Summary
MicroRNAs predominantly decrease gene expression; however, specific mRNAs are translationally upregulated in quiescent (G0) mammalian cells and immature Xenopus laevis oocytes by an FXR1a-associated microRNP (microRNA-protein complex) that lacks the microRNP repressor, GW182. Their mechanism in these conditions of decreased mTOR signaling and therefore, reduced canonical (cap-and-poly(A)-tail-mediated) translation, remains undiscovered. Our data reveal that mTOR inhibition in human THP1 cells enables microRNA-mediated activation. Activation requires shortened/no poly(A)-tail targets; polyadenylated mRNAs are partially activated upon PAIP2 overexpression, which interferes with poly(A)-bound PABP, precluding PABP-enhanced microRNA-mediated inhibition and canonical translation. Consistently, inhibition of PARN deadenylase prevents activation. P97/DAP5, a homolog of canonical translation factor, eIF4G, which lacks PABP- and cap binding complex-interacting domains, is required for activation and thereby, for the oocyte immature state. P97 interacts with 3′-UTR-binding FXR1a-associated microRNPs and with PARN, which binds mRNA 5′ caps, forming a specialized complex to translate recruited mRNAs in these altered canonical translation conditions.
Introduction
MicroRNAs are small non-coding RNAs that associate with Argonaute (AGO) proteins and target mRNAs in a sequence-specific manner to regulate their gene expression (Jonas and Izaurralde, 2015). MicroRNPs (microRNA-protein complexes) primarily cause deadenylation and translation repression through a repressor, GW182, and involve canonical translation factors (Moretti et al., 2012; Meijer et al., 2013; Jonas and Izaurralde, 2015). Our data revealed that in quiescent (G0) mammalian cells, such as in human THP1 leukemic cells, and in immature folliculated Xenopus laevis oocytes, select mRNAs are translationally activated (Vasudevan et al., 2007; Mortensen et al., 2011) by a microRNA-associated protein complex that contains AGO2 and an isoform of an RNA binding protein, Fragile-X-Mental-Retardation-Syndrome-Related protein 1a (FXR1a, FXR1a-microRNP), but lacks repressive GW182. Overexpression of FXR1a promotes translation and FXR1a-microRNP selects targets in the nucleus, such as TNFα mRNA in serum-starvation induced G0, and Myt1 mRNA in immature oocytes (Truesdell et al., 2012; Mortensen et al., 2011). Others have observed microRNA-mediated activation of TNFα and other specific mRNAs in distinct conditions (Lin et al., 2011; Tserel et al., 2011; Zhang et al., 2014; Iwasaki and Tomari, 2009). The mechanism of microRNA-mediated activation remains to be uncovered.
In G0 cells and immature oocytes, canonical translation is compromised in part due to poly(A) tail shortening, and low mechanistic/mammalian Target of Rapamycin (mTOR) kinase activity that reduces phosphorylation and increases activity of the inhibitor of the cap-binding complex, eukaryotic translation initiation factor 4E binding protein (eIF4EBP/4EBP) (Thoreen et al., 2012; Seal et al., 2005; Radford et al., 2008). Yet ongoing translation is observed (Mortensen et al., 2011; Loayza-Puch et al., 2013), suggesting that non-canonical translation mechanisms operate on specific mRNAs here and could be utilized for microRNA-mediated activation. mRNA poly(A) tails can be short in immature oocytes (Radford et al., 2008) and the deadenylase, poly(A)-specific ribonuclease, PARN (Korner et al., 1998), shows increased poly(A) tail shortening in serum-starved G0 cells (Seal et al., 2005). PARN activity is stimulated upon mRNA cap binding, which is enhanced in G0 (Seal et al., 2005). Consistently, we found that deadenylated mRNAs show microRNA-mediated activation in G0-like oocytes (Mortensen et al., 2011). Importantly, microRNAs have been shown to activate translation in Drosophila embryo extracts (Iwasaki and Tomari, 2009) and other conditions and RNAs (Zhang et al., 2014) in the absence of canonical cap and poly(A) tail. These data indicate a non-canonical translation mechanism that connects mRNAs recruited by FXR1a-microRNP with ribosomes.
Here we investigated the mechanism of translation activation by microRNAs in oocytes and human THP1 G0 cells. We find microRNA-mediated activation of targets upon reducing mTOR activity in THP1 cells, where—similar to serum-starved G0—the essential activator, FXR1, is altered, and canonical cap-dependent translation is compromised due to reduced 4EBP phosphorylation by mTOR (Loayza-Puch et al., 2013; Thoreen et al., 2012; Seal et al., 2005). Consistent with these conditions of poly(A) shortening (Seal et al., 2005; Radford et al., 2008), our data reveal that activation targets have shortened poly(A) tails. Such mRNAs avoid binding Poly(A) binding protein (PABP), which may not be required as PABP promotes canonical translation that is impaired in G0 (Thoreen et al., 2012; Seal et al., 2005), and can enhance microRNP-mediated inhibition (Moretti et al., 2012; Jonas and Izaurralde, 2015). FXR1a-microRNP interacts with Death-associated protein 5 (p97/DAP5), a homolog of canonical translation factor, eIF4G. P97 lacks eIF4G domains to interact with cap binding eIF4E, and PABP, but like eIF4G, recruits eIF3, and thereby, 40S ribosome subunits, and mediates non-canonical translation (Levy-Strumpf et al., 1997) of deadenylated mRNAs to which it is recruited by FXR1a-microRNP. Activation needs PARN, which interacts with p97 and FXR1a-microRNP, and shows increased binding to mRNA caps in G0, where the canonical cap binding complex (eIF4E-eIF4G) interaction is impaired by dephosphorylated 4EBP (Seal et al., 2005). Our data reveal miR16 and miR369-3p targets that are associated with FXR1a-microRNP and need FXR1, PARN and p97 for translation in G0. These results reveal a distinct mechanism of translation by FXR1a-microRNP, which connects specific targets with shortened poly(A) tails, and 5′ caps bound by PARN, with a specialized translation factor, p97, to promote non-canonical translation under these conditions of decreased canonical translation.
Results
Unadenylated target mRNA shows microRNA-mediated activation in immature oocytes
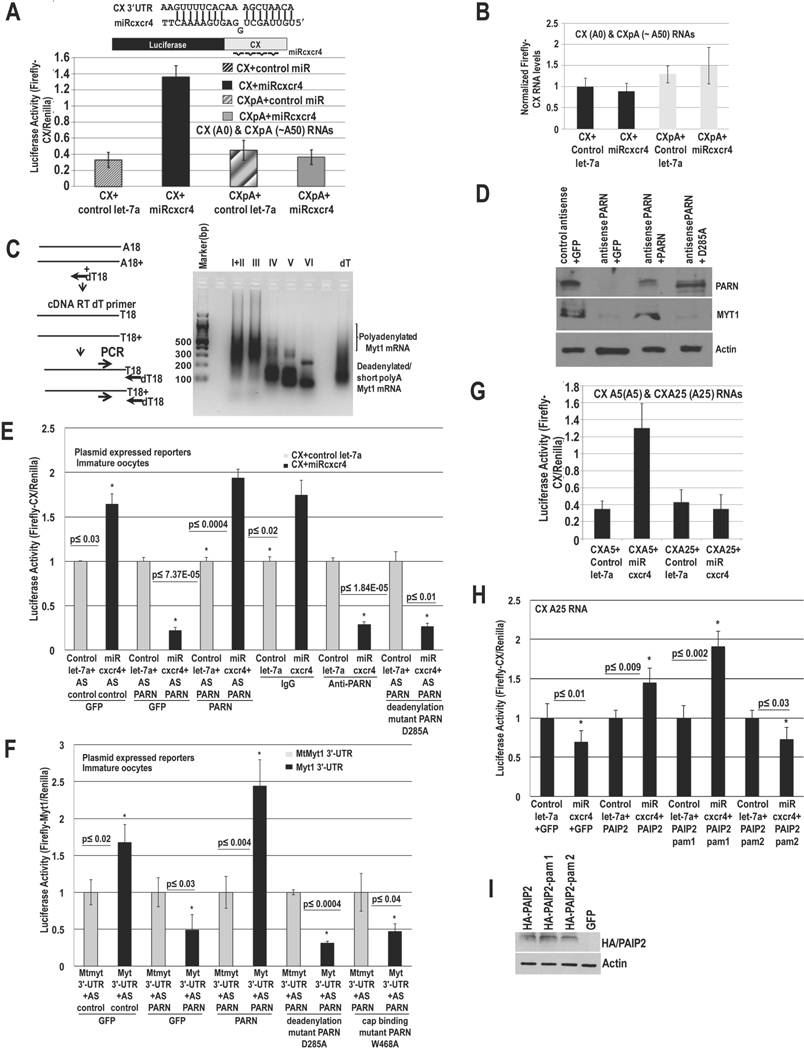
Many mRNAs display a short poly(A) tail (≤25As) in immature oocytes (Radford et al., 2008), where we observed microRNA-mediated activation with deadenylated mRNAs (Mortensen et al., 2011). We tested unadenylated and adenylated mRNAs for microRNA-mediated activation in stage IV immature oocytes. CX Firefly luciferase mRNA, with 4 target sites for a synthetic microRNA, miRcxcr4, was in vitro transcribed and injected into the nucleus as capped, unadenylated (CX A0) or capped, polyadenylated mRNA (CXpA with ~50As, Fig. S1A), along with Renilla for normalization, and miRcxcr4 or control microRNA, let-7a (Fig. 1A). The unadenylated reporter showed activation with miRcxcr4 compared to let-7a (Fig. 1A), while the polyadenylated reporter did not. RNA levels do not change (Fig. 1B). These data suggest that unadenylated mRNAs can be translationally activated by microRNAs in these conditions. Poly(A) tail (RACE-PAT) assay (SI extended procedures) of endogenous target, Myt1 mRNA, shows poly(A) tail shortening (Fig. 1C) in oocyte stages IV–VI where MYT1 protein is generated (Mortensen et al., 2011), consistent with the need for unadenylated mRNAs for activation (Fig. 1A–B).
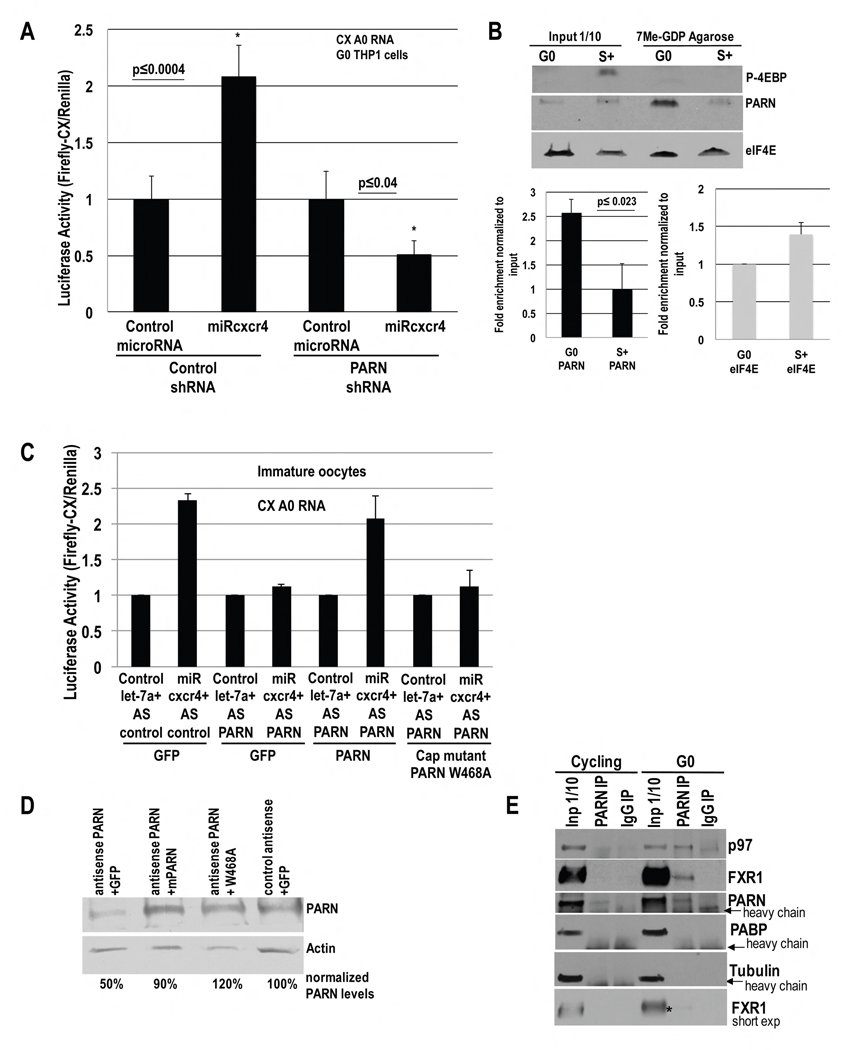
Fig. 1. MicroRNA-mediated activation requires poly(A) shortened mRNAs and PARN to avoid PABP in immature oocytes.
A. Luciferase assay of in vitro transcribed capped CX mRNAs (CX=Firefly Luciferase reporter with 4 target sites for artificial microRNA, miRcxcr4, in its 3′-UTR), without/with a poly(A) tail (CX(A0)=CX A0=no poly(A) tail, CXpA=~A50, Fig. S1A), which were injected into stage IV oocyte nuclei with Renilla mRNA and miRcxcr4 or control let-7a. B. qRT-PCR of RNAs in A. C. RACE-PAT assay of endogenous target Myt1 mRNA in immature oocyte stages; oligo dT/RNase H treated RNA=deadenylated RNA control. D.–E. Oocytes were injected with: control antisense (AS control)+GFP, PARN antisense (AS PARN)+GFP, PARN antisense+ PARN (mouse PARN cDNA insensitive to antisense against Xenopus PARN) and PARN antisense+PARN D285A deadenylase inactive mutant, followed by: D. Western blot of PARN and endogenous target MYT1, and E. these oocytes, as well as oocytes injected with control IgG or anti-PARN antibody, were injected with CX and Renilla plasmids and miRcxcr4 or let-7a, followed by Luciferase analysis. PAT assays of reporters (Fig. S1Bi–ii) and endogenous Myt1 mRNA (Fig. S1Biii–iv) reveal altered poly(A) tails; RNA levels do not correlate with translation/poly(A) changes (Fig. S1C–D). F. Oocytes were injected with antisense and rescue plasmids as noted and then with Firefly Luciferase plasmids with the 3′-UTR of endogenous miR16 target, Myt1, or with the 3′-UTR miR16 target site mutated (MtMyt1 3′-UTR) and Renilla, followed by Luciferase assay. G. CXA5 and CXA25 mRNAs (CX mRNAs with A5 and A25, Fig. S1A), Renilla mRNA and miRcxcr4 or let-7a were injected into oocytes, followed by Luciferase assay to test activation with poly(A) tails less than or greater than a PABP binding site (~20 As). H. Oocytes were injected with GFP, HA-tagged PAIP2, PAIP2-pam 1or PAIP2-pam 2 (pam 1 deleted) cDNAs and with CXA25 mRNA, Renilla and miRcxcr4 or let-7a, followed by Luciferase assay to test if PAIP2 enables activation with CXA25. I. Western blot of samples in H. Actin=loading control. Graphs=average of ≥ 3 experiments; error bars=standard error of mean (SEM) or standard deviation (A, G); p≤0.05 or indicated. See Fig. S1.
Inhibition of PARN prevents activation
PARN deadenylase is active in G0 (Seal et al., 2005) and oocytes (Korner et al., 1998) where we observe activation with unadenylated targets (Fig. 1A–B). To test whether PARN is needed for activation, we used antisense depletion or immunoneutralization with an antibody that abrogates PARN activity (Korner et al., 1998), and analyzed activation of CX reporter, injected as a CMV expressed plasmid along with Renilla and miRcxcr4 or control let-7a. PARN antisense (Fig. 1D, PARN depletion) and immunoneutralization cause oocyte maturation (Kim and Richter, 2007) and prevented activation of CX mRNA by miRcxcr4 compared to let-7a (Fig. 1E). Introducing mouse PARN mRNA (PARN, resistant to antisense against Xenopus PARN sequence) prior to antisense depletion of endogenous PARN, restored activation of CX mRNA by miRcxcr4 (Fig. 1D–E), with no related change in RNA levels (Fig. S1C).
To test whether the deadenylase activity of PARN was needed, a point mutant D285A (Ren et al., 2002) that lacks deadenylase activity was created in antisense-resistant mouse PARN cDNA. Unlike wild type PARN, introducing D285A PARN mRNA prior to depletion of endogenous PARN did not restore activation of CX mRNA by miRcxcr4 compared to let-7a (Fig. 1D–E). Since total RNA levels do not correlatively change (Fig. S1C), poly(A) tail (PAT) assays (SI Extended Procedures) were used to check if deadenylation by PARN was affecting poly(A) tail lengths of plasmid derived CX mRNA. CX mRNA poly(A) tails are longer upon PARN depletion, compared to A20 control mRNA (Fig. S1Bi lanes 5 versus 4 versus 2), or upon antibody-mediated inhibition (Fig. S1Bii lanes 12, 6 versus 11, 5), and is not rescued by D285A PARN (Fig. S1Bii lanes 13) compared to control antisense (Fig. S1Bii lane 7), or rescue of PARN antisense oocytes with wildtype PARN (Fig. S1Bii lane 9)—where poly(A) tails are short, similar to a control sample treated with oligo dT/RNAse H to remove the poly(A) tail (Fig. S1Bi lane 6, Fig. S1Bii lane 15). These results suggest that upon PARN inhibition, poly(A) tails are not shortened and microRNA-mediated activation does not occur (Fig. 1D–E), correlating with Fig. 1A, where adenylated mRNAs do not show activation.
Similar results were obtained with a Luciferase reporter with the 3′-UTR of endogenous miR16 target, Myt1 mRNA (Fig. 1F). Endogenous MYT1 protein also decreased upon PARN depletion and was restored by antisense-resistant PARN but not by D285A (Fig. 1D), without related change in RNA levels (Fig. S1D). Consistently, Myt1 poly(A) tail is longer upon PARN depletion or inhibition, compared to rescue with antisense-resistant PARN, IgG, deadenylated dT/RNAse H control, or A20 control (Fig. S1Biii lanes 3 or 4 versus 6 or 5; S1Biv lanes 2 versus 3 or 1). These data indicate that PARN is required in part, for shortening mRNA poly(A) tails to enable activation. Increased repression upon PARN depletion (Fig. 1E) may reflect long poly(A) tails (Fig. S1B) that can bind PABP and promote repressive GW182 recruitment (Moretti et al., 2012). These results indicate that activation requires PARN deadenylase.
PAIP2 enables activation of adenylated mRNAs
Poly(A) tails recruit PABP/Xenopus embryonic PAB (ePAB, PABP found in oocytes (Voeltz et al., 2001)), which enables poly(A) mediated canonical translation that is decreased in G0-like conditions (Thoreen et al., 2012). PABP also promotes repressive GW182-microRNP recruitment (Jonas and Izaurralde, 2015; Moretti et al., 2012). We tested if an mRNA with a poly(A) tail shorter than a PABP binding site (~20 As protected, 12 As binding site (Sachs et al., 1986)), would not bind PABP and would show activation, similar to unadenylated mRNAs (Fig. 1A). Translation of CX mRNA with A0 or A5 tails is upregulated in response to miRcxcr4 (Fig. 1G, S1A, CXA5, Fig. 1A, CX A0) but mRNAs with A25 tail (Fig. 1G, S1A, CXA25) and greater (Fig. 1A, CXpA, ~50As) prevent activation, likely due to binding of ePAB. ePAB is a stable protein that cannot be efficiently depleted by antisense but can be decoyed by Poly(A) binding protein Interacting Protein 2 (PAIP2), which disrupts PABP/ePAB interactions with poly(A) tail and with the cap complex associated factor, eIF4G (Yanagiya et al., 2010). Expression of PAIP2 and the pam 1 domain of PAIP2 that interferes with PABP interaction with eIF4G and poly(A), partially rescues activation of CXA25 (Figs. 1H–I, PAIP2, PAIP2-pam 1); PAIP2 with deletion of pam 1 domain (Figs. 1H–I, PAIP2-pam 2) does not rescue activation of CXA25. These data argue that poly(A) tail shortened mRNA (Fig. 1A–G, S1B–D) is needed for activation, to preclude PABP/ePAB (Fig. 1H–I), and its interactions with the canonical cap binding complex and poly(A) tail.
P97 is required for activation
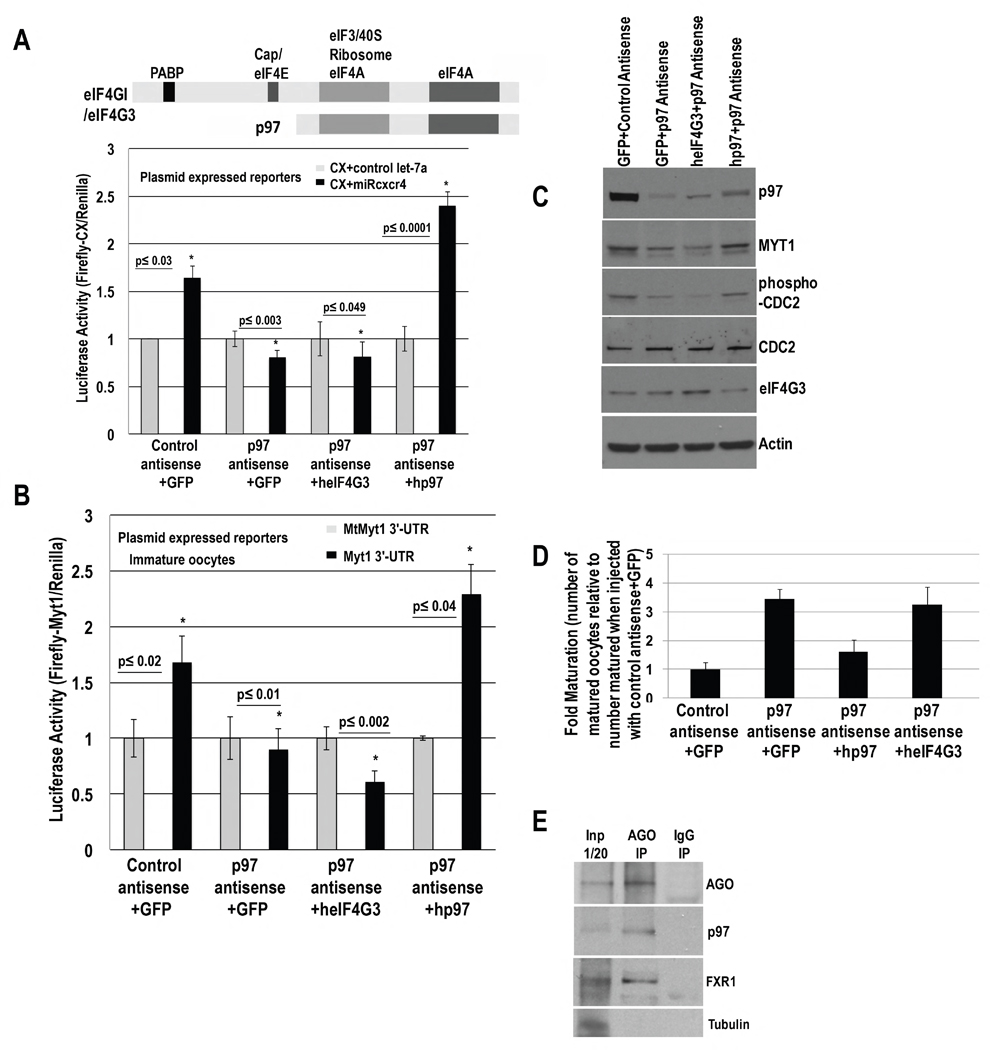
Activation would involve non-canonical factors to recruit ribosomes in these conditions of reduced canonical cap and poly(A) mediated translation. In an antisense screen against candidate factors in oocytes, antisense depletion of DAP5/p97 abrogated microRNA-mediated activation of CX reporter (Fig. 2A,C), compared to control antisense. P97 is an eIF4G homolog that lacks domains to interact with PABP and cap binding protein but recruits eIF3-40S ribosome subunits for non-canonical translation (Levy-Strumpf et al., 1997). RNA levels do not reflect the decrease in Luciferase activity upon p97 depletion (Fig. S2A). When oocytes depleted of p97 were injected with human p97 (resistant to antisense against Xenopus p97 sequence) plasmid, microRNA-mediated activation was rescued, compared to complementation with GFP or eIF4G3, another eIF4G homolog (Fig. 2A, C). Similar results were obtained with a Luciferase reporter with the 3′-UTR of endogenous miR16 target, Myt1 mRNA (Fig. 2B). These data indicate that p97 mediates microRNA-directed activation in oocytes.
Fig. 2. P97 mediates activation and is involved in the immature oocyte state.
P97, eIF4GI, eIF4G3 (eIF4G3/eIF4GII, distinct from eIF4G2/DAP5/p97) domains. Oocytes were injected with control or p97 antisense and GFP, human p97 or eIF4G3 rescue cDNA plasmids for 6–7 hr and then with: A. CX and Renilla plasmids, and miRcxcr4 or control let-7a or B. Myt1 3′-UTR or mtMyt1 3′-UTR Firefly Luciferase reporters and Renilla plasmids. C. Western blot of p97, MYT1, and immaturity marker, phospho-CDC2 (MYT1 phosphorylates CDC2), upon p97 depletion and rescue in A. 57% and 61% decrease in MYT1, and 65% and 64% decrease in phospho-CDC2, were observed with p97 depletion and upon lack of rescue with eIF4G3; 94% rescue of MYT1 and 84% rescue of phospho-CDC2 was observed with human p97 compared to control antisense. Actin=loading control. RNA levels do not change significantly (Fig. S2A–B). D. Maturation upon p97 depletion and rescue (samples in A) by scoring for Germinal Vesicle Breakdown (GVBD). Y axis=fold maturation, which is the number of matured oocytes in samples injected with: p97 antisense+GFP, p97 antisense+human p97 (hp97), or p97 antisense+human eIF4G3 (heIF4G3), compared to number of oocytes matured when injected with control antisense+GFP (set to 1). E. Immunoprecipitation (IP) with AGO antibody and control IgG from RNAse treated oocyte extracts; Western blot for p97, FXR1 and AGO; Tubulin=negative control. Graphs=average of ≥ 3 experiments; error bars=SEM; p≤0.05 or indicated. See Fig. S2.
P97 is involved in oocyte immaturity
MicroRNA-mediated activation regulates MYT1 expression, which is required for the immature oocyte state (Mortensen et al., 2011). Since p97 is involved in microRNA-mediated activation, it would regulate the immature state. We analyzed maturation by scoring for germinal vesicle breakdown (GVBD) in oocytes injected with control or p97 antisense, and complemented with GFP, human p97 or eIF4G3 plasmids. We find increased maturation in p97 depleted oocytes; immaturity is restored upon rescue with human p97 but not with GFP or eIF4G3 (Fig. 2D). Consistently, endogenous MYT1 decreases upon p97 depletion (Fig. 2C) without related change in RNA levels (Fig. S2B), and results in decrease of phospho-CDC2, a marker of immaturity (Mortensen et al., 2011). MYT1 and phospho-CDC2 levels are rescued by human p97 (Fig. 2C), indicating that p97 contributes to the immature oocyte state.
P97 interacts with AGO
To test whether the microRNP interacts with p97, AGO immunoprecipitation analysis was performed in RNase treated extracts (Truesdell et al., 2012). AGO antibody co-immunoprecipitates p97 and FXR1 (Fig. 2E), indicating that FXR1-microRNP can interact with p97.
Deadenylated mRNAs exhibit microRNA-mediated activation in G0 THP1 cells
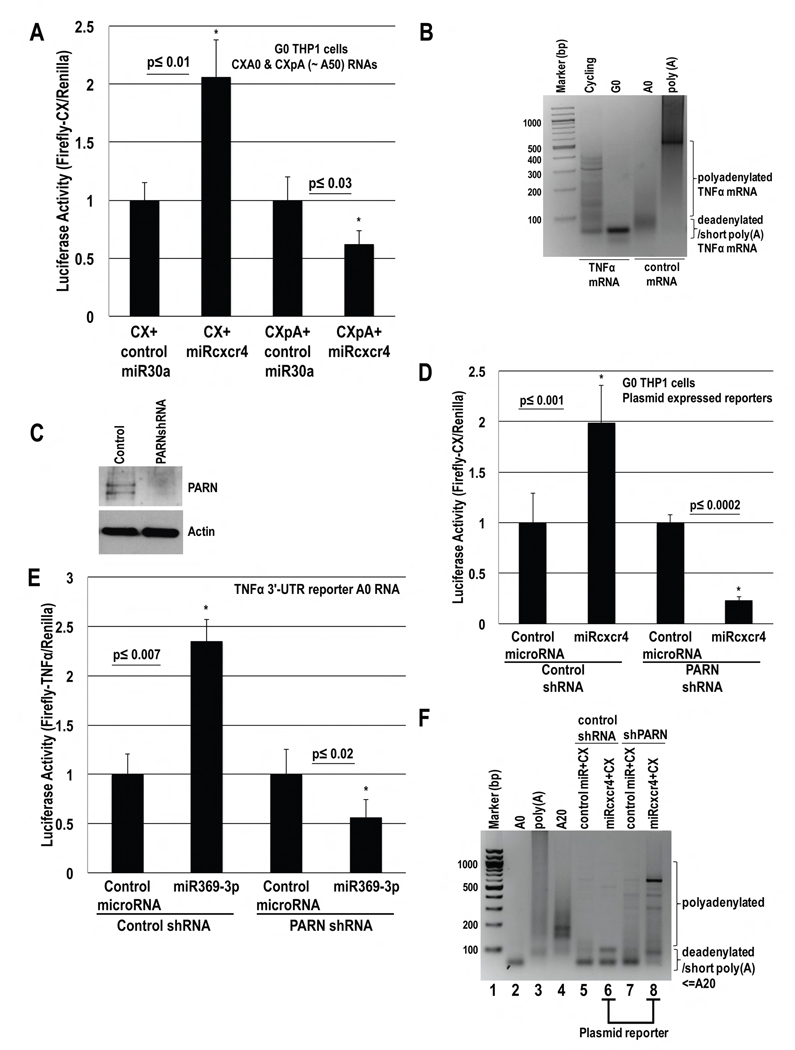
We tested capped, unadenylated CXA0 and polyadenylated CXpA (~50As) mRNAs (blocked at the 3′-end with cordycepin to prevent degradation) for activation in human THP1 G0 cells (Fig. 3A). These reporters, Renilla, and control miR30a or miRcxcr4, were nucleofected into THP1 cells, which were serum-starved for 42 hr prior to Luciferase analysis. Only unadenylated mRNA showed activation with miRcxcr4 compared to control miR30a (Fig. 3A). PAT assay revealed that endogenous activation target TNFα mRNA, has a shorter poly(A) tail in THP1 G0 compared to cycling cells (Fig. 3B), indicating that targets of microRNA-mediated activation require shortened poly(A) tails in THP1 G0 as in oocytes (Figs. 1A, S1B).
Fig. 3. MicroRNA-mediated activation requires poly(A) shortened mRNAs and PARN in human THP1 G0 Cells.
A. In vitro transcribed capped CX mRNAs, without/with a poly(A) tail (CXA0, CXpA, Fig. S1A), & cordycepin to protect 3′ ends, were nucleofected with Renilla plasmid, and miRcxcr4 or control miR30a, into THP1 cells, and then serum-starved for 42 hr before Luciferase analysis. B. PAT assay of TNFα mRNA in cycling and G0 THP1 cells, indicates shortened poly(A) tails in G0 compared to control A0 and poly(A) mRNAs. C.–E. THP1 cells nucleofected with control or PARN shRNA plasmids, along with: D. CX and Renilla plasmids and doxycycline inducible miRcxcr4 or miR30a plasmids, or E. Firefly Luciferase A0 RNA with the 3′-UTR of TNFα, Renilla plasmid, and miR369-3p or control miR30a. After 30 hr of shRNA expression, cells were serum-starved for 42 hr followed by: C. Western blot for PARN, D.–E. Luciferase assay. F. PAT assay of CX mRNA (samples in D) shows shortened poly(A) tail (<A20) in control shRNA cells (lane 6), and extra forms greater than 20As upon shPARN/PARN depletion (lane 8), compared to control A0, A20 and poly(A) mRNAs (lanes 2–4). Poly(A) forms are more evident in the miRcxcr4 sample (lane 8) than in the control microRNA sample. PAT assay of endogenous TNFα and CD209 (Fig. S3Di–ii; Western, RNA levels Fig. 7Fi, S3C, S7Gi), shows poly(A) tails upon PARN depletion. Actin=loading control. Graphs=average of ≥ 3 experiments; error bars=SEM; p≤0.05 or indicated. See Fig. S3.
PARN knockdown prevents activation
To test whether PARN is needed for activation in G0 THP1 cells as in oocytes (Fig. 1D–F), control or PARN shRNA plasmids, CX Firefly Luciferase and Renilla plasmids as well as doxycycline inducible miRcxcr4 or control miR30a plasmids were nucleofected into THP1 cells. After 2 days of shRNA expression and PARN depletion (Fig. 3C), microRNAs were induced with doxycycline and cells were serum-starved for 2 days to induce G0. While control shRNA G0 cells show activation of CX reporter by miRcxcr4 compared to miR30a, PARN depleted cells do not (Fig. 3D); this effect is not due to RNA levels (Fig. S3A). Similar loss of activation (Fig. 3E) is observed with a Luciferase reporter with the 3′-UTR of miR369-3p target TNFα (Vasudevan et al., 2007), and with endogenous TNFα protein (Fig. 7Fi) upon PARN depletion, with no change in RNA levels (Fig. S3B–C).
Since loss of activation is observed with adenylated mRNAs (Fig. 3A), PAT assay was used to uncover if loss of deadenylation by PARN led to long poly(A) tails on plasmid derived CX and endogenous TNFα mRNAs. Consistent with oocyte results (Fig. 1D–F, S1B), PARN knockdown G0 cells show longer poly(A) tails on CX mRNA (Fig. 3F lanes 8)—compared to control shRNA cells (Fig. 3F lane 6) that show poly(A) tails shorter than control A20 mRNA (Fig. 3F lane 4); this effect is not due to RNA levels (Fig. S3A). Similar results were observed with endogenous TNFα mRNA, with extended poly(A) tails upon PARN depletion (Fig. S3Di lanes 6 versus 4; Fig. S3C RNA levels do not change). These results indicate that activation requires PARN in THP1 G0 cells.
P97 interacts with FXR1 and AGO2 in THP1 cells
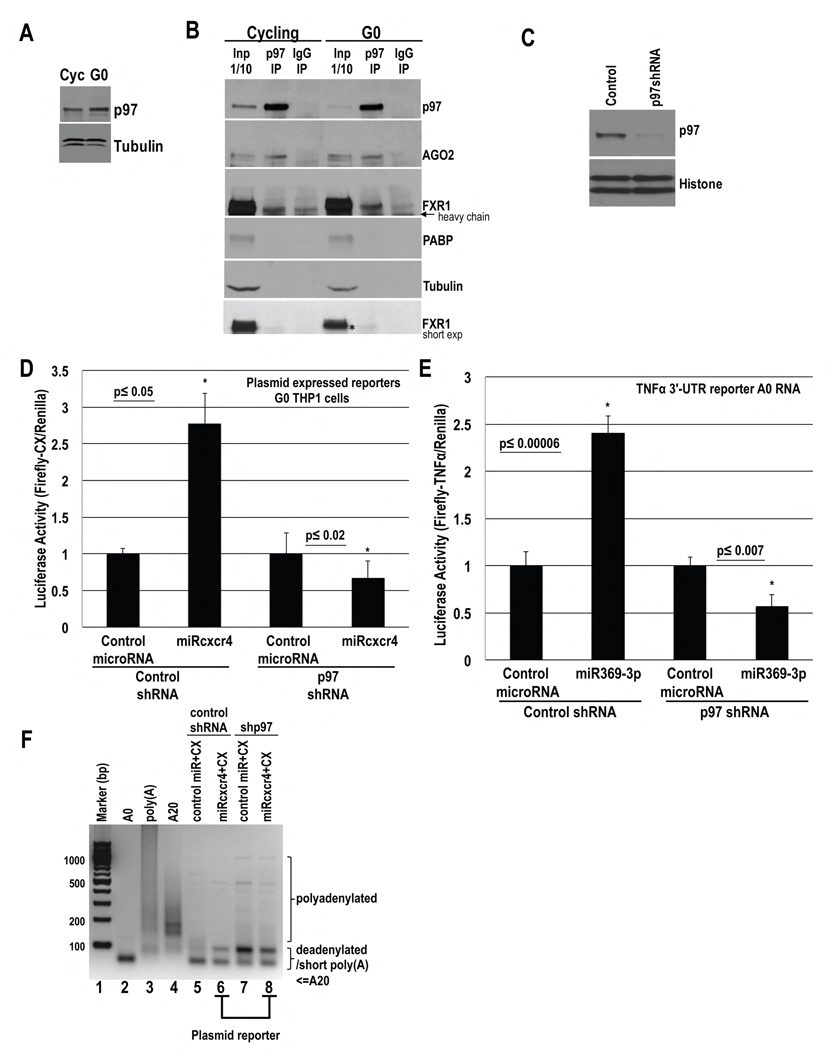
p97 is present in THP1 (Fig. 4A). To test whether p97 interacts with FXR1a-microRNP in THP1 cells, we analyzed p97 immunoprecipitates from in vivo formaldehyde-crosslinked, RNase-treated extracts (Truesdell et al., 2012). P97 interacts with AGO2 and FXR1 (Fig. 4B), but not PABP that associates with repressive GW182-microRNPs and is involved in canonical translation that is reduced in G0. These data indicate that FXR1a-microRNP may promote translation of associated mRNAs via interaction with the non-canonical translation factor, p97.
Fig. 4. P97 interacts with FXR1 and AGO2 and is required for activation.
A. Western blot of p97 in serum-grown (Cycling, Cyc) and 2 day serum-starved (G0) THP1 cells. B. IP of p97 and control IgG from in vivo crosslinked, pre-cleared, RNase treated extracts followed by Western blot of p97, AGO2 and FXR1. PABP, Tubulin=negative controls.* marks FXR1 band immunoprecipitating with p97. C.–E. THP1 cells were nucleofected with control or p97 shRNA plasmids and D. CX and Renilla plasmids, and miRcxcr4 or miR30a, or E. Firefly Luciferase A0 mRNA with the 3′-UTR of TNFα, Renilla plasmid and miR30a or miR369-3p. After 30 hr, cells were serum-starved for 2 days followed by C. Western blot of p97 and D.–E. Luciferase assay. F. PAT assay of CX mRNA (samples in D) with few faint higher bands upon shp97/p97 depletion (lane 8), compared to control cells (lane 6). Tubulin, Histone H3=loading control. Graphs=average of 3 experiments; error bars=SEM; p≤0.05 or indicated. See Fig. S4.
P97 is required for activation in THP1 G0 Cells
To test whether activation requires p97, control or p97 shRNA plasmids, CX Firefly Luciferase and Renilla plasmids, as well as doxycycline inducible miRcxcr4 or control miR30a plasmids were nucleofected into THP1 cells. After 2 days of shRNA expression and p97 depletion (Fig. 4C), microRNAs were induced with doxycycline and cells were serum-starved for 2 days. Cells treated with control shRNA show activation of CX mRNA by miRcxcr4 compared to control miR30a (Fig. 4C–D). P97 depletion resulted in loss of CX mRNA activation by miRcxcr4 (Fig. 4C–D), without RNA level changes (Fig. S4A), indicating that p97 is required for activation of CX mRNA. Similar activation was observed with control shRNA, and not with p97 shRNA, with a Luciferase reporter with the TNFα 3′-UTR in the presence of miR369-3p (Fig. 4E), as well as with endogenous TNFα protein (Fig. 7Fi), without RNA level changes (Fig. S4B–C). These data suggest that activation in G0 requires p97.
PARN interacts with p97 and FXR1, and shows increased binding to the cap in G0, which is required for activation
If PARN is only needed for deadenylation (Fig. 1D–F, S1B, 3C–F, S3D), then A0 mRNA lacking a poly(A) tail would show activation in PARN depleted cells. If PARN has an additional role in activation apart from deadenylation, then A0 mRNA will not show activation in PARN depleted cells because the other function is still absent.
We tested A0 CX mRNA in PARN depleted G0 cells: while control shRNA treated cells show activation with A0 CX mRNA, PARN depleted cells do not show activation with A0 CX mRNA by miRcxcr4 (Fig. 5A), with no change in RNA levels (Fig. S5A). This was also observed with an A0 Luciferase reporter with the TNFα 3′-UTR (Fig. 3E). These data suggest that PARN is needed in activation, for an additional role apart from mRNA deadenylation. PARN binding to mRNA caps increases in G0 as the canonical cap complex is inhibited by dephosphorylated 4EBP (Seal et al., 2005). PARN may affect activation via increased cap binding in G0.
Fig. 5. PARN interacts with p97 and FXR1, and shows increased binding to the cap in G0, which is required for activation.
A. Luciferase activity in THP1 cells nucleofected with CX A0 mRNA, PARN or control shRNA and Renilla plasmids, and miRcxcr4 or miR30a, for 30 hr, followed by serum-starvation for 42 hr. B. Cap column purification and Western blot of PARN, eIF4E and phospho-4EBP in G0 and serum-grown (S+) cells. PARN and eIF4E enrichment on cap column graphed. C. Oocytes were injected as noted with PARN antisense, antisense resistant PARN or W468A cap binding PARN mutant; then injected with CX A0 mRNA, Renilla, and let-7a or miRcxcr4, followed by Luciferase assay. D. Western blot of PARN in samples in C. E. IP of PARN and control IgG from in vivo crosslinked, pre-cleared, RNase treated THP1 extracts followed by Western blot of p97, FXR1 and PARN. PABP, Tubulin=negative controls. * marks FXR1 band immunoprecipitating with PARN. Graphs=average of ≥ 3 experiments; error bars=SEM; p≤0.05 or indicated. See Fig. S5.
Purification over cap column (7-Methyl GDP agarose) revealed a 2.5-fold increase in PARN binding to the cap column in G0 compared to serum-grown cells (Fig. 5B, Western blot & quantitation) while eIF4E binding decreased (25%), consistent with previous studies (Seal et al., 2005). To test whether cap binding by PARN is required for activation, we tested CX A0 mRNA with a cap binding defective PARN mutant, W468A (Nilsson et al., 2007), in oocytes depleted of endogenous PARN. Wildtype PARN rescues activation of CX A0, but W468A failed to rescue activation in PARN depleted oocytes (Fig. 5C–D), implicating a role for cap binding by PARN for activation. Similar results were obtained with plasmid expressed, Luciferase reporter with 3′-UTR of endogenous miR16 target, Myt1 (Fig. 1F, W468A), indicating that cap binding by PARN is required on plasmid derived, endogenously transcribed, target mRNA and not just synthetic CX A0 RNA. These data indicate that in addition to its deadenylation role (Fig. 1D–F, D285A, 3C–F), cap binding by PARN is required for activation.
mRNA cap binding by PARN could enable access for p97, recruited by the 3′-UTR binding FXR1a-microRNP (Fig. 4B)—to the 5′ end, where p97 can initiate non-canonical translation. Consistently, p97 and FXR1a co-immunoprecipitate with PARN from in vivo crosslinked, RNase treated extracts (Fig. 5E). These data indicate that in G0, where the cap binding complex is inhibited by dephosphorylated 4EBP, PARN binds mRNA 5′ caps and interacts with p97, associated with 3′-UTR binding FXR1a-microRNP.
Inhibition of mTOR in serum-grown cells leads to microRNA-mediated activation
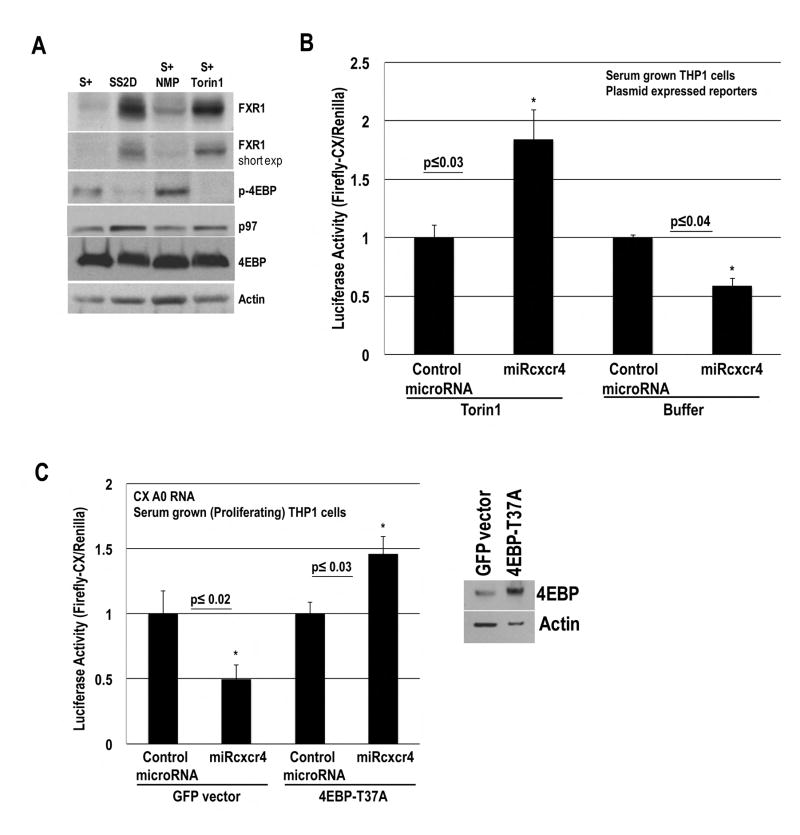
To investigate whether low mTOR activity, observed in G0 (Thoreen et al., 2012; Seal et al., 2005), enables microRNA-mediated activation, we inhibited mTOR with the inhibitor, Torin1 (Thoreen et al., 2012), in serum-grown THP1 cells, where microRNA-mediated repression is observed. CX Firefly Luciferase and Renilla plasmids, along with doxycycline inducible control miR30a or miRcxcr4 plasmids, were nucleofected into THP1 cells. The microRNAs were induced with doxycycline, and the cells were treated with buffer (NMP) or Torin1 in serum-media. Western blot shows that Torin1 compared to buffer treated serum-grown cells exhibit dephosphorylation of 4EBP and increased FXR1, similar to 2 day serum-starved G0 cells (Fig. 6A) and immature oocytes (Fig. S6D), where activation is observed (these changes are not seen in the first 18–24 h of serum-starvation or in mature oocytes, where activation is not observed (Vasudevan et al., 2007; Mortensen et al., 2011)). FXR1a overexpression leads to microRNA-mediated activation in low density, serum-grown cells (Truesdell et al., 2012), indicating that Torin1 treated serum-grown cells could show activation. Serum-grown cells treated with buffer show the expected repression of CX translation in response to miRcxcr4 (Fig. 6B). In contrast, serum-grown cells treated with Torin1, reveal 1.8 fold activation of CX translation in response to miRcxcr4, (Fig. 6B), with no increase in RNA levels (Fig. S6B).
Fig. 6. Inhibition of mTOR-mediated phosphorylation of 4EBP leads to activation.
A. Western blot of FXR1, 4EBP, phospho-4EBP, p97 and Actin in serum-grown, 2 day serum-starved or serum-grown cells treated with NMP buffer or Torin1. 9 fold and 4 fold increase in FXR1 are observed in serum-starved G0 and in Torin1-treated cells, with 4 fold decrease in phospho-4EBP in both conditions, compared to serum-grown or serum-grown+NMP buffer treated cells. B. Luciferase activity of CX plasmid, nucleofected with Renilla and inducible miRcxcr4 or miR30a plasmids in THP1 serum-grown cells, treated with doxycycline to induce microRNAs and with NMP buffer or Torin1. C. Luciferase activity of CX A0 mRNA, nucleofected with Renilla plasmid, miRcxcr4 or miR30a, and GFP or eIF4EBP-T37A plasmids in THP1 serum-grown cells. 4EBP Western blot. Graphs=average of ≥ 3 experiments; error bars=SEM; p≤0.05 or indicated. See Fig. S6.
To validate that the effects of mTOR inhibition (G0, Torin1) on activation are through loss of mTOR directed phosphorylation and inhibition of 4EBP, we tested a constitutively dephosphorylated mutant 4EBP, 4EBP-T37A (similar to the form in G0 (Gingras et al., 1999)). If mTOR-inhibited G0 and Torin1 conditions mediate activation by dephosphorylated 4EBP, then 4EBP-T37A should mimic the dephosphorylated form of G0 and promote activation even in serum grown conditions. 4EBP-T37A or GFP control, along with CX Firefly mRNA, Renilla, and miR30a or miRcxcr4 were nucleofected into serum-grown cells. Similar to Torin1 treated serum-grown cells, expression of 4EBP-T37A—but not GFP—in serum-grown cells lead to 1.47 fold activation of CX reporter in the presence of miRcxcr4 (Fig. 6C), with no increase in RNA levels (Fig. S6C). These data suggest that low mTOR activity in THP1 G0 and immature oocytes, mediates microRNA-directed activation in part via dephosphorylated 4EBP—which leads to decreased canonical translation as well as increased cap binding and deadenylation by PARN—and enables non-canonical translation of mRNAs recruited by FXR1a-microRNP that associates with PARN and p97.
Identification of activation targets in G0 THP1 cells
To identify activation targets, we conducted proteomic analysis by Tandem-Mass Tag (TMT) mass spectrometry (SI extended procedures, Table S1) in G0 cells with and without FXR1 depletion. FXR1 knockdown was used in order to globally identify activation targets of more than one microRNA—since FXR1a is required for activation by FXR1a-microRNP in G0 (Vasudevan et al., 2007) and does not associate with repressive GW182 in these cells (Fig. S5D–E). Stable inducible FXR1 and control shRNA THP1 cell lines were used to deplete FXR1 (Fig. 7Ai) and identify targets with decreased protein levels (Fig. S7Ai, Tables S1, S2, S7), which included immune genes (CD209, miR16 target, Fig. 7) and other categories (Table S3, Fig. S7Aii).
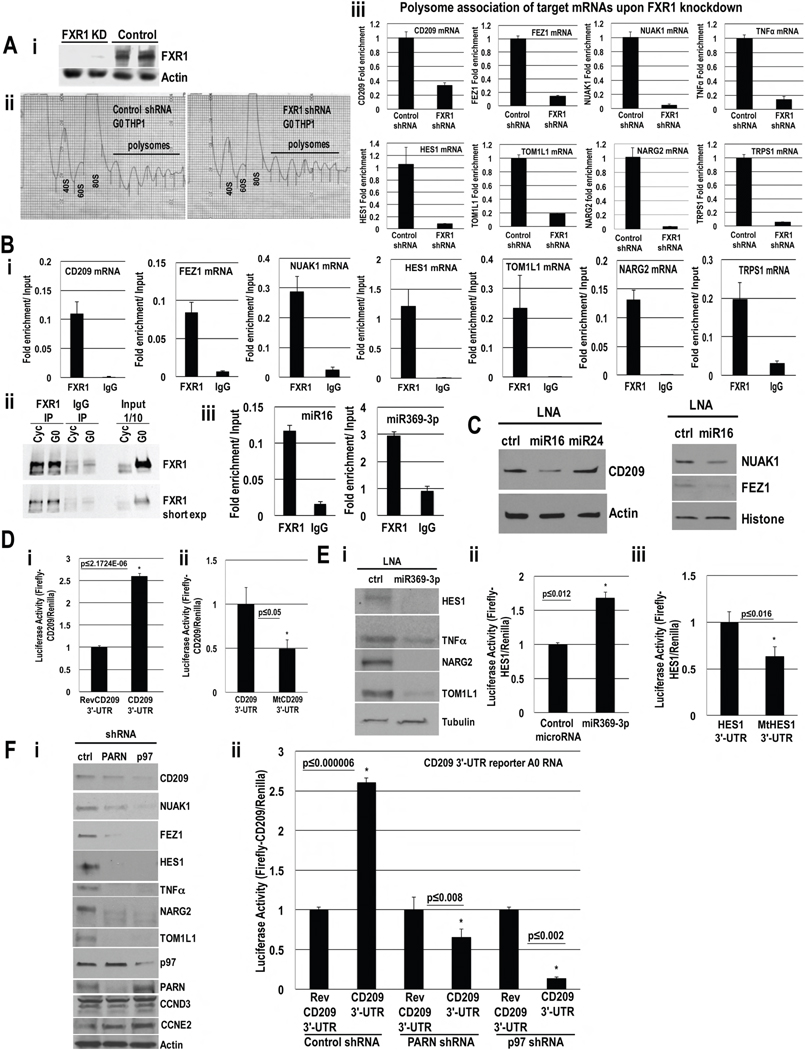
Fig. 7. Identification of activation targets in THP1 G0 Cells.
A. i. Western blot of FXR1 in control or FXR1 shRNA cells. ii. Polysome profiles of control and FXR1 shRNA G0 cells. iii. qRT-PCR of polysomal fraction mRNAs to test candidates (Table S7) for polysome association upon FXR1 knockdown; no decrease in total RNA levels (Fig. S7C). B. Associated RNAs in FXR1 and IgG IP relative to input in G0 cells of i. miR16 (Table S4) and miR369-3p (Table S5) targets. ii. Western blot of FXR1 IP. iii. miR16 and miR369-3p association with FXR1; negative controls, Fig. S7Di. C. Western blot of miR16 targets (Table S4, S7, Fig. S7B) upon miR16 inhibition with an LNA inhibitor or control LNAs in G0. RNA levels do not decrease (Fig. S7E). D. Luciferase activity in G0 cells nucleofected with Renilla plasmid, Firefly Luciferase A0 mRNAs with: CD209 3′-UTR and i. reverse CD209 3′-UTR (REV) ii. miR16 target site mutated, mtCD209 3′-UTR, and serum-starved for 42 hr. RNA levels do not change (Fig. S7Fi). E. i. Western blot of miR369-3p targets (Table S5, S7, Fig. S7B) upon miR369-3p inhibition with an LNA inhibitor or control LNA. RNA levels do not change (Fig. S7E). Luciferase activity in G0 cells nucleofected with Renilla plasmid, Firefly Luciferase A0 mRNAs with HES1 3′-UTR and ii. miR369-3p or control miR30a, iii. miR369-3p target site mutated, mtHES1 3′-UTR, and serum-starved for 42 hr. RNA levels do not change (Fig. S7Fii). F. i. Western blot of miR16 and miR369-3p activation targets and negative controls (CCND3 (top band), CCNE2) in G0 with control, PARN and p97 depletion. RNA levels do not change (Fig. S3C, S4B, S7Gi). ii. Luciferase activity in G0 cells nucleofected with Firefly Luciferase A0 mRNAs with CD209 3′-UTR or reverse CD209 3′-UTR (REV), Renilla and control, PARN or p97 shRNA plasmids for 30 hr, and serum-starved for 42 hr. RNA levels do not change (Fig. S7Gii). Actin, Histone, Tubulin=loading controls. Graphs=average of ≥ 3 experiments; error bars=SEM; p≤0.05 or indicated. See Fig. S7, Tables S1–S7.
Since miR16 is present in the nucleus where activation targets are recruited (Truesdell et al., 2012) and associates with FXR1 (Fig. 7Bii–iii), miR16 may mediate activation, apart from repression with GW182-microRNPs. As a second screen, top candidates that decreased upon FXR1 knockdown (Fig. S7A–B, Table S1, S2, S7), which are predicted by TargetScan and MiRror programs (Friedman et al., 2009; Friedman and Linial, 2013) as miR16 targets (Table S4, S7), were selected. Proteomic differences could be due to protein stability or translation or RNA levels. As a third screen, to check for regulation at the translation level, the candidates were tested for decreased polysome association by qRT-PCR in polysomal fractions from polysome profiles of FXR1 knockdown compared to control shRNA cells (Fig. 7Aii). The targets show decreased polysome association upon FXR1 knockdown (Fig. 7Aiii), without decrease in total RNA levels (Fig. S7C, some RNA levels increase possibly due to indirect stabilization when not translated upon FXR1 knockdown) and thus require FXR1 for polysome association. Proteomic data was also normalized for RNA levels, identified by microarray, to provide normalized protein data. Top targets such as CD209, FEZ1 and NARG2 (Table S7), are affected at the protein level after normalization for RNA levels (Table S2), and along with their need for FXR1 for polysome association (Fig. 7Aiii) indicate that FXR1 mediates their translation. MicroRNA target candidates with known expression in G0—TNFα (Vasudevan et al., 2007), HES1 (Coller et al., 2006), and NUAK1 (Sun et al., 2013) (Table S7)—that are secreted (TNFα) and thus not analyzed as the analysis excluded extracellular media, or are not resolved by mass spectrometry that is limited to ~8000 genes, also require FXR1 for polysome association (Fig. 7Aiii, S7C). Fourth, FXR1a-microRNP recruitment of targets (Truesdell et al., 2012), directs these mRNAs for activation while targets for repression are not recruited by FXR1a. Thus, to rule out candidates that decreased upon FXR1 knockdown due to indirect effects, the targets were screened for association with FXR1 (Fig. 7Bi–ii), and with AGO2 (Fig. S7Diii–iv, Table S7).
Candidate targets of activation were validated in 3 ways (Table S7, SI extended procedures). First, the requirement for the targeting microRNA, miR16, for expression was tested using an LNA inhibitor against miR16. Western blots reveal that the targets (CD209, FEZ1, NUAK1) decreased upon miR16 inhibition (Fig. 7C) compared to control inhibitors, with no decrease in RNA levels (Fig. S7E, some RNA levels increase possibly due to indirect stabilization when not translated due to LNA inhibition). Second, 3′-UTR Luciferase reporters of miR16 target CD209 were tested. CD209 3′-UTR reporter showed activation (Fig. 7D) with no change in RNA levels (Fig. S7Fi), compared to reporters with reversed 3′-UTR or mutations in the 3′-UTR miR16 target site, indicating that the miR16 target site of the mRNA was needed for activation by endogenous miR16. Third, targets were tested for their requirement for p97 and PARN by Western blot of endogenous proteins and Luciferase assay of CD209 3′-UTR reporter. P97 and PARN depletion decreased endogenous target levels (Fig. 7Fi) and Luciferase activity of CD209 3′-UTR reporter (Fig. 7Fii), but not RNA levels (Fig. S7Gi–ii), indicating that the targets need p97 and PARN for their translation.
To check activation by a second microRNA, we tested predicted targets of miR369-3p from FXR1-regulated proteins (Tables S1, S5, S7), as miR369-3p associates with FXR1 (Fig. 7Biii) and translates TNFα in G0 (Vasudevan et al., 2007). MiR369-3p candidate mRNAs—including TNFα, mass spectrometry-identified TOM1L1 and NARG2 (Table S5), and HES1, a G0-expressed candidate (Coller et al., 2006)—associate with FXR1 (Fig. 7Bi) and need FXR1 for polysome association (Fig. 7Aiii). Decreased target protein is observed with miR369-3p LNA inhibitor (Fig. 7Ei), with no decrease in RNA levels (Fig. S7E), validating that miR369-3p is needed for their translation. HES1 3′-UTR reporter assay (Fig. 7Eii–iii) revealed the need for an intact miR369-3p target site for activation, with no change in RNA levels (Fig. S7Fii). PARN and p97 depletion decreased TNFα 3′-UTR Luciferase activity (Fig. 3E, 4E) but not RNA levels (Fig. S3B, S4C), and decreased endogenous target proteins (Fig. 7Fi) but not RNA levels (Fig. S3C, S4B, S7Gi)—revealing that the targets need PARN and p97 for their translation.
Discussion
We previously showed that FXR1a-microRNP activates translation of specific mRNAs in G0 mammalian cells and G0-like Xenopus laevis immature oocytes (Truesdell et al., 2012). These conditions decrease canonical translation; yet specific mRNAs are translated (Loayza-Puch et al., 2013; Mortensen et al., 2011; Radford et al., 2008), indicating that activation involves a non-canonical mechanism to connect specific mRNAs with ribosomes. Our studies reveal the mechanism of microRNA-mediated activation, where poly(A) shortened mRNAs are recruited by FXR1a-microRNPs for non-canonical translation, via PARN and p97, in distinct, low mTOR activity conditions, where canonical translation is decreased.
The poly(A) tail is an enhancer of translation initiation; but unadenylated mRNAs such as Histone mRNAs (Marzluff et al., 2008) and viral RNAs (HCV (Jopling et al., 2005) are translated. We find unadenylated mRNA is required for activation by microRNAs (Figs. 1A–C, 3A–B) in these conditions where canonical cap and poly(A) tail mediated translation is inhibited by dephosphorylated 4EBP. Consistently, microRNAs have been shown to activate translation of unadenylated mRNAs in the absence of canonical cap and poly(A) tail roles (Iwasaki and Tomari, 2009; Zhang et al., 2014).
mRNAs with tails longer than a PABP site (Sachs et al., 1986) are not stimulated by microRNAs (Figs. 1A, 1G, 3A). The poly(A) tail recruits PABP, which promotes canonical translation that is reduced by dephosphorylated 4EBP in these conditions (Thoreen et al., 2012; Seal et al., 2005). PABP also enhances mRNA interactions with repressive GW182 (Jonas and Izaurralde, 2015; Beilharz et al., 2009; Moretti et al., 2012). Consistently, an endogenous activation target, TNFα, has a shortened poly(A) tail in G0 (Fig. 3B), likely to avoid PABP; accordingly, TNFα mRNA does not associate with PABP in G0 (Fig. S5B–C). Spermatid mRNAs are translated upon PABP inhibition by PAIP2 (Yanagiya et al., 2010), which disrupts PABP interactions with poly(A) tail and eIF4G. Consistently, activation of adenylated mRNAs is partially rescued by PAIP2 expression (Fig. 1H–I). These data indicate that activation selects mRNAs that avoid PABP—preventing PABP-mediated canonical translation that is inhibited in these conditions, as well as PABP-enhanced repressive GW182 effects—and are recruited instead by an FXR1a-microRNP that lacks GW182 in these cells (Fig. S5D–E).
Canonical translation is reduced here in part, due to dephosphorylated 4EBP that interferes with interaction of the cap binding factor, eIF4E with eIF4G (Thoreen et al., 2012), and due to poly(A) shortening by PARN (Figs. 1D–F, S1B, 3A–F, S3D) (Seal et al., 2005; Radford et al., 2008), which excludes PABP (Figs. 1G–I, S5B–C) and its interaction with eIF4G; eIF4G binds eIF3, which recruits 40S ribosome subunits. Activation would need non-canonical translation factors to recruit ribosomes to specific mRNAs—to replace the impaired cap binding eIF4E mediated recruitment of eIF4G that brings in eIF3–ribosomes. P97, a homolog of canonical eIF4G, can recruit eIF3–40S subunits but lacks eIF4G domains that interact with eIF4E and PABP (Levy-Strumpf et al., 1997) and thus causes non-canonical translation of such poly(A)-shortened mRNAs. P97 is required for activation (Fig. 2A–B, 4C–E) and thereby, for oocyte immaturity (Fig. 2C–D), revealing a physiological role for this mechanism. P97 associates with AGO and FXR1 (Fig. 2E, 4B), indicating that FXR1a-microRNP may connect its recruited targets to 40S ribosome subunits via interactions with p97 and thus promote translation.
PARN deadenylase is active in G0 (Seal et al., 2005) and oocytes (Korner et al., 1998) where activation needs unadenylated targets (Fig. 1A–C, 3A–B). Consistently, PARN deadenylase activity is needed (Fig. 1D–F, D285A) and PARN depletion leads to long poly(A) tails (Fig. S1B, 3F, S3D) that do not show activation (Fig. 1A, 3A). In THP1 G0, poly(A) shortened mRNAs are not decayed immediately and are translated (Fig. 3, S3), as in immature oocytes, where deadenylated mRNAs are stable due to decreased decay (Voeltz et al., 2001).
Deadenylation is not the only role of PARN needed, as unadenylated mRNA failed to show activation upon PARN depletion (Fig. 5A), indicating additional roles. PARN binding to mRNA caps increases in G0 (Fig. 5B) where canonical cap binding complex is inhibited by dephosphorylated 4EBP (Seal et al., 2005) (Fig. 6A, and Fig. S6D, immature oocytes); consistently, deadenylation (Fig. 1D–F (D285A), 3F, S3D), as well as cap binding by PARN is needed for activation (Fig. 5C–D, 1F, W468A). Also, p97 and FXR1 interact with PARN (Fig. 5E). These data suggest that PARN could connect p97—that is recruited by the 3′-UTR binding FXR1a-microRNP on specific mRNAs—with the 5′ mRNA cap, which may connect the 5′ and 3′ ends in place of the impaired, conventional poly(A)-cap link (PABP-eIF4E/eIF4G) in these reduced canonical translation conditions (Fig. S7H).
Proteomic studies with FXR1 depletion revealed miR16 (CD209, FEZ1, NUAK1) and miR369-3p (HES1, TOM1L1, NARG2) activation targets (Fig. 7, S7, Tables S1–S5, S7) that need FXR1 for polysome association (Fig. 7Aiii), are associated with FXR1 (Fig. 7Bi, and with AGO2, Fig. S7Diii–iv), need their targeting microRNAs (Fig. 7C–E), PARN and p97 (Fig. 7F) for translation. These results show activation with more than one microRNA that are associated with FXR1 (Fig. 7Biii). Other targets (Tables S1, 4, 5) need to be validated likewise for activation to rule out other regulatory processes (Bhattacharyya et al., 2006). Our data reveal cell state/signaling factor (HES1, TOM1L1) and immune genes (CD209, Table S3, Fig. S7Aii) as activation targets in THP1 G0, providing insights into G0 expression.
Specific mRNA recruitment by FXR1 in the nucleus (Truesdell et al., 2012) is essential for activation in G0; targets not recruited by FXR1a-microRNP are not activation targets, and can be recruited for repression by GW182-microRNPs that do not interact with FXR1 in these cells (Fig. S5D–E). CCND3 and CCNE2, targets of repression of miR16 (Fig. S7Dii), are not affected by (Tables S1, S4, S7) or associated with FXR1 (Fig. S7Di)—and thus are not activation targets. Consistently, their levels are not affected by depletion of factors needed for activation: p97 or PARN (Fig. 7Fi).
Our studies reveal essential features that promote microRNP-mediated activation in G0 and oocytes: (1) nucleofection/nuclear-injected mRNA for nuclear recruitment (Truesdell et al., 2012), (2) poly(A) minus target (avoiding PABP, Figs. 1, 3), (3) mRNA recruitment by FXR1a-microRNP (Truesdell et al., 2012) that is devoid of GW182 (Fig. S5D–E), and PABP in these cells (Fig. 4B, 5E, S5B–C), (4) specific low mTOR activity conditions: immature oocytes, G0 THP1 (serum-starved for greater than 18–24 hr (Vasudevan et al., 2007)), THP1 with inhibited mTOR or dephosphorylated 4EBP (Fig. 6, S6), (5) reduced canonical translation by dephosphorylated 4EBP (Fig. 6, S6D), (6) cap-bound PARN (Fig. 5B–D, 1F), and (7) increased FXR1 (Fig. 6A, S6D). These characteristics provide markers for activation by FXR1a-microRNP to overcome the variability of G0 heterogeneity. Whether these features are adequate in other cells remain to be tested. Our studies suggest a non-canonical translation mechanism where activation targets, recruited by FXR1a-microRNP, have shortened poly(A) tails and are translated via PARN and p97, in conditions with reduced canonical translation (Fig. S7H).
Experimental Procedures
Cells
THP1 cells were grown at 3–5 × 105 cells/ml in 10% FBS and RPMI 1640 (Vasudevan et al., 2007) and serum-starved at 1 × 105 cells/ml for ≥2 days for G0 or 42 hr (RNA transfection). Torin1 in N-methyl-2-pyrrolidone (NMP) and rapamycin (Cayman chemical) were added at 250nM and 100nM for 2 days.
Quantitative RT-PCR Analyses
cDNA synthesis was performed using Random Primers (Invitrogen) and the cDNA was subjected to PCR amplification with 60°C annealing and 40–45 cycles for different cDNA preparation yields. Primers used in oocytes were described previously (Mortensen et al., 2011; Truesdell et al., 2012). Primers to exogenously introduced Firefly and Renilla reporters, as well as endogenous mRNAs and reporters, used for oocyte and mammalian cell samples, are listed in Table S6.
Oocyte Protein and RNA Analyses
All extracts (RNA and protein) were prepared after collagenase treatment followed by manual defolliculation and examination to ensure complete defolliculation of oocytes (Moon et al., 2006; Mortensen et al., 2011) to prevent contamination of the samples with follicular material. The samples for immunoprecipitation were treated with RNase A/T1 and Turbo DNase (Ambion). For all gels, equal numbers of (20) oocytes, isolated as described (Moon et al., 2006; Mortensen et al., 2011), were run in each lane.
Luciferase Assay
Extract preparation and luciferase assays were performed as described in the manufacturer’s protocol (Promega). The average ratios of Luciferase values from at least three biological replicate experiments are depicted with the SD or SEM as error bars. The Luciferase activity was normalized to total nucleic acid values, representing relative cell numbers, or to total oocyte numbers.
Cap affinity purification
Binding of PARN to a cap column from G0 and serum-grown THP1 cells was performed as described previously (Seal et al., 2005; Tahmasebi et al., 2014). Cell lysates were pre-treated with micrococcal nuclease, prepared in buffer A (20mM HEPES/KOH pH 7.5, 50mMKCl, 1mM NaF, β-glyerophosphate, 0.1 mM EDTA, 1% NP40, 10% glycerol, 2mM DTT and EDTA-free protease inhibitor cocktail from Roche), pre-cleared using anti-V5 agarose (2,000rpm for 10 min), diluted upto 500μl with buffer A and incubated for 30 min at 4°C with m7GDP-agarose (Axxora) that had been blocked (0.5X SSPE, 0.05% tween, 0.1% PVP) for 1 hr before use. After incubation, beads were washed 5 times with buffer A, resuspended in SDS-PAGE loading buffer, and analyzed by Western blotting.
RNA
Capped mRNAs were transcribed using the T7 ultra mMESSAGE mMACHINE transcription system (Ambion/Applied Biosystems), subjected to a G-50 spin column or gel purified; they were quantitated on the NanoDrop Spectrophotometer. Except for control poly(A) mRNA, CXpA, CXA25, A20, and A5, all Luciferase reporters were transcribed without a poly(A) tail (A0) and had no known polyadenylation sequence. CXpA polyadenylation and cordycepin (Sigma and Cayman chemical) addition to the 3′end was carried out as described in the same system except the ATP was replaced with cordycepin in the latter case. CXA25 was transcribed using the above system from a PCR product of the CX plasmid primed with a T7 primer to the 5′ T7 site on the plasmid and to the 3′end of the CX reporter sequence using the primer, (T)25CTAGAAGGCACAGTCGAGGC, to add A25 to the 3′end. A20 and A5 were synthesized similarly. T7 transcription was performed in the presence of Anti-Reverse Cap Analogs (ARCA) to produce ARCA-capped mRNAs, as described for the T7 ultra mMESSAGE mMACHINE transcription system (Ambion/Applied Biosystems).
Polysome analysis
Polysome analysis (Fig. 7Aii), followed by qRT-PCR analysis (Fig. 7Aiii) of polysome associated mRNAs, in FXR1 knockdown and control shRNA cells was conducted as described previously (Gandin et al., 2014; Lee et al., 2014). 15% and 50% (w/v) sucrose solutions were prepared in buffer A (10 mM Tris-HCl, pH 7.4, 100 mM KCl, 5 mM MgCl2, 100 μg/ml cycloheximide and 2 mM DTT). Sucrose density gradients were prepared as previously described (Vasudevan and Steitz, 2007; He and Green, 2013). FXR1 knockdown and shRNA control cells were induced with 1μg/ml of doxycycline continually to cause knockdown of FXR1 (Fig. 7Ai); the cells were grown in serum for 3 days to induce the shRNAs, followed by 2 days of serum-starvation to induce G0. Just before harvesting, the cells were treated with 100 μg/ml cycloheximide at 37°C for 5 min. Collected cells were washed with cold PBS containing cycloheximide and then lysed in buffer A containing 1% Triton X-100 and 40 units/μl murine RNase Inhibitor (NEB). Cleared cell lysates were loaded on sucrose gradients followed by ultracentrifugation (Beckman Coulter Optima L90) for 2 hours at 38,000 rpm at 4°C in an SW40 rotor. Samples were fractionated by density gradient fractionation system (Isco). The heavy polysome (>3 polysomes (Preiss et al., 2003; Fu et al., 2012)) fractions were pooled, lysed in proteinase K buffer (Truesdell et al., 2012) and total RNA was extracted with 3 volumes of Trizol (Invitrogen). The decrease in polysome associated mRNAs in FXR1 knockdown compared to control shRNA cells was analyzed by qRT-PCR analysis (Fig. 7Aiii) of candidate targets in polysome associated mRNA samples, normalized to tRNA-lys and to total target RNA levels.
Xenopus laevis oocytes
Oocytes were harvested as described from human chorionic gonadotropin (hcG)-stimulated frogs by partial ovariectomy (Moon et al., 2006; Mortensen et al., 2011) and manipulated as described in SI Extended Procedures. Pre-stimulated frogs were obtained from NASCO (WI). The oocyte removal and animal handling protocols adhered strictly to the institutionally approved guidelines and regulations, and were reviewed, approved and renewed annually by the Subcommittee on Research Animal Care (SRAC), which serves as the Institutional Animal Care and Use Committee (IACUC) for MGH. Additional Oocyte, Plasmids, RNA, protein analyses are described in SI Extended Procedures.
GEO Accession GSE77512 (RNA levels microarray)
Supplementary Material. (Supplementary Information, SI)
Acknowledgments
The study is funded by Cancer Research Institute, D. & M-E Ryder, Leukemia & Lymphoma Society, Smith Family Foundation & NIGMS GM100202 grants to SV. SL is funded by Fund for Medical Discovery fellowship. We thank Partners Healthcare Center for Personalized Genetic Medicine facility for microarray data, M. Serra & J.M. Traverso for plasmids, O. Le Tonqueze for stable cell lines, J.A. Steitz & N. Sonenberg for comments, E. Wahle & M. Wormington for anti-PARN.
Footnotes
Author Contributions
SIAB conducted the research; SST, SL, SK, AC & RDM contributed data; AY made reagents; EJ & RIS did bioinformatics; MB & WH did proteomics; SIAB & SV analyzed data; SV supervised the research & wrote the manuscript.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Beilharz TH, Humphreys DT, Clancy JL, Thermann R, Martin DI, Hentze MW, Preiss T. microRNA-mediated messenger RNA deadenylation contributes to translational repression in mammalian cells. PLoS One. 2009;4:e6783. doi: 10.1371/journal.pone.0006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-Mediated Translational Repression in Human Cells Subjected to Stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol. 2006;4:e83. doi: 10.1371/journal.pbio.0040083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman Y, Linial M. miRror2.0: a platform for assessing the joint action of microRNAs in cell regulation. J Bioinform Comput Biol. 2013;11:1343012. doi: 10.1142/S0219720013430129. [DOI] [PubMed] [Google Scholar]
- Fu S, Fan J, Blanco J, Gimenez-Cassina A, Danial NN, Watkins SM, Hotamisligil GS. Polysome profiling in liver identifies dynamic regulation of endoplasmic reticulum translatome by obesity and fasting. PLoS Genet. 2012;8:e1002902. doi: 10.1371/journal.pgen.1002902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Sikstrom K, Alain T, Morita M, McLaughlan S, Larsson O, Topisirovic I. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J Vis Exp. 2014;87 doi: 10.3791/51455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SL, Green R. Polysome analysis of mammalian cells. Methods Enzymol. 2013;530:183–92. doi: 10.1016/B978-0-12-420037-1.00010-5. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Tomari Y. Argonaute-mediated translational repression (and activation) Fly. 2009 Jul-Sep;3(3):204–206. [PubMed] [Google Scholar]
- Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev. 2007;21:2571–2579. doi: 10.1101/gad.1593007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner CG, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Truesdell SS, Bukhari SI, Lee JH, Letonqueze O, Vasudevan S. Upregulation of eIF5B controls cell-cycle arrest and specific developmental stages. Proc Natl Acad Sci U S A. 2014;111:E4315–E4322. doi: 10.1073/pnas.1320477111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Strumpf N, Deiss LP, Berissi H, Kimchi A. DAP-5, a novel homolog of eukaryotic translation initiation factor 4G isolated as a putative modulator of gamma interferon-induced programmed cell death. Mol Cell Biol. 1997;17:1615–1625. doi: 10.1128/mcb.17.3.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Liu LZ, Addison JB, Wonderlin WF, Ivanov AV, Ruppert JM. A KLF4-miRNA-206 Autoregulatory Feedback Loop Can Promote or Inhibit Protein Translation Depending upon Cell Context. Mol Cell Biol. 2011;31:2513–2527. doi: 10.1128/MCB.01189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza-Puch F, Drost J, Rooijers K, Lopes R, Elkon R, Agami R. p53 induces transcriptional and translational programs to suppress cell proliferation and growth. Genome Biol. 2013;14:R32. doi: 10.1186/gb-2013-14-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- Moon KH, Zhao X, Yu YT. Pre-mRNA splicing in the nuclei of Xenopus oocytes. Methods Mol Biol. 2006;322:149–163. doi: 10.1007/978-1-59745-000-3_11. [DOI] [PubMed] [Google Scholar]
- Moretti F, Kaiser C, Zdanowicz-Specht A, Hentze MW. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat Struct Mol Biol. 2012;19:603–608. doi: 10.1038/nsmb.2309. [DOI] [PubMed] [Google Scholar]
- Mortensen RD, Serra M, Steitz JA, Vasudevan S. Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA-protein complexes (microRNPs) Proc Natl Acad Sci U S A. 2011;108:8281–8286. doi: 10.1073/pnas.1105401108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson P, Henriksson N, Niedzwiecka A, Balatsos NA, Kokkoris K, Eriksson J, Virtanen A. A multifunctional RNA recognition motif in poly(A)-specific ribonuclease with cap and poly(A) binding properties. J Biol Chem. 2007;282:32902–32911. doi: 10.1074/jbc.M702375200. [DOI] [PubMed] [Google Scholar]
- Preiss T, Baron-Benhamou J, Ansorge W, Hentze MW. Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nat Struct Biol. 2003;10:1039–1047. doi: 10.1038/nsb1015. [DOI] [PubMed] [Google Scholar]
- Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta. 2008;1779:217–229. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren YG, Martinez J, Virtanen A. Identification of the active site of poly(A)-specific ribonuclease by site-directed mutagenesis and Fe(2+)-mediated cleavage. J Biol Chem. 2002;277:5982–5987. doi: 10.1074/jbc.M111515200. [DOI] [PubMed] [Google Scholar]
- Sachs AB, Bond MW, Kornberg RD. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- Seal R, Temperley R, Wilusz J, Lightowlers RN, Chrzanowska-Lightowlers ZM. Serum-deprivation stimulates cap-binding by PARN at the expense of eIF4E, consistent with the observed decrease in mRNA stability. Nucleic Acids Res. 2005;33:376–387. doi: 10.1093/nar/gki169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Gao L, Chien HY, Li WC, Zhao J. The regulation and function of the NUAK family. J Mol Endocrinol. 2013;51:R15–R22. doi: 10.1530/JME-13-0063. [DOI] [PubMed] [Google Scholar]
- Tahmasebi S, Alain T, Rajasekhar VK, Zhang JP, Prager-Khoutorsky M, Khoutorsky A, Dogan Y, Gkogkas CG, Petroulakis E, Sylvestre A, Ghorbani M, Assadian S, Yamanaka Y, Vinagolu-Baur JR, Teodoro JG, Kim K, Yang XJ, Sonenberg N. Multifaceted Regulation of Somatic Cell Reprogramming by mRNA Translational Control. Cell Stem Cell. 2014;14:606–16. doi: 10.1016/j.stem.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truesdell SS, Mortensen RD, Seo M, Schroeder JC, Lee JH, Letonqueze O, Vasudevan S. MicroRNA-mediated mRNA Translation Activation in Quiescent Cells and Oocytes Involves Recruitment of a Nuclear microRNP. Sci Rep. 2012;2:842. doi: 10.1038/srep00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tserel L, Runnel T, Kisand K, Pihlap M, Bakhoff L, Kolde R, Peterson H, Vilo J, Peterson P, Rebane A. MicroRNA expression profiles of human blood monocyte-derived dendritic cells and macrophages reveal miR-511 as putative positive regulator of Toll-like receptor 4. J Biol Chem. 2011;286:26487–26495. doi: 10.1074/jbc.M110.213561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Ongkasuwan J, Standart N, Steitz JA. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 2001;15:774–788. doi: 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagiya A, Delbes G, Svitkin YV, Robaire B, Sonenberg N. The poly(A)-binding protein partner Paip2a controls translation during late spermiogenesis in mice. J Clin Invest. 2010;120:3389–3400. doi: 10.1172/JCI43350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, Zhang H, Guo P, Sun H, Guo L, Zhang Y, Fu XD. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.