Abstract
Over the past 8 years, the discovery of 11 new human polyomaviruses (HPyVs) has revived interest in this DNA tumor virus family. Although HPyV infection is widespread and largely asymptomatic, one of these HPyVs, Merkel cell polyomavirus (MCV), is a bona fide human tumor virus. JC virus (JCV), BK virus, HPyV7, and trichodysplasia-spinulosa virus (TSV) can cause nonneoplastic diseases in the setting of immunosuppression. Few specific reagents are available to study the biology of the newly discovered HPyVs. We developed a pan-HPyV-screening method using a cocktail of 3 antibodies that, when combined, recognize T antigen proteins of all HPyVs. We validated detection characteristics of the antibody cocktail by immunoblotting and immunohistochemistry and screened 1,184 cases, including well-defined diseases and tumor tissue microarrays. This assay robustly detected MCV, TSV, JCV, and HPyV7 in etiologically related diseases. We further identified WU polyomavirus in a case of chronic lymphocytic lymphoma-associated bronchitis. Except for scattered, incidentally infected cells in 5% of lung squamous cell carcinomas and colon adenocarcinomas, a broad panel of tumor tissues was largely negative for infection by any HPyV. This method eliminates known HPyVs as suspected causes of cancers investigated in this study. Pan-HPyV survey can be applied to identify diseases associated with recently discovered polyomaviruses.
A screen for human polyomaviruses in 1,184 tumor samples finds no evidence for a role of polyomaviruses in the development of these cancers.
Introduction
All human polyomaviruses (HPyVs) share fundamental features of genome organization and structure but can differ in tissue tropism and disease association (1, 2). Infection with HPyVs is mostly asymptomatic and widespread in the general population. These viruses are part of the normal microbial flora but, in the context of immune suppression, can cause a spectrum of diseases as the sequela of unchecked viral replication or unbalanced expression of early versus late viral genes. HPyV diseases run the spectrum from inflammatory, to hyperplastic, to neoplastic disorders and include BK virus–related (BKV-related) nephropathy (PVAN) (3), JC virus–related (JCV-related) progressive multifocal leukoencephalopathy (PML) (4), trichodysplasia spinulosa (TS) (5), HPyV7-related epithelial hyperplasia (6), and Merkel cell carcinoma (MCC) (7). Since the discovery of JCV and BKV in 1971, 11 new polyomavirus species have been identified. Much remains unknown about the newly discovered HPyVs. Given the increasing use of immunosuppressive treatments due to transplantation and acquired or primary immune deficiency, reactivation of these normally commensal viruses may result in new disease syndromes.
HPyVs are small nonenveloped double-stranded DNA viruses with 4.8- to 5.3-kb genomes divided into early, late, and noncoding control regions (1, 2). The early region encodes for large T (LT) and small T (sT) regulatory proteins and can also encode for alternative frame (8) and splice variants of LT proteins. The late region comprises structural genes that produce viral capsid proteins VP1, VP2, and VP3. Some polyomaviruses (PyVs) also encode a microRNA that targets the early transcript and thus modulates the expression levels of LT protein (9, 10). All HPyV T antigens are potential oncoproteins based on their conserved tumor suppressor–targeting domains. Enormous resources and efforts have been spent in searching for polyomavirus-induced tumors and diseases (particularly for nonhuman simian virus 40 [SV40]) by PCR-based methods (11–21). However, results have been controversial and inconclusive due to the limitations of this technique: although PCR is simple and sensitive, it is also prone to contamination, does not provide localization information, and does not distinguish between coincidental passenger infections in the tumor milieu and a true causal association. Only Merkel cell polyomavirus (MCV) has been established to cause human cancer among the polyomaviruses, and there is a need for an assay that can rapidly and accurately assess whether other polyomaviruses play a role in tumors.
Immunohistochemistry (IHC) is a well-established and powerful technique that provides information about the localization and quantitation of target protein epitopes. Detection of viral antigen by IHC can frequently define severity and extent of an infection. We developed a pan-polyomavirus immunohistochemistry test (P-PIT). By analyzing the reactivity of several PyV antibodies, we found that the combination of the PAb416 commercial antibody (22), 2t2 (23), and Xt7 (24) is sufficient to robustly detect the early proteins of all HPyVs not only in immunoblotting of cellular lysates, but also in tissue specimens. We were able to identify a case of WU virus–associated (WUV-associated) bronchitis in a lung biopsy from a patient with chronic lymphocytic leukemia (CLL) by rolling circle amplification (RCA) that was found to be reactive for PAb416 in an initial diagnostic pathology evaluation. We demonstrate that the use of this cocktail of 3 antibody P-PIT can be applied easily to tissue arrays or a large selection of patient tissue samples to screen for known HPyVs and has the potential to reveal new members of the polyomavirus family.
Results
Validation of the reactivity of PyV T antigen-specific antibodies.
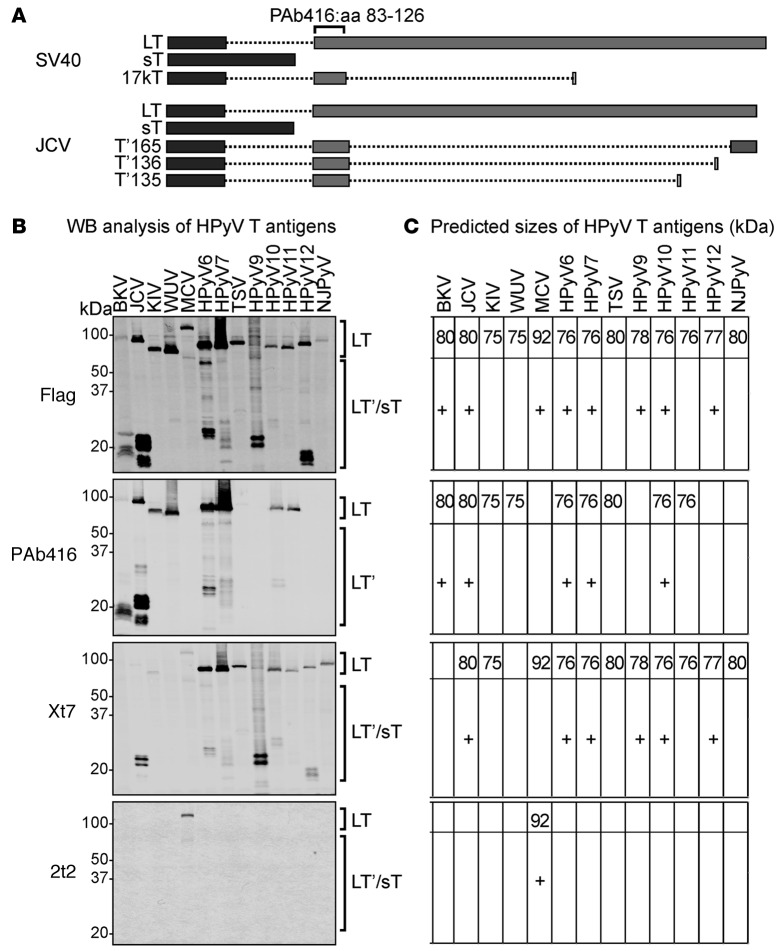
In order to evaluate the reactivity for available PyVs T antigen antibodies, early regions of all HPyVs were either synthesized or PCR amplified and cloned into an expression vector with an N-terminal flag tag. Following sequence confirmation, each construct was transfected into HEK293 cells. Cells lysates were subjected to SDS-PAGE and immunoblotted with various antibodies. The mouse IgG2a mAb designated PAb416 is a commercial antibody generated against SV40 T antigen (22) that recognizes a conserved epitope at aa 83–126 within exon 2 (Figure 1A). It is widely used for the clinical diagnosis of JCV and BKV infection in human tissues. The Xt7 mAb was generated against purified HPyV10 sT antigen (24). The 2t2 mAb was developed against the NRKEREALCKLLEIAPN peptide epitope of the common J-domain present in both MCV LT and sT proteins (23) and was used in preference to the CM2B4 antibody (25), which recognizes an epitope present only on the MCV LT antigen.
Figure 1. Detection of human polyomavirus early region proteins in HEK293 cells.
(A) Splicing arrangement of simian virus 40 (SV40) and JC virus (JCV) T antigens. Rectangles indicate the coding sequences: dark gray rectangles represent exon 1 and the light gray rectangles represent exon 2. Dotted lines represent the intron sequences. The PAb416 diagnostic antibody recognizes an epitope within aa 83–126 of exon 2 that is conserved in most of the human polyomaviruses (HPyVs). Different splicing events in JCV T antigen give rise to truncated forms of large T (LT), including T′165, T′136, and T′135. (B) Early gene expression of HPyVs was analyzed by Western blotting (WB) using anti-Flag, PAb416, Xt7, or 2t2. Observed sizes of LT from different HPyVs vary between 75 and 125 kDa. HPyV T antigen splice variants are between 15 and 60 kDa (HPyV T antigen splice variants are indicated by a plus sign in C). PAb416 antibody detects early proteins from BKV, JCV, KIV, WUV, HPyV6, HPyV7, TSV, HPyV10, and HPyV11. Xt7 antibody detects early region proteins from HPyV6, HPyV7, TSV, HPyV9, HPyV10, HPyV11, HPyV12, and NJPyV and detects KIV, MCV, and JCV splice forms weakly. 2t2 antibody detects MCV LT and the 57 kDa T. (C) The predicted sizes of HPyV T antigens (in kDa) are shown for respective immunoblots. sT, small T; BKV, BK virus; JCV, JC virus; KIV, KI virus; WUV, WU virus; MCV, Merkel cell polyomavirus; TSV, trichodysplasia-spinulosa virus; NJPyV, New Jersey polyomavirus.
We were able to detect all HPyV LTs and their T antigen isoforms. Immunoblotting with anti-Flag antibody revealed LT protein expression from all HPyV early regions, since all these isoforms have an intact N-terminal Flag tag (Figure 1B). JCV, BKV, and MCV LT proteins migrate slower than the previous predicted size (ref. 25 and Figure 1C). We detected known alternative splice variants of T antigen for BKV, JCV, MCV but also found similar splicing events in WUV, HPyV6, HPyV7, HPyV9, HPyV10, and HPyV12.
The diagnostic PAb416 antibody detects LT and LT splice isoforms, but not sT, which lacks the epitope-binding site (Figure 1A). As expected, PAb416 immunoblotting detects LT and truncated LT of BKV and LT, T′165, T′136, and T′135 of JCV (Figure 1, B and C). In addition, it is also reactive for KI virus (KIV), WUV, HPyV6, HPyV7, trichodysplasia-spinulosa virus (TSV), HPyV10, and HPyV11 LT. PAb416 revealed alternatively spliced isoforms of LT for HPyV6, HPyV7, and HPyV10.
Although Xt7 was generated using HPyV10 sT as immunogen, it also detects LT and other splice forms consistent with a recognition epitope in a shared sT/LT exon. Xt7 is reactive for HPyV6, HPyV7, TSV, HPyV9, HPyV10, HPyV11, HPyV12, and New Jersey polyomavirus (NJPyV) but is also weakly reactive for JCV, KIV, and MCV. The 2t2 epitope is unique and thus only detects MCV LT and the 57 kDa T. Simultaneous incubation with all 3 antibodies showed a pattern similar to anti-Flag immunoblotting (data not shown).
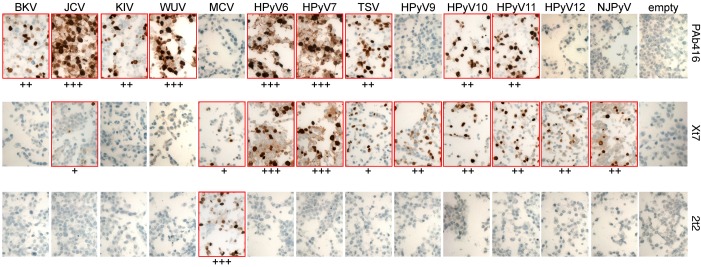
Immunohistochemical staining results were consistent with immunoblotting studies. The staining with PAb416, Xt7, and 2t2 antibodies for various T antigens was predominantly nuclear (dark brown) (Figure 2). For JCV, KIV, WUV, HPyV6, HPyV7, HPyV9, HPyV10, and HPyV11, T antigen expression was also detected in the cytoplasm (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/jci.insight.85562DS1). Control cells transfected with empty vector showed low levels of cytoplasmic background staining with Xt7. Immunostaining using all 3 antibodies detected T antigens of all HPyVs. The combination had an additive advantage in generating a more robust signal strength as compared to the use of single antibodies for HPyVs in which more than one antibody was reactive (JCV, MCV, HPyV6, HPyV7, TSV, HPyV10, and HPyV11) (data not shown).
Figure 2. Immunohistochemical characterization of HEK293 cells expressing HPyV early proteins.
Cell pellet microarray sections were stained with PAb416, Xt7, or 2t2 overnight at 4°C and counterstained with hematoxylin. Positive staining (outlined in red) intensity is graded from 0 to 3 (+, weak; ++, moderate; +++, strong). Original magnification, ×20. BKV, BK virus; JCV, JC virus; KIV, KI virus; WUV, WU virus; MCV, Merkel cell polyomavirus; HPyV, human polyomavirus; TSV, trichodysplasia-spinulosa virus; NJPyV, New Jersey polyomavirus.
Validation of pan-HPyV screening for known disease-associated HPyVs.
The optimal condition for the P-PIT-screening method was developed using JCV-positive PML, MCV-positive or -negative MCC, HPyV7-positive skin rash, and TSV-positive TS tissue sections. Different antigen retrieval buffer conditions and varying antibody dilutions and incubation periods were evaluated in the optimization. The combination of all 3 antibodies detected immunohistochemical positivity for JCV, MCV, HPyV7, and TSV in PML, MCC, skin rash, and TS, respectively. We did not observe any nonspecific staining in several control tissues, including MCV-negative MCC (Figure 3). As a further control, we used an MCC tissue microarray (TMA) comprising duplicate or triplicate cores from each different case. We performed separate immunostaining with the pan-HPyV cocktail (Figure 4A) and the CM2B4 MCV LT antibody (Figure 4B). We excluded all cytokeratin 20–negative (CK20-negative) cases from the TMA, since recent literature in the field suggests that the vast majority of CK20-negative cases diagnosed in the past as MCCs are not usually infected with MCV (26). The immunostaining results showed exact concordance between the pan-HPyV cocktail and the CM2B4 antibody: 20 of 25 CK20-positive MCC cases were immunopositive with both assays, the remaining 5 cases were immunonegative with both (Table 1).
Figure 3. Application of the pan–human polyomavirus survey to known HPyV-associated diseases.
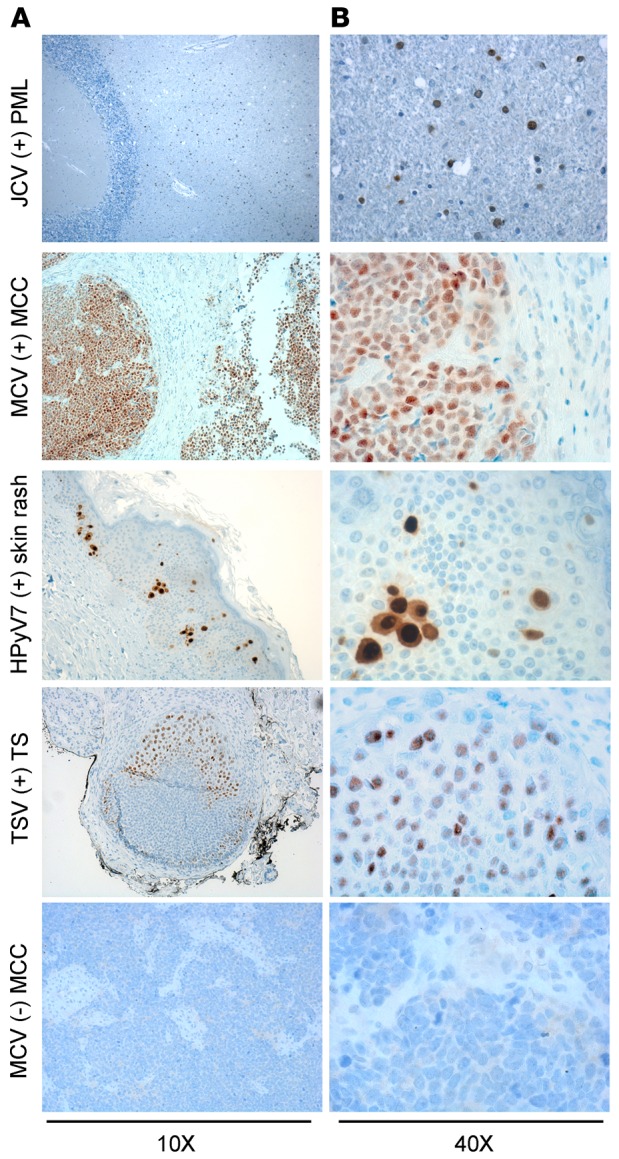
Formalin-fixed paraffin-embedded tissue sections from JC virus–related (JCV-related) progressive multifocal leukoencephalopathy (PML), Merkel cell polyomavirus–positive (MCV-positive) or -negative Merkel cell carcinoma (MCC), human polyomavirus 7–related (HPyV7-related) skin rash lesion, and trichodysplasia-spinulosa virus-related (TSV-related) trichodysplasia spinulosa (TS) were stained with triple antibody combination overnight and counterstained with hematoxylin. JCV T antigen is detected mostly in oligodendrocytes. Nuclear MCV T antigen is detected in the whole tumor section. HPyV7 T antigen is expressed in the nuclei and cytoplasm of the hyperplastic epithelium. TSV T antigen is expressed in the affected hair follicle cells. Original magnification, ×10 (A); ×40 (B).
Figure 4. Application of pan-human polyomavirus screening to different tissue microarrays.
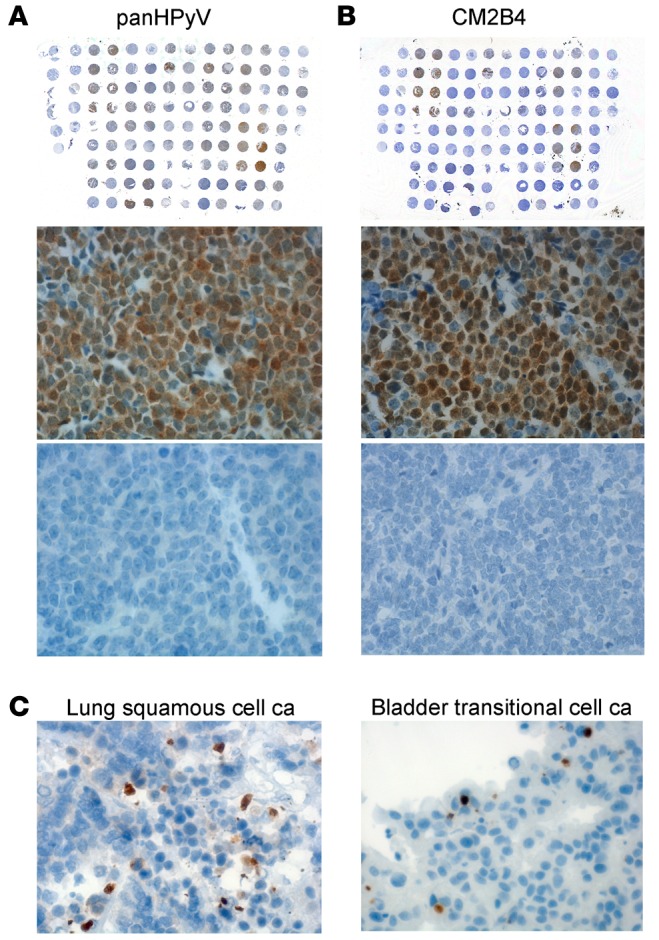
A Merkel cell carcinoma (MCC) tissue microarray (TMA) was either immunostained with (A) a combination of PAb416, Xt7, and 2t2 or with (B) CM2B4. The same Merkel cell polyomavirus–positive (MCV-positive) (middle row) and -negative (bottom row) MCC case samples from each slide are shown for both stainings. Original magnification, ×40 (middle and bottom); ×1 (top). (C) Representative examples for passenger infection cases observed in lung squamous cell carcinoma (ca) and bladder transitional cell ca (brown). Original magnification, ×40.
Table 1. Immunohistochemistry staining results for CK20-positive MCC tumor microarray with CM2B4 or pan-HPyV.
After validating the sensitivity and specificity of the P-PIT-screening method, we analyzed TMA sections from different tumor types (Table 2). We applied the screening test to lung (236 cases), oral cavity (65 cases), stomach (36 cases), colon (94 cases), bladder (137 cases), kidney (65 cases), skin (50 cases), and breast (85 cases) TMAs (Table 2) and did not find any cases in which the staining pattern would suggest a direct involvement of viral proteins in tumorigenesis, as defined by nuclear staining in the majority of tumor cells juxtaposed to negative reactivity in infiltrated normal tissue. However, we did observe that 1.5% of bladder transitional cell carcinoma cases and 5% of lung squamous cell carcinoma and colon adenocarcinoma cases had scattered and rare cells, showing convincing nuclear positivity suggestive of incidental or passenger infection in rare cells by HPyVs (Figure 4C).
Table 2. Tumor TMAs.
Absence of HPyV T antigen expression in human cancers reported to be associated with SV40.
The presence of SV40 DNA has been episodically reported in malignant mesotheliomas, brain tumors, osteosarcomas, non-Hodgkin lymphomas, and breast and colon cancers (18–21). In contrast, there have been epidemiological and molecular studies that suggest a lack of SV40 involvement in human disease (reviewed in refs. 1 and 21). We performed P-PIT on tumor TMAs representing brain neoplasia (261 cases, including 4 choroid plexus tumors and 11 ependymomas), malignant mesothelioma (88 cases), and non-Hodgkin lymphoma (38 cases) (Table 2). We found no evidence of HPyV T antigen expression in any of the tissues tested.
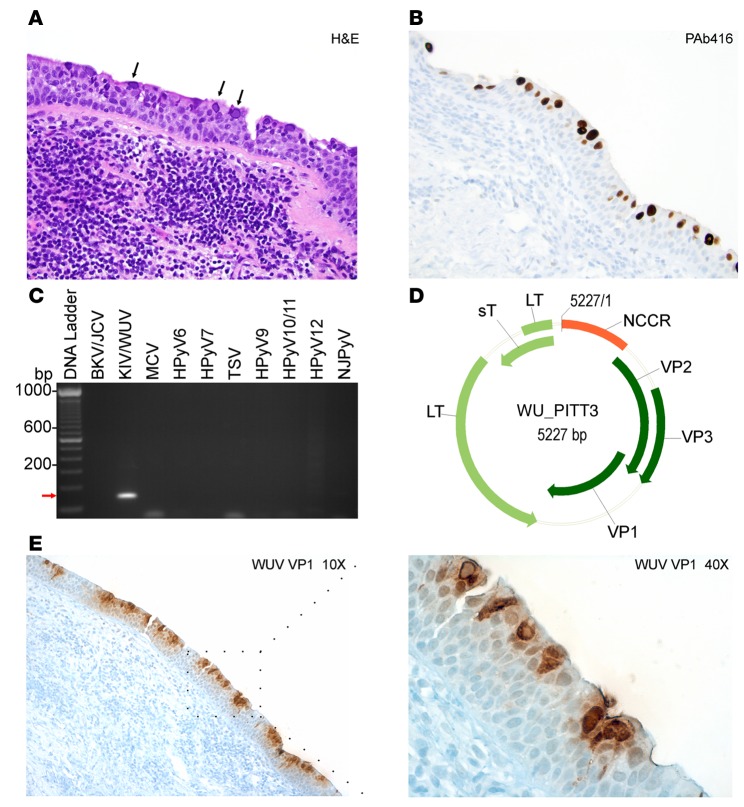
Detection of WUV-associated acute bronchitis in a patient with pulmonary CLL.
We were able to identify and localize extensive infection by WUV in respiratory epithelial cells in a patient with bronchitis and co-occurrence of pulmonary CLL. A 61-year-old male patient with CLL presented with a dry, persistent nonproductive cough, with patchy ground-glass opacities in both lungs. By H&E, a bronchial biopsy showed viral bronchitis mimicking squamous dysplasia, overlying a submucosal infiltrate of CLL/small lymphocytic lymphoma, characterized by dense lymphocytic infiltrates dominated by CD20-positive B cells and fewer CD3-positive T cells. The bronchial mucosa had abundant numbers of enlarged cells with enlarged nuclei with glassy, homogenous, central inclusions, which, in some cells, were associated with marginal displacement of the chromatin to the nuclear membrane (Figure 5A). Immunostaining of lung and bronchial tissues for cytomegalovirus, herpes simplex virus type 1 and 2, varicella-zoster virus, and adenoviruses were negative; however, PAb416 staining was positive (Figure 5B).
Figure 5. Detection and molecular characterization of WU virus isolated from a chronic lymphocytic leukemia patient.
(A) Hematoxylin and eosin (H&E) staining of a lung tissue biopsy sample shows intranuclear inclusion bodies (black arrows). (B) PAb416 staining correlates with H&E staining and shows nuclear staining of the respiratory epithelium (brown). (A and B) Original magnification, ×20. (C) Specific primers for different human polyomaviruses (HPyVs) were tested on rolling circle amplification products. KI virus/WU virus–specific (KIV/WUV-specific) amplification is indicated with a red arrow. BKV, BK virus; JCV, JC virus; MCV, Merkel cell polyomavirus; TSV, trichodysplasia-spinulosa virus; NJPyV, New Jersey polyomavirus. (D) Genome map of the WUV isolate WU_PITT3. The early region encodes for large T (LT) and small T (sT) antigens. The late region comprises viral capsid protein VP1, VP2, and VP3. The noncoding control region (NCCR) contains the putative origin of replication and promoters. (E) Immunohistochemistry staining of WUV with VP1 antibody shows nuclear and cytoplasmic staining of a large number of lumenally located cells of the dysplastic respiratory epithelia (brown). Original magnification, ×10 (left); ×40 (right).
To identify which polyomavirus is the causative agent for bronchitis, we extracted DNA from formalin-fixed paraffin-embedded samples for traditional PCR using HPyV-specific primers (Supplemental Table 1 and Figure 5C). Initial PCR amplification showed positive amplification of an approximately 180-bp product using a primer pair that identified both KIV and WUV. Subsequent sequencing analysis identified sequence match to WUV from the amplified product. To obtain the whole-genome sequence of the isolate, we performed RCA followed by PCR using overlapping primers (Supplemental Table 2). Extracted DNA products were sequenced and analyzed using Blast NCBI software (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The complete genome sequence of the new isolate was designated WU_PITT3 (Figure 5D) and deposited under genBank accession number KU049032.
In comparison to the WU reference S1 strain (EF444550.1), WU_PITT3 had 57 nucleotide-mismatch polymorphisms and a 2-bp gap (nucleotide position 2813 and 4536). The majority of polymorphisms are in the VP region. Interestingly, sT and LT coding regions had only 4 and 2 mismatch polymorphisms, respectively (data not shown). The WU_PITT3 isolate shows 99% sequence identity with the previously reported EU711055 (27) and GU296365.1 (28) isolates, carrying single missense polymorphisms in VP1 and sT coding regions, respectively (Table 3).
Table 3. Sequence comparison of WU_PITT3 and other WUV strains.
Differential expression of early and late proteins in HPyV-associated diseases.
We further assessed WUV protein expression using a specific antibody directed against the WU VP1 virion protein (Figure 5E and ref. 29). Strong VP1 reactivity was detected in both the nuclei and cytoplasm of a large number of cells lining the respiratory tract, most of which are located on the lumenal surface of the dysplastic epithelia (Figure 5E). These results indicate that infected cells contained actively replicating WUV expressing both early and late WUV proteins. The peripheral location of immunohistochemically positive cells may suggest that fully differentiated, ciliated respiratory epithelial cells are most permissive to WUV lytic replication.
We performed immunohistochemical staining on MCC, skin rash, and TS tissue sections with respective virus-specific VP1 antibodies and compared the extent and localization of these viral proteins to that of T antigen expression (Figure 6). We verified that MCV-positive tumors do not express any late gene products (Figure 6A, MCV VP1). HPyV7 VP1 staining using 6V32 mAb (6) revealed strong cytoplasmic and intercellular reactivity and some nuclear reactivity as well as HPyV7 T antigen staining (Figure 6B). In contrast, TSV T antigen expression was relatively more abundant compared to that of TSV VP1 in the TS tissue section (Figure 6C, TSV VP1). There were more cells that expressed TSV T antigen alone compared to cells with both TSV T antigen and VP1 expression. These results demonstrate the relevance of targeting T antigen expression, which is expressed in all pathologies associated with known HPyV-related diseases, in a screening assay. In contrast, MCC, which exemplifies an HPyV-related neoplasm, does not express late proteins such as VP1.
Figure 6. Comparison of T antigen and VP1 immunostaining of known human polyomaviruses.
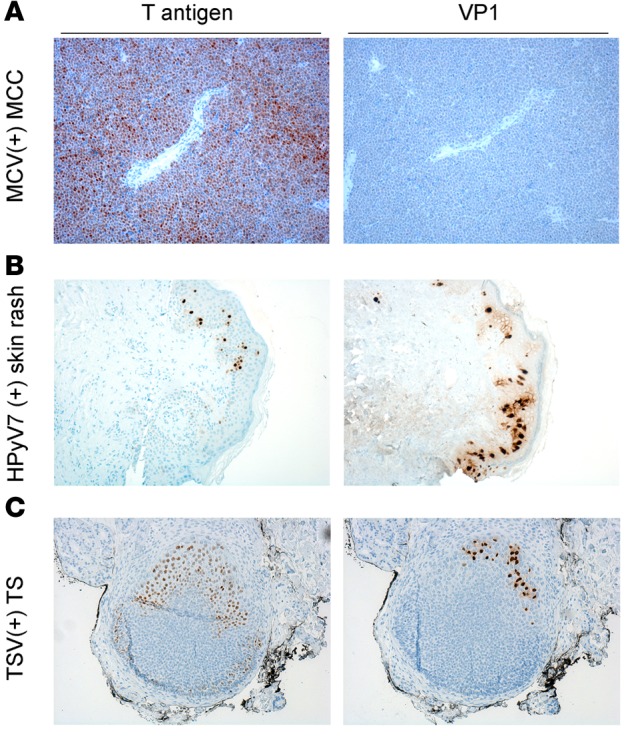
T antigen and virus-specific VP1 staining in (A) Merkel cell polyomavirus (MCV-positive) Merkel cell carcinoma (MCC), (B) human polyomavirus 7–positive (HPyV7-positive) skin rash, and (C) trichodysplasia-spinulosa virus-positive (TSV-positive) trichodysplasia spinulosa (TS) tissue. For T antigen staining, a triple antibody combination of PAb416, Xt7, and 2t2 was used. For VP1 staining, virus-specific antibodies CM9B2, 6V32, and TSV-VP1 were used for MCV, HPyV7, and TSV, respectively. MCV-positive tumors positive for T antigen do not express VP1. HPyV7-positive skin rash shows cytoplasmic, intercellular, and some nuclear VP1 expression and scattered T antigen expression in the epidermis. TSV T antigen is more broadly expressed in contrast to VP1 in the TS lesion. Original magnification, ×10.
Discussion
We developed P-PIT, an IHC-based pan-HPyV-screening method to facilitate the detection of PyV early gene products in tissue samples or arrays. The combination of 3 different PyV-specific antibodies enabled detection of overexpressed T antigens from all HPyVs reported to date by immunoblotting and IHC methods. Flag immunoblotting of HPyV early region constructs overexpressed in HEK293 cells revealed LT expression from each HPyV in addition to that of alternatively spliced T isoforms. The variability in the ratios of expression levels between LT and other T antigen isoforms has been observed in polyomaviruses and can result from diverse factors, including cell context, splicing efficiency of overexpressed constructs, and whether expression occurs in the course of natural infection (2, 30, 31). At present, we do not know which RNA cis elements and/or transacting host factors are involved in the regulation of alternative splicing.
The detection specificities for each antibody combine to provide full coverage of all HPyV T antigens. PAb416 and Xt7 overlap in their detection of JCV, HPyV6, HPyV7, TSV, HPyV10, and HPyV11 T antigens, and using the triple antibody combination increases the chances of IHC detection of these HPyVs, even if their T antigens are expressed at low levels. At the same time, the broad specificity of the antibodies suggests that detection is through conserved antigenic epitopes, which may potentially facilitate the discovery of new polyomaviruses. Once the pan-HPyV staining reveals a positive result, sequential single antibody staining along with PCR, if feasible, can narrow down or identify the probable HPyV involved (Figure 7). Validation of this immunohistochemical staining on tissue biopsies from known HPyV-associated diseases confirms the specificity and sensitivity of this method. Further, immunohistochemical visualization provides critical information on the localization, cell tropism, and extent of specific HPyV infections.
Figure 7. Diagram of single antibody staining, following a reactive pan–human polyomavirus survey.
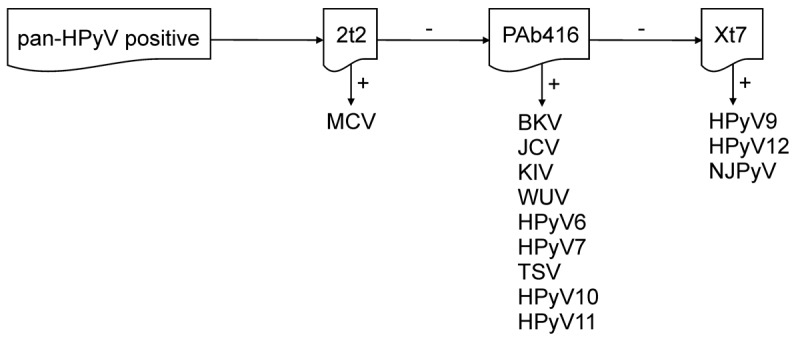
HPyV, human polyomavirus; MCV, Merkel cell polyomavirus; BKV, BK virus; JCV, JC virus; KIV, KI virus; WUV, WU virus; TSV, trichodysplasia-spinulosa virus; NJPyV, New Jersey polyomavirus.
With the exception of MCV in MCC, the involvement of other HPyVs in tumorigenesis has been debated during the last decade, with no clear evidence or progress on causality (1). This is in large part due to the lack of reliable diagnostic tests and limitations of the screening methods used in the past. The pan-HPyV IHC staining applied to an MCC TMA underscores the sensitivity and specificity of this assay, with results correlating to CM2B4 staining, a test used for the diagnosis of MCV-positive MCC. We screened over 100 lung (adenocarcinoma and squamous cell) carcinomas, bladder carcinomas, and brain tumors and between 50 and 100 oral cavity, gastric, colon, kidney, skin, breast carcinoma, non-Hodgkin lymphoma, and malignant melanoma cases with no evidence of HPyV involvement in the pathogenesis of these neoplasms. Rare cells showing HPyV infection in some of these cases are likely to represent incidental or passenger infection. This occurred more frequently in squamous cell lung carcinoma tissues as compared to lung adenocarcinoma tissues, and the detection of scattered positive cells also occurred in some colon and bladder carcinomas. These tumor subtypes may offer a more permissive host environment for HPyV infection. Alternatively, the epithelial surfaces from which these lung, colon, and bladder tumors arise may represent the natural niche for specific HPyVs. Both KIV and WUV were initially isolated from respiratory secretions (27–29), and BKV as well as JCV have been determined to be detectable from the urogenital system (11–13). Nevertheless, our results suggest that HPyVs do not play a driver role in the cancers we tested. In contrast to studies suggesting the involvement of SV40 in human neoplasia, we found no evidence of the expression of SV40 early proteins in ependymomas, choroid plexus and other brain tumors, mesotheliomas, and non-Hodgkin lymphomas.
All HPyVs have the potential to cause nonneoplastic disease in the context of immunosuppression. As reported previously, peripheral blood mononuclear cell (PBMC) DNA samples collected from 83 healthy individuals were negative for MCV DNA; however, MCV was detected in 9.5% of the PBMCs of 21 HIV/AIDS patients without MCC (25). Similarly, in HIV-positive individuals, JCV and BKV DNA was found in 22.8% and 51.1% of the PBMC samples, respectively, a positivity rate higher than that found in healthy individuals (32). Reactivation of various other HPyVs has been reported following organ transplantation and immunosuppressive therapy (33, 34).
PML, PVAN, and TS appear to be the consequence of active virus replication, which occurs in the context of immune suppression. In contrast, MCV, the first polyomavirus linked to human cancer, is not caused by viral reactivation and replication but rather by clonal integration of the viral genome with restricted expression of only the early region T antigen proteins (10). Late capsid proteins are not expressed in MCC, while early region sT and LT expression is indispensable for transformation and T antigen expression is required in every tumor cell (25, 35). We conclude that HPyV T antigen detection in scattered and rare cells most likely indicates a passenger infection of a restricted nature. Recently, BKV was reported to be associated with a high-grade urothelial tumor (36). In this singular case, the BKV genome was integrated into the host genome, which eliminated BKV VP1 but not T antigen expression, supporting this model of HPyV tumorigenesis.
Further application of the pan-HPyV-screening method was validated by the detection of WUV-associated bronchitis in a patient with CLL. Since its discovery in 2010, no clear link between WUV and a human disease has been found, although low levels WUV DNA can be detected in screens of respiratory tissues by PCR (37). PCR-based studies of 30 lung cancer patients compared to healthy individuals indicated no direct association between WUV infection and lung adenocarcinoma (38). However, recently, a lung transplant patient with Job’s syndrome and a bone marrow transplant recipient with severe acute respiratory distress syndrome were reported to have WUV pulmonary infection (29, 39). In the present study, we have identified a case of WUV-associated bronchitis with the demonstration of both early and late viral antigens in intact dysplastic lung epithelia. The WUV genome sequenced from the present case shows similarity with two previously published WUV isolates (27, 28) but has multiple polymorphisms in comparison to the reference WUV genome. It unknown at this time whether these differences relate to increased virulence in the context of immune suppression.
In summary, the pan-HPyV-screening method we established enables detection of T antigens and thus not only allows detection of lytic infection but also has the potential to uncover HPyV-related oncogenic processes in which late antigens are not expressed. The P-PIT screening assay will be valuable for addressing the perennial controversy of HPyV-related cancer and other disease associations.
Methods
Cell lines and plasmids.
HEK293 cells (ATCC) were maintained DMEM (Cellgro, 10-013) supplemented with 10% FBS (Sigma-Aldrich).
In order to generate HPyV early region expression constructs, template plasmids comprising the early regions of BKV, JCV, HPyV6, HPyV7, and HPyV10 (40) (provided by C. Buck, National Cancer Institute, Bethesda, Maryland, USA) and KIV and WUV (Addgene, plasmid 37093 and 37094) were amplified with Phusion Hot Start Flex DNA polymerase (NEB) or Platinum Taq DNA polymerase (Invitrogen) using primer pairs comprising an N-terminal flag tag sequence, SalI restriction endonuclease site, and a C-terminal SacII restriction endonuclease site. Early regions of TSV, HPyV9, HPyV11, HPyV12, and NJPyV, with an N-terminal flag tag and flanking restriction sites for subcloning, were synthesized (GenScript GU989205, HQ696595, JX463183, JX308829, and KF954417 were used for the synthesis). Amplified PCR products and isolated DNA were digested with restriction endonucleases and subcloned into a modified pcDNA6 vector (Invitrogen) linearized with XhoI and SacII restriction endonuclease enzymes (NEB) and transformed into NEB Stable competent cells (NEB) following ligation reaction with T4 ligase (NEB). MCV genomic LT (41) was subcloned into a pCMVTag2B vector, which has an N-terminal flag tag. Following restriction digestion and sequencing analysis, DNA isolated from bacteria was used to transfect HEK293 cells with Lipofectamine 2000 (Invitrogen).
Hybridoma production.
The 6V32 mAb (6) and 2t2 (23) and Xt7 (24) hybridoma cells were gifts from C. Buck (National Cancer Institute). General characteristics of the mAbs have been previously described (42). 6V32 mAb was raised against virus-like particles composed of HPyV6 VP1 and cross-reacts with HPyV7 VP1. Xt7 and 2t2 hybridoma cells were grown to maximum confluence in CD Hybridoma medium (Gibco) supplemented with Hybridoma Fusion and Cloning Supplement (Roche) and glutamine (Sigma-Aldrich). Hybridoma supernatant was collected, filtered (0.22-μm filter, Nalgene), and subsequently concentrated using Amicon filtration system (Millipore).
The mAb CM9B2 was generated by standard methods by immunizing mice with KLH-derived DKGKAPLKGPQKASQKES peptide from the MCV VP1 antigen (43).
The open reading frame of the TSV-VP1 gene was subcloned into pEU-His-TEV-vector (CellFree Sciences). Full-length His-TsPyV-VP1 protein was synthesized using the robotic synthesizer (Protemist XE; CellFree Sciences) as described previously (44). The cell-free translation reaction mixture was lysed by the addition of 8 M urea and then mixed with Ni-sepharose High Performance beads (GE Healthcare) in the presence of 20 mM imidazole. The beads were washed 3 times with washing buffer (20 mM Tris-HCl, pH 7.5, 500 mM NaCl) containing 40 mM imidazole. His-VP1 proteins were then eluted with washing buffer containing 8 M urea and 500 mM imidazole and concentrated using Amicon Ultra centrifugal filters (Millipore). The mAbs specific for TSV-VP1 (clone 14) and cross-reactive (clone 12) for both TSV and JCV VP1 (data not shown) were generated using a previously described hybridoma technology (45). In brief, 300 μg of His-tagged full-length VP1 protein was injected into the footpads of BALB/c mice using keyhole limpet hemocyanin as an adjuvant. Four weeks later, spleen cells were isolated and fused to the myeloma cell line SP2/O using polyethylene glycol 1500 as previously described (46). Isotype determination was performed with the mouse mAb isotyping test kit (Bio-Rad) according to the manufacturer’s protocol.
Genomic DNA isolation, RCA, and sequencing.
To isolate DNA from formalin-fixed paraffin sections, tissue samples were collected in a microfuge tube and incubated in 1 ml of Xylene on a rotator for 15 minutes. Following centrifugation at 16,000 g for 3 minutes, the pellet was rinsed with 1 ml of 100% ethanol, followed by 1 ml of 70% ethanol at room temperature for 5 minutes. The air-dried pellet was resuspended in 500 μl of digestion buffer (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 25 mM EDTA, pH 8.0, and 0.5% sodium dodecyl sulfate [SDS]). 25 μl of Proteinase K (Promega, 20 mg/ml) was added to the lysate and incubated overnight at 55°C for full digestion. Following phenol/chloroform extraction, DNA was precipitated with 0.5 volume of ammonium acetate (7.5 M), 2.5 volume of ice-cold ethanol, and glycogen overnight at –20°C. After centrifugation at 16,000 g for 1 hour at 4°C, the pellet was rinsed with 70% ethanol, air dried, and dissolved in small volumes of 0.1 M Tris-EDTA buffer at room temperature for 3 hours.
Extracted DNA was treated with PreCR repair mix (NEB) prior to RCA using TempliPhi (GE Healthcare) according to the manufacturer’s recommendations. 0.5 to 1 μl of extracted DNA or 1 μl of RCA template was used for the PCR reaction and supplemented with Phusion DNA polymerase (NEB) and virus-specific primer pairs (Supplemental Tables 1 and 2). PCR products extracted from agarose gel (Qiagen) were used for sequencing analysis (McLAB).
Western blot.
HEK293 cells were lysed in radioimmunoprecipitation assay buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP40, 0.1% SDS, 0.5% Na deoxycholate, and protease inhibitors (Complete cocktail with EDTA, Roche). Lysates were kept on ice for 20 minutes, sonicated, and then centrifuged at 17,500 g for 10 minutes at 4°C. Protein concentration was estimated by DC protein assay kit (Bio-Rad). Lysates were mixed with 2x Laemmli loading dye and denatured at 100°C for 10 minutes. Proteins were separated by SDS-PAGE and transferred onto a Hybond-C nitrocellulose membrane (GE Healthcare). Following blocking with 5% nonfat dry milk in TBS, membranes were incubated with PAb416 (clone Ab-2, Millipore, DP02, 1:1,000), Xt7 (1:2 hybridoma supernatant), 2t2 (1:2 hybridoma supernatant), or anti-Flag (clone M2, Sigma-Aldrich, F1804, 1:3,000) in TBS supplemented with 5% BSA overnight at 4°C. Following incubation with IRD-conjugated secondary antibody (Li-Cor, 1:15,000) in blocking buffer, proteins were visualized by Li-Cor Odyssey imaging system.
IHC.
HEK293 cells were transfected with various expression constructs. 48 hours after transfection, cells were harvested and pellets were fixed in 10% neutral-buffered formalin and processed with HistoGel (Thermo Scientific) for routine histology. Formalin-fixed paraffin-embedded cell pellets, tumor tissues, and TMA sections (oral cavity, stomach, colon, bladder, kidney, skin, brain, mesothelial, and lymphoma TMAs were obtained from US Biomax Inc.) were stained according to a previously published protocol (25). Briefly, slides were deparaffinized in xylene and rehydrated in a series of ethanol solutions. Endogenous peroxidase activity was blocked by incubation of the slides with 3% hydrogen peroxide for 15 minutes. Epitope retrieval was performed using 1 mM EDTA buffer, pH 8.0, at 125°C for 3 minutes and 90°C for 15 seconds in an antigen retrieval chamber (Decloaking chamber, Biocare medical). After blocking (Protein block, serum free, Dako), primary antibodies PAb416 (1:100), 2t2 (1:5 hybridoma supernatant or approximately 8 μg/ml concentrated mAb), Xt7 (1:5 hybridoma supernatant or approximately 8 μg/ml concentrated mAb), CM2B4 (1:10 hybridoma supernatant or 0.6 μg/ml mAb), MCV VP1 (CM9B2, 2.5 μg/ml), 6V32 (1:100, approximately 2 μg/ml), TSV VP1 (1:100 hybridoma supernatant), or mouse mAb WUV VP1 (NN-Ab06, 1:2,000, provided by David Wang, Washington University, St. Louis, Missouri, USA) diluted in antibody buffer (1% BSA, 0.1% gelatin, 0.5% Triton-X, 0.05% sodium azide in PBS, pH 7.4) were applied to each section overnight at 4°C in a humidified chamber. Following extensive rinsing in TBS, sections were incubated with mouse Envision Polymer (Dako) for 30 minutes at room temperature, reacted with diaminobenzidine (DAB, Dako) or with 3-amino-9-ethylcarbazole high-sensitivity substrate (Dako), and counterstained with hematoxylin (Dako). Unless otherwise indicated, DAB chromogen was used as chromogen.
Study approval.
This project did not involve human subjects, as defined by US federal regulations [46.102(f)]. Informed consent was not required for the experiments performed in this study, as all human samples were obtained as deidentified tissue and are covered by University of Pittsburgh IRB exemption protocols PRO07100312, PRO07110141, and PRO07100376.
Author contributions
TT, YC, and PSM designed research, analyzed data, and wrote the manuscript. TT and SAY conducted experiments and acquired and analyzed data. JH, YM, MFF, LPS, RB, and AR provided critical tissues, TMAs, and antibody reagents.
Supplementary Material
Acknowledgments
The authors thank Chris Buck, Rachel Schowalter, Masahiro Shuda, Clayton Wiley, and David Wang for providing reagents and for critical reading of the manuscript. We thank Louis Hardin for help with the manuscript. This work was supported by NIH National Cancer Institute grants R01CA136806, CA136363, and CA170354 and American Cancer Society Professorships to P.S. Moore and Y. Chang. This project used the University of Pittsburgh Cancer Institute Tissue and Research Pathology Services and Li-COR Imaging Facility, which is supported in part by NIH grant P30CA047904.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2016;1(2):e85562. doi:10.1172/jci.insight.85562.
Contributor Information
Tuna Toptan, Email: toptant@gmail.com.
Samuel A. Yousem, Email: yousemsa@upmc.edu.
Jonhan Ho, Email: hoj@upmc.edu.
Yuki Matsushima, Email: matusima-y@city.kawasaki.jp.
Laura P. Stabile, Email: stabilela@upmc.edu.
Rohit Bhargava, Email: rbhargava@mail.magee.edu.
Akihide Ryo, Email: aryo@yokohama-cu.ac.jp.
Patrick S. Moore, Email: psm9@pitt.edu.
Yuan Chang, Email: yc70@pitt.edu.
References
- 1.DeCaprio JA, Garcea RL. A cornucopia of human polyomaviruses. Nat Rev Microbiol. 2013;11(4):264–276. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White MK, Gordon J, Khalili K. The rapidly expanding family of human polyomaviruses: recent developments in understanding their life cycle and role in human pathology. PLoS Pathog. 2013;9(3):e85562. doi: 10.1371/journal.ppat.1003206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1(7712):1253–1257. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- 4.Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1(7712):1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 5.van der Meijden E, Janssens RWA, Lauber C, Bavinck JNB, Gorbalenya AE, Feltkamp MCW. Discovery of a new human polyomavirus associated with Trichodysplasia Spinulosa in an immunocompromized patient. PLoS Pathog. 2010;6(7):1–10. doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho J, et al. Human polyomavirus 7-associated pruritic rash and viremia in transplant recipients. J Infect Dis. 2014;211(10):1560–1565. doi: 10.1093/infdis/jiu524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319(5866):1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter JJ, et al. Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes. Proc Natl Acad Sci U S A. 2013;110(31):12744–12749. doi: 10.1073/pnas.1303526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo GJ, Chen CJ, Sullivan CS. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology. 2009;383(2):183–187. doi: 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Bauman Y, et al. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe. 2011;9(2):93–102. doi: 10.1016/j.chom.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Abend JR, Jiang M, Imperiale MJ. BK virus human cancer: Innocent until proven guilty. Semin Cancer Biol. 2009;19(4):252–260. doi: 10.1016/j.semcancer.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pino L, Rijo E, Nohales G, Frances A, Ubre A, Arango O. Bladder transitional cell carcinoma and BK virus in a young kidney transplant recipient. Transpl Infect Dis. 2013;15(1):E25–E27. doi: 10.1111/tid.12042. [DOI] [PubMed] [Google Scholar]
- 13.Khalili K, Del Valle L, Otte J, Weaver M, Gordon J. Human neurotropic polyomavirus, JCV, and its role in carcinogenesis. Oncogene. 2003;22(33):5181–5191. doi: 10.1038/sj.onc.1206559. [DOI] [PubMed] [Google Scholar]
- 14.Zheng H, et al. Oncogenic role of JC virus in lung cancer. J Pathol. 2007;212(3):306–315. doi: 10.1002/path.2188. [DOI] [PubMed] [Google Scholar]
- 15.Brouchet L, et al. Detection of oncogenic virus genomes and gene products in lung carcinoma. Br J Cancer. 2005;92(4):743–746. doi: 10.1038/sj.bjc.6602409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niv Y, Vilkin A, Levi Z. Patients with sporadic colorectal cancer or advanced adenomatous polyp have elevated anti-JC virus antibody titer in comparison with healthy controls: a cross-sectional study. J Clin Gastroenterol. 2010;44(7):489–494. doi: 10.1097/MCG.0b013e3181d7a347. [DOI] [PubMed] [Google Scholar]
- 17.Rollison DE, Sexton WJ, Rodriguez AR, Kang LC, Daniel R, Shah KV. Lack of BK virus DNA sequences in most transitional-cell carcinomas of the bladder. Int J Cancer. 2007;120(6):1248–1251. doi: 10.1002/ijc.22494. [DOI] [PubMed] [Google Scholar]
- 18.Weiss AF, Portmann R, Fischer H, Simon J, Zang KD. Simian virus 40-related antigens in three human meningiomas with defined chromosome loss. Proc Natl Acad Sci U S A. 1975;72(2):609–613. doi: 10.1073/pnas.72.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss AF, Zang KD, Birkmayer GD, Miller F. SV 40 related papova-viruses in human meningiomas. Acta Neuropathol. 1976;34(2):171–174. doi: 10.1007/BF00684667. [DOI] [PubMed] [Google Scholar]
- 20.Vilchez RA, Butel JS. SV40 in human brain cancers and non-Hodgkin’s lymphoma. Oncogene. 2003;22(33):5164–5172. doi: 10.1038/sj.onc.1206547. [DOI] [PubMed] [Google Scholar]
- 21.Qi F, Carbone M, Yang H, Gaudino G. Simian virus 40 transformation, malignant mesothelioma and brain tumors. Expert Rev Respir Med. 2011;5(5):683–697. doi: 10.1586/ers.11.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harlow E, Crawford LV, Pim DC, Williamson NM. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39(3):861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Li J, Schowalter RM, Jiao J, Buck CB, You J. Bromodomain protein Brd4 plays a key role in Merkel cell polyomavirus DNA replication. PLoS Pathog. 2012;8(11):e85562. doi: 10.1371/journal.ppat.1003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta T, Robles MT, Schowalter RM, Buck CB, Pipas JM. Expression of the Small T antigen of Lymphotropic Papovavirus is sufficient to transform primary mouse embryo fibroblasts. Virology. 2015;487:112–120. doi: 10.1016/j.virol.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shuda M, et al. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int J Cancer. 2009;125(6):1243–1249. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miner AG, et al. Cytokeratin 20-negative Merkel cell carcinoma is infrequently associated with the Merkel cell polyomavirus. Mod Pathol. 2015;28(4):498–504. doi: 10.1038/modpathol.2014.148. [DOI] [PubMed] [Google Scholar]
- 27.Babakir-Mina M, et al. Identification of the novel KI and WU polyomaviruses in human tonsils. J Clin Virol. 2009;46(1):75–79. doi: 10.1016/j.jcv.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Bialasiewicz S, Whiley DM, Lambert SB, Nissen MD, Sloots TP. Detection of BK, JC, WU, or KI polyomaviruses in faecal, urine, blood, cerebrospinal fluid and respiratory samples. J Clin Virol. 2009;45(3):249–54. doi: 10.1016/j.jcv.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Siebrasse EA, et al. WU polyomavirus in respiratory epithelial cells from lung transplant patient with Job syndrome. Emerg Infect Dis. 2015;21(1):103–106. doi: 10.3201/eid2101.140855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bollag B, Kilpatrick LH, Tyagarajan SK, Tevethia MJ, Frisque RJ. JC virus T′ 135, T′ 136 and T′ 165 proteins interact with cellular p107 and p130 in vivo and influence viral transformation potential. J Neurovirol. 2006;12(6):428–442. doi: 10.1080/13550280601009553. [DOI] [PubMed] [Google Scholar]
- 31.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest. 2011;121(9):3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Degener A, et al. Detection of JC and BK viral genome in specimens of HIV-1 infected subjects. New Microbiol. 1997;20(2):115–122. [PubMed] [Google Scholar]
- 33.Chen Y, et al. Asymptomatic reactivation of JC virus in patients treated with natalizumab. N Engl J Med. 2009;361(11):1067–1074. doi: 10.1056/NEJMoa0904267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mengel M, et al. Incidence of polyomavirus-nephropathy in renal allografts: influence of modern immunosuppressive drugs. Nephrol Dial Transplant. 2003;18(6):1190–1196. doi: 10.1093/ndt/gfg072. [DOI] [PubMed] [Google Scholar]
- 35.Houben R, et al. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010;84(14):7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kenan DJ, Mieczkowski PA, Burger-Calderon R, Singh HK, Nickeleit V. The oncogenic potential of BK-polyomavirus is linked to viral integration into the human genome. J Pathol. 2015;237(3):379–389. doi: 10.1002/path.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Astegiano S, et al. Prevalence and clinical impact of polyomaviruses KI and WU in lung transplant recipients. Transplant Proc. 2010;42(4):1275–1278. doi: 10.1016/j.transproceed.2010.03.107. [DOI] [PubMed] [Google Scholar]
- 38.Teramoto S, et al. Detection of KI polyomavirus and WU polyomavirus DNA by real-time polymerase chain reaction in nasopharyngeal swabs and in normal lung and lung adenocarcinoma tissues. Microbiol Immunol. 2011;55(7):525–530. doi: 10.1111/j.1348-0421.2011.00346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siebrasse EA, Nguyen NL, Willby MJ, Erdman DD, Menegus MA, Wang D. Multiorgan WU polyomavirus infection in bone marrow transplant recipient. Emerg Infect Dis. 2016;22(1):24–31. doi: 10.3201/eid2201.151384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buck CB, Phan GQ, Raiji MT, Murphy PM, McDermott DH, McBride AA. Complete genome sequence of a tenth human polyomavirus. J Virol. 2012;86(19):e85562. doi: 10.1128/JVI.01690-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shuda M, et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci U S A. 2008;105(42):16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. [on February 9, 2016];National Cancer Institute Center for Cancer Research Web site. http://home.ccr.cancer.gov/Lco/BuckLabAntibodies.htm
- 43.Feng H, et al. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS One. 2011;6(7):e85562. doi: 10.1371/journal.pone.0022468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsunaga S, et al. Wheat germ cell-free system-based production of hemagglutinin-neuraminidase glycoprotein of human parainfluenza virus type 3 for generation and characterization of monoclonal antibody. Front Microbiol. 2014;5(208):1–11. doi: 10.3389/fmicb.2014.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura K, Nozaki N, Saijo M, Kikuchi A, Ui M, Enomoto T. Identification of the nature of modification that causes the shift of DNA topoisomerase II beta to apparent higher molecular weight forms in the M phase. J Biol Chem. 1994;269(40):24523–24526. [PubMed] [Google Scholar]
- 46.Kimura K, Nozaki N, Enomoto T, Tanaka M, Kikuchi A. Analysis of M phase-specific phosphorylation of DNA topoisomerase II. J Biol Chem. 1996;271(35):21439–21445. doi: 10.1074/jbc.271.35.21439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.