Abstract
Intratumor heterogeneity (ITH) leads to an underestimation of the mutational landscape portrayed by a single needle biopsy and consequently affects treatment precision. The extent of colorectal cancer (CRC) genetic ITH is not well understood in Chinese patients. Thus, we conducted deep sequencing by using the OncoGxOne™ Plus panel, targeting 333 cancer-specific genes in multi-region biopsies of primary and liver metastatic tumors from three Chinese CRC patients. We determined that the extent of ITH varied among the three cases. On average, 65% of all the mutations detected were common within individual tumors. KMT2C aberrations and the NCOR1 mutation were the only ubiquitous events. Subsequent phylogenetic analysis showed that the tumors evolved in a branched manner. Comparison of the primary and metastatic tumors revealed that PPP2R1A (E370X), SETD2 (I1608V), SMAD4 (G382T), and AR splicing site mutations may be specific to liver metastatic cancer. These mutations might contribute to the initiation and progression of distant metastasis. Collectively, our analysis identified a substantial level of genetic ITH in CRC, which should be considered for personalized therapeutic strategies.
Introduction
Colorectal cancer (CRC) is one of the leading causes of death worldwide, accounting for an estimated 1.2 million new cases and 600,000 deaths every year [1]. CRC is the third most prevalent cancer and the fifth leading cause of cancer deaths in China. The incidence of CRC has also increased in the Chinese population in recent years [2]. Although considerable progress has been made regarding CRC treatment, including targeted therapy, the five-year relative survival of patients with distant metastases is only 12.5% [3]. Therefore, an extensive understanding of the molecular biology of CRC is highly desirable. These new insights would subsequently improve the treatment of CRC.
Intratumor heterogeneity (ITH) leads to an underestimation of the mutational landscape portrayed by a single needle biopsy and consequently affects treatment precision. With the development of next-generation sequencing (NGS), ITH has recently been elucidated in substantial detail in several cancer types [4–7], including CRC [8, 9]. Kim et al. [8] conducted multi-region biopsies of primary and liver metastatic regions from five CRCs through whole-exome sequencing and observed that only 19.8%–53.9% of the mutations in a given sample were universal. They also identified APC, KRAS, and TP53 mutations in all regional biopsies. The sequencing coverage of their study is relative low, which may lead to overlook the low frequency mutation. Whereas in Kogita study, [9] they only targeted 50 cancer genes, and hence the identified the extent of CRC genetic ITH was limited. In addition the extent of CRC genetic ITH has not been established in Chinese patients.
We conducted deep coverage sequencing by using the SureSelect Target Enrichment Kit and OncoGxOne™ Plus panel of DNA from multi-region biopsies of primary and liver metastatic tumors of three Chinese CRC patients to characterize CRC ITH in Chinese patients. We determined the mutation landscape and identified the substantial level of genetic ITH in CRC.
Materials and Methods
Patients and samples
Three patients with multi-regional primary colon carcinoma surgically resected in the First People’s Hospital of Yunnan Province between October 2013 and April 2014 were enrolled in the study. Pathological tumor staging was conducted in accordance with the TNM classification (Table 1). This study was approved by the ethics committee of The First People's Hospital of Yunnan Province (2014YXLH029). All patients in the study provided written informed consent for the use of resected tissue.
Table 1. Basic patient characteristics.
| Sample | Age | Gender | Location | Cell Type | Differentiation | T stage | N stage | M stage |
|---|---|---|---|---|---|---|---|---|
| P1 | 62 | male | colon | adenoca | Moderately | 2 | 1 | 0 |
| P2 | 60 | female | colon | adenoca | Moderately | 2 | 2 | 0 |
| P3 | 71 | female | colon | adenoca | Hight and Moderately | 3 | 2 | 2 |
DNA isolation
The Formalin-fixed, paraffin-embedded (FFPE) specimens were subjected to histological review, only those containing sufficient tumor cells (at least 70% tumor cells), as determined by hematoxylin and eosin staining, and matched normal tissues were selected for DNA isolation. Genomic DNA was isolated by using the QIAamp® DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s procedure with slight modifications. Genomic DNA samples were quantified by using the NanoDrop 2000 spectrophotometer (Agilent, Santa Clara, CA, USA). The isolated DNA was stored at −80°C until analysis.
Microsatellite testing
The microsatellite instability (MSI) status was determined for each case by using five mononucleotide or dinucleotide microsatellites markers (BAT25, BAT26, D17S250, D2S123, and D5S346) [10]. Primers were 5′-labeled with HEX, FAM, or TET (Sangon Biotech Co., Ltd., Shanghai, China). All microsatellite loci were amplified from matched normal and tumor DNA through fluorescence multiplex polymerase chain reaction (PCR). Then, the PCR products were sequenced on the ABI 3730XL automated sequencer by using a fragment analysis software (Gene Scan, Perkin Elmer, Waltham, MA, USA). Microsatellite marker stability was analyzed by using the GeneMapper software. The MSI status was classified as MSI-high if ≥30% of markers were unstable, MSI-low if <30% of markers were unstable, and no shifts or additional peaks for microsatellite stable (MSS) colon cancer.
Target enrichment and sequencing
Sample sequence enrichment and library preparation was conducted by using the SureSelect Target Enrichment Kit (Agilent, Santa Clara, CA, USA) and OncoGxOne™ Plus cancer panel (GENEWIZ, Inc., South Plainfield, NJ, USA) according to the manufacturer’s instructions. OncoGxOne Plus, a targeted oncology panel, contains 333 cancer-specific genes (including all known cancer driver genes and 64 targeted and chemotherapy related genes). Briefly, 200 ng of genomic DNA from each sample was fragmented by acoustic shearing on a Covaris instrument. Fractions of 150–200 bp were ligated to SureSelect Adaptor Oligo and PCR-amplified for 10 cycles. For target enrichment, the entire library was hybridized by using the SureSelect Oligo Capture Kit for 16 or 24 h at 65°C. Biotinylated hybrids were captured, and the enriched libraries were completed under 16 PCR cycles. The resulting purified libraries were pooled and sequenced by using the Illumina® MiSeq™ NGS instrument for 2 × 150 paired-end sequencing reads (Illumina, San Diego, CA, USA) according to the manufacturer’s protocols. The genes in the OncoGxOne™ Plus cancer panel are listed in S1 Table.
Bioinformatics analysis
Sequencing data were accessed through the MiSeq Reporter. Data quality was checked by using FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/) and then aligned to the human reference genome (hg19) by using the Burrows–Wheeler alignment tool to generate a bam file [11]. Variant calling was conducted by using the Genome Analysis Toolkit, by default [12]. Raw variant calls were filtered out by using allele frequencies > 5% and altered reads>7X. Then, the resulting variants were annotated by the Illumina Variant Studio version 2.2.3 (Illumina, San Diego, CA, USA) and ANNOVAR [13]. Known single-nucleotide polymorphisms were excluded by using variants inthedbSNP 137 (hg19) [14] and SNPs presented in the 1000 Genomes data. The potential germinal mutations were sequenced in matched normal tissues.
Phylogenetic analysis
We inferred ancestral relationships between primary tumors and metastases of Patient 3 by mutation profiles. Neighbor trees were constructed as previously described by Kim et al. [8], with some modifications. We used phytools [15] to calculate neighbor distances and neighbor algorithm implemented in the PHYLIP [16] software package to infer the phylogenetic tree.
Results
Overview of the NGS data
Three CRC patients (P1, P2, and P3) were included in this study. Then, 10 samples from 3 patients were sequenced, with a median coverage of 250 reads. We identified ~8,000 somatic single-nucleotide variations (SNVs) (4,444 to 8,348) and ~850 (457 to 1,154) somatic insertions and deletions (indels) in the 10 biopsy samples from the 3 CRC cases (Table 2).
Table 2. Summary of somatic SNVs and small indels.
| Category | P1-1 b | P1-2 | P2-1 | P2-2 | P2-3 | P3-1 | P3-2 | P3-3 | L1 | L2 |
|---|---|---|---|---|---|---|---|---|---|---|
| A:SNV | ||||||||||
| downstream | 31 | 46 | 47 | 49 | 45 | 31 | 36 | 31 | 30 | 33 |
| intergenic | 1258 | 4046 | 1334 | 2054 | 1314 | 973 | 2181 | 1648 | 922 | 4169 |
| intronic | 1728 | 2137 | 2217 | 2932 | 2483 | 1592 | 1656 | 1562 | 1656 | 1701 |
| Within_non_coding_gene | 596 | 725 | 606 | 674 | 622 | 502 | 533 | 480 | 489 | 543 |
| upstream | 28 | 51 | 32 | 54 | 41 | 38 | 35 | 24 | 53 | 55 |
| upstream;downstream | 24 | 38 | 22 | 20 | 25 | 19 | 24 | 18 | 18 | 24 |
| UTR3 | 522 | 579 | 632 | 740 | 678 | 559 | 571 | 555 | 594 | 590 |
| UTR5 | 104 | 128 | 116 | 125 | 122 | 125 | 124 | 123 | 129 | 124 |
| stop_gaineda | 7 | 7 | 5 | 4 | 5 | 4 | 5 | 6 | 4 | 8 |
| nonsynonymous_CODINGa | 262 | 271 | 238 | 244 | 241 | 257 | 265 | 250 | 258 | 264 |
| synonymous_CODING | 308 | 316 | 284 | 292 | 287 | 285 | 288 | 282 | 286 | 291 |
| splicinga | 4 | 4 | 2 | 2 | 2 | 4 | 4 | 4 | 5 | 5 |
| B:indel | ||||||||||
| downstream | 6 | 11 | 9 | 12 | 9 | 10 | 10 | 9 | 7 | 8 |
| intergenic | 58 | 177 | 107 | 124 | 101 | 69 | 100 | 72 | 62 | 135 |
| intronic | 226 | 530 | 532 | 632 | 541 | 366 | 391 | 341 | 305 | 316 |
| Within_non_coding_gene | 32 | 60 | 56 | 64 | 60 | 52 | 53 | 45 | 52 | 63 |
| upstream | 3 | 10 | 6 | 10 | 6 | 5 | 7 | 4 | 6 | 8 |
| UTR3 | 107 | 248 | 240 | 263 | 236 | 216 | 240 | 209 | 193 | 203 |
| UTR5 | 11 | 28 | 26 | 31 | 31 | 28 | 29 | 26 | 31 | 31 |
| frameshift_codinga | 10 | 11 | 12 | 13 | 13 | 14 | 13 | 12 | 12 | 11 |
| nonframeshift_codinga | 4 | 4 | 4 | 4 | 4 | 3 | 3 | 3 | 3 | 3 |
| splicinga | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
a Mutations from these categories might alter protein.
b P1-1 and P1-2 are primary-tumor regions of colectomy specimen from patient 1; P2-1~P2-3 are primary-tumor regions of colectomy specimen from patient 2; P3-1~P3-3 are primary-tumor regions of colectomy specimen from patient 3, L1 and L2 are two regions of the excised liver metastasis lesion from patient 3.
Patient 1
A 62-year-old male was diagnosed with MSS colon cancer, with a 3.5 cm × 3 cm × 1.2 cm tumor. Two regions (P1-1 and P1-2) of the primary tumor exhibited a moderately differentiated adenocarcinoma, and the pathological stage of the tumor was T2N1M0. The patient received no prior treatment.
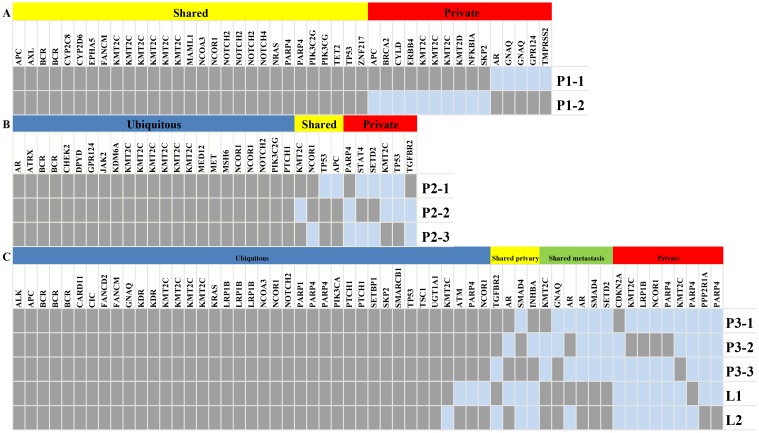
The nonsynonymous somatic point mutations and indels that lead to protein alterations are summarized in S2 Table. Their distributions in different regions across the tumor were mapped (Fig 1A). By comparison of the mutation spectra between primary regions, we determined that 24% (11/45) of the mutations were specific to P1-1, whereas 13% (6/45) of the mutations were specific to P1-2. The remaining 63% of the mutations were shared by both regions. By comparing our result with the 127 significantly mutated genes identified by Kandoth et al. [17], we determined that P1-1 and P1-2 contained APC (428_429del), KIT (A755T), KMT2C (R909K, C391X, G315S D348N, P309S, and .Y816_I817delinsX), NCOR1 (S63L), NRAS (G12D), and PIK3CG (K344Q) mutations. P1-1 contained seven additional mutations, namely, APC (Q706X), KMT2D (3860_3861del), KMT2C (S338L, K306fs, and a splicing site mutation) BRCA2 (T3030fs), and ERBB4 splicing site mutation. P1-2 contained another AR (57_58del) mutation.
Fig 1. Genetic ITH in three Patient.
A Genetic ITH in Patient 1. The regional distribution of 45 somatic variants in 3 primary tumor regions (P1-1 and P1-2); B Genetic ITH in Patient 2. The regional distribution of 33 somatic variants in 3 primary tumor regions (P2-1, P2-2, and P2-3); C Genetic ITH in Patient 3. The regional distribution of 58 somatic variants in 3 primary and 2 metastatic regions. The heat map indicates the presence (gray) or absence (dark blue) of a mutation in each region. The color bars above the heat map specify the categories of mutations.
Patient 2
A 60-year-old female was diagnosed with MSS colon cancer, with a 3.8 cm × 3.5 cm × 1.2 cm tumor. Three regions (P2-1, P2-2, and P2-3) of the primary tumor exhibited a moderately differentiated histology and right hepatic lobe metastasis. Multiple lymph node metastases (pN2) were detected, and the pathological stage of the tumor was T2N4M0. The patient received no prior treatment.
For Patient 2, targeted resequencing of DNA from the three regions of primary tumor was conducted. This process revealed 25 nonsynonymous point mutations and 8 indels (S2 Table). Their distributions in different regions across the tumor were mapped (Fig 1B) as described by Gerlinger et al. [5]. We distinguished the 33 mutations into 23 ubiquitous mutations (those that presented in all regions in each CRC), 6 private mutations (those that presented in only one region), and the rest called shared mutations. On average, a single biopsy exhibited 28 somatic mutations, accounting for 85% of all the mutations observed in this tumor. The mutation landscapes of the different regions were highly similar to one another.
We identified several significantly mutated genes, including ATRX (P571A), CHEK2 (P358S), KDM6A (A1038T), NCOR1 (N208K, S63L), KMT2C (A746S, G892R, P309S, R909K, D348N, and Y816_I817delinsX), and AR (57_59del), which were ubiquitous mutations. The TGFBR2 (E125fs) was specific to P2-1, SETD2 (I1608V) mutation was specific to P2-2, TP53 (T218P), KMT2C (K339N) was specific to P2-3. APC (L693fs) and TP53 (R89W) was shared by P2-2 and P2-3. NCOR1 (K85N) was shared by P2-2 and P2-1. KMT2C (Q755X) is shared by P2-1 and P2-3.
Patient 3
A 71-year-old female was diagnosed with MSS-high and moderately differentiated colon cancer, with a 9 cm × 5 cm × 3 cm primary tumor and a 1.5 cm × 1 cm × 0.5 cm right hepatic lobe metastasis. Three regions (P3-1, P3-2, and P3-3) of the primary cancer and two metastatic regions were resected, and the pathological stage of the tumor was T3N2M2. The patient received no prior treatment. For this patient, targeted resequencing was conducted on the DNA from the three regions (P3-1, P3-2, and P3-3) of the primary tumor and two metastatic regions. The process revealed 47 nonsynonymous point mutations and 11 indels (S2 Table). Their distributions in different regions across the tumor were mapped (Fig 1C).
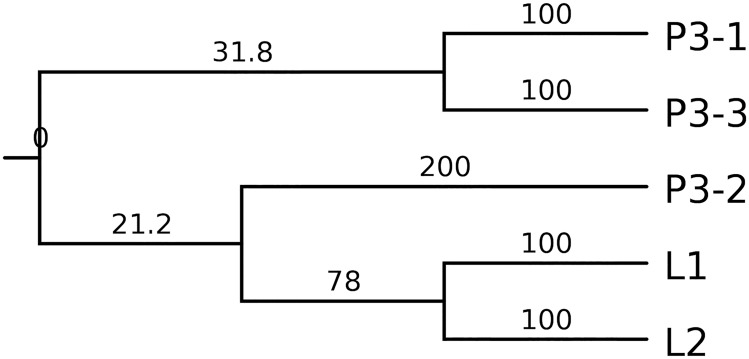
We constructed a phylogenetic tree (Fig 2), which showed a branched, rather than a linear, tumor evolution, to represent the evolutionary history of the CRC. P3-2 mutation spectra were more similar to L1 and L2, indicating that the origins of distant metastases could be traced to one of the primary sites.
Fig 2. Phylogenetic relationship between multi-regional biopsies inferred from somatic mutations.
Branch lengths in the tree are proportional to the number of nonsynonymous mutations in the corresponding branches.
Overall, target sequencing identified 45 somatic mutations per biopsy, accounting for 78% of all mutations observed in this tumor. 60% of all the mutations detected by multi-region sequencing in the colectomy specimen were ubiquitous mutations present in all regions. When we analyzed the primary cancer, an average of 45 somatic mutations were detected per biopsy, which accounted for 87% of all the mutations identified in this tumor. For the metastatic sites, a single biopsy revealed 92% of all the mutations detected in this tumor.
In the significantly mutated genes, we determined that KRAS (G13D), PIK3CA (E545), TP53 a splicing site mutation, APC (E854fs), KMT2C (E141G, K339N, P309S, and Y816_I817delinsX), SETBP1 (Q1558L), and NCOR1 (S63L) were ubiquitous mutations, whereas CDKN2A (D74A), SMAD4 (W168X), NCOR1 (N208K), KMT2C (G315C and K306fs) and INHBA (V58I) were specific to the primary cancer. PPP2R1A (E370X), SETD2 (I1608V), SMAD4 (G382T), and AR splicing site mutations were specific to liver metastatic cancer. However, these mutations were absent in the primary cancer regions. The KRAS, TP53, and PIK3CA mutations, which have often been used as molecular biomarkers in CRC, also presented in all the regions.
Metastatic tumor-specific mutations
By comparing the mutation landscape between primary and metastatic regions, we detected that several mutations were specific to the metastatic regions. SMAD4 is a key transducer of transforming growth factor-β superfamily signaling, which regulates cell proliferation, differentiation, and apoptosis [18]. Loss of Smad4 is correlated with CRC metastasis [19–21]. Meanwhile, AR belongs to a family of nuclear receptors that act as transcription factors. AR has been shown to regulate cell migration by suppressing the nuclear factor kappa B/matrix metallopeptidase 9 pathway and further inhibiting hepatocellular carcinoma metastasis [22, 23]. AR splice variants are known to promote tumor metastasis [24]. The PPP2R1A gene encodes the structural subunit of the PP2A enzyme, which is a highly conserved serine/threonine phosphatase that serves a broad spectrum of biological roles, including the negative regulation of signal transduction, cell cycle progression [25], and gene expression. PPP2R1A facilitates tumor cell–lymphatic endothelial cell interactions during melanoma cell metastasis [26].
Discussion
The multi-region mutation landscape analysis of the three CRC tumors provided substantial evidence of ITH, with the extent of ITH varying among the three cases. In Patient 1, we sequenced two spatially separated sites of primary cancer. Their mutational spectra exhibited a 63% overlap. In Patient 2, a single biopsy exhibited 28 somatic mutations on average, accounting for 85% of all the mutations observed in this tumor. By contrast, in Patient 3, 78% of all the mutations were observed in this tumor. Of all the somatic mutations revealed by multi-region sequencing, 30% to 40% were heterogeneous and thus cannot be detected in every sequenced region. Kim et al. [8] determined that 46%–80% of the mutations were undetectable across all the regional biopsies in their cohort. The level of genetic ITH in their samples was higher than that in ours. The sequencing results of the primary cancers of Patients 1 and 2 were more similar to each other than those of the samples from the aforementioned study. These findings imply that single tumor biopsies cannot accurately represent a tumor’s genomic mutational landscape, which significantly affects targeted therapy.
We constructed a phylogenetic tree by using the data from Patient 3. The tree indicated that CRC evolved in a branched, rather than a linear, manner. The branched evolution model has been validated in many types of cancers [27], including CRC [8]. Traditionally, CRC is considered to evolve in a linear manner [28]. Recent studies showed that both evolution models exist in CRC [29].
Branched tumor evolution underlines the necessity to target mutations located in the main trunk of the phylogenetic tree. We determined that KMT2C and NCOR1 (S63L) were mutated in all regions compared with the significantly mutated genes reported by Kandoth et al. [17]. KMT2C is a member of the human trithorax/mixed-lineage leukemia family [30] and is involved in histone modification. KMT2C is frequently altered in many types of cancer, including liver cancer [31], cutaneous squamous cell carcinoma [32], and esophageal squamous cell cancer [33]. In our research, we determined that each patient has at least seven mutations in KMT2C, the clinical significance of which remains to be validated. We also observed a NCOR1 missense mutation. NCOR1 is a 270 k nuclear receptor corepressor [34] that participates in ligand-dependent transcriptional repression by the estrogen receptor-α [35]. KMT2C and NCOR1 may represent two potential targetable genes in the setting of heterogeneity.
KRAS, TP53, and PIK3CA are the most significantly mutated genes in CRC [36]. However, only Patient 3 had these mutations in our cohort. These mutations were identified in all regions obtained from the patient and showed high concordance between matched primary and metastatic CRC. This result is consistent with that of Brannon et al. [37]. The primary cancer mutation spectra accurately represent those of the metastatic lesions in terms of currently available predicted CRC biomarkers.
PPP2R1A (E370X), SETD2 (I1608V), SMAD4 (G382T), and AR splicing site mutations were determined to be potential metastatic tumor-specific mutations. We suppose that these mutations could drive CRC metastasis. Given the limited sample size, the frequency of these mutations in CRC metastases must be assessed in a larger cohort. Further functional studies are necessary to address their roles in CRC metastasis.
Genomic analysis from single needle biopsies may underestimate the mutational landscape of heterogeneous tumors. ITH may partly explain the poor validation of CRC biomarkers due to sampling bias. Reconstructing the tumor clonal structure and identifying the ubiquitous alterations located in the trunk of the phylogenetic tree may contribute to the discovery of more effective biomarkers and therapeutic approaches.
In conclusion, we identified the substantial extent of ITH in CRC. In the context of this phenomenon, we determined that CRC evolved in a branched manner and several molecular events may contribute to CRC progression. We investigated only three cases because of financial constraints. Further studies with large sample sizes should be conducted to confirm these findings.
Supporting Information
(PDF)
(XLSX)
Data Availability
Sequencing data are available from the Sequence Retrieve Archive (SRA) database (accession number SRP065361).
Funding Statement
This work was supported by Foundation of Medical Leading Talent of Yunnan Province (No. L-201205) KHW, Foundation of Yunnan Institute of Digestive Disease (No. 2014NS122) KHW, Department of Science and Technology of Yunnan Province—Kunming Medical University (NO. 2015FB100) HFZ, National Natural Science Foundation of China (81460007) XYK. http://www.nsfc.gov.cn/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61(2):69–90. 10.3322/caac.20107 . [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu L, et al. Report of incidence and mortality in China cancer registries, 2009. Chinese Journal of Cancer Research. 2013;25(1):10–21. 10.3978/j.issn.1000-9604.2012.12.04 PMC3555299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R, DeSantis C, Jemal A. Colorectal cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64(2):104–17. 10.3322/caac.21220 [DOI] [PubMed] [Google Scholar]
- 4.Kim TM, Jung SH, Baek IP, Lee SH, Choi YJ, Lee JY, et al. Regional biases in mutation screening due to intratumoural heterogeneity of prostate cancer. The Journal of pathology. 2014;233(4):425–35. 10.1002/path.4380 . [DOI] [PubMed] [Google Scholar]
- 5.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366(10):883–92. Epub 2012/03/09. 10.1056/NEJMoa1113205 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nature Communications. 2014;5 10.1038/ncomms3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–9. 10.1126/science.1256930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim TM, Jung SH, An CH, Lee SH, Baek IP, Kim M, et al. Subclonal genomic architectures of primary and metastatic colorectal cancer based on intratumoral genetic heterogeneity. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015. Epub 2015/05/17. 10.1158/1078-0432.ccr-14-2413 . [DOI] [PubMed] [Google Scholar]
- 9.Kogita A, Yoshioka Y, Sakai K, Togashi Y, Sogabe S, Nakai T, et al. Inter- and intra-tumor profiling of multi-regional colon cancer and metastasis. Biochemical and biophysical research communications. 2015;458(1):52–6. 10.1016/j.bbrc.2015.01.064 . [DOI] [PubMed] [Google Scholar]
- 10.Berg KD, Glaser CL, Thompson RE, Hamilton SR, Griffin CA, Eshleman JR. Detection of microsatellite instability by fluorescence multiplex polymerase chain reaction. The Journal of molecular diagnostics: JMD. 2000;2(1):20–8. Epub 2001/03/29. 10.1016/s1525-1578(10)60611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. Epub 2009/05/20. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43(5):491–8. Epub 2011/04/12. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38(16):e164 Epub 2010/07/06. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherry ST, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001;29(1):308–11. 10.1093/nar/29.1.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S, Zhou J, Zhang X, Zhou X, Zhu M, Zhang Y, et al. Mutation Analysis of the APC Gene in a Chinese FAP Pedigree with Unusual Phenotype. ISRN gastroenterology. 2011;2011:909121 10.5402/2011/909121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Retief JD. Phylogenetic analysis using PHYLIP. Methods in molecular biology (Clifton, NJ). 2000;132:243–58. Epub 1999/11/05. . [DOI] [PubMed] [Google Scholar]
- 17.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–9. 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alazzouzi H, Alhopuro P, Salovaara R, Sammalkorpi H, Jarvinen H, Mecklin JP, et al. SMAD4 as a prognostic marker in colorectal cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11(7):2606–11. Epub 2005/04/09. 10.1158/1078-0432.ccr-04-1458 . [DOI] [PubMed] [Google Scholar]
- 19.Losi L, Bouzourene H, Benhattar J. Loss of Smad4 expression predicts liver metastasis in human colorectal cancer. Oncol Rep. 2007;17(5):1095–9. Epub 2007/03/29. . [PubMed] [Google Scholar]
- 20.Li X, Liu B, Xiao J, Yuan Y, Ma J, Zhang Y. Roles of VEGF-C and Smad4 in the lymphangiogenesis, lymphatic metastasis, and prognosis in colon cancer. Journal of gastrointestinal surgery: official journal of the Society for Surgery of the Alimentary Tract. 2011;15(11):2001–10. 10.1007/s11605-011-1627-2 . [DOI] [PubMed] [Google Scholar]
- 21.Xiao D-S, Wen J-F, Li J-H, Wang K-S, Hu Z-L, Zhou J-H, et al. Effect of DPC4 Gene on Invasion and Metastasis of Colorectal Carcinoma Cells. Acta Biochimica et Biophysica Sinica. 2006;38(12):883–92. 10.1111/j.1745-7270.2006.00233.x [DOI] [PubMed] [Google Scholar]
- 22.Ma WL, Hsu CL, Yeh CC, Wu MH, Huang CK, Jeng LB, et al. Hepatic androgen receptor suppresses hepatocellular carcinoma metastasis through modulation of cell migration and anoikis. Hepatology (Baltimore, Md). 2012;56(1):176–85. Epub 2012/02/10. 10.1002/hep.25644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang BG, Du T, Zang MD, Chang Q, Fan ZY, Li JF, et al. Androgen receptor promotes gastric cancer cell migration and invasion via AKT-phosphorylation dependent upregulation of matrix metalloproteinase 9. Oncotarget. 2014;5(21):10584–95. Epub 2014/10/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong D, Sethi S, Li Y, Chen W, Sakr WA, Heath E, et al. Androgen receptor splice variants contribute to prostate cancer aggressiveness through induction of EMT and expression of stem cell marker genes. The Prostate. 2015;75(2):161–74. Epub 2014/10/14. 10.1002/pros.22901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bluemn EG, Spencer ES, Mecham B, Gordon RR, Coleman I, Lewinshtein D, et al. PPP2R2C loss promotes castration-resistance and is associated with increased prostate cancer-specific mortality. Molecular cancer research: MCR. 2013;11(6):568–78. Epub 2013/03/16. 10.1158/1541-7786.mcr-12-0710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christianson DR, Dobroff AS, Proneth B, Zurita AJ, Salameh A, Dondossola E, et al. Ligand-directed targeting of lymphatic vessels uncovers mechanistic insights in melanoma metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(8):2521–6. Epub 2015/02/11. 10.1073/pnas.1424994112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGranahan N, Swanton C. Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution. Cancer cell. 2015;27(1):15–26. 10.1016/j.ccell.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 28.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–67. 10.1016/0092-8674(90)90186-I [DOI] [PubMed] [Google Scholar]
- 29.Naxerova K, Brachtel E, Salk JJ, Seese AM, Power K, Abbasi B, et al. Hypermutable DNA chronicles the evolution of human colon cancer. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(18):E1889–98. Epub 2014/04/23. 10.1073/pnas.1400179111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruault M, Brun ME, Ventura M, Roizès G, De Sario A. MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukaemia. Gene. 2002;284(1–2):73–81. 10.1016/S0378-1119(02)00392-X [DOI] [PubMed] [Google Scholar]
- 31.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nature genetics. 2012;44(7):760–4. http://www.nature.com/ng/journal/v44/n7/abs/ng.2291.html. 10.1038/ng.2291 [DOI] [PubMed] [Google Scholar]
- 32.Pickering CR, Zhou JH, Lee JJ, Drummond JA, Peng SA, Saade RE, et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20(24):6582–92. 10.1158/1078-0432.CCR-14-1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song Y, Li L, Ou Y, Gao Z, Li E, Li X, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509(7498):91–5. 10.1038/nature13176 [DOI] [PubMed] [Google Scholar]
- 34.Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377(6548):397–404. [DOI] [PubMed] [Google Scholar]
- 35.Merrell KW, Crofts JD, Smith RL, Sin JH, Kmetzsch KE, Merrell A, et al. Differential recruitment of nuclear receptor coregulators in ligand-dependent transcriptional repression by estrogen receptor-alpha. Oncogene. 2011;30(13):1608–14. Epub 2010/11/26. 10.1038/onc.2010.528 . [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brannon AR, Vakiani E, Sylvester BE, Scott SN, McDermott G, Shah RH, et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome biology. 2014;15(8):454 Epub 2014/08/29. 10.1186/s13059-014-0454-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(XLSX)
Data Availability Statement
Sequencing data are available from the Sequence Retrieve Archive (SRA) database (accession number SRP065361).