Abstract
Immune repertoires are collections of lymphocytes that express diverse antigen receptor gene rearrangements consisting of Variable (V), (Diversity (D) in the case of heavy chains) and Joining (J) gene segments. Clonally related cells typically share the same germline gene segments and have highly similar junctional sequences within their third complementarity determining regions. Identifying clonal relatedness of sequences is a key step in the analysis of immune repertoires. The V gene is the most important for clone identification because it has the longest sequence and the greatest number of sequence variants. However, accurate identification of a clone’s germline V gene source is challenging because there is a high degree of similarity between different germline V genes. This difficulty is compounded in antibodies, which can undergo somatic hypermutation. Furthermore, high-throughput sequencing experiments often generate partial sequences and have significant error rates. To address these issues, we describe a novel method to estimate which germline V genes (or alleles) cannot be discriminated under different conditions (read lengths, sequencing errors or somatic hypermutation frequencies). Starting with any set of germline V genes, this method measures their similarity using different sequencing lengths and calculates their likelihood of unambiguous assignment under different levels of mutation. Hence, one can identify, under different experimental and biological conditions, the germline V genes (or alleles) that cannot be uniquely identified and bundle them together into groups of specific V genes with highly similar sequences.
Keywords: high throughput sequencing, gene identification
1. Introduction
The diversity of the immune T cell receptor (TCR) and B cell receptor (BCR or antibody, Ig) repertoires allows T cells and B cells to respond to a wide variety of pathogens and establish protective immunity. Repertoire diversity is generated in a combinatorial fashion. Each antigen receptor is a tetramer made up of two heterodimers; each heterodimer consists of a heavy and a light chain. The variable portions of these chains (V regions) arise by recombination of individual members of variable (V), diversity (D only in the case of heavy chains), and joining (J) gene segments [1, 2]. V(D)J recombination of individual coding elements from the V, D, and J genes and junctional modifications result in considerable combinatorial diversity [3]. Lymphocytes are subjected to additional rounds of selection during the immune response, and B cells can undergo further diversification via somatic hypermutation of their antibodies [4].
Quantifying repertoire diversity is important in studies of inflammation [5], reaction to disease [6], vaccination, autoimmunity [7] and cancer [8]. By finding the dominant clones in a given repertoire or by studying the distribution of V gene usage, researchers can gain a better understanding of how the human immune system responds to a particular antigen or is perturbed by or during disease. Our ability to study the repertoire is greatly enhanced by the advent of high-throughput sequencing technologies [9]. However the resulting deluge of data has its own issues. Sequences are often partial and the error rates are significant [10]. Moreover, the sheer amount of data means that there is very little possibility to do quality control and analysis without the aid of computational means.
The clone (also referred to as clonotype) is the unit of selection of the immune response. Clones are collections of sequences that are associated with B cells that derive from a common precursor cell. To properly understand repertoire diversity, we first need to separate the sequences into clones [11]. Unique sequence variants are insufficient for this purpose because they often represent sequencing errors and even sequences with larger numbers of nucleotide differences than predicted by sequencing error may be clonally related. To assign sequence membership into clones, the V, (D) and J genes within each rearrangement need to be associated with their corresponding germline (unarranged) gene segments. Currently, there are several programs that have been developed to perform this function. The two most commonly used are IMGT’s (ImMunoGeneTics) High V-Quest, which uses local alignment to find the best match between the sequence and V, D, and J gene segments [12], and IgBLAST, which breaks the sequence into a k-letter word list, scans the database for possible matching words, and evaluates the significance [13]. Good performance is also achieved by a three-dimensional dynamic programming algorithm for V(D)J segments called SoDA [14] and by applied statistical models, such as the hidden Markov model (HMM), used by iHMMune-align to obtain the optimized parameters fitting to the rearranged antibody [15]. IMGT’s High V-Quest, and iHMMune-align will give multiple identifications when they do not have a conclusive identification. IgBLAST, on the other hand, will always give multiple identifications. (Features of each identification method are provided in Table 1).
Table 1.
Comparison of existing methods
| Program | Algorithm | Stand alone version |
Give multiple identifications? |
Control of germline database |
|---|---|---|---|---|
| IMGT/High V-Quest | Local alignment | No | Yes (no quality score) | No |
| IgBLAST | BLAST searches performed against a user-selected germline V gene database | Yes | Yes | Yes |
| SoDA | Local alignment and 3D dynamic programming | Yes | No | Yes |
| iHMMune-align | Hidden Markov model | Yes | Yes | Yes |
| Conserved Anchor method | Hamming distance after finding conserved anchors | Yes | Yes (when tied) | Yes |
Despite their many strengths, these methods do not take into account the a priori similarity of some germline V genes. Some V genes are more similar to each other than others. Thus there is some a priori non-uniform rate at which certain V genes can be confused with others due to point mutations, deletions or other errors [16]. In fact, some highly similar V genes (such as VH4-30-02 and VH4-30-04) are indistinguishable from each other even if unmutated. This problem is conpounded by two major issues of the high-throughput sequencing technology: (1) it generates short, partial sequences of the Ig genes and (2) it has a significant error rate [10]. Because of this in many high-throughput experiments there is less than the full component of positions with which to differentiate between germline V genes and even more of them are a priori indecipherable. None of the above methods takes into account this source of confusion. Even if they do score all possible good hits, they do not first calculate the likelihood that two germline V genes would show similar scores.
Here we therefore present two innovations: (1) a rapid heavy chain alignment method based on highly stable anchoring positions in V genes that are identical across all germline genes [12, 17] and seldom survive when mutated ([18, 19] and see below) and (2) a general framework of assessing confidence in V gene identification. Using the latter, we have calculated the mutation distances between V genes from the compiled IMGT list of the different human and murine germline V genes [12] and determined those V genes whose germline source cannot be discriminated at different V gene lengths and mutation levels.
2. Method and materials and calculation
2.1 Identifying which germline V genes are a priori too similar to discriminate
To identify which germline V genes can and which cannot be distinguished from each other, we use an alignment method to take a first pass at their identification. After the first round of identification, we make an initial estimate of the distribution of V gene lengths and mutation levels. Based on this estimation, we calculate the likelihood of two germline V genes/alleles being confused by chance for the estimated V gene lengths and levels of mutation (see Calculation). Those germline genes whose probability of being confused when assigned is above our pre-defined threshold (P > 0.01 in the examples shown here) will be identified as giving a mixed V gene identification (so-called “V-ties”). We use a sample-based estimate of mutation/ sequence length as we wish to compare clones and sequences across an entire experiment. This requires the assumption that in a single experiment or sample the mutation pattern/level and V gene length are consistent among sequences. We can then predict the V-ties we expect to find in the experiment while retaining a consistent set of common germline associations that we can use for clonal assignment and clonal diversity analysis throughout the experiment. If some sequences are suspected to be uncharacteristically mutated and thus skewing the estimated level of repertoire error/noise, they can be removed from analysis and V-ties can be reassigned.
It is important to note that the assignment of germline V-ties is a specific one as the potential confusion of germline V genes assigned will always be the same specific small subset of all the V genes. The amount of V gene sequence positions we observe changes which germlines will be V-ties. Lack of sequence information can be divided into two types: (1) Partial sequence reads and (2) mutation/sequencing error. Tables showing the list of potentially confused V genes given specific V gene identifications at 100, 150, 200 nucleotide and full sequences length with 0.05, 0.15 and 0.30 mutation frequencies are found in the supplemental materials (Supplemental Table 1–4).
The precision of V gene identification we consider for the sake of clonal identification is usually at the level of the gene or in some cases, if mutation rates are low and we have full sequences, the allele. Using this method, we can consider a set of unique sequences or clones and assign to each the germline gene for which we have adequate confidence. Some V genes can be fully differentiated at the gene level, some at the allele level and some, for a given dataset of specific sequencing quality or level of mutation, can only be assigned at the level of V-ties with one or two other potential germline V genes. The issue we are pinpointing here is one of germline similarity. Some germline genes/ alleles are so similar that when we query their mutant progeny we cannot discriminate between them with adequate confidence as random error may confuse them. Thus re-sequencing (if it does not remove error) will not change the type of V-ties we identify. PCR error (and selection) can skew the distribution such that the more mutated but more “false germline like” sequence will be more prevalent.
A set of Matlab codes for calculating V-ties can be found on-line at: https://github.com/DrexelSystemsImmunologyLab/ConservedIdentification.git. It can take germline aligned and V(D)J gene associated sequences from any identification method and calculate V-ties base on these identifications.
2.2. Description of the conserved anchor method of germline association
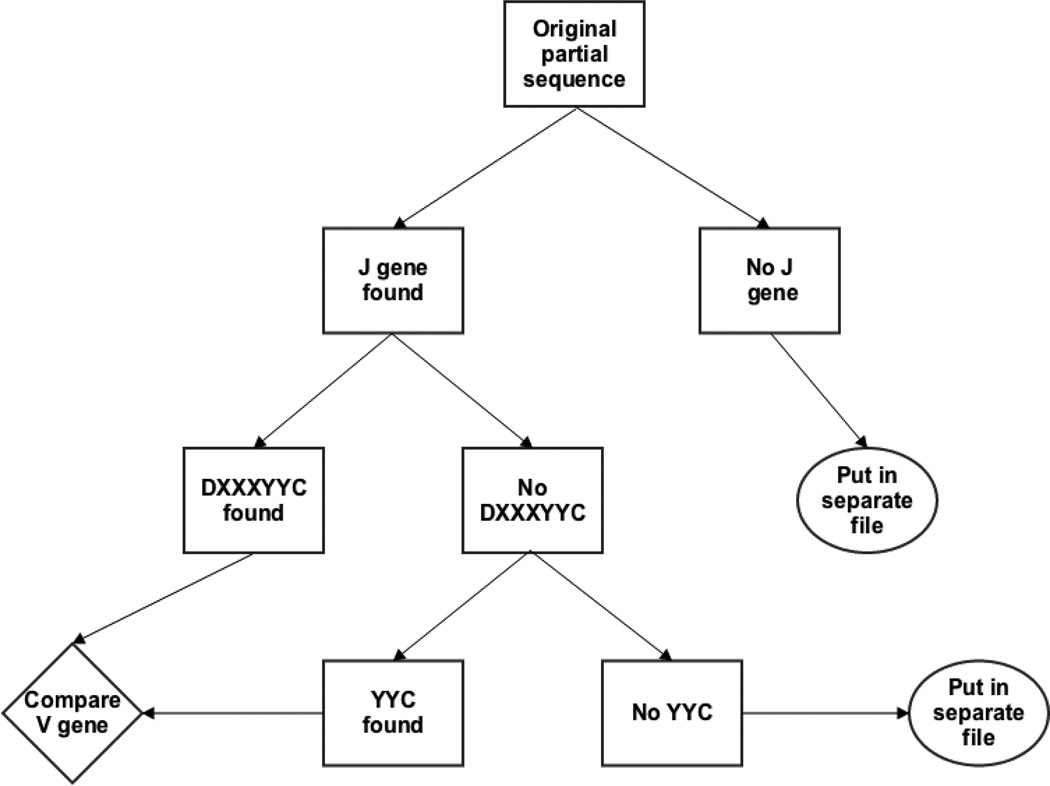
In addition to providing an assessment of our confidence in germline VH gene assignment, we describe herein a novel method of VH gene identification and alignment to IMGT numbering. Our method utilizes consistencies of VH gene structure to make alignment much faster without any loss in accuracy. We show here how it applies to human B cell VH genes and show how it can be modified for use on human VL genes and on murine VH and VL genes. Our human VH germline identification method starts with JH gene identification and then continues to anchor the VH gene and align it. First, we find JH genes by exact match of nucleotides. The nucleotides we use for the JH gene are shown in Table 2 and are located at positions 46 to 63 of the JH gene alignment according to IMGT numbering [20]. If no match is found, the nucleotide strings used will be reduced by one codon from the 3’ end and new strings are then used to find the match. This process is repeated until we can find a match. However, a minimum of twelve matching nucleotides is required to ensure the accuracy of matching. If still no match is found, then the reverse complement of original sequence is used and the aforementioned steps are repeated to find matches. If no match is found in either the original sequence or its reverse complement, then the sequence is put in a separate file (Figure 1). Second, we pinpoint the position of the human VH gene using the highly conserved amino acid sequence ‘DXXXXXC’ which starts with an aspartic acid (D) residue at position 98 (by IMGT numbering) and ends with a Cysteine (C) residue at position 104. These positions are highly conserved at both the amino acid and the nucleotide level as the D is encoded by the nucleotides ‘GAC’ at position 292 to 294 in all but two alleles of functional and non-partial human heavy chain genes while the C is encoded by TGT at positions 310–312 in all but 5 alleles of one gene (Table 3). In these 5 alleles, the V gene ends out of frame with a TG at positions 310–311.
Table 2.
Nucleotides used to identify human JH genes, at positions 46–63 by IMGT numbering
| J gene | Nucleotide |
|---|---|
| IGHJ1/4/5 | TGGTCACCGTCTCCTCAG |
| IGHJ2 | TGGTCACTGTCTCCTCAG |
| IGHJ3 | TGGTCACCGTCTCTTCAG |
| IGHJ6 | CGGTCACCGTCTCCTCAG |
Figure 1. Workflow of germline association process.
(1) Search partial sequences for germline J gene signature. Sequences with no J gene found are put to a separate file. (2) Sequences with identified J gene are checked for germline V anchor DXXXYYC. (3) Counting from the anchor positions, sequences are compared to all germlines and minimally distant germline(s) is/are assigned. (4) Sequences without this anchor will be checked for second anchor YYC and similarly compared. (5) Sequences without any anchor sites are put to a separate file. (For further details see text, Methods Section 2.2)
Table 3.
Nucleotide variations on ‘TATTACTGT’ & ‘GAC’ in human VH genes and alleles
| Incomplete YYC | |||
| Name | AA position | NT sequence | AA sequence |
| IGHV2-70*02 | 102–104 | TATTACTG | YY |
| IGHV2-70*03 | 102–104 | TATTACTG | YY |
| IGHV2-70*04 | 102–104 | TATTACTG | YY |
| IGHV2-70*06 | 102–104 | TATTACTG | YY |
| IGHV2-70*07 | 102–104 | TATTACTG | YY |
| IGHV2-70*08 | 102–104 | TATTACTG | YY |
| Different nucleotide or AA sequences | |||
| Name | AA position | NT sequence | AA sequence |
| IGHV2-70*13 | 102–104 | TATTATTGT | YYC |
| IGHV3-20*01 | 102–104 | TATCACTGT | YHC |
| IGHV4-31*10 | 102–104 | GACTACTGT | DYC |
| IGHV4-34*11 | 102–104 | TATTGCTGT | YCC |
| IGHV4-4*01 | 102–104 | TATTGCTGT | YCC |
| IGHV7-4-1*05 | 102–104 | TGTTACTGT | CYC |
| IGHV3-30*05 | 98 | GGC | G |
| IGHV4-31*05 | 98 | GCG | A |
We identify GACNNNNNNNNNNNNNNNTGT in the sequence. If the sequences lack the GAC nucleotide or it’s synonymous mutation or if we find multiple ‘DXXXXXC’ we search for the nucleotides encoding YYC, at amino acid positions 102–104. These too are highly conserved at the nucleotide level and will most commonly take on the form of TATTACTGT (Table 3). If neither of the primary nucleotide motifs encoding DXXXXXC or YYC is found, we will search for other nucleotide combinations that can encode them (Figure 1). If after these steps still no V gene anchor is found, the sequence is put into another separate rejection file for sequences with an identified J and no V.
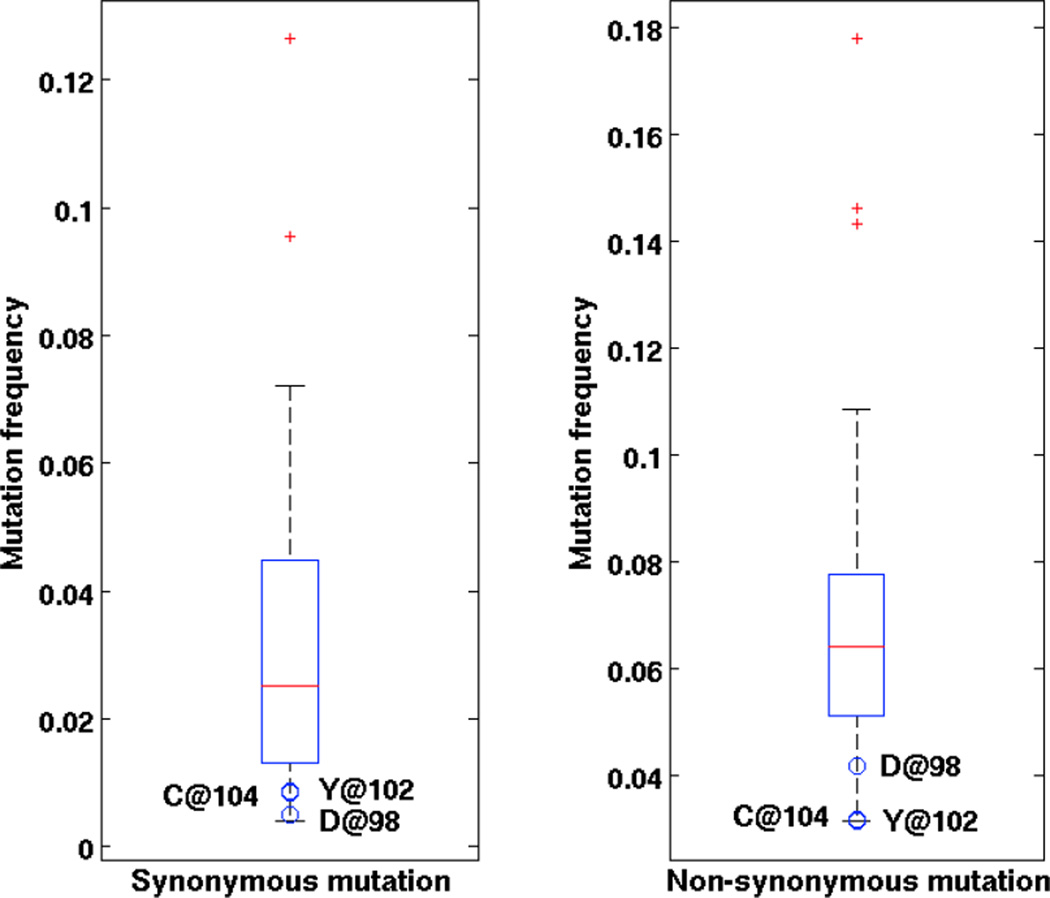
To test how frequently these positions were mutated, we sent a set of 150,000 sequences (as described in section 2.4) [21] to be aligned using High V-Quest [12]. This analysis resulted in 92,491 unique sequences. We found ‘D’ mutated synonymously 457 (0.49%) times and non-synonymously 3850 (4.16%) times, while ‘C’ is found 785 (0.85%) and 2921 (3.16%) times respectively (Table 4). As we would expect from negative selection and random error these ratios of mutation either match or fall below the ratios of 8 to 1 and 7 to 1 expected for D or C (under uniform patterns of mutation or error and remembering that C can mutate to stop). The combination of both anchoring sites ‘DXXXXXC’ was found mutated 732 (0.54%) times. The other possible source of confusion, in which DXXXXXC occurs more than once in a V gene sequence happened 1293 times. But in all these cases only one of the pair had the second anchor YYC. The first ‘Y’ at position 102 was found mutated synonymously 807 times and non-synonymously 2934 times, and the second ‘Y’ at position 103 was found 1921 and 7989 was ‘YHC’ (IGHV3-20*01) at those positions and 383 whose germline was ‘YCC’ (IGHV4-34*11) at those positions (Table 4). The combination of ‘YYC’ was found mutated simultaneously 292 times. The mutation frequencies of D at position 98, Y at position 102 and C at position 104 are the lowest among all positions (Figure 2). It is important to note that the above data are from high-throughput sequencing data of DNA, and include sequencing errors. In similar experiment, studying mutated B cell receptors taken from human lymph nodes, the genes were sequenced from barcoded mRNA, where consensus alignments were used to create the sequences of B cell receptors and most if not all of the sequencing errors were fixed [19]. In this barcoded data set [19] we found the nucleotides encoding C mutated 79 times synonymously and zero times non-synonymously in 3,017 sequences with copy number greater than one.. This leads us to predict that, with proper sequencing error correction, the amount of unique sequences lost by relying upon the Anchor method, should drop below the 2% level we observe here (see Results section 4.3).
Table 4.
Numbers of synonymous and non-synonymous mutations of the anchor positions in 92,491 unique DNA sequences identified using High V-Quest [21] and 3,017 mRNA sequences with copy number >1 [19].
| DNA | mRNA | |||||||
|---|---|---|---|---|---|---|---|---|
| Position | 98 | 102 | 103 | 104 | 98 | 102 | 103 | 104 |
| Amino acid | D | Y | Y (H/C) | C | D | Y | Y (H/C) | C |
| Nucleotide | GAC | TAT | TAC | TGT | GAC | TAT | TAC | TGT |
| Synonymous | 457 | 807 | 1921 | 785 | 14 | 99 | 178 | 79 |
| Non-synonymous | 3850 | 2934 | 7381 | 2921 | 54 | 0 | 0 | 0 |
Figure 2. Boxplots of mutation frequency of amino acid positions 76–105 in [21].
Red line indicates median. Blue box indicates 25% and 75% quartile. Whiskers indicate the furthest data not considered outlier. Red dots indicate outliers. Blue circles indicate mutation frequencies of D, Y and C at position 98, 102 and 104. (A) Synonymous mutations; (B) Non-synonymous mutations.
After the relative position of the sequence and germline are determined by the anchor(s), all the alleles of all V genes in the IMGT database are compared with the sequences. The allele(s) with the fewest mismatches will be assigned as the germline source of the sequence. If multiple germline genes are identified as being equally distant from the query sequence, they will be both labeled as possible germline sources. It is important to note that such a confusion of identity can happen with any two germline genes but is much rarer than the appearance of V-ties and implies either a lack of information or high mutation levels. The method described here does not identify insertion/deletions in sequence. However, we apply an insertion/deletion control after the identification. If there are more than 9 mutations in a sliding window of 15 nucleotides, we label the sequences as having potential insertion/deletion(s) and identify their germline source using local alignment. A set of Matlab codes for the Anchoring method of germline association can be viewed online at: https://github.com/DrexelSystemsImmunologyLab/ConservedIdentification.git.
Similar sequence anchor points are found in human light chains and TCR, and in the murine V genes for BCRs and TCRs (Table 5). In human and murine light chains, the dominant codon encoding ‘D’ at position 98 is ‘GAT’ instead of ‘GAC’ in heavy chains and TCR. However, in murine TCR α chains the ‘D’ is at IMGT position 100 and ‘YYC’ at position 104. In TCR there is no clearly dominant amino acid combiantion at positions 102–104. They have relatively equal codon usage encoding ‘YYC’, ‘YLC’ and ‘YFC’ at these positions. The anchor variations for these chains are found in the supplemental materials (Supplemental Tables 5–12).
Table 5.
Anchors in other BCR and TCR genes. The default amino acid at position 98 is D. The default amino acids at position 102–104 are YYC. Leucine (L) and Phenylalanine (F) are very common variants at position 103.
| D at 98 | YYC at 102–104 | ||||||
|---|---|---|---|---|---|---|---|
| Number of alleles |
V gene type |
Total No. of germline genes |
With NT variation |
With AA variation |
With NT variation |
With AA variation (YLC/YFC) |
With AA change at both positions |
| Human | □ | 49 | 2 | 1 | 6 | 5 | 0 |
| κ | 61 | 0 | 0 | 29 | 2 | 0 | |
| λ | 65 | 20 | 0 | 31 | 4 | 0 | |
| α | 88 | 25 | 3 | 0 | 78 (24/48) | 0 | |
| β | 106 | 34 | 28 | 0 | 106 (51/41) | 0 | |
| Mouse | Heavy | 258 | 22 | 3 | 99 | 50 | 1 |
| κ | 120 | 20 | 0 | 74 | 31 | 0 | |
| α | 206 | 40 | 0 | 0 | 165 (36/124) | 0 | |
| β | 46 | 17 | 16 | 0 | 46 (19/21) | 0 | |
2.3. Simulated validation of the consistent mis-identification of V-ties
To validate our method of V gene alignment and germline origin identification, we used simulated mutant sequences. The method for generating these sequences is described in Section 2.4. These simulated sets of mutant sequences were compared to a reference set of all functional and non-partial alleles [12] (see Supplemental Table 13). We compared our identification of the simulated dataset using the anchor method with those from two commonly used V gene identification tools: IMGT’s High V-Quest and NCBI’s IgBLAST. We used the downloadable IgBLAST (version 2.2.28) with the same set of germlines as in our method. High V-Quest uses the entire germline dataset in IMGT of which our set is a subset.
High V-Quest uses a global pairwise alignment without insertions or deletions [12] and outputs multiple V gene identifications without a metric for preference (insertion/deletions are fixed at later stage). IgBLAST makes a k-letter word list (k=9 for V gene identification by default), scans the database for possible matching words and evaluates the significance [13] (Table 1). In this way they generate a list of possible V gene identifications with their significance. For this reason we consider the top five hits as identifications from IgBLAST. To compare both methods, we also ran IgBLAST with different word sizes: the default word size 9 and the minimum word size allowed of 4, which we determined would give optimal identification results.
We determined if identification occurred correctly at the gene level only, not at the level of alleles. We have divided the results into three categories to evaluate the performance of each method.
Category 1: the gene is distinguished and the unique identification is correct
Category 2: the correct gene is identified along with the expected confusing gene (as described in section 2.1).
Category 3: other, unpredicted misidentifications
2.4 Germline and mutant sequences used
(i) Germline sequences analyzed for V-ties: All germlines and alleles analyzed are from the IMGT database version 3.1.2, also in Supplemental Table 14. The exact identification of V-ites will depend on list of known germline genes that is queried.
(ii) Human IgH rearrangements: Peripheral blood B cell DNA was enriched using a dual step PCR based amplicon capture as described previously [21].
(iii) Simulated sequences: Simulated sequences were made with V genes randomly mutated with a 0.02, 0.05, 0.10, 0.15 and 0.3 mutation frequencies, uniformly spread across the sequence, but not the anchoring points. We generated 500 mutated sequences for each of the 192 known functional and non-partial human VH gene alleles or a total of 96,000 sequences. We did not mutate the anchor sites described in 2.2. This is because we are attempting to test the miss-assignment of expected sets of germlines that form V-ties. The mutation of anchor sites does not in any way change the likelihood of confusing germline V genes as the anchor sites are identical in all germline V genes and thus have no power in determining them. These sequence datasets can be provide in fasta format on request.
3. Calculating the likelihood of confusing two V genes
We calculated the similarities of the BCR and TCR V genes and alleles by how many nucleotide differences they have at certain sequence lengths. The probability p of two specific V alleles, of the same or different V genes, to be confused at a particular length and mutation frequency is given by the following equation:
Where p is the hypergeometric probability of each value of x (from K/2 to K) using the corresponding size of the population, M, number of items with the desired characteristic in the population, K, and number of samples drawn, N. In this distribution, if we assume that mutations have equal probability of targeting at each position, M stands for the length of alignment, K the differences between two genes/alleles and N is the number of estimated mutations in the sequence. The estimated mutation number N is calculated from the average alignment length L and r is the average fraction of sequence positions that are mutated. 0.33 is the probability of one nucleotide mutating into others, assuming equal chance (no bias). Although this form of calculation ignores known patterns of mutation it allows us to generalize the method across species and include species where a good model of mutation does not exist. Also we do not set a prior probability for V gene or allele usage. The knowledge of exact germline gene usage across the population in humans is still very limited and is at present beyond the scope of this analysis.
4. Results
4.1. Germline V gene similarity
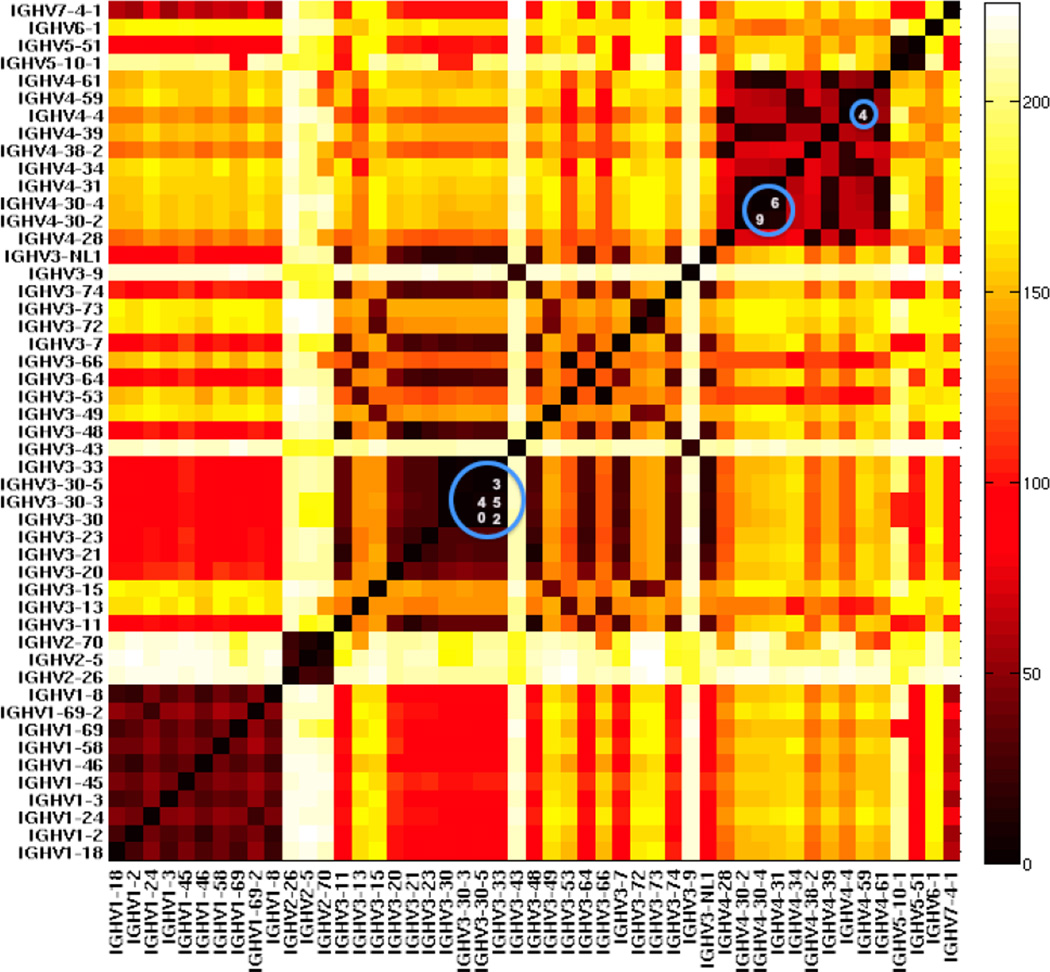
We counted the number of different nucleotides between any two human heavy chain variable alleles when counting from the 3’ end (Figure 3). We found genes from the same families were often highly similar to each other, especially in the VH1 and VH4 families. In addition, certain genes in the VH3 family are very similar. Certain alleles in IGHV3-30 and IGHV3-33 only have two nucleotide differences. IGHV3-30-5*01 and IGHV3-30*18 have exactly the same nucleotides. We also calculated the differences within BCR light chains and both β and α TCR V genes (Supplemental Figures 1–4).
Figure 3. Heat map of the minimal sequence difference of each human germline VH gene pair [12] at full sequence length.
Minimal nucleotide differences among all alleles pairs between genes. Distances range from 0 (black) through red (100) and yellow (200) to white (>200). The comparison begins at the 3' end of each full-length VH gene. The blue numbers in the circles represent the numbers of mismatched nucleotides in the most similar pairs. These pairs of VH genes cannot be discriminated from each other at almost any length or mutation frequency.
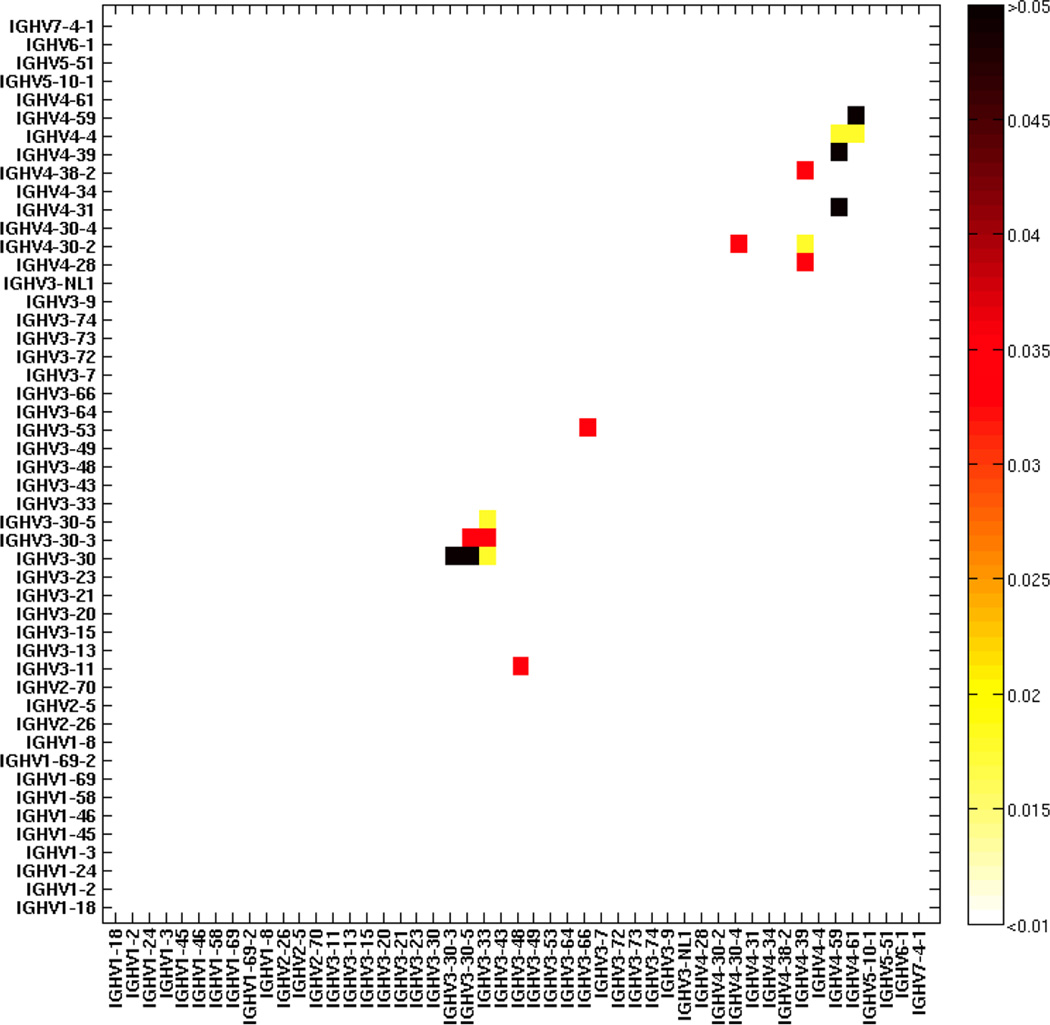
Using the hypergeometric calculation described in the Methods and Calculations sections, we found that the likelihood of failed V gene assignment increases, as one would predict, with higher mutation frequencies and shorter read lengths (Figure 4 and Supplementary Zip file). As shown in Figure 4, we calculated at 150 nucleotide length and 0.05 mutation frequency, that some VH genes (Supplemental Table 2) have more than 0.01 probability of being confused with each other and thus mutants from these germline V genes cannot be definitively distinguished. We would call these germline V genes at this sequence length/mutation level V-ties. Mutated sequences that are assigned either of these germline V genes should instead be assigned the V-ties they belong to (Supplemental Table 2).
Figure 4.
The probabilities of confusing the germline association of a mutated sequence between any two germline VH genes at 150 nucleotide length and 0.05 mutation frequency (calculated as described in Calculations). Likelihoods range from white p<0.01 to black p>0.05).
We have generated sets of tables delineating exactly which VH genes cannot be unequivocally uniquely identified at 0.05, 0.15 and 0.3 mutation frequencies and 100, 150, 200 and full-sequence lengths (Supplemental Tables 1–4 and Supplementary Zip file). We have done so for human BCRs and TCRs (Supplementary Zip file). While TCRs do not mutate, in many cases very short reads are used and there can be a sequencing error of ~ 1–3% [22]. We therefore only created such tables with 0.03 error frequencies. TCR α chain V gene germlines can be clearly distinguished even at 100 nucleotide length. The set of V genes that cannot be distinguished for TCR β chain can be seen in Supplemental Table 15. We have also calculated the probabilities of gene pairs confusing with each other at aforementioned mutation levels and sequence lengths for human BCR and TCRs so one can set his/her own threshold instead of 0.01 used in this paper (Supplementary Zip file). Finally we supply a Matlab code (https://github.com/DrexelSystemsImmunologyLab/ConservedIdentification.git) that can filter sets of genes with identified germline sequences so that the V genes that cannot be uniquely identified are explicitly identified the appropriate germline V-ties are assigned.
4.2. Identifying V-ties with the Anchor method, with High V-Quest and IgBLAST
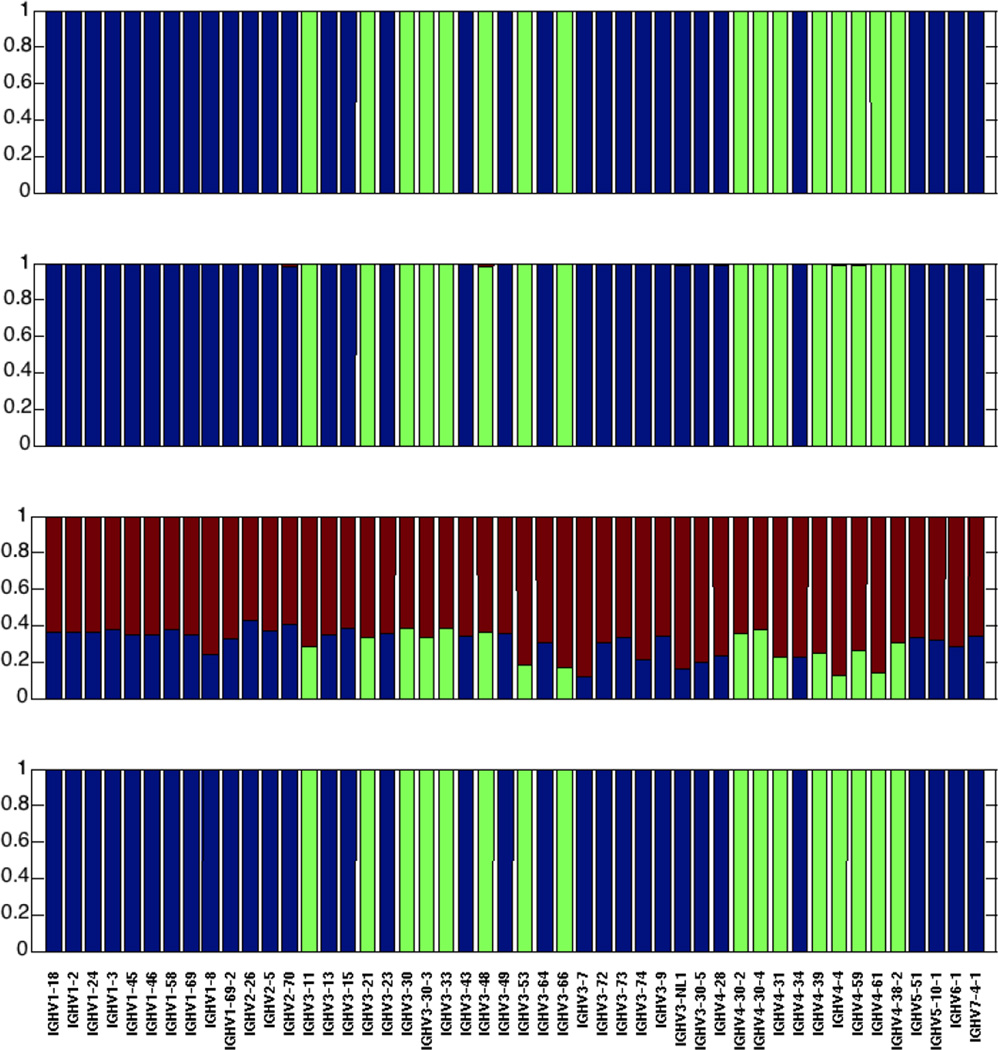
To see if V-ties appeared where they were predicted to appear we compared the V assignments using two standard algorihems for V gene assignment and our novel Anchor method. To do so we used simulated sequences (mutated as described in Methods) as only with those could we know a-priori their actual germline source. To our surprise we observed that not all the methods did a good job of identifying germlines. IgBLAST at its default settings has very poor VH gene identification. However, all three methods can give reasonable results by optimizing the sequence feature parameters (Figure 5 at 150 nucleotide length and 0.15 mutation frequency and Supplementary Figures 5–15 for 100, 150, 200 nucleotide and full length and with 0.05. 0.15 and 0.30 mutation frequencies).
Figure 5. Comparison of human VH identification results using three different sequence identification methods at n=150 nucleotide read length and 0.15 mutation frequency.
From top to bottom (A) The Anchor method (this paper); (B) High V-Quest; (C) IgBLAST using the default word length (9 characters); (D) IgBLAST using the minimal word length (4 characters). In all cases the blue color represents the fraction of correctly identified sequences in which a VH gene is uniquely identifiable; green is the fraction of identifications when a gene is not distinguishable from at least one other gene (using the calculation described in the Calculation and Methods section) and either the real gene or the one we predict to confuse it with are identified; red is the fraction of incorrect identifications.
Most importantly, in terms of our predication of V-ties, for all methods, whenever sequences are partial length or somatically mutated, a consistent subset of genes will be misidentified in the way we predicted (category 2 identification – green bars in Figure 5 and see Calculations and Methods section). It is important to note that such misidentifications could not be distinguished from correct identifications or other types of error in a non-simulated set of sequences. Thus, as we suggest in Methods, the only solution for these consistently miss-assigned germline V genes is to combine them with their appropriate confounding germline V genes (Figure 5). These sequences should not be confused with sequences that are equidistant from two different germline genes by their mutation count. The germline V genes identified as V-ties are clearly identified at the level of the specific V-tie, and will most often be identified as being closest to one specific germline V of those associated by the V-tie. However, their chance of being randomly identified or misclassified as a specific other V gene is significant (considered to be p > 0.01 in our Calculations and Methods section) and either the real gene or the one we predict to confuse it with are identified; red is the fraction of other incorrect identifications that cannot be explained by V-ties.
4.3 Comparing the Anchoring method to other alignment methods
Computational efficiency
To test the efficiency of our germline association method, we selected 10,000 sequences (299±6 nucleotides in length that have a partial VH gene sequence, all of the junctional sequences and a partial JH gene sequence) with an estimated 0.03 mutation frequency [21] (see Section 2.4). It took our method 35.54 seconds to finish the identification while IgBLAST needed 347.58 seconds using the default word size of 9 nucleotides and 3,877.59 seconds using the minimum word size (4 nucleotides). This analysis shows that our method is less computationally intensive than IgBLAST, especially when using the more accurate minimum word size there. Although High V-Quest is clearly the standard high-throughput alignment program used in our field it does not have a stand-alone version and jobs need to be queued on the IMGT server. This makes that and not the processing speed the relevant time limiting step in using it. Thus from what we can compare we can conclude that the Anchoring method outperforms IgBLAST (×10 compared to the default word size and ×100 when using the more accurate minimal word size).
Sequence loss
To test to what extent the use of the Anchor positions causes us to lose sequence data, we compared a single IMGT High V-Quest run of 150,000 sequences to our alignment of the same set of genes. The sequences were taken at random from a set of 1.8 million B cell heavy chain V(D)J gene sequences sequenced from human blood [21]. From these 150,000 sequences, we were able to identify 141,496 (94.33%) using the V Anchor method discussed in Section 2.2, while with High V-Quest we identified 146,821 (97.88%). Discounting duplicated sequences, we identified 88,209 unique sequences using the Anchor method described here and High V-Quest identified 92,490. 87,958 of these unique sequences were in complete agreement with respect to their copy number and V identity between two methods. There were also a few sequences identified only by one method. The V Anchor method identified 154 unique sequences and High V-Quest 4,460 unique sequences that the other method did not.
It is important to note that much of the extra diversity that was only identified by High V-Quest (5% more sequences) is probably due to sequencing error. To remove unique sequence types generated through sequencing error we next considered only unique sequences with copy number >1 [11]. The removal of singleton sequences indeed improves our level identification in comparison to High V-Quest. Both methods agree on the identity of 15,252 unique sequences of copy number >1 and High V-Quest identifies only 357 additional sequences (or 2% more). There are also a few genes that are identified by both methods where they do not agree as to the V gene assignment and/or the copy number. The discrepancies between the two methods are interesting as they reveal the minor issues with each method. One reason for these differences is that we are comparing partial sequences of unequal length. With High V-Quest, alignments are performed not only to functional V genes but also to pseudogenes and incomplete V genes in the IMGT database. Hence there are 15 unique sequences to which the Anchor method assigns specific V genes that IMGT High V-Quest considers to most closely resemble partial sequences or pseudogenes. In addition there are 13 unique sequences that IMGT’s High V-Quest assigns to a pseudogene whose component sequences we considered to be two different V genes. Finally there are 4 sequences (out of 150,000) that were identified as containing indels by High V-Quest and did not pass our threshold (Supplemental Table 16–18). In all instances it is hard to categorically state which method was correct. However, these examples pinpoint potential limitations in our method of insertion/deletion detection and in High V-Quest user operability, which does not allow the user to select which subset or version of the database of germline genes they wish to compare to the mutant sequences.
5. Discussion
The BCR and TCR repertoires play critical roles in immune function and pathogenesis [23, 24]. One of the first steps in studying immune responses is to study how immune repertories shift in response to antigens, vaccines and pathogens. Identification of germline genes that comprise the building blocks of the antigen receptor is a crucial first step in repertoire analysis. Unfortunately, this step is difficult because germline genes can be highly similar and can undergo somatic mutation (only in the case of BCRs) and be subject to sequencing error (BCR and TCR). High-throughput sequencing methods generate large numbers of sequences at a low cost, providing a way to essentially map the immune repertoire, but can use short read lengths and have high sequence error rates. For this reason it is now critical to categorize, as much as possible, the reason for uncertainty in germline gene assignment.
As we have shown in the Results, at all sequence lengths, the V genes from the same gene family are quite similar and can have differences as low as one or two nucleotides. Some alleles of different V genes are even identical to each other over short lengths. For example, IGHV3-30*07 and IGHV3-33*04 are identical in the last 119 nucleotides. As the sequence read length increases, V genes can be better differentiated, but even with full-length sequence data if the mutation frequency is above 0.1, some germline V genes are effectively indistinguishable.
This raises the problem that some germline V genes cannot be well discriminated. However, they are not unknowable as they are similar only to other specific V genes and can be discriminated from most other germline V genes. In our method, after the first round of identification, we estimate the expected variability in our data. Based on the alignment length and mutation frequency of the first round identification, we can calculate the likelihood of error due to mutation (or other sequence changes) using the simple hypergeometric test described in Calculations. V genes that we calculate as being impossible to distinguish at a given length/mutation/error rate will be identified as a single germline source. When we construct clones in later steps these V genes will be put in the same clone as they are indistinguishable. In this way, we can have more reliable identifications and know at what level of categorization (family, gene or allele) the identifications are definitive. This is especially useful in studies of inflammation wherein B cellclones can be highly mutated.
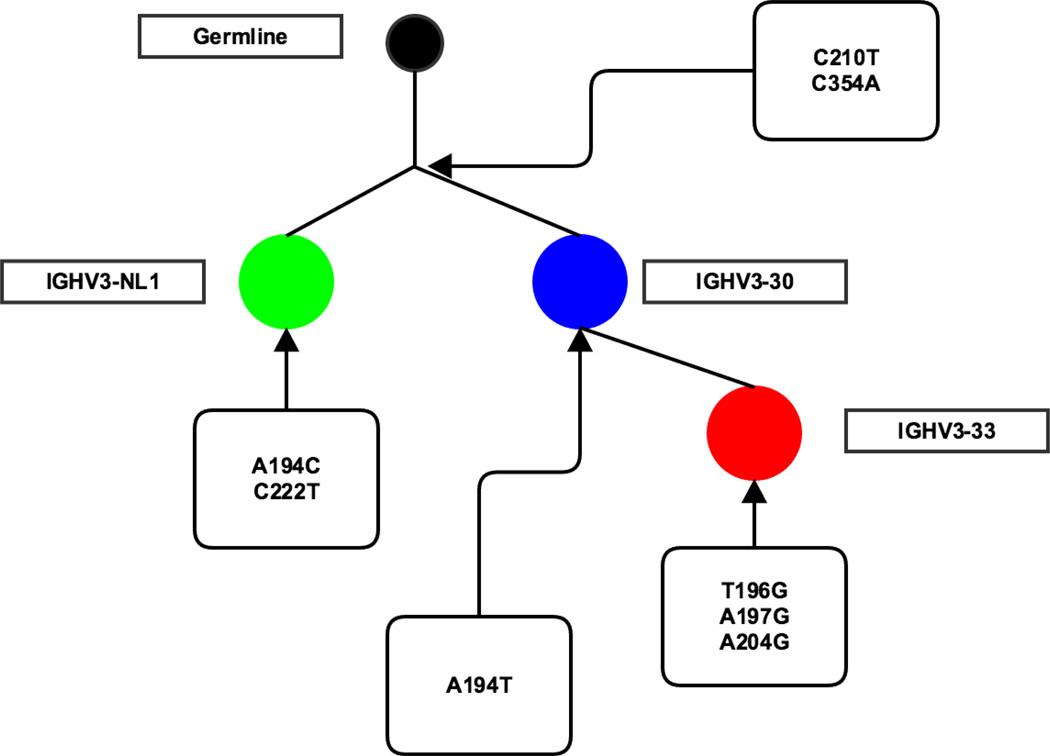
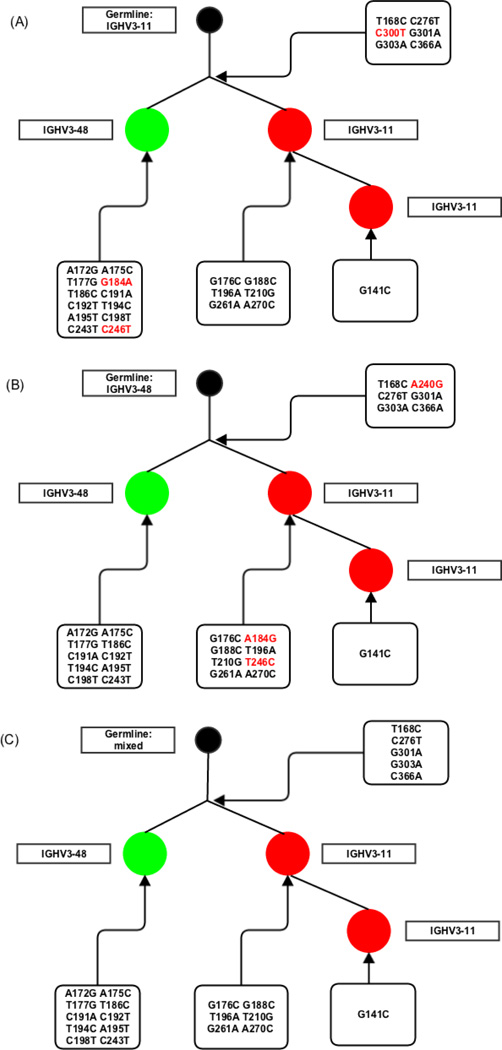
Figure 6 shows four sequences [21] that would not be associated into a single clone if we did not consider V-ties. Applying V-ties, they were identified as IGHV3-30, IGHV3-33 and/or IGHV3-NL1, and put in the same clone, as these genes have a p> 0.01 to be confused at the length (90.45 nucleotides) and mutation frequency (0.02) found in this dataset. Despite the low mutation rate in the sample in general, these sequences had two mutations (C210T and C354A) in common and the same CDR3 (CARDRASCPDYW) confirming that they probably belonged to the same clone, which would have been missed if standard V gene alignment practices had been followed [11]. Figure 7 shows another example where allowing for a more ambiguous V gene assignment helps to identify the full clone. The identical CDR3 and five common mutations from the germline sequences (T168C, C276T, G301A, G303A and C366A) found in all three sequences suggest they are in the same clone. However, if we had not considered the inherent ambiguity in assigning them to IGHV 3-11 or 3-48, we would have considered them to comprise two separate clones and have assigned them up to 3 erroneous mutations. By giving them the hybrid V-ties assignment we consider only mutations we are sure of and correctly assign these sequences to a single clone (Figure 7c).
Figure 6. Part of a phylogenetic tree of a clone from naïve B cells in human.
Each circle represents a sequence. Different colors show what V gene the sequences were originally identified. The germline was made by ignoring the different positions of tied V genes. The mutations on each branching level are shown. The numbers in each circle show how many additional mutations each sequence has.
Figure 7. Phylogenetic tree of a clone from a plasmablast dataset of a lupus patient.
Each circle represents a sequence. Different colors and tags show what V gene the sequences were originally identified. The germline was made by (A) filling in IGHV3-11*01 germline sequence, (B) filling in IGHV3-48*01 germline sequence or (C) ignoring the different positions of these V genes. The mutations of each hypothetical and real node are shown. The additional mutations in (A) and (B) compared to (C) are shown in red.
Beyond explicit indication of the specificity of V gene identification, our germline identification Anchor method performs as well or better than existing human germline identification and clonal assignment methods. Specifically, at high mutation levels IgBLAST (with the default word size) does not work well. As shown in the Results, as mutation frequency increases, the performance of IgBLAST quickly worsens (Figure 5 and Supplemental Figures 5–15). This error rate can be corrected by shortening the word size but then computing time balloons to 100-fold longer than the Anchor method described here. High V-Quest and the Anchor method have equally reliable V gene identifications. The Anchor method allows for command line alignment of sequences and control over the members of the germline V gene database used to compare with the query sequences. High V-Quest identifies ~ 2% more unique sequences than the Anchor method. Lack of control of the gene database used to compare with the query sequences is problematic when we have knowledge of our input beforehand. For example we know the V genes are all functional when we extract the sequence from immunized patient blood, but if we compare sequences to non-functional germline genes as well we could assign them as germline source if mutant sequences happened to exhibit more similarity to them and not their germline source. Finally, the V Anchor method allows us to ensure that all sequences are aligned in terms of IMGT numbering even if we are uncertain of their exact germline source. In this way we can both retain information on CDR3 structure and at least some measure of intra-clonal diversity analysis can be achieved even if we are not sure of the precise germline source of the clone.
In summary, we have developed a new methodology (Anchor method) to rapidly identify the originating germline genes of rearranged antibody and TCR sequences, based on conserved sites. In addition, we have analyzed the similarity of V genes at different lengths, and created matrices of small V gene groups (V-ties) that cannot be discriminated because of sequence similarity of their germline genes for different levels of sequence information. The exact identification of V-ties will depend on the list of known germline genes that is queried. Herein we have identified V ties for the current, most widely used human and murine IMGT heavy chain germline V gene lists. The exact groups of V-ties we show here derive from the genes in these lists. The V-ties would change if we added new V gene alleles and of course will also change when we look at the immune systems of other species. However, the methodology and consequences of our analysis remain the same: We will identify some germline genes/ alleles that cannot be uniquely associated or discriminated when querying their mutant progeny.
To allow scientists to identify the V-ties in their data, given the set of relevant germline genes in the repertoires they are analyzing, we have created a simple set of programs (in Matlab). These programs implement the Anchor alignment method and post process any BCR alignment results, from High V-Quest or IgBLAST or our own conserved site Anchor method, so as to assign V genes to their appropriate V-ties under different sequencing lengths and levels of mutation.
Supplementary Material
Acknowledgements
The authors would like to thank Gregory Schwartz and Aaron Rosenfeld for their insightful comments and helpful arguments. Research reported in this publication was supported by the National Institute Of Allergy And Infectious Diseases of the National Institutes of Health under Award Number P01AI106697 and by NIH P30-CA016520. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alt Frederick W, Baltimore David. Joining of Immunoglobulin Heavy Chain Gene Segments: Implications from a Chromosome with Evidence of Three D-JH fusions. Proceedings of the National Academy of Sciences. 1982;79(13):4118–4122. doi: 10.1073/pnas.79.13.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrie Howard T, Livak Ferenc, Burtrum Douglas, Mazel Svetlana. T Cell Receptor Gene Recombination Patterns and Mechanisms: Cell Death, Rescue, and T Cell Production. The Journal of Experimental Medicine. 1995;182(1):121–127. doi: 10.1084/jem.182.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tonegawa Susumu. Somatic Generation of Antibody Diversity. Nature. 1983;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 4.Grimaldi Christine M, Hicks Ruthmarie, Diamond Betty. B Cell Selection and Susceptibility to Autoimmunity. The Journal of Immunology. 2005;174(4):1775–1781. doi: 10.4049/jimmunol.174.4.1775. [DOI] [PubMed] [Google Scholar]
- 5.Goronzy Jörg J, Weyand Cornelia M. Ageing, Autoimmunity and Arthritis: T-cell Senescence and Contraction of T-cell Repertoire Diversity – Catalysts of Autoimmunity and Chronic Inflammation. Arthritis Research and Therapy. 2003;5(5):225–234. doi: 10.1186/ar974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abe Jun, Kotzm Brian L, Melssner Cody, Melish Marian E, Takahashi Masato, Fulton David, Romagne Francois, Malissen Bernard, Leung Donald Y. Characterization of T Cell Repertoire Changes in Acute Kawasaki Disease. The Journal of Experimental Medicine. 1993;177(3):791–796. doi: 10.1084/jem.177.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hershberg Uri, Meng Wenzhao, Zhang Bochao, Haff Nancy, St Clair E William, Cohen Philip L, McNair Patrice D, Li Ling, Levesque Marc C, Luning Prak Eline T. Persistence and Selection of an Expanded B-cell Clone in the Setting of Rituximab Therapy for Sjögren’s Syndrome. Arthritis Research & Therapy. 2014;16(1):R51. doi: 10.1186/ar4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houghton Alan N. Cancer Antigens: Immune Recognition of Self and Altered Self. The Journal of Experimental Medicine. 1994;180(1):1–4. doi: 10.1084/jem.180.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berglund Eva C, Kiialainen Anna, Syvänen Ann-Christine. Next-generation Sequencing Technologies and Applications for Human Genetic History and Forensics. Investig Genet. 2011;2(2011):23. doi: 10.1186/2041-2223-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Lin, Li Yinhu, Li Siliang, Hu Ni, He Yimin, Pong Ray, Lin Danni, Lu Lihua, Law Maggie. Comparison of Next-Generation Sequencing Systems. BioMed Research International. 2012;2012 doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hershberg Uri, Luning Prak Eline T. The Analysis of Clonal Expansions in Normal and Autoimmune B Cell Repertoires. Philosophical Transactions of the Royal Society B. 2015;307(1676):20140239. doi: 10.1098/rstb.2014.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brochet Xavier, Lefranc Marie-Paule, Giudicelli Véronique. IMGT/VQUEST: the Highly Customized and Integrated System for IG and TR Standardized V-J and V-D-J Sequence Analysis. Nucleic Acids Research. 2008;36(suppl 2):W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Jian, Ma Ning, Madden Thomas L, Ostell James M. IgBLAST: an Immunoglobulin Variable Domain Sequence Analysis Tool. Nucleic Acids Research. 2013;(2013):gkt382. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volpe Joseph M, Cowell Lindsay G, Kepler Thomas B. SoDA: Implementation of a 3D Alignment Algorithm for Inference of Antigen Receptor Recombinations. Bioinformatics. 2006;22(4):438–444. doi: 10.1093/bioinformatics/btk004. [DOI] [PubMed] [Google Scholar]
- 15.Gaëta Bruno A, Malming Harald R, Jackson Katherine JL, Bain Michael E, Wilson Patrick, Collins Andrew M. iHMMune-align: Hidden Markov Model-based Alignment and Identification of Germline Genes in Rearranged Immunoglobulin Gene Sequences. Bioinformatics. 2007;23(13):1580–1587. doi: 10.1093/bioinformatics/btm147. [DOI] [PubMed] [Google Scholar]
- 16.Kepler Thomas B. Reconstructing a B-cell Clonal Lineage. I. Statistical Inference of Unobserved Ancestors. F1000Res. 2013;2 doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz Gregory W, Hershberg Uri. Conserved Variation: Identifying Patterns of Stability and Variability in BCR and TCR V Genes with Different Diversity and Richness Metrics. Physical Biology. 2013;10(3):035005. doi: 10.1088/1478-3975/10/3/035005. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz Gregory W, Hershberg Uri. Germline Amino Acid Diversity in B Cell Receptors is a Good Predictor of Somatic Selection Pressures. Frontiers in Immunology. 2013;4 doi: 10.3389/fimmu.2013.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stern Joel NH, Yaari Gur, Vander Heiden Jason A, Church George, Donahue William F, Hintzen Rogier Q, Huttner Anita J, Laman Jon D, Nagra Rashed M, Nylander Alyssa, Pitt David, Ramanan Sriram, Siddiqui Bilal A, Vigneault Francois, Kleinstein Steven H, Hafler David A, O’Connor Kevin C. B Cells Populating the Multiple Sclerosis Brain Mature in the Draining Cervical Lymph Nodes. Science Translational Medicine. 2014;6(248):248ra107–248ra107. doi: 10.1126/scitranslmed.3008879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefranc Marie-Paule, Pommie Christelle, Ruiz Manuel, Giudicelli Veronique, Foulquier Elodie, Truong Lisa, Thouvenin-Contet Valerie, Lefranc Gerard. IMGT Unique Numbering for Immunoglobulin and T cell Receptor Variable Domains and Ig Superfamily V-like Domains. Developmental & Comparative Immunology. 2003;27(1):55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 21.Meng Wenzhao, Jayaraman Sahana, Zhang Bochao, Schwartz Gregory W, Daber Robert D, Hershberg Uri, Garfall Alfred L, Carlson Christopher S, Luning Prak Eline T. Trials and Tribulations with VH Replacement. Frontiers in Immunology. 2014;5 doi: 10.3389/fimmu.2014.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall Brendan, Schulz Ruth, Zhou Min, Mellor Andrew. Alternative Splicing and Hypermutation of a Nonproductively Rearranged TCR Alpha-chain in a T cell Hybridoma. The Journal of Immunology. 1999;162(2):871–877. [PubMed] [Google Scholar]
- 23.Mauri Claudia, Bosma Anneleen. Immune Regulatory Function of B cells. Annual Review of Immunology. 2012;30(2012):221–241. doi: 10.1146/annurev-immunol-020711-074934. [DOI] [PubMed] [Google Scholar]
- 24.Kronenberg Mitchell, Siu Gerald, Hood Leroy E, Shastri Nilabh. The Molecular Genetics of the T-cell Antigen Receptor and T-cell Antigen Recognition. Annual Review of Immunology. 1986;4(1):529–591. doi: 10.1146/annurev.iy.04.040186.002525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.