Abstract
Lung cancer is the leading cause for cancer-related death, however, the pathogenesis mechanism is poorly understood. Although the rhotekin (RTKN) gene has been reported to encode an effector for the Rho protein that has critical roles in regulating cell growth, the role of RTKN in lung cancer has not been investigated. In clinical lung cancer patient tumor samples, we identified that the RTKN gene expression level was significantly higher in tumor tissues compared to that of the adjacent normal tissues. To investigate the molecular mechanisms of RTKN in lung cancer, we established RTKN stable knock-down A549 and SPC-A-1 lung adenocarcinoma cell lines using lentiviral transfection of RTKN shRNA and evaluated the antitumor effects. The results showed that RTKN knock-down inhibited lung adenocarcinoma cell viability, induced S phase arrest and increased cell apoptosis. In addition, RTKN knock-down inhibited lung cancer cell invasion and adhesion. Further analysis showed that the S phase promoting factors cyclindependent kinase (CDK)1 and CDK2 levels were decreased in RTKN knock-down cells, and that the DNA replication initiation complex proteins Minichromosome maintenance protein complex (MCM)2 and MCM6 were decreased as well in RTKN knock-down cells. These results indicated that the RTKN protein was associated with lung cancer in clinic samples and exerted anticancer activity in lung adenocarcinoma cells through inhibiting cell cycle progression and the DNA replication machinery. These findings suggest that RTKN inhibition may be a novel therapeutic strategy for lung adenocarcinoma.
Keywords: rhotekin, lung cancer, adenocarcinoma, cell viability
Introduction
Lung cancer is the leading cause of cancer-related death worldwide, with an increasing mortality each year (1). Non-small cell lung cancer (NSCLC) accounts for 80–85% of all lung cancers. NSCLC subtypes include adenocarcinoma, squamous cell carcinoma and large cell carcinoma. The majority of patients diagnosed with NSCLC are diagnosed at advanced stages with local or distant metastases. Standard NSCLC treatment includes chemotherapy and surgery which have severe side effects and limited efficacy (2). Targeted therapy, which specifically attacks cancer cells with designated molecular targets, has emerged as a promising strategy due to its high efficacy and reduced side effects (3,4). Several drugs targeting key oncogenesis signaling molecules have been developed and showed efficacy for specific patient groups, such as erlotinib targeting epidermal growth factor (EGF) (5), bevacizumab targeting vascular endothelial growth factor (VEGF) (6) and crizotinib targeting anaplastic lymphoma kinase (ALK) (7). However, due to the complexity of pathogenic pathways in individual patients, it is urgent to uncover the largely unknown molecular origins of lung cancer and provide new targets for lung cancer therapy.
The rhotekin (RTKN) gene encodes a scaffold protein which interacts with active GTP-bound Rho proteins and interferes with the conversion to inactive GDP-bound Rho proteins (8). Rho proteins regulate critical cell functions including cell growth and transformation, cytokinesis, transcription, and smooth muscle contraction. Rho signaling pathway dysregulation was implicated in several forms of cancer (9). Although the RTKN gene has been reported to be associated with several cancer types such as bladder cancer, gastric cancer and breast cancer (10–12), the role of RTKN in lung cancer has not been investigated.
Cancer cells are characterized by uncontrolled proliferation (13). Cell cycle progression and DNA replication are essential events for cell proliferation (14). Cell cycle was finely tuned by a number of factors including cyclins and cyclin-dependent kinases (CDKs) (15). CDK1 is a catalytic subunit of the M-phase promoting factor (MPF), which promotes G1/S and G2/M transitions of eukaryotic cells (16). CDK2 is part of a cyclin-dependent protein kinase complex. CDK2 activity is essential during G1/S transition (17). Minichromosome maintenance protein complex (MCM) is involved in the initiation of DNA replication. The complex formed by MCM2, 4, 6, and 7 was shown to regulate the helicase activity of the pre-replication complex (18,19).
Here we report that the RTKN gene expression level was significantly higher in tumor tissue of lung cancer patients. Further analysis in RTKN stable knock-down A549 and SPC-A-1 lung adenocarcinoma cells indicated that RTKN knock-down exhibited antitumor activity as evidenced by decreased cancer cell viability, induction of cell cycle arrest, increased apoptosis, and decreased invasion and migration. Detailed analysis showed that RTKN knock-down decreased the cell cycle regulators CDK1 and CDK2 expression, as well as the DNA replication modulators MCM2 and MCM6 expression.
Materials and methods
Clinical patient samples
Primary tissues were collected from patients who received surgery for lung cancer at our institution. All the patients had given their informed consent. Dissected samples were frozen immediately after surgery and stored at −80°C until needed.
Cell culture
A549, H460, H128, H1299, SPC-A-1 and SK-MES human lung adenocarcinoma cell lines were obtained from the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen) and maintained in an incubator with a humidified atmosphere of 95% air and 5% CO2 at 37°C.
Establishment of stable RTKN knock-down cell lines
A549 and SPC-A-1 stable knock-down cell lines were constructed using the pLKO.1-EGFP vector-based lentiviral transduction system. shRNA target RTKN and non-target control shRNA (NC) were synthesized and cloned into the pLKO.1-EGFP vector using AgeI and EcolI restriction sites. pLKO.1-EGFP-RTKN or pLKO.1-EGFP-NC and the packing vector psPAX2 and pMD2G were co-transfected with into 293T cells. Lentivirus particles were produced in 293T cells. A549 and SPC-A-1 cells were transfected by the lentiviruses and selected using the EGFP green fluorescence marker. Three shRNA sequences targeting the RTKN gene are: RTKN-F1: CCGGG AACTGCGGTTAGAGCTGTATCTCGAGATACAGCTCT AACCGCAGTTCTTTTTC and AATTGAAAAAGAACTG CGGTTAGAGCT-GTATCTCGAGATACAGCTCTAACCG CAGTTC; RTKN-F2: CCGGGAAGCAG-TGCTGTGATGA AATCTCGAGATTTCATCACAGCACTGCTTCTTTTTC and AATTGAAAAAGAAGCAGTGCTGTGATGAAATCT CGAGATTTCATCACAGCACTGCTTC; and RTKN-F3: CCGGAAGAACCCTTGGAGCAAACATCTCGAGATGTT TGCTCCAAGGGTTCTTTTTTTC and AATTGAAAAAA AGAACCCTTGGAGCAAACATCTCGAGATGTTTGCTC CAAGGGTTCTT.
Quantitative RT-PCR
Total RNA was extracted using TRIzol reagent, reverse transcribed to cDNA, and quantified by real-time PCR using SYBR Green Universal Master Mix (all reagents were from Thermo Fisher Scientific, Waltham, MA, USA). Results were normalized by using human GAPDH mRNA levels as internal control. Relative mRNA levels were expressed as fold change to control. The primer pairs used are as follows: RTKN (forward) GCCGCTGCTTACTATTGC and (reverse) GTGCTTCCCGACTTTCTG; GAPDH (forward) CACCCACTCCTCCACCTTTG and (reverse) CCA CCACCCTGTTGCTGTAG; CDK1 (forward) ACCATACCC ATTGACTAAC and (reverse) ATAAGCACATCCTGAAGAC; CDK2 (forward) CCAGGAGTTACTTCTATGCCTGA and (reverse) TTCATCCAGGGGAGGTACAAC; MCM2 (forward) CTACCAGCGTATCCGAATC and (reverse) GTTG AGGGAGCCATCATAG; and MCM6 (forward) CCAAACA TCTGCCGAAATC and (reverse) TCAGTGTCCCTGTAAA GTC.
Western blotting
Cells were lysed with RIPA lysis buffer containing protease inhibitor cocktails (both from Thermo Scientific). Protein concentrations were determined using BCA protein assay kit (Thermo Scientific). Equal amounts of lysates (20-40 µg of protein) were resolved with 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein blots were transferred to a nitrocellulose membrane and probed with the corresponding primary antibodies. The membrane was then incubated with appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies, and the protein expression was detected by Immobilon Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA, USA). Antibodies used were all from Abcam (Cambridge, MA, USA): RTKN (Ab154954), CDK1 (Ab18), CDK2 (Ab6538), MCM2 (Ab4461), MCM6 (Ab190948), and GAPDH (Ab8254).
Cell viability assay
Cell viability was determined using the Cell Counting Kit-8 (CCK-8) assay kit (Dojindo Molecular Technologies, Japan) following the manufacturer's instructions. Briefly, 5,000 cells were seeded into each well of 96-well plates in triplicates. Cells were cultured for 24 h and then 10 µl of the CCK-8 solution was added to each well and incubated for 1 h. The absorbance at 450 nm was measured using a microplate reader. Results were calculated as percentage to cells numbers at 0 h.
Cell cycle analysis
Cells were cultured for 24 h and then suspended and fixed with ethanol. After an overnight incubation at −20°C, cells were washed with PBS and incubated with 1 mg/ml RNase A at 37°C for 30 min. The cells were then incubated with 50 µg/ml propidium iodide (PI) on ice for 10 min in the dark. Cell cycle distribution was analyzed using FACSCalibur instrument (BD Biosciences, San Jose, CA, USA) equipped with CellQuest software.
Apoptosis
Cell Apoptosis was determined using the Annexin V-APC apoptosis detection kit (BD Biosciences) following the manufacturer's instructions. Briefly, cells were detached by trypsin, harvested and stained with Annexin V-APC for 10 min at room temperature. Cell were then incubated with 50 µg/ml PI on ice and analyzed using flow cytometry on the FACSCalibur instrument. Annexin V positive cells were considered the apoptotic fraction.
Tumor invasion assay
Cancer cell invasion was evaluated using Matrigel and Transwells in triplicates. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) without serum 24 h prior to seeding. Transwells (24-well plates) were rinsed with PBS and then each Transwell was coated with 80 µl Matrigel at 37°C for 30 min. Cells were detached with trypsin and resuspended with DMEM containing 1% FBS. Cells (1×105) in 0.5 ml suspension were seeded in each Transwell. DMEM (0.75 ml) with 10% FBS was added into each well of the 24-well plate under the Transwell. Cells were incubated at 37°C for 48 h and then fixed with 4% formaldehyde. The cells were stained with 0.5% crystal violet solution, rinsed with PBS and air dried. Residue cells in Transwell were wiped away and the migrated cells were visualized under a microscope. Cell numbers from triplicate wells in each group were counted for statistical analysis.
Cell adhesion assay
Cells were detached by trypsin and resuspended. Cells (1×105) were plated in each well of fibronectin coated 12-well plates and culture in growth medium for 1 h at 37°C in the incubator for evaluation of adhesion. Cell culture medium was discarded and cells were washed with PBS twice. Cells were then fixed with 5 ml of 4% formaldehyde for 15 min and washed with PBS. Cells were stained with Giemsa staining buffer for 30 min and unbound dye was rinsed with water. The plates were air dried and cell images were captured under a microscope. Cells in 3 triplicate wells were counted for each group.
Data analysis
All the experiments were performed in triplicates. Results are expressed as mean ± SD. The statistical difference between multiple treatments and control was analyzed using one-way ANOVA. Differences between two groups were analyzed by Student's t-test. A p-value of <0.05 was considered statistically significant.
Results
The RTKN levels are elevated in lung cancer cells in vivo and in vitro
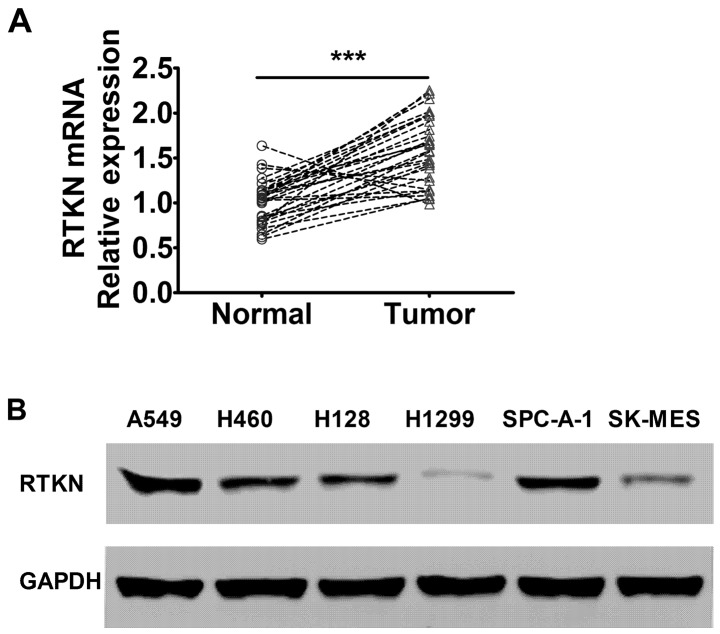
To study the expression pattern of RTKN gene in lung cancer, tumor and matched benign tissue from 30 non-small cell lung cancer cases were collected and evaluated by RT-PCR analysis. The results showed that RTKN gene mRNA level was significantly elevated in lung cancer tissue compared with matched benign tissue, indicating that the RTKN gene may be associated with the development of lung cancer (Fig. 1A). We further detected the RTKN expression levels in different lung adenocarcinoma cell lines. The results showed that RTKN level was elevated in several lung adenocarcinoma cell lines with A549 and SPC-A-1 showing the highest levels among the cell lines evaluated (Fig. 1B).
Figure 1.
RTKN is overexpressed in lung cancer tumor tissue and cell lines. (A) RTKN mRNA level was determined by RT-PCR in lung tumor tissue and matched adjacent normal tissue. ***P<0.001. (B) RTKN protein level was detected in lung cancer cell lines by western blotting using anti-RTKN and anti-GAPDH antibodies.
The RTKN knock-down inhibits lung adenocarcinoma cell viability and induces cell cycle arrest and apoptosis
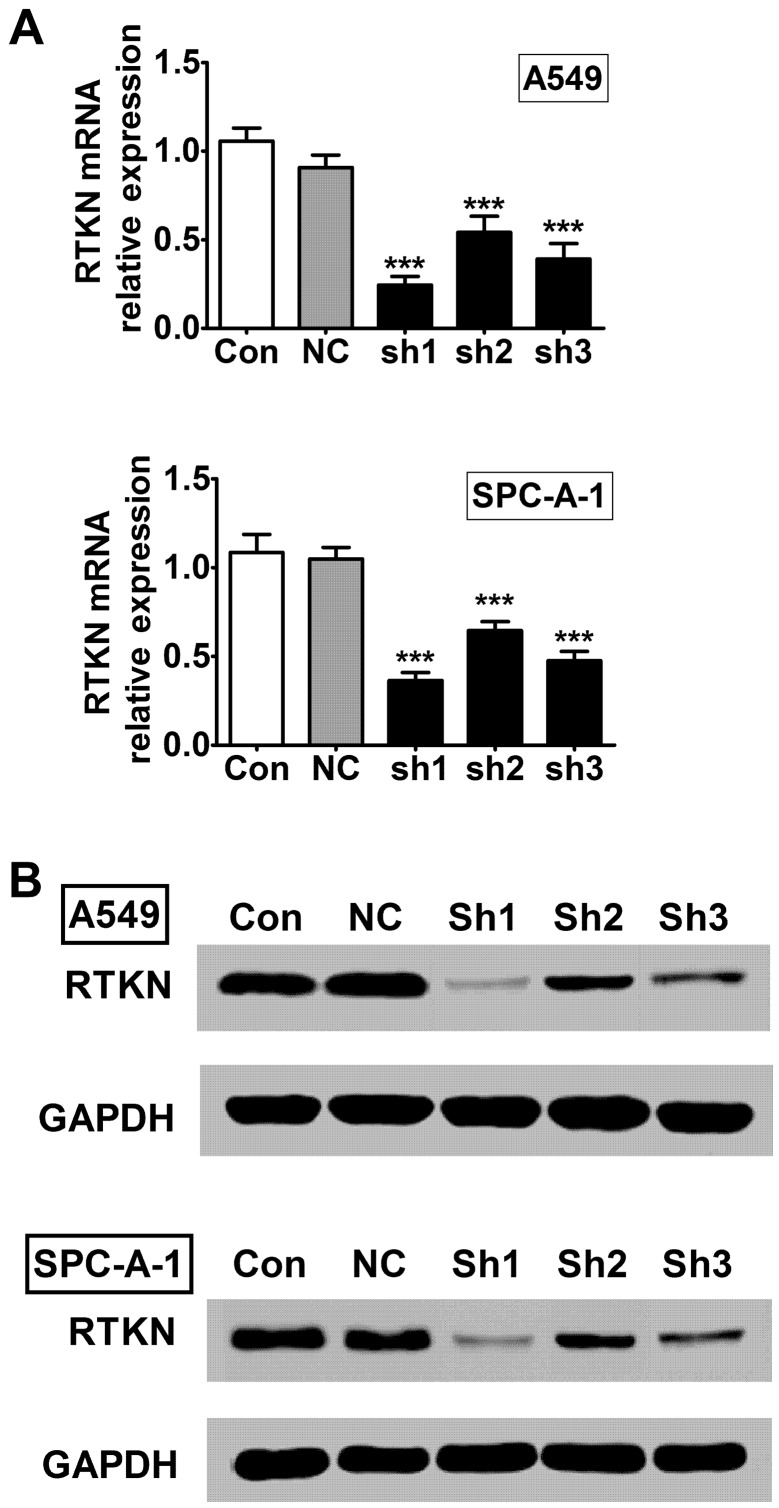
To further study the role of RTKN in lung cancer, we established RTKN stable knock-down A549 and SPC-A-1 lung adenocarcinoma cell lines using lentiviral system. We designed three shRNA constructs (sh1, sh2, and sh3) and detected their knock-down efficiencies on RTKN mRNA and protein levels in A549 and SPC-A-1 cells. The first construct (sh1) showed the best inhibiting efficiency in both cell lines compared with other two constructs (Fig. 2), therefore it was selected for further experiments.
Figure 2.
The RTKN stable knock-down A549 and SPC-A-1 lung adenocarcinoma cells lines. A549 and SPC-A-1 cells were transfected with lentivirus containing three constructs of RTKN shRNA (sh1, sh2, and sh3) or non-target controls shRNA (NC). Cells without transfection were used as control (CON). (A) RTKN mRNA levels were determined by RT-PCR. (B) RTKN protein levels were examined by western blotting. ***P<0.001.
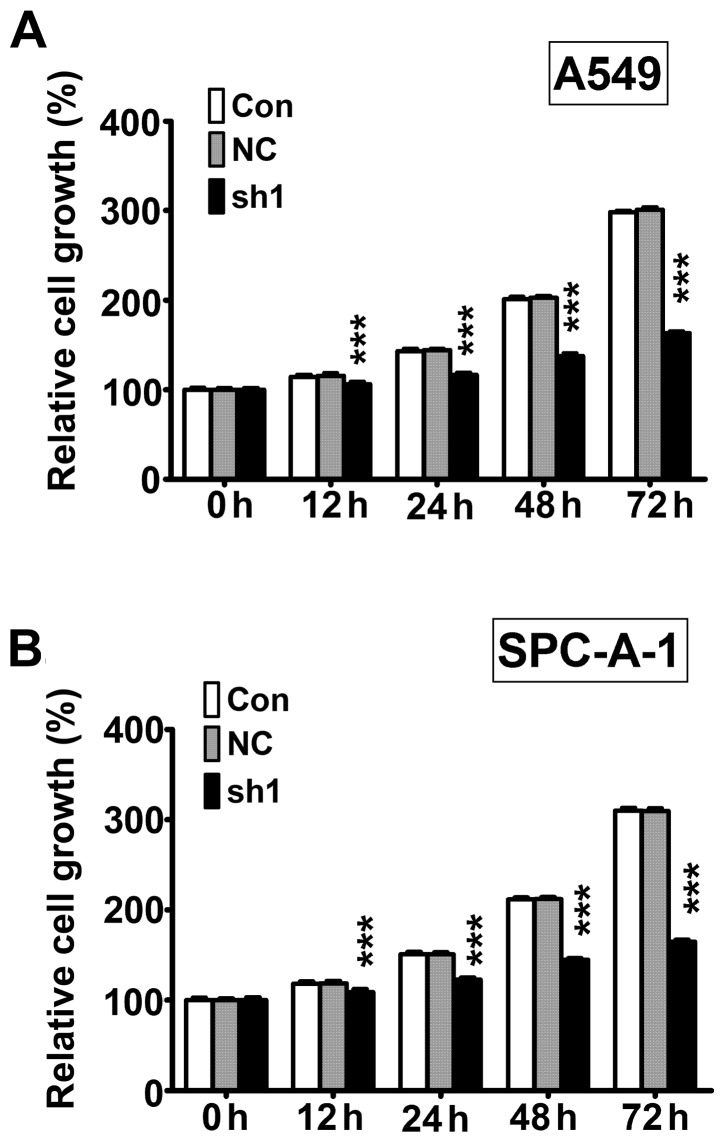
Next, we measured cell viability using CCK-8 assay. The results showed that cell viability was significantly lower after 12, 24, 48, and 72 h in RTKN knock-down (sh1) cells compared with non-transfected control cells (Con) and non-target scramble control shRNA transfected cells (NC) in both A549 (Fig. 3A) and SPC-A-1 (Fig. 3B) cell lines.
Figure 3.
The RTKN knock-down inhibits cell viability in lung cancer cells. (A) A549 and (B) SPC-A-1 cells were cultured for 24 h and cell viability were determined using CCK-8 assay. Results are expressed as percentage to cells at 0 h. ***P<0.001.
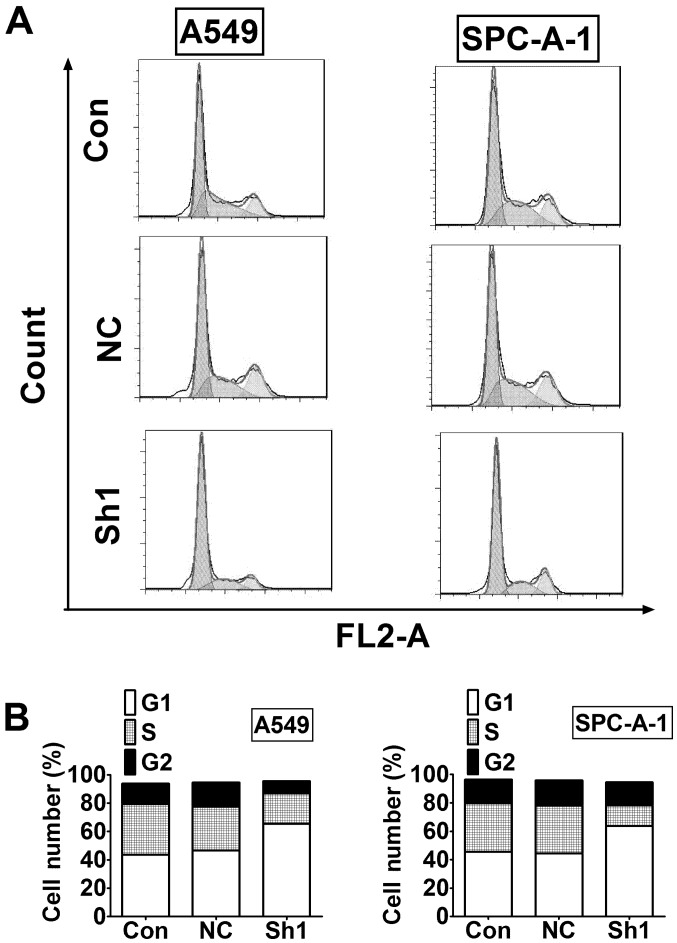
To explore the mechanism of reduced cell viability after RTKN inhibition, we examined cell cycle distribution using flow cytometry. The results showed that G1 phase was greatly increased in RTKN knock-down cells compared to control cells in both A549 and SPC-A-1 cells (Fig. 4).
Figure 4.
The RTKN knock-down induces G1 phase arrest in lung cancer cells. (A) Cell cycle was analyzed by flow cytometry. (B) Cell cycle distribution in A was quantified using CellQuest software.
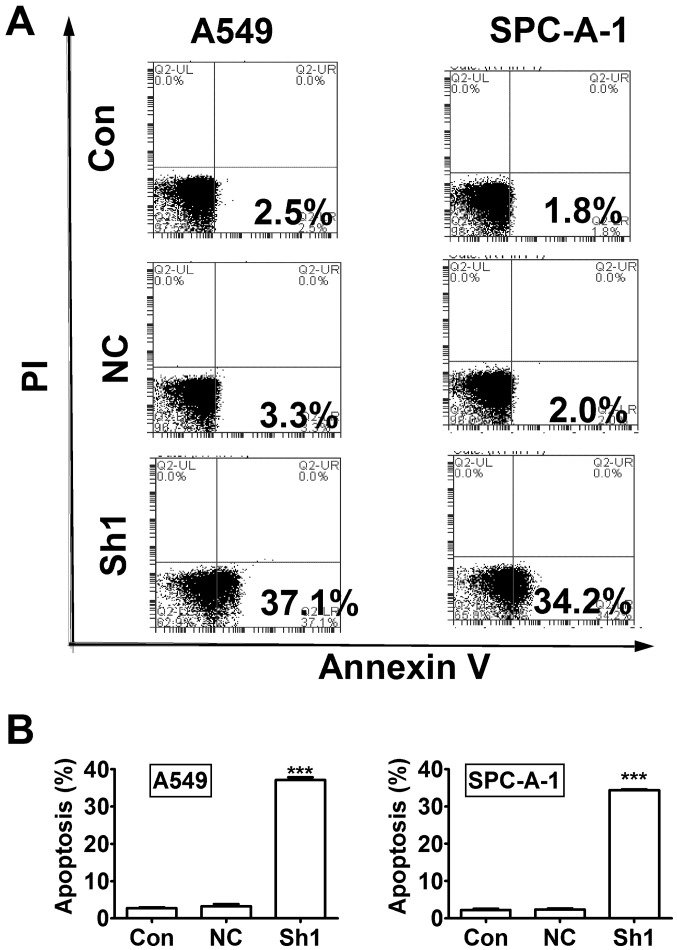
Next, we evaluated cell apoptosis using Annexin V-APC staining and flow cytometry. The results showed significant increase of apoptosis percentage in RTKN knock-down A549 and SPC-A-1 cells compared to control knock-down cells (Fig. 5, P<0.001).
Figure 5.
The RTKN knock-down induces apoptosis in lung cancer cells. (A) Cell apoptosis was analyzed by Annexin V staining and flow cytometry. (B) Annexin V stained cells were considered apoptotic and apoptosis rate was calculated. ***P<0.001.
The RTKN knock-down blocks signaling pathways in cell growth
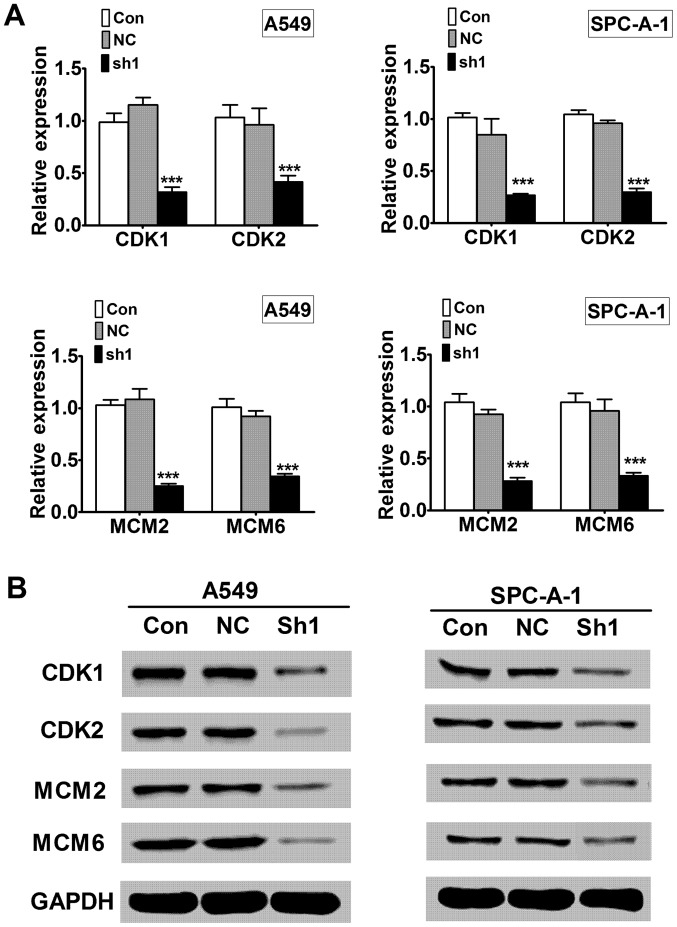
To investigate the underlying mechanism of RTKN shRNA induced growth inhibition, we examined signaling molecules in cell cycle progression (CDK1 and CDK2) and DNA replication (MCM2 and MCM6). Quantitative RT-PCR showed that RTKN knock-down decreased CDK1, CDK2, MCM2, and MCM6 mRNA levels compared to the control knock-down (Fig. 6A, P<0.001). Western blot analysis showed that the protein levels of CDK1, CDK2, MCM2 and MCM6 were also decreased in RTKN knock-down cells compared to control knock-down cells (Fig. 6B).
Figure 6.
The RTKN knock-down inhibits signaling pathways in cell growth. (A) mRNA levels of CDK1, CDK2, MCM2, and MCM6 were analyzed using RT-PCR. Results were expressed as fold change to non-transfected control cells (Con). (B) Protein levels of CDK1, CDK2, MCM2, and MCM6 were analyzed using western blotting with indicated antibodies. GAPDH was used as a loading control. ***P<0.001.
The RTKN knock-down inhibits lung cancer cell invasion and adhesion
To further investigate the role of RTKN in lung cancer cells, we examined A549 and SPC-A-1 lung cancer cell invasion and adhesion after RTKN knock-down. The Transwell tumor invasion assay showed that migrated cells were significantly decreased in RTKN knock-down A549 and SPC-A-1 cells compared to control knock-down cells (Fig. 7, P<0.001). Next, we investigated the role of RTKN in cell adhesion.
Figure 7.
The RTKN knock-down inhibits lung cancer cell invasion. (A) A549 and SPC-A-1 lung cancer cell invasion was evaluated using Transwell assay. Representative images of migrated cells are shown. (B) Migrated cells from three independent wells of each group were counted and analyzed. ***P<0.001.
The results showed a significant reduction in cell adhesion in RTKN knock-down A549 and SPC-A-1 cells compared to control knock-down cells (Fig. 8, P<0.001). Taken together, the results indicated that the RTKN knock-down decreased lung cancer cell invasion and adhesion.
Figure 8.
The RTKN knock-down inhibits lung cancer cell adhesion. (A) Cell adhesion was determined as described in Material and methods. Representative images of adhered cells are shown. (B) Adhered cells from three independent wells of each group were counted and analyzed. ***P<0.001.
Discussion
Non-small cell lung cancer is the major type of lung cancer with adenocarcinoma as the main subtype. Targeted cancer therapy showed high efficacy and low toxicity compared with traditional chemotherapy. Identifying specific tumor genesis pathways could promote personalized cancer therapy and greatly improve cancer treatment outcome. We studied tumor tissue from lung cancer patients and found that the RTKN gene was highly expressed in tumor tissue compared with benign tissue. We further investigated the role of RTKN in lung cancer and underlying mechanisms using RTKN stable knockdown lung adenocarcinoma cell lines. Our results showed that knock-down of RTKN exhibited antitumor activity in lung cancer cells.
Rho GTPase is a master regulator of cytoskeleton in multiple cell functions such as cell migration, adhesion and cytokinesis. Upon binding GTP, Rho exerts its functions through downstream Rho effectors, such as ROCK, mDia, Citron, PKN, Rhophilin and Rhotekin (RTKN) (9,20–22). The RTKN gene was only identified recently and its physiological functions remain largely unknown (8,23). RTKN has been reported to be associated with several cancer types such as bladder cancer (12), gastric cancer (11), and breast cancer (10).
However, the role of RTKN in lung cancer and its molecular mechanisms have not been reported. We identified RTKN as a potential oncogenic factor in lung cancer, as evidenced by the findings that the RTKN knock-down inhibited lung cancer cell viability, invasion and adhesion.
Suppression of apoptosis and disregulation of cell viability are key characteristics of cancer cells which enables uncontrolled expansion and invasion (14). Our results demonstrated increased cell apoptosis in RTKN knock-down lung cancer cells compared to control knock-down cells, therefore indicating antitumor activity of RTKN inhibition. Cell cycle progression was finely tuned by cyclin complexes, in which cyclin-dependent kinases play important roles (15). CDK1 and CDK2 are key regulators of G1-S transition (24). Our results showed that cells were arrested in G1 phase in RTKN knockdown cells. Consistently, further analysis revealed that CDK1 and CDK2 levels are decreased in RTKN knock-down cells, indicating that RTKN affected cell cycle regulatory proteins and cell growth. DNA replication is the major event in cell viability (25). MCM complex is required for DNA replication and recruited to the origin recognition complex (ORC) during late mitosis/early G1 phase (18,19). Our results revealed that MCM2 and MCM6 levels are decreased in RTKN knockdown lung cancer cells, indicating that RTKN affected the DNA replication machinery and thus cell proliferation.
Further experiments are needed to determine the protein levels through western blotting and the cellular distribution through immunohistochemistry staining for RTKN in human cancer samples. Also, it is important to establish animal tumor models to further validate the antitumor effects of RTKN inhibition or knockdown in vivo.
In summary, our results from both clinical specimens and cultured lung cancer cells revealed that the RTKN level was elevated in lung cancer and that RTKN inhibition exerts antitumor effects in lung cancer cells. These finding suggested that RTKN may be a potential therapeutic target in lung cancer treatment.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Peters S, Adjei AA, Gridelli C, Reck M, Kerr K. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23(Suppl 7):vii56–vii64. doi: 10.1093/annonc/mds226. [DOI] [PubMed] [Google Scholar]
- 3.Kumar M, Ernani V, Owonikoko TK. Biomarkers and targeted systemic therapies in advanced non-small cell lung cancer. Mol Aspects Med. 2015 Jul 14; doi: 10.1016/j.mam.2015.06.009. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black A, Morris D. Personalized medicine in metastatic non-small-cell lung cancer: Promising targets and current clinical trials. Curr Oncol. 2012;19(Suppl 1):S73–S85. doi: 10.3747/co.19.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pérez-Soler R, Chachoua A, Hammond LA, Rowinsky EK, Huberman M, Karp D, Rigas J, Clark GM, Santabárbara P, Bonomi P. Determinants of tumor response and survival with erlotinib in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22:3238–3247. doi: 10.1200/JCO.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 6.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, Lilenbaum R, Johnson DH. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 7.Pender A, Popat S. The efficacy of crizotinib in patients with ALK-positive non-small cell lung cancer. Ther Adv Respir Dis. 2015;9:97–104. doi: 10.1177/1753465815577738. [DOI] [PubMed] [Google Scholar]
- 8.Reid T, Furuyashiki T, Ishizaki T, Watanabe G, Watanabe N, Fujisawa K, Morii N, Madaule P, Narumiya S. Rhotekin, a new putative target for Rho bearing homology to a serine/threonine kinase, PKN, and rhophilin in the rho-binding domain. J Biol Chem. 1996;271:13556–13560. doi: 10.1074/jbc.271.23.13556. [DOI] [PubMed] [Google Scholar]
- 9.Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem J. 2000;348:241–255. doi: 10.1042/bj3480241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen M, Bresnick AR, O'Connor KL. Coupling S100A4 to Rhotekin alters Rho signaling output in breast cancer cells. Oncogene. 2013;32:3754–3764. doi: 10.1038/onc.2012.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu CA, Wang MJ, Chi CW, Wu CW, Chen JY. Overexpression of rho effector rhotekin confers increased survival in gastric adenocarcinoma. J Biomed Sci. 2004;11:661–670. doi: 10.1007/BF02256132. [DOI] [PubMed] [Google Scholar]
- 12.Fan J, Ma LJ, Xia SJ, Yu L, Fu Q, Wu CQ, Huang XH, Jiang JM, Tang XD. Association between clinical characteristics and expression abundance of RTKN gene in human bladder carcinoma tissues from Chinese patients. J Cancer Res Clin Oncol. 2005;131:157–162. doi: 10.1007/s00432-004-0638-8. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 14.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 15.Norbury C, Nurse P. Animal cell cycles and their control. Annu Rev Biochem. 1992;61:441–470. doi: 10.1146/annurev.bi.61.070192.002301. [DOI] [PubMed] [Google Scholar]
- 16.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 17.Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 18.Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 19.Lygerou Z, Nurse P. Cell cycle. License withheld - geminin blocks DNA replication. Science. 2000;290:2271–2273. doi: 10.1126/science.290.5500.2271. [DOI] [PubMed] [Google Scholar]
- 20.Sahai E, Marshall CJ. ROCK and Dia have opposing effects on adherens junctions downstream of Rho. Nat Cell Biol. 2002;4:408–415. doi: 10.1038/ncb796. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol. 1999;1:136–143. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- 22.Ishizaki T, Naito M, Fujisawa K, Maekawa M, Watanabe N, Saito Y, Narumiya S. p160ROCK, a Rho-associated coiled-coil forming protein kinase, works downstream of Rho and induces focal adhesions. FEBS Lett. 1997;404:118–124. doi: 10.1016/S0014-5793(97)00107-5. [DOI] [PubMed] [Google Scholar]
- 23.Fu Q, Yu L, Liu Q, Zhang J, Zhang H, Zhao S. Molecular cloning, expression characterization, and mapping of a novel putative inhibitor of rho GTPase activity, RTKN, to D2S145-D2S286. Genomics. 2000;66:328–332. doi: 10.1006/geno.2000.6212. [DOI] [PubMed] [Google Scholar]
- 24.Morgan DO. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 25.Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]