Abstract
BRCA mutated ovarian cancers respond better to platinum-based therapy and to the recently approved PARP-inhibitors. There is the need for efficient and timely methods to detect both somatic and germline mutations using formalin-fixed paraffin-embedded (FFPE) tissues and commercially available technology. We used a commercial kit exploring all exons and 50bp exon-intron junctions of BRCA1 and BRCA2 genes, and semiconductor next-generation sequencing (NGS) on DNA from 47 FFPE samples of high-grade serous ovarian cancers. Pathogenic mutations were found in 13/47 (28%) cancers: eight in BRCA1 and five in BRCA2. All BRCA1 and two BRCA2 mutations were germline; three BRCA2 mutations were somatic. All mutations were confirmed by Sanger sequencing. To evaluate the performance of the NGS panel, we assessed its capability to detect the 6,953 variants described for BRCA1 and BRCA2 in ClinVar and COSMIC databases using callability analysis. 6,059 (87.1%) variants were identified automatically by the software; 829 (12.0%) required visual verification. The remaining 65 (0.9%) variants were uncallable, and would require 15 Sanger reactions to be resolved. Thus, the sensitivity of the NGS-panel was 99.1%. In conclusion, NGS performed with a commercial kit is highly efficient for detection of germline and somatic mutations in BRCA genes using routine FFPE tissue.
Keywords: BRCA1-BRCA2, ovarian carcinoma, next generation sequencing, PARP inhibitor, olaparib
INTRODUCTION
Ovarian cancer is the most deadly tumour of the female reproductive system with 238,700 new cases and 151,900 deaths worldwide [1-3], of which 65,500 new cases and 42,700 deaths in Europe in 2012 [4]. In Italy, ovarian cancer accounts for 30% of all tumours of the female genital apparatus, and in 2013 there were 4,800 new cases and 37,829 prevalent cases [5].
BRCA1 and BRCA2 are among the most frequently mutated genes in high-grade ovarian serous carcinoma, which is responsible for the vast majority of ovarian cancer deaths [6, 7]. BRCA1 and BRCA2 genes are key partners of the homologous recombination (HR) DNA repair system, together with ATM, BARD1, NBN and other genes [6]. Indeed, germline and somatic mutations in HR genes occur in about 30% of patients with ovarian carcinoma, of which up to 75% are in BRCA1 and BRCA2 genes [6, 8].
Patients carrying a germline or somatic BRCA1/BRCA2 mutation have been associated with a better prognosis and a better response to platinum-based therapy [8-11]. A particular class of drugs, poly(ADP-ribose) polymerase-inhibitors (PARPi), has been shown to be effective for targeted treatment of cancers harbouring BRCA1 or BRCA2 mutations [12-18]. The PARP-1 protein is critical to the repair of single-strand DNA breaks. In cells with defective HR, such as the BRCA mutations carriers, PARP-1 inhibition is synthetic lethal and results in cell cycle arrest and subsequent apoptosis [15]. On December 2014 the European Medicines Agency (EMA) and the U.S.A. Food and Drug Administration (FDA) approved the PARPi olaparib [13-15] for treatment of BRCA1/BRCA2 mutated ovarian cancer.
Investigating BRCA mutational status in ovarian cancer patients has thus a key role, not only for the identification of familial cancer predisposition but also to address therapeutic choices. Germline testing of BRCA is widespread in medical genetics laboratories, but this approach excludes patients with somatic BRCA mutations, i.e. only present in cancer cells, from the opportunity to avail of PARPi therapies. Testing BRCA on formalin-fixed paraffin-embedded (FFPE) samples would permit the simultaneous assessment of both somatic and germline mutations using an easily accessible material that is routinely available in any pathology laboratory worldwide.
High throughput next-generation sequencing (NGS) technologies permit fast multiplex testing on small quantities of DNA with budding applications in cancer diagnostics as they improve both the capacity and the cost-effectiveness of mutational analysis compared with Sanger [19-23]. To assess the feasibility of using NGS in routine diagnostic activity for BRCA analysis, we have investigated BRCA1 and BRCA2 mutations in 47 high-grade serous tumours of the ovary, using a commercially available kit and semiconductor NGS on FFPE tissue samples.
RESULTS
The results of NGS targeted sequencing are reported in Table 1 and an example is shown Figure 1. DNA from all samples was successfully amplified in multiplex PCR and an adequate library for NGS was obtained. The mean read length was 112 base pairs and a mean coverage of 3,507x was achieved, with 99.6% target bases covered more than 100x.
Table 1. Pathogenic mutations in BRCA1 and BRCA2 detected by next-generation sequencing of 47 ovarian cancers.
| Case | BRCA1 | BRCA2 | Mutation type | Germline-somatic | dbSNP ID | ClinVar class |
|---|---|---|---|---|---|---|
| 3506 | c.5329dupC p.Gln1777ProfsTer74 | - | Frameshift | Germline | rs397507247 | Pathogenic |
| 3513 | c.676delT p.Cys226ValfsTer8 | - | Frameshift | Germline | rs80357941 | Pathogenic |
| 3508 | c.1687C>T p.Gln563Ter | - | Nonsense | Germline | rs80356898 | Pathogenic |
| 3521 | c.2405_2406delTG p.Val802GlufsTer7 | - | Frameshift | Germline | rs80357706 | Pathogenic |
| 3528 | c.2405_2406delTG p.Val802GlufsTer7 | - | Frameshift | Germline | rs80357706 | Pathogenic |
| 3489 | c.3767_3768delCA p.Thr1256ArgfsTer10 | - | Frameshift | Germline | rs730881440 | Pathogenic |
| 3505 | c.5125_5127delGTT p.Val1709del | - | In-frame deletion | Germline | rs80358344 | Pathogenic |
| 3520 | c.5309C>T p.Pro1770Leu | - | Missense | Germline | - | -* |
| 3512 | - | c.2813delC p.Ala938GlufsTer22 | Frameshift | Somatic | - | -** |
| 3523 | - | c.6202dupA p.Ile2068AsnfsTer10 | Frameshift | Germline | rs397507833 | Pathogenic |
| 3514 | - | c.6574delA p.Met2192TrpfsTer14 | Frameshift | Germline | - | -** |
| 3501 | - | c.7069_7070delCT p.Leu2357ValfsTer2 | Frameshift | Somatic | rs80359636 | Pathogenic |
| 3516 | - | c.8614G>T p.Glu2872Ter | Nonsense | Somatic | - | -** |
This variant is not recorded on CinVar, but a variant on the very same codon, ClinVar variant :c.5309C>G (p.Pro1770Arg, rs80357462) is recorded as Pathogenic.
These variants are not recorded in either dbSNP or ClinVar, however they cause a premature stop codon, which is a feature of pathogenic mutations.
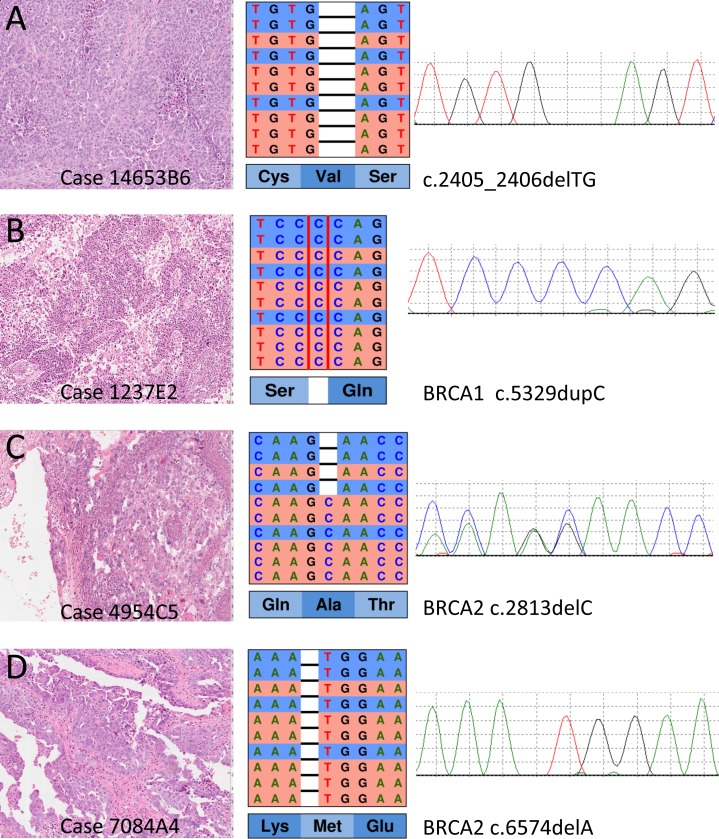
Figure 1. Representative examples of mutations detected at next generation sequencing with the HR1 kit and validated by Sanger sequencing.
On the left, the histological section of the primary ovarian cancer (Hematoxylin and eosin stain) from which DNA has been prepared after microdisection of the most cellular areas. In the middle, the representation of the results of next-generation sequencing where the reads (red for forward and blue for reverse) are aligned to the reference genome as provided by the Integrative Genomics Viewer (IGV v.2.3, Broad Institute) software. On the right, the representation of Sanger sequencing results for each cancer to validate the mutations (forward strand was used in A. B. and D.; reverse strand was used in C.). The BRCA1 mutations in A, B and the BRCA2 mutation in D are homozygous in cancer tissue as shown in both IGV and Sanger representations; these mutations were heterozygous in germline DNA of the respective patients. The BRCA2 mutation in C was heterozygous in tumour tissue, and its somatic nature was determined by its absence in matching normal DNA (not shown).
Pathogenic variants
Pathogenic mutations in BRCA genes were found in 13 of the 47 (28%) ovarian cancers: eight were in BRCA1 and five in BRCA2 (Table 1).
Of the eight mutations in BRCA1 gene, five were frame-shifts resulting in a premature stop codon, one was a nonsense mutation, one was an in-frame single codon deletion and one was a missense mutation. Frame-shift, nonsense and in-frame deletion mutations are recorded as pathogenic in the ClinVar database; the only missense mutation found (c.5309C>T; p.Pro1770Leu) has not yet been recorded, but falls in the same codon where a pathogenic missense mutation (c.5309C>G; p.Pro1770Arg) is recorded in ClinVar (Table 1). All of the BRCA1 mutations were germline as assessed by the analysis of DNA from matched normal tissue.
Of the five BRCA2 mutations, four were frame-shifts (three deletions and one insertion) resulting in a premature stop codon and one was a nonsense point mutation. Three of the BRCA2 mutations were somatic and two were germline as assessed by the analysis of matched normal tissue DNA (Table 1). One of the somatic (c.7069_7070delCT) and one of the germline (c.6202dupA) mutations are recorded as pathogenic in the ClinVar database. The remaining two somatic and one germline BRCA2 mutations have not yet been recorded but were considered pathogenic as they cause a premature stop codon, a feature of pathogenic mutations.
All the above variants were confirmed at Sanger sequencing (Figure 1), therefore our estimated specificity was 100%.
Additional variants
In the BRCA1 gene, an additional germline variant, c.3119G>A (p.Ser1040Asn), was found in two cases. This variant is recorded as benign/likely benign in the ClinVar database by eight submitters, as uncertain by one and as pathogenic by one; its frequency in the global population is 1% according to the 1000Genomes project database [24]. In the BRCA2 gene, two germline single nucleotide polymorphisms (SNP) were also detected in 29 patients: 24 harboured the c.1114A>C (p.Asn372His) SNP, three the c.865A>C (p.Asn289His) SNP, and two patients had both. The first SNP, c.1114A>C (p.Asn372His), has been recorded as benign in the ClinVar database by four submitters and as pathogenic by one; its frequency in the global population is 25% according to the 1000Genomes project database. The second SNP, c.865A>C (p.Asn289His), has been recorded as benign by seven submitters; its frequency in the global population is 7%.
Sensitivity of targeted NGS
To evaluate the performance of the NGS panel, we assessed its capability to detect the 6,953 variants described for BRCA1 and BRCA2, comprising 6,106 germline variants in the ClinVar database and 1,071 somatic mutations in COSMIC database, of which 224 overlap (Table 2).
Table 2. Somatic and Germline BRCA mutation callability analysis of HR1 Next-Generation kit vs Sanger sequencing.
| GENE | Coding region (bp) | ClinVar – COSMIC Variants* | Total variants* | N. Sanger needed$ | Callability ° of HR1 Next-Generation Sequencing and Variant Caller software | |||
|---|---|---|---|---|---|---|---|---|
| AutomaticCalls | Called by IGV** | Uncallable*** | Sensitivity (%) | |||||
| BRCA1 | 5,659 | 2,522 - 398 | 2,841 | 55 | 2,520 | 320 | 1 | 99.9 |
| BRCA2 | 10,262 | 3,584 - 673 | 4,112 | 77 | 3,539 | 509 | 64 | 98.4 |
| Total | 15,921 | 6,106 - 1,071 | 6,953 | 132 | 6,059 | 829 | 65 | 99.1 |
Callability analyis using the Torrent Variant Caller.
Germline variants listed in the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/) and mutations in COSMIC database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/). The number of total variants is less than the sum of variants from both databases (7177) because 224 variants overlap.
Estimate based on one reaction per 150 bp (one per exon if <150 bp) using DNA from formalin-fixed paraffin-embedded tissue
Visual verification of sequences with Integrative Genomics Viewer (IGV v.2.3, Broad Institute) software.
Mutations within homopolymer stretches, artefact-prone regions of the genes, or not covered by the NGS panel. The number of Sanger to solve these 65 blind spots would be 1 for BRCA1 and 14 for BRCA2.
Analysis of callable/uncallable/poorly mapped loci was performed to discriminate between variants which can be automatically detected by the software from those that are hindered by sequencing errors due to homopolymers or PCR amplification artefacts. The procedure is described in detail in the Methods section. This analysis showed that the HR1 NGS-kit obtained a clear sequence of the DNA regions harbouring 6059 (87.1%) of these variants that were automatically identified by the Variant Caller Plugin software (Torrent Suite Software v4.6; Life Technologies), while the regions harbouring the remaining 894 (12.9%) variants were challenging for automatic detection. Of these, 829 (12.0%) variants were automatically identified by the software but with low confidence due to their proximity to homopolymer stretches or artefact-prone regions and thus required confirmation/correction by visual inspection of the region using the Integrative Genomics Viewer (IGV) v2.3 (Broad Institute). In detail, the number of variants that required visual inspection to be confirmed was 320 for BRCA1 and 509 for BRCA2. The remaining 65 (0.9%) variants were uncallable. Of these 65, 13 (0.2%) were single/double base insertion or deletions located within homopolymer stretches, 1 for BRCA1 and 12 for BRCA2, and both software and visual inspection were insufficient to discern between an artefact and a true alteration. The remaining 52 (0.7%) variants reside in regions of BRCA2 that are difficult to amplify by current NGS library approaches for FFPE tissue, and thus could not be amplified in HR1 panel. Uncallable variants would require 15 Sanger sequencing reactions to be resolved (Table 2). In conclusion, the sensitivity of BRCA1 and BRCA2 NGS sequencing was 99.1%.
DISCUSSION
The assessment of BRCA mutational status in ovarian cancer patients plays a double role. The first is the identification of familial cancer predisposition; the second is to address therapeutic choices.
BRCA1 and BRCA2 germline mutations are known risk factors for ovarian cancer [2, 7, 25-33], and it has been reported that up to 44% of patients without a family history of ovarian cancer harbour a germline BRCA1 or BRCA2 mutation, and as such may act as first alert for descendants [10].
The better response to platinum and the recent approval of PARP-inhibitors for therapy of ovarian cancers harbouring mutations in BRCA genes calls for methods able to detect not only germline but also somatic mutations in these genes.
Molecular tests to analyse BRCA mutations are generally carried out on germline DNA from blood samples to identify hereditary mutations as part of risk assessment programs. However, in order to identify somatic mutations present in neoplastic cells to permit the patient to benefit from drugs such as olaparib, it is necessary to analyze DNA from the tumour tissue.
The analysis of BRCA on diagnostic tissue is complex due to: 1) the type of possible mutations in these genes; 2) the fact that these mutations may be found in any part of these genes which are very large and as such require the entire gene to be sequenced; and 3) the limited quantity and low quality of DNA available from routine diagnostic FFPE tissue is limited. It is therefore important to be able to sequence the entire BRCA genes using minimal amounts of DNA to identify pathogenic mutations relevant to patient treatment.
In this study, we tested a commercially available NGS sequencing panel on small quantities of DNA purified from FFPE tissue to evaluate the use of these technologies for diagnostic applications.
The analysis of 47 high grade serous ovarian cancers identified 13 (28%) pathogenic mutations, 8 BRCA1 and 5 BRCA2. All BRCA1 and two BRCA2 mutations were germline while three BRCA2 mutations were somatic. BRCA1 and BRCA2 mutations were mutually exclusive. All 13 BRCA variants were confirmed by Sanger sequencing, demonstrating an estimated specificity of 100%.
To evaluate sensitivity of the NGS panel, we assessed its capability to detect all 6,953 germline or somatic variants described for BRCA1 and BRCA2 in the ClinVar and COSMIC databases using the analysis of callable loci (details in Methods section). A total of 6,059 (87.1%) of these variants were automatically identified by the Torrent Variant Caller software. A further 829 (12.0%) were imprecisely identified by the software and required confirmation/correction by visual inspection of the region. This was due to the mutations being either inside or in close proximity of homopolymer stretches or PCR amplification artefacts. The remaining 65 (0.9%) variants were uncallable resulting in the sensitivity of the HR1 NGS-panel at 99.1%, as it would miss 65/6,953 variants. These could be resolved, however, with 15 Sanger sequencing reactions.
In conclusion, our study shows that next-generation sequencing performed with a commercial kit (HR1, 4Bases) is highly efficient for detection of germline and somatic mutations in BRCA1 and BRCA2 genes using DNA from routinely available FFPE tissue.
MATERIALS AND METHODS
Cases
The study series comprised 47 samples of high grade serous ovarian carcinomas, diagnosed according to WHO classification criteria [2], from patients who underwent surgical resection between 2010 and 2015 (mean age 61±12 years, median 62 years) at the Department of Obstetrics and Gynaecology of the University Hospital Trust of Verona. Five patients were diagnosed at stage IV, 35 at stage IIIC, 4 at stage I and 3 at stage IC, according to FIGO staging system [34].
Ethics
The samples were acquired from the Integrated University Hospital Trust of Verona Pathology archives under the amended Program 1885, that permits the acquisition of FFPE samples by the ARC-Net (Applied Research on Cancer) biobank of the University of Verona and the Hospital Trust of Verona following the re-consent of patients or the anonymization of samples under protocol 52438 Prog. 1885 approved 23/11/2010. The amendment also addresses the regulatory issue of data protection and disclosure in genomic studies. Subsequent approval for this study was presented and approved under protocol 44541 on 29/09/2015.
DNA extraction and qualification
FFPE samples of ovarian cancers were enriched for neoplastic cellularity to a minimum of 70% by manual microdissection of 10 consecutive 4-μm sections. DNA was purified using the QIAamp FFPE Tissue Kit (Qiagen) and qualified as previously reported [35, 36]. Briefly, DNA was quantified using Qubit (LifeTechnologies) platforms, and its quality was further evaluated by NanoDrop (Life Technologies) and PCR analysis using the BIOMED 2 PCR multiplex protocol [36].
Deep sequencing of multiplex PCR amplicons
The HR1 kit (4bases SA, Switzerland) was used. This kit explores all exons of BRCA1 (n=24; NM_007300.3) and BRCA2 (n=27; NM_000059.3) and 50 bp exon-intron junctions. The kit is composed of three multiplex PCR primer pools, and uses 30 nanograms of DNA, ten per primer pool, for multiplex PCR amplification, followed by ligation of a specific barcode-sequence to each sample for identification The quality of the obtained libraries was evaluated by the Agilent 2100 Bioanalyzer on-chip electrophoresis (Agilent Technologies) as previously described [35]. Emulsion PCR for clonal amplification of libraries was performed with the Ion OneTouch OT2 System and the Hi-Q OT2 200 Kit (Life Technologies); libraries were processed in batches of eight samples per emulsion. Sequencing of the libraries was performed on Personal Genome Machine (PGM, Life Technologies) using the Ion 318 Chip and the Ion PGM Hi-Q Sequencing 200 Kit (Life Technologies).
Data analysis and variant calling
Data analysis, including alignment to the hg19 human reference genome and variant calling, was done using the Torrent Suite Software v4.6 (Life Technologies). Filtered variants were annotated using a custom pipeline based on vcflib (https://github.com/ekg/vcflib), SnpSift [37], the Variant Effect Predictor (VEP) software [38] and NCBI RefSeq database. Alignments were visually verified with the Integrative Genomics Viewer (IGV) v2.3 [39].
Analysis of callable loci
Analysis of callable/uncallable/poorly mapped loci was performed using the Torrent Variant Caller, to discriminate between variants which can be automatically detected from those that are hindered by sequencing errors due to homopolymers or PCR amplification artefacts. SNPs and small (<100 bp) INDELs spanning the coding regions of BRCA1 and BRCA2 were retrieved from the ClinVar and COSMIC databases in VCF format, converted to a Hotspots file and used to guide variant calling. In this way, the variant caller is forced to analyse a given hotspot coordinate; if there is no mutation, the software outputs that the position is “reference”; otherwise it outputs the mutation detected. If there are problems in the sequence at that position, the software outputs a “no call” value indicating if variant calling failed due to strand bias, low quality of bases, noise in the sequence, poor mapping. All the “no call” positions were further inspected by visual verification of the alignment file to ascertain whether the “no call' status was due to artefacts or homopolymer misalignment. This analysis was carried out for each of the 47 samples and the frequency of “no call” status for each hotspot coordinate was recorded.
DNA Sanger sequencing
Mutations of BRCA1 and BRCA2 were validated by Sanger sequencing (primer sequences available upon request). PCR products were purified using Agencourt AMPure XP magnetic beads (Beckman Coulter) and labelled with BigDye® Terminator v3.1 (Applied Biosystems). Agencourt CleanSEQ magnetic beads (Beckman Coulter) were used for post-labeling DNA fragment purification, and sequence analysis was performed on the Applied Biosystems 3130xl Genetic Analyzer.
Acknowledgments
We thank Giada Bonizzato, Sonia Grimaldi, and Paola Merlini of the ARC-Net biobank.
Abbreviations
- EMA
European Medicines Agency
- FDA
Food and Drug Administration
- FFPE
formalin-fixed paraffin embedded
- FIGO
International Federation of Gynecology and Obstetrics
- HR
homologous recombination
- IGV
Integrative Genomics Viewer
- NGS
Next-Generation Sequencing
- PARP
poly(ADP-ribose) polymerase
- PARPi
poly(ADP-ribose) polymerase-inhibitors
- PCR
polymerase chain reaction
- PGM
personal genome machine
- SNP
single nucleotide polymorphism
Footnotes
GRANT SUPPORT
Italian Cancer Genome Project grant from the Italian Ministry of Research (FIRB - RBAP10AHJB), Associazione Italiana Ricerca sul Cancro (AIRC grant n. 12182), and Seventh Framework European Community grant (Cam-Pac, Grant agreement no: 602783).
CONFLICTS OF INTEREST
4bases provided kits free of charge.
Authors' contribution
RTL, CB, AS: ideation and planning of the study; AP, EP: histopathological evaluation according to WHO classification; BR: sample microdissection; NS: preparation and quality control of DNA; MS, AM: next-gen sequencing including data analysis and interpretation; GTu: Sanger sequencing; CL, IC: sample choice and preparation, collection and assembly of clinical-pathological data; AS, MF, GTo: clinical-pathological data analysis and interpretation of results related to clinical implications of the peculiar tumour type; all authors participated in writing and approved the final, submitted manuscript.
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Kurman RJ, Carcangiu ML, Herrington CS, Young RH. WHO Classification of Tumours of Female Reproductive Organs. Lyon: International Agency for Research on Cancer (IARC); 2014. [Google Scholar]
- 3.Tewari KS, Monk BJ. The 21st century handbook of clinical ovarian cancer. Berlin Heidelberg New York: Springer; 2015. [Google Scholar]
- 4.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 5.AIOM-AIRTUM 2015 The numbers of cancer in Italy ( http://www.aiom.it/C_Common/Download.asp?file=/$Site$/2015_I_Numeri_del_cancro.pdf)
- 6.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilton JL, Geisler JP, Rathe JA, Hattermann-Zogg MA, DeYoung B, Buller RE. Inactivation of BRCA1 and BRCA2 in ovarian cancer. J Natl Cancer Inst. 2002;94:1396–1406. doi: 10.1093/jnci/94.18.1396. [DOI] [PubMed] [Google Scholar]
- 8.Pennington KP, Walsh T, Harrell MI, Lee MK, Pennil CC, Rendi MH, Thornton A, Norquist BM, Casadei S, Nord AS, Agnew KJ, Pritchard CC, Scroggins S, Garcia RL, King MC, Swisher EM. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20:764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dann RB, DeLoia JA, Timms KM, Zorn KK, Potter J, Flake DD, 2nd, Lanchbury JS, Krivak TC. BRCA1/2 mutations and expression: response to platinum chemotherapy in patients with advanced stage epithelial ovarian cancer. Gynecol Oncol. 2012;125:677–682. doi: 10.1016/j.ygyno.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Alsop K, Fereday S, Meldrum C, deFazio A, Emmanuel C, George J, Dobrovic A, Birrer MJ, Webb PM, Stewart C, Friedlander M, Fox S, Bowtell D, Mitchell G. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol. 2012;30:2654–2663. doi: 10.1200/JCO.2011.39.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolton KL, Chenevix-Trench G, Goh C, Sadetzki S, Ramus SJ, Karlan BY, Lambrechts D, Despierre E, Barrowdale D, McGuffog L, Healey S, Easton DF, Sinilnikova O, Benitez J, Garcia MJ, Neuhausen S, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307:382–390. doi: 10.1001/jama.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, De Greve J, Lubinski J, Shanley S, Messiou C, A'Hern R, Tutt A, Ashworth A, Stone J, Carmichael J, Schellens JH, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 13.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott C, Meier W, Shapira-Frommer R, Safra T, Matei D, Macpherson E, Watkins C, Carmichael J, Matulonis U. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 14.Lee JM, Hays JL, Annunziata CM, Noonan AM, Minasian L, Zujewski JA, Yu M, Gordon N, Ji J, Sissung TM, Figg WD, Azad N, Wood BJ, Doroshow J, Kohn EC. Phase I/Ib study of olaparib and carboplatin in BRCA1 or BRCA2 mutation-associated breast or ovarian cancer with biomarker analyses. J Natl Cancer Inst. 2014;106:dju089. doi: 10.1093/jnci/dju089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tewari KS, Eskander RN, Monk BJ. Development of Olaparib for BRCA-Deficient Recurrent Epithelial Ovarian Cancer. Clin Cancer Res. 2015;21:3829–3835. doi: 10.1158/1078-0432.CCR-15-0088. [DOI] [PubMed] [Google Scholar]
- 16.Wagner LM. Profile of veliparib and its potential in the treatment of solid tumors. Onco Targets Ther. 2015;8:1931–1939. doi: 10.2147/OTT.S69935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oza AM, Cibula D, Benzaquen AO, Poole C, Mathijssen RH, Sonke GS, Colombo N, Spacek J, Vuylsteke P, Hirte H, Mahner S, Plante M, Schmalfeldt B, Mackay H, Rowbottom J, Lowe ES, et al. Olaparib combined with chemotherapy for recurrent platinum-sensitive ovarian cancer: a randomised phase 2 trial. Lancet Oncol. 2015;16:87–97. doi: 10.1016/S1470-2045(14)71135-0. [DOI] [PubMed] [Google Scholar]
- 18.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott CL, Meier W, Shapira-Frommer R, Safra T, Matei D, Fielding A, Spencer S, Dougherty B, Orr M, Hodgson D, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 19.Bria E, Pilotto S, Amato E, Fassan M, Novello S, Peretti U, Vavala T, Kinspergher S, Righi L, Santo A, Brunelli M, Corbo V, Giglioli E, Sperduti I, Milella M, Chilosi M, et al. Molecular heterogeneity assessment by next-generation sequencing and response to gefitinib of EGFR mutant advanced lung adenocarcinoma. Oncotarget. 2015;6:12783–95. doi: 10.18632/oncotarget.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luchini C, Capelli P, Fassan M, Simbolo M, Mafficini A, Pedica F, Ruzzenente A, Guglielmi A, Corbo V, Scarpa A. Next-generation histopathologic diagnosis: a lesson from a hepatic carcinosarcoma. J Clin Oncol. 2014;32:e63–66. doi: 10.1200/JCO.2012.47.5855. [DOI] [PubMed] [Google Scholar]
- 21.Mafficini A, Amato E, Fassan M, Simbolo M, Antonello D, Vicentini C, Scardoni M, Bersani S, Gottardi M, Rusev B, Malpeli G, Corbo V, Barbi S, Sikora KO, Lawlor RT, Tortora G, et al. Reporting Tumor Molecular Heterogeneity in Histopathological Diagnosis. PLoS One. 2014;9:e104979. doi: 10.1371/journal.pone.0104979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scarpa A, Sikora K, Fassan M, Rachiglio AM, Cappellesso R, Antonello D, Amato E, Mafficini A, Lambiase M, Esposito C, Bria E, Simonato F, Scardoni M, Turri G, Chilosi M, Tortora G, et al. Molecular typing of lung adenocarcinoma on cytological samples using a multigene next generation sequencing panel. PLoS One. 2013;8:e80478. doi: 10.1371/journal.pone.0080478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simbolo M, Mafficini A, Agostini M, Pedrazzani C, Bedin C, Urso ED, Nitti D, Turri G, Scardoni M, Fassan M, Scarpa A. Next-generation sequencing for genetic testing of familial colorectal cancer syndromes. Hered Cancer Clin Pract. 2015;13:18. doi: 10.1186/s13053-015-0039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majdak EJ, De Bock GH, Brozek I, Perkowska M, Ochman K, Debniak J, Milczek T, Cornelisse CJ, Jassem J, Emerich J, Limon J, Devilee P. Prevalence and clinical correlations of BRCA1/BRCA2 unclassified variant carriers among unselected primary ovarian cancer cases - preliminary report. Eur J Cancer. 2005;41:143–150. doi: 10.1016/j.ejca.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 26.Rafnar T, Benediktsdottir KR, Eldon BJ, Gestsson T, Saemundsson H, Olafsson K, Salvarsdottir A, Steingrimsson E, Thorlacius S. BRCA2, but not BRCA1, mutations account for familial ovarian cancer in Iceland: a population-based study. Eur J Cancer. 2004;40:2788–2793. doi: 10.1016/j.ejca.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Sarantaus L, Vahteristo P, Bloom E, Tamminen A, Unkila-Kallio L, Butzow R, Nevanlinna H. BRCA1 and BRCA2 mutations among 233 unselected Finnish ovarian carcinoma patients. Eur J Hum Genet. 2001;9:424–430. doi: 10.1038/sj.ejhg.5200652. [DOI] [PubMed] [Google Scholar]
- 28.Stratton JF, Gayther SA, Russell P, Dearden J, Gore M, Blake P, Easton D, Ponder BA. Contribution of BRCA1 mutations to ovarian cancer. N Engl J Med. 1997;336:1125–1130. doi: 10.1056/NEJM199704173361602. [DOI] [PubMed] [Google Scholar]
- 29.Yazici H, Glendon G, Yazici H, Burnie SJ, Saip P, Buyru F, Bengisu E, Andrulis IL, Dalay N, Ozcelik H. BRCA1 and BRCA2 mutations in Turkish familial and non-familial ovarian cancer patients: a high incidence of mutations in non-familial cases. Hum Mutat. 2002;20:28–34. doi: 10.1002/humu.10090. [DOI] [PubMed] [Google Scholar]
- 30.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schrader KA, Hurlburt J, Kalloger SE, Hansford S, Young S, Huntsman DG, Gilks CB, McAlpine JN. Germline BRCA1 and BRCA2 mutations in ovarian cancer: utility of a histology-based referral strategy. Obstet Gynecol. 2012;120:235–240. doi: 10.1097/AOG.0b013e31825f3576. [DOI] [PubMed] [Google Scholar]
- 32.Pruthi S, Gostout BS, Lindor NM. Identification and Management of Women With BRCA Mutations or Hereditary Predisposition for Breast and Ovarian Cancer. Mayo Clin Proc. 2010;85:1111–1120. doi: 10.4065/mcp.2010.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Excellence NIfHaC NICE guidelines [CG164] 2013 [Google Scholar]
- 34.Prat J, Oncology FCoG. Staging classification for cancer of the ovary, fallopian tube, and peritoneum. Int J Gynaecol Obstet. 2014;124:1–5. doi: 10.1016/j.ijgo.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Simbolo M, Gottardi M, Corbo V, Fassan M, Mafficini A, Malpeli G, Lawlor RT, Scarpa A. DNA qualification workflow for next generation sequencing of histopathological samples. PLoS One. 2013;8:e62692. doi: 10.1371/journal.pone.0062692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zamo A, Bertolaso A, van Raaij AW, Mancini F, Scardoni M, Montresor M, Menestrina F, van Krieken JH, Chilosi M, Groenen PJ, Scarpa A. Application of microfluidic technology to the BIOMED-2 protocol for detection of B-cell clonality. J Mol Diagn. 2012;14:30–37. doi: 10.1016/j.jmoldx.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 37.Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, Lu X. Using Drosophila melanogaster as a Model for Genotoxic Chemical Mutational Studies with a New Program, SnpSift. Front Genet. 2012;3:35. doi: 10.3389/fgene.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]