Abstract
Porcine epidemic diarrhea virus (PEDV) is the cause of an economically important swine disease. Previous studies suggested that PEDV does not elicit a robust IFN response, but the mechanism(s) used to evade or block this innate immune response was not known. In this study, we found that PEDV infection blocked synthetic dsRNA-induced IFN-β production by interfering with the activation of interferon regulatory factor 3 (IRF3). We identified PEDV replicase encoded papain-like protease 2 (PLP2) as an IFN antagonist that depends on catalytic activity for its function. We show that levels of ubiquitinated proteins are reduced during PEDV infection and that PEDV PLP2 has deubiquitinase (DUB) activity that recognizes and processes both K-48 and K-63 linked polyubiquitin chains. Furthermore, we found that PEDV PLP2 strongly inhibits RIG-I- and STING-activated IFN expression and that PEDV PLP2 can be co-immunoprecipitated with and deubiquitinates RIG-I and STING, the key components of the signalling pathway for IFN expression. These results show that PEDV infection suppresses production of IFN-β and provides evidence indicating that the PEDV papain-like protease 2 acts as a viral DUB to interfere with the RIG-I- and STING-mediated signalling pathway.
Introduction
Porcine epidemic diarrhea virus (PEDV) is a member of the genus Alphacoronavirus, family Coronaviridae included in the order Nidovirales together with the families Arteriviridae and Roniviridae. Despite the availability of vaccines, the disease caused by PEDV has broken out frequently in many swine-raising countries, resulting in considerable economic loss (Pensaert & de Bouck, 1978). Accumulating evidence suggests that PEDV encodes a defensive mechanism(s) to evade the antiviral activities of IFN. PEDV suppresses production of type I IFN (IFN-α/β) in infected Vero cells and alveolar macrophages (Charley et al., 2006; Laude et al., 1993). Furthermore, PEDV diminishes IFN-α/β production induced by infection with transmissible gastroenteritis coronavirus (TGEV) or transfection with double-stranded RNA (dsRNA) (Albina et al., 1998; Miller et al., 2004). However, the mechanisms used by PEDV to suppress the production of type I IFN remain unclear.
The innate immune system is the first line of defence that protects the host against viral infection (O’Neill & Bowie, 2010). Viral infections are sensed by pattern-recognition receptors (PRRs) of the innate immune system, such as toll-like receptors, retinoid acid-inducible gene (RIG)-I-like helicase (RLH) family members RIG-I and melanoma differentiation associated protein 5 (MDA-5), that recognize pathogen-associated molecular patterns (PAMPs) and then trigger an antiviral response (Wilkins & Gale, 2010). Upon engagement with viral nucleic acids, these PRRs transduce signals to the downstream kinase with the help of different adaptor proteins (MAVS/IPS-1/VISA/Cardif for RIG-I, TRIF for TLR3 and MyD88 for TLR7/8/9). Transcription factors IFN regulatory factor-3 (IRF3), nuclear factor κB (NF-κB) and ATF-2/c-jun are co-ordinately activated, stimulating the expression of type I interferons (IFN-α/β). Type I IFNs induce the activation of STAT transcription factors that induce the expression of hundreds of IFN-stimulated genes (ISGs) which establish an antiviral state in surrounding cells, thereby limiting viral replication and spread (Schindler et al., 2007).
Coronaviruses (CoV), including PEDV, are positive strand RNA viruses that replicate in the cytoplasm of infected cells and produce a nested set of double-stranded RNA intermediates during viral RNA synthesis (Sawicki et al., 2007). Despite the generation of dsRNA intermediates, it has been reported that CoV infection generally does not induce high levels of IFN production (Clementz et al., 2010; Devaraj et al., 2007). In order to combat the antiviral effects of IFN-α/β, many viruses, such as coronavirus, have evolved distinct strategies to inhibit IFN signalling pathways for their survival (Haller & Weber, 2007). Human coronavirus, severe acute respiratory syndrome coronavirus (SARS-CoV), encodes at least six innate immune antagonists, including non-structural protein 1 (nsp1) (Narayanan et al., 2008), the papain-like protease domain in nsp3 (Devaraj et al., 2007), nucleocapsid protein (Kopecky-Bromberg et al., 2007; Lu et al., 2011), membrane protein (Siu et al., 2009) and the products of ORF6 and ORF3b (Kopecky-Bromberg et al., 2007). Our previous work found that human NL63 CoV (HCoV-NL63) (van der Hoek et al., 2005) also encodes a papain-like protease, termed PLP2, which has deubiquitinase (DUB) activity and antagonizes IFN induction (Chen et al., 2007; Clementz et al., 2010). Our recent studies indicated that NL63 and SARS-CoV papain-like proteases (PLPs) antagonize STING- (also known as MITA/ERIS/MYPS) mediated antiviral innate immune signalling through disruption of STING dimer and deubiquitination of RIG-I and STING regulators of the IFN expression pathway (Sun et al., 2012). A recent study also showed that DUB activity is conserved in all members of the arterivirus family and that both arteri- and nairovirus DUBs inhibit RIG-I mediated innate immune signalling (van Kasteren et al., 2012).
In the present study, we investigated the mechanisms used by PEDV to interact with the host antiviral innate immune response. Our results show that PEDV infection prevents IFN-β expression in Vero cells and blocks ubiquitination of cellular proteins. We demonstrated that the core domain of PEDV PLP2 acts as an IFN antagonist and is a coronaviral DUB. This study represents a first step in elucidating the role of papain-like protease in PEDV pathogenesis and provides new insights for the development of novel vaccines to control PEDV outbreaks.
Results
PEDV infection fails to activate IFN-β and interrupts poly(I : C)-mediated IFN-β induction
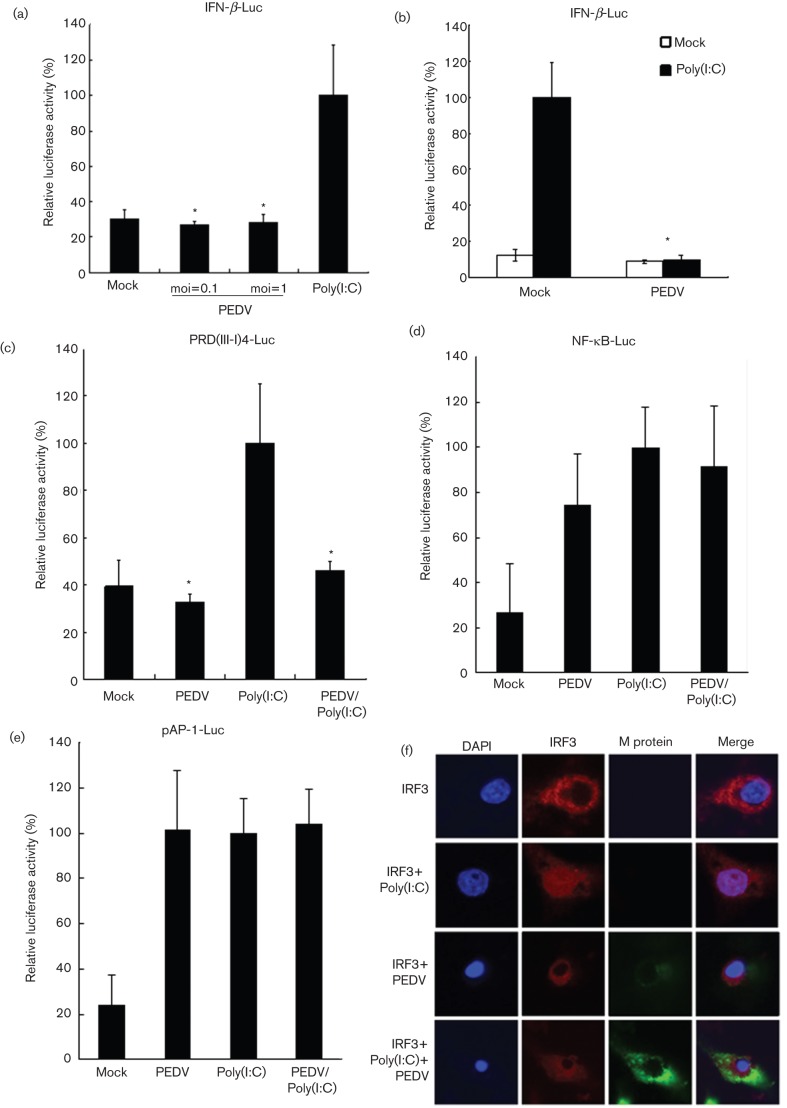
To investigate the activation of IFN-β in PEDV-infected cells, Vero E6 cells were co-transfected with IFN-β-Luc and pRL-TK reporter plasmids and then infected with PEDV. After 12 h of infection, cell lysates were prepared and IFN-β promoter-driven luciferase activity was assessed. Transfection with poly(I : C) was used as positive control to test whether Vero E6 cells can recognize dsRNA to activate IFN-β promoter activity. As shown in Fig. 1(a), IFN-β promoter-driven luciferase activity was barely detectable in PEDV-infected cells in comparison to a strong reporter signal in cells transfected with poly(I : C), indicating that PEDV infection failed to activate IFN-β promoter activity. In order to examine whether PEDV inhibits dsRNA-induced IFN-β promoter activity, mock- and PEDV-infected Vero E6 cells were cotransfected with poly(I : C) and IFN-β-Luc, and IFN-β promoter activity was analysed. As shown in Fig. 1(b), the IFN-β promoter was activated 10-fold when mock-infected cells were transfected with poly(I : C), whereas the activation of IFN-β promoter induced by poly(I : C) transfection was significantly inhibited in PEDV infected cells. The results indicated that PEDV infection interrupts dsRNA-mediated IFN-β induction.
Fig. 1.
PEDV does not induce IFN-β production and blocks dsRNA-induced IFN-β promoter activation. (a) PEDV does not induce IFN-β production. Vero E6 cells were cotransfected with IFN-β-Luc and an internal control plasmid pRL-TK, followed by PEDV infection at an m.o.i. of 0.1 or 1. Poly(I : C) was used as a positive control. Cells were harvested and subjected to a Dual-luciferase assay after PEDV infection or poly(I : C) transfection. The results were expressed as mean relative luciferase (firefly luciferase activity divided by Renilla luciferase activity) with standard deviation from repeated experiments carried out in triplicate. (b) PEDV blocks dsRNA-induced IFN-β promoter activation. Vero E6 cells were infected with PEDV at an m.o.i. of 0.1. Cells were then cotransfected with IFN-β-Luc and pRL-TK. Twenty-four hours later, cells were transfected with or without poly(I : C). Cells were subjected to Dual-luciferase assay as described in (a). (c–e) PEDV inhibits dsRNA-induced activation of IRF3 but not of NF-κB and AP-1. Vero E6 cells were infected or mock-infected with PEDV at an m.o.i. of 0.1, and then cotransfected with pRL-TK and PRD(III-I)4-Luc (c), pNF-κB-Luc (d), or pAP-1-Luc (e), respectively. Twenty-four hours later, cells were transfected with or without poly(I : C). Luciferase activities were measured as described in (a). Asterisks indicate statistical significance (P<0.05). (f) PEDV inhibits dsRNA-induced IRF3 nuclear translocation. Vero E6 cells were transfected with IRF3 expression construct. After 12 h, cells were infected or mock-infected with PEDV at an m.o.i. of 0.1. Another 12 h later, cells were transfected or mock-transfected with poly(I : C). Fluorescence was examined by using a confocal microscope.
IFN-β transcription requires the activation of transcription factors NF-κB, IRF3 and AP-1 and their subsequent binding to the IFN-β promoter (Bovolenta et al., 1995; Thanos & Maniatis, 1995). To investigate the mechanisms used by PEDV to inhibit host antiviral IFN-β expression, the transcriptional activity of NF-κB, IRF3 and AP-1 was analysed using the luciferase assay for identifying the exact transcription factor involved in the inhibition by PEDV on the IFN-β promoter. We found that IRF3-dependent IFN-β activation was significantly inhibited by PEDV infection, and PEDV infection inhibits poly(I : C) activated IRF3-dependent IFN-β activation (Fig. 1c). In contrast, NF-κB and AP-dependent IFN-β expression were normally activated by PEDV infection (Fig. 1d, e). IRF3, a cytoplasmic protein, migrates to the nucleus and binds to the PRDIII and PRDI sites of IFN-β promoter to initiate IFN-β transcription upon viral infection (Fitzgerald et al., 2003). To determine whether PEDV prevents IRF3 migration from the cytoplasm to the nucleus, Vero E6 cells were transiently transfected with IRF3 expression construct and the subcellular localization of the protein was analysed using confocal microscopy. As shown in Fig. 1(f), IRF3 was located exclusively in the cytoplasm in mock-infected Vero E6 cells, but it rapidly translocated to the nucleus when those cells were transfected with poly(I : C). In contrast, nuclear IRF3 translocation did not occur in PEDV-infected cells. Moreover, PEDV prevented the nuclear translocation of IRF3 induced by poly(I : C) (Fig. 1f). Taken together, PEDV infection inhibits the activation of IRF3, but not NF-κB and AP-1. These observations collectively suggest that PEDV suppresses IFN-β transcription by interfering with the IRF3-mediated IFN expression signalling pathway.
PEDV PLP2 is an IFN antagonist
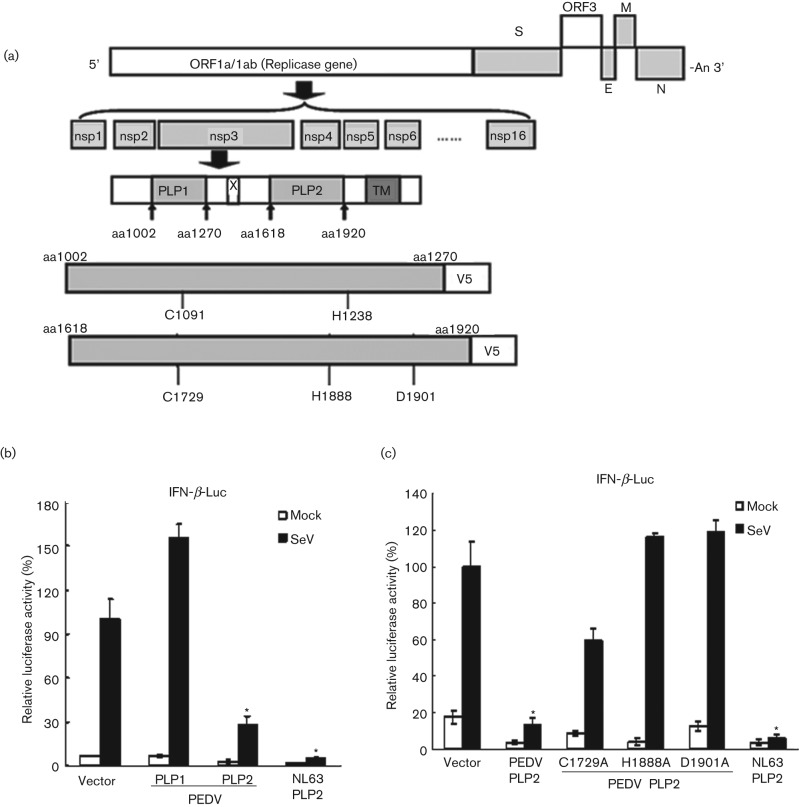
Our previous work demonstrated that the papain-like proteases of human coronaviruses, SARS-CoV and NL63-CoV, act as IFN antagonists (Clementz et al., 2010; Devaraj et al., 2007; Sun et al., 2012). To determine if the PLP of PEDV is an IFN antagonist that may contribute to inhibition of type I IFN expression during PEDV infection, the PLP1 and PLP2 core domains were constructed and the catalytic mutants (C1729A, H1888A, D1901A) were generated as indicated in Fig. 2(a). HEK293T cells were transfected with the plasmids encoding PEDV PLP1, PLP2 or its corresponding catalytic mutants separately together with IFN-β luciferase and Renilla luciferase reporters for 24 h. The cells were then infected with Sendai virus to activate the RIG-I-dependent IFN-β expression pathway. We observed that the IFN-β promoter activated by Sendai virus was inhibited in the presence of PEDV PLP2, which was similar to the previously reported IFN antagonist of NL63 PLP2 (Clementz et al., 2010; Sun et al., 2012). In contrast, the core domain of PEDV PLP1 showed negligible inhibition activity of the IFN-β promoter (Fig. 2b). These results suggest that PEDV PLP2, but not PLP1, is an interferon antagonist.
Fig. 2.
PEDV PLP2 is an IFN antagonist. (a) Schematic diagram illustrating PEDV PLP1 and PLP2 constructs used in this study. The PEDV papain-like protease domains (PLP1 and PLP2), their boundary sites (arrowheads) and the presence of the predicted transmembrane (TM) domains within nsp3 are indicated. The catalytic core domains of PLP1 and PLP2 and catalytic sites of each domain are indicated. (b) PEDV PLP2, but not PLP1, is an IFN antagonist. HEK293T cells were co-transfected with either PLP1 or PLP2 at amounts of 200 ng, IFN-β-Luc and pRL-TK. HCoV NL63 PLP2 was used as a positive control as an inhibitor of IFN-β expression. Cells were then mock infected or infected with Sendai virus to activate the IFN expression pathway. Luciferase activities were assayed as described in (a). (c) PEDV PLP2 inhibits IFN-β expression depending on its protease activity. HEK293T cells were co-transfected with 200 ng of PEDV PLP2 and catalytic mutants C1729A, H1888A and D1901A, with IFN-β-Luc and pRL-TK, followed by the assay which was performed similarly to that in (b). Asterisks indicate statistical significance (P<0.05).
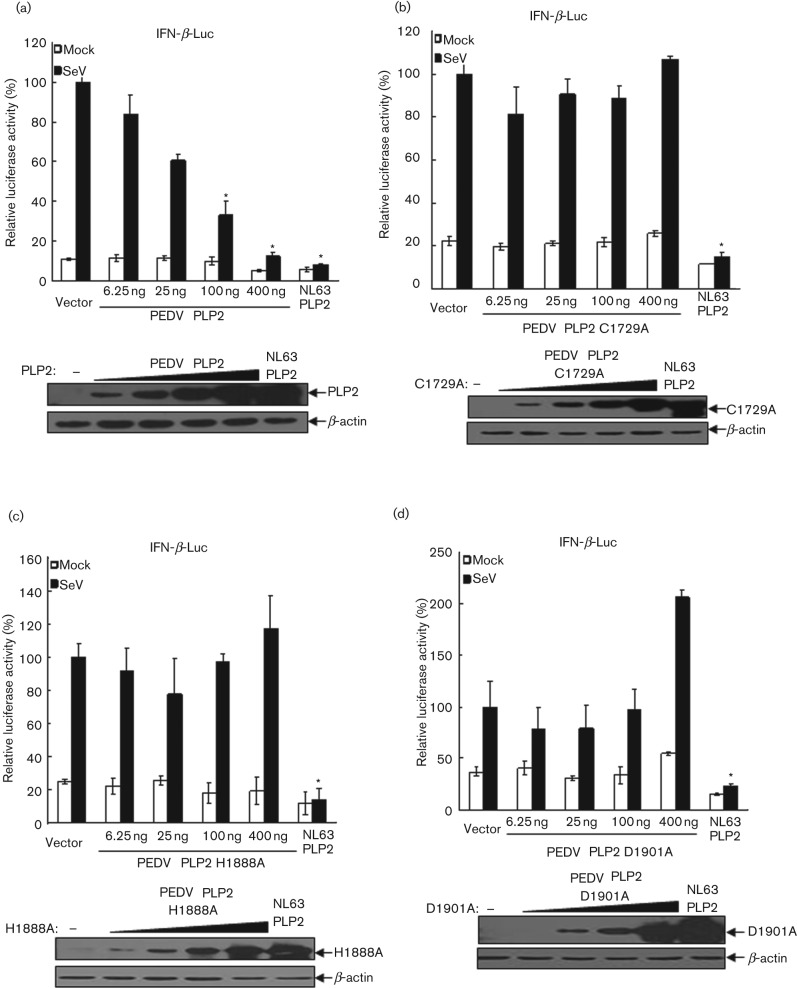
To determine if catalytic activity is essential for PEDV PLP2-mediated inhibition of IFN expression, we performed a sequence alignment with other coronavirus PLP domains and identified the conserved catalytic triad of PLP2, and catalytic mutants (C1729A, H1888A, D1901A) were generated. PEDV PLP2 or catalytic mutants of PLP2 (C1729A, H1888A, D1901A) were transfected with the IFN-β Luc and pRL-TK plasmids into HEK293T cells, which were then infected with Sendai virus to activate IFN-β promoter activity. We observed that the PLP2 mutants with mutation at the catalytic sites of H1888 and D1901 almost completely lost IFN antagonistic activity compared to that of wt PLP2, despite the PLP2 mutant with C1729A showing reduced inhibition of IFN-β promoter activity (Fig. 2c). Furthermore, PLP2 exhibits a clear dose-dependent inhibition of IFN promoter activity (Fig. 3a), whereas the PLP2 mutants (C1729A, H1888A, D1901A) completely lost IFN antagonistic activity at the concentrations tested (Fig. 3b–d). Taken together, these results demonstrated that PEDV PLP2 is an IFN antagonist which is dependent on an intact catalytic triad.
Fig. 3.
PLP2 inhibits IFN-β expression in a dose-dependent manner. HEK293T cells were cotransfected with increasing amounts of PEDV PLP2 (a) and the catalytic mutants C1729A (b), H1888A (c) and D1901A (d), together with reporters of IFN-β-Luc and pRL-TK, followed by the assay which was performed similarly to that in Fig. 2(b, c). Asterisks indicate statistical significance (P<0.05).
PEDV PLP2 has DUB activity
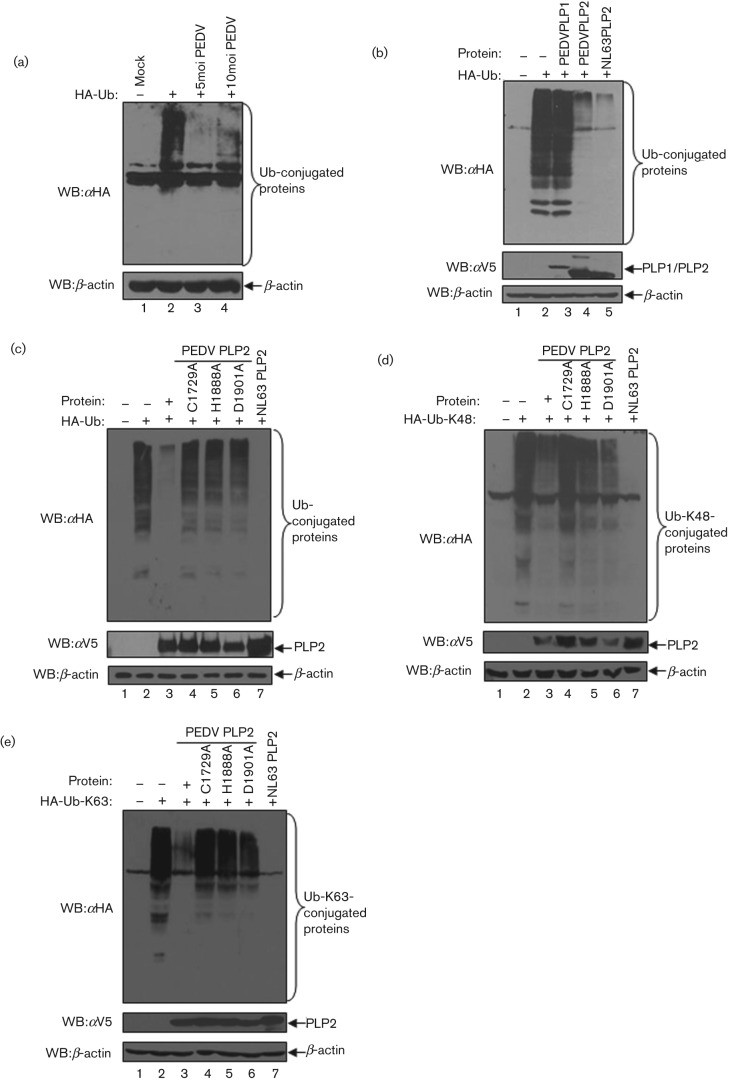
Our previous work demonstrated that the papain-like proteases of human coronaviruses, SARS-CoV and NL63-CoV, are coronaviral DUBs (Barretto et al., 2005; Clementz et al., 2010; Devaraj et al., 2007; Sun et al., 2012). Here, we asked if PEDV infection exhibits any DUB activity against host cellular proteins. First, Vero E6 cells were transfected with pcDNA HA-Ub, the cells were infected or mock-infected with PEDV at an m.o.i. of 5 for another 24 h. Cell lysates were then prepared and the extent of ubiquitinated proteins was assessed via Western blotting assay with anti-HA antibodies. We found that the level of ubiquitin (Ub)-conjugated proteins was reduced dramatically in the cells infected with PEDV (Fig. 4a, lanes 3 and 4), indicating that PEDV encodes some proteins which have deubiquitinating activity, or PEDV infection disrupts the ubiquitination machinery in host cells to block the DUB activity.
Fig. 4.
PEDV PLP2 has DUB activity that is dependent on its catalytic activity. (a) PEDV has deubiquitinating activity. Vero E6 cells were transfected with HA-tagged ubiquitin and then mock infected or infected with PEDV at an m.o.i. of 5 or 10. The HA-tagged Ub-conjugated proteins were assayed with anti-HA antibody by Western blotting (top panel). β-Actin was detected from whole-cell lysates (WCL) as a loading control. (b) PEDV PLP2, but not PLP1, has DUB enzyme activity. HEK293T cells were transfected with HA-tagged Ub and either PEDV PLP1 or PLP2. HCoV NL63 PLP2 was used as a positive control of coronaviral DUB activity. Proteins were extracted and analysed by Western blotting with an anti-HA antibody to visualize HA-tagged Ub-conjugated proteins (top panel) and anti-V5 antibody (middle panels) to visualize PLP construct expression. β-Actin was detected by Western blotting as protein loading control (bottom panel). (c–e) PLP2 removes both K48- and K63-linked polyubiquitin chains. HEK293T cells were transfected with HA-tagged Ub (c), HA-Ub-K48 (d) or HA-Ub-K63 (e) with PEDV PLP2 and the catalytic mutants C1729A, H1888A and D1901A. HCoV NL63 PLP2 was used as a positive control for DUB activity. HA-tagged Ub-conjugated proteins were assayed as described above.
To investigate if PEDV papain-like protease core domains, PLP1 and PLP2, have DUB activity, PEDV PLP1 and PLP2 were separately transfected into HEK293T cells, together with pcDNA HA-Ub. We found that expression of PLP2 resulted in a dramatic reduction in the level of Ub-conjugated proteins (Fig. 4b, lane 4), which was similar to NL63-CoV PLP2 as reported previously (Clementz et al., 2010). In contrast, PLP1 did not show any significant reduction of HA-Ub conjugates (Fig. 4b, lane 3).
The two most common types of ubiquitinated protein are linked through ubiquitin lysine 48 (K48) and lysine 63 (K63) (Pickart & Fushman, 2004). To investigate whether PEDV PLP2 has selectivity for ubiquitinated substrates with K48 or K63 linkages, HEK293T cells were transfected with PLP2 or the corresponding catalytic mutants (C1729A, H1888A and D1901A) together with pcDNA-HA-Ub (Fig. 4c), pcDNA-HA-Ub K48 (Fig. 4d) or pcDNA-HA-Ub K63 (Fig. 4e) and the extent of ubquitinated products was assessed by Western blotting. We found that PEDV PLP2 exhibits strong global DUB activity with significant reduction in the level of Ub-, K48- and K63-conjugated proteins (Fig. 4c–e, lane 3). However, the catalytic mutants (C1729A, H1888A and D1901A) of PLP2 completely lost DUB activity against all types of ubiquitinated proteins (Fig. 4c–e, lanes 4–6). These results demonstrate that PEDV PLP2 has potent DUB activity which is dependent on its catalytic activity. Furthermore, PEDV PLP2 DUB exhibits hydrolytic activity toward ubiquitinated branched proteins, for both K48 and K63 linkages.
PEDV PLP2 negatively regulates RIG-I and STING-mediated IFN-β expression
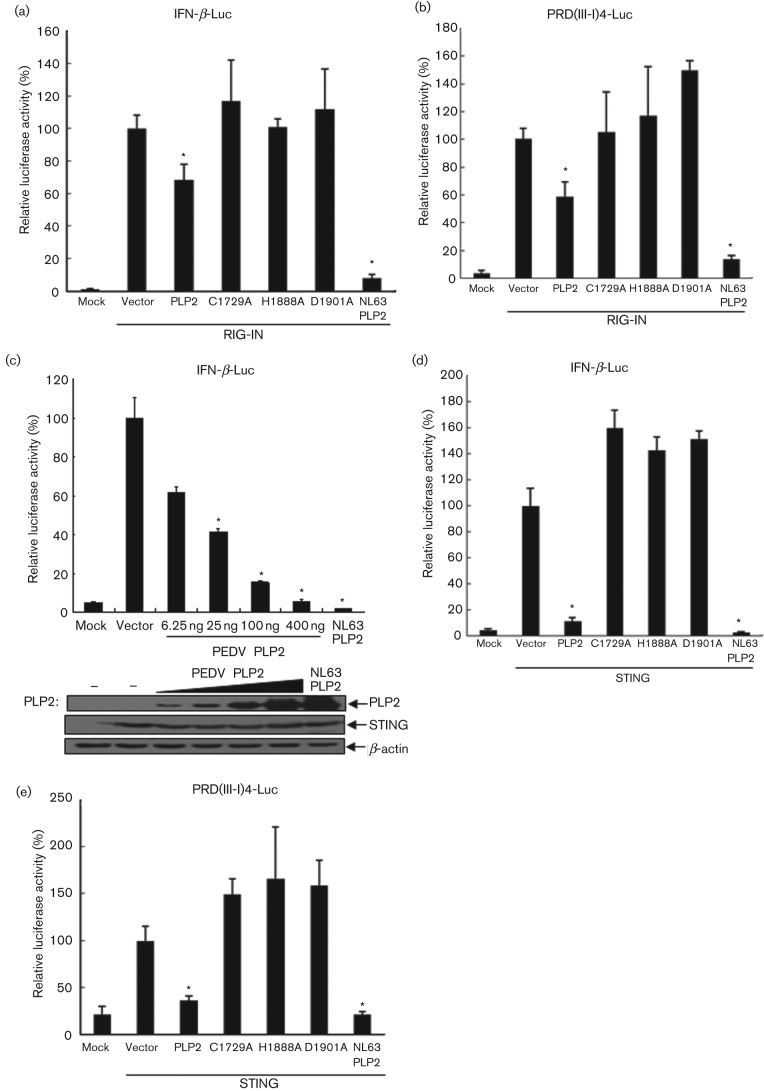
We previously discovered that human coronavirus (NL63 and SARS) PLPs negatively regulate innate antiviral immune response by disrupting STING-mediated IFN induction (Sun et al., 2012). Based on these findings, we hypothesized that PEDV PLP2 inhibits the IFN expression pathway through a similar mechanism. To test this hypothesis HEK293T cells were transfected with PEDV PLP2 or the corresponding catalytic mutants (C1729A, H1888A, D1901A) together with Flag-RIG-IN, the N-terminal helicase domain of RIG-I as its constitutively active mutant, or Flag-STING, and IFN-β-Luc or PRD(III-I)4-luc reporters; NL63 PLP2 was used as a positive control for IFN antagonist (Charley et al., 2006; Zeng et al., 2010). IFN-β and IRF3 promoter-driven luciferase activity was detected 24 h later. As expected, RIG-IN activated IFN-β and IRF3 promoter activity were inhibited significantly by PEDV PLP2, and this inhibitory activity of PEDV PLP2 seemed to be catalytic-dependent (Fig. 5a, b). In addition, PEDV PLP2 possessed dose-dependent inhibitory function on STING activated IFN-β promoter (Fig. 5c). In addition, STING activated IFN-β and IRF3 expression were also inhibited significantly by PEDV PLP2 in a catalytic-dependent manner (Fig. 5d, e). Overall, these data indicate that PEDV PLP2 strongly inhibits RIG-I and STING-activated IFN-β expression.
Fig. 5.
PEDV PLP2 inhibits RIG-I and STING-activated IFN-β expression. (a, b) PEDV PLP2 inhibits RIG-I-activated IFN expression. HEK293T cells were cotransfected with IFN-β-Luc (a) or PRD(III-I)4-Luc (b) together with PEDV PLP2 and the catalytic mutants C1729A, H1888A and D1901A, with RIG-IN to activate the IFN expression pathway, followed by the assay which was performed similarly to that in Fig. 2(b, c). (c) PEDV PLP2 inhibits STING-activated IFN expression. HEK293T cells were cotransfected with increasing amounts of PEDV PLP2 along with reporters of IFN-β-Luc and STING for activation of IFN-β expression, followed by treatments that were performed similarly to those in Fig. 2(b, c). (d, e) PEDV PLP2 inhibits STING-activated IFN expression which is dependent on its protease activity. HEK293T cells were cotransfected with STING and either PLP2 or catalytic mutants (C1678A, H1836A and D1849A), and IFN-β-Luc (d) or PRD(III-I)-Luc (e), followed by the assay which was performed similarly to that in Fig. 2(b, c). Asterisks indicate statistical significance (P<0.05).
PEDV PLP2 negatively regulates IFN-β expression by removing ubiquitinated conjugates from RIG-I and STING
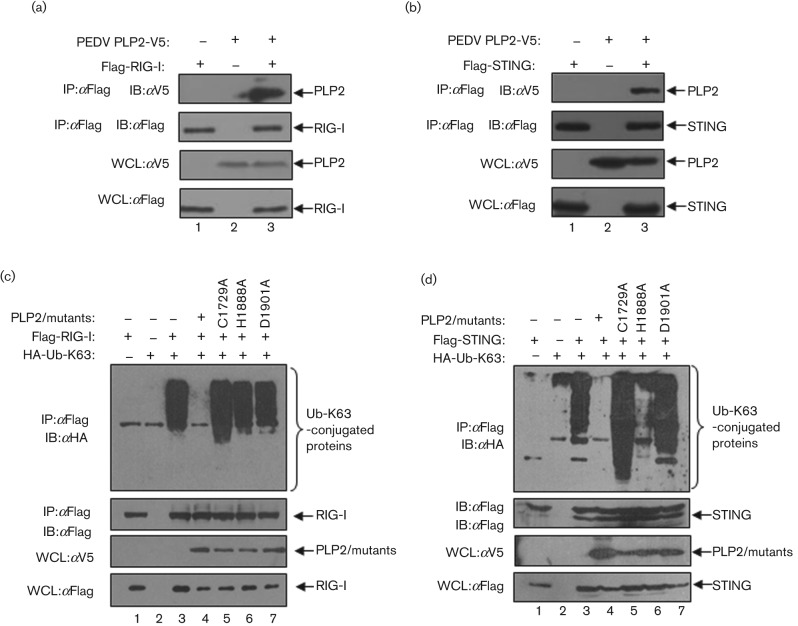
Modification of signalling molecules, such as RIG-I and STING, by ubiquitination plays a critical role in activation of the IFN response (Bhoj & Chen, 2009; Bibeau-Poirier & Servant, 2008; Isaacson & Ploegh, 2009). Here, we asked if PEDV PLP2 can recognize and deubiquitinate key regulators of RIG-I and STING in the IFN signalling pathway. HEK293T cells were transfected with PEDV PLP2 together with Flag-RIG-I or Flag-STING for 24 h, and the interaction of PLP2 and RIG-I or STING was assessed via co-immunoprecipitation and Western blotting assay. The results show that PEDV PLP2 is detected in association with RIG-I, as well as STING (Fig. 6a, b, lane 3). Next, we wanted to determine if PEDV PLP2 can recognize and deubiquitinate RIG-I and STING, since our previous report demonstrated that NL63 PLP2 blocks ubiquitination of several signalling molecules in the IFN expression pathway, such as RIG-I, STING and TBK1 (Sun et al., 2012). HEK293T cells were transfected with PEDV PLP2 or the corresponding catalytic mutants, together with HA-Ub-K63 and Flag-tagged versions of either RIG-I or STING; cell lysates were subjected to immunoprecipitation and immunoblotting to determine the ubiquitination status of the immunoprecipitated proteins (Fig. 6c, d). We found that there was a dramatic reduction in the amount of ubiquitinated RIG-I (Fig. 6c, lane 4) and STING (Fig. 6d, lane 4) in cells expressing PEDV PLP2. We also investigated the role of PEDV PLP2 catalytic activity in mediating deubiquitination of RIG-I and STING. Cells were transfected with HA-Ub-K63 and either wt or catalytic mutants of PEDV PLP2; as expected, we did not detect any reduction in the level of ubiquitinated RIG-I and STING in the presence of the C1729A, H1888A and D1901A mutants of PLP2 (Fig. 6c, d, lanes 5–7). These results suggest that PEDV PLP2 DUB activity contributes to deubiquitination of RIG-I and STING. One another possibility is that PEDV PLP2 interacts with RIG-I and STING, which blocks access of RIG-I and STING to the host ubiquitination machinery.
Fig. 6.
PEDV PLP2 interacts with and deubiquitinates RIG-I and STING. (a, b) PEDV PLP2 interacts with RIG-I and STING. HEK293T cells were transfected with Flag-tagged RIG-I (a) or STING (b) with V5-tagged PLP2, and co-immunoprecipitation experiments were performed with the indicated antibodies. The co-immunprecipitates were blotted with anti-V5 (top panel) and reprobed with anti-Flag (second panel) for RIG-I or STING detection. The input lysates were blotted with anti-V5 to detect PLP2 (third panel) and anti-Flag to detect RIG-I or STING (bottom panel). (c, d) PEDV PLP2 deubiquitinates RIG-I and STING in a catalytic-dependent manner. HEK293T cells were co-transfected with Flag-tagged RIG-I (c) or STING (d) together with HA-tagged Ub-K63, V5-tagged PLP2 or the catalytic mutants of C1729A, H1888A and D1901A. The lysates were immunoprecipitated with anti-Flag antibody and the immunoprecipitates were blotted with anti-HA to detect Ub-K63-conjugated RIG-I (c) or STING (d) (top panel) and reprobed with anti-Flag for RIG-I or STING detection (second panel). The input lysates were blotted with anti-V5 to detect PLP2 and the catalytic mutants (third panel) and anti-Flag to detect RIG-I and STING (bottom panel).
Discussion
In this study, we investigated the mechanism of IFN antagonism imposed by the PLPs of PEDV. We show that PEDV infection does not stimulate the production of IFN-β in Vero E6 cells. Moreover, dsRNA-induced IFN-β expression is inhibited by PEDV infection, indicating that PEDV encodes an interferon antagonist. Importantly, we provide evidence that PEDV PLP2 can act as an IFN antagonist, which may contribute to the inhibitory effect of PEDV on the IFN-β pathway in cells. In addition, we provide data showing that, like human SARS-CoV PLpro and NL63-CoV PLP2, PEDV PLP2 is a viral DUB enzyme which acts on both K48- and K63-linked Ub chains. Overall, our results are consistent with the idea that negative regulation of host antiviral innate immunity by PLP/DUB enzyme is a general characteristic of many human and animal coronaviruses.
One of the most intriguing results from this study is the finding that both the IFN antagonist and DUB activity of PEDV PLP2 are dependent on its protease activity, since mutation of the catalytic sites of PLP2 resulted in the loss of IFN antagonist and DUB activity, suggesting a DUB/protease-dependent mechanism for type I IFN antagonism. PEDV PLP2 has an IFN antagonism profile that is remarkably different from that of NL63-CoV PLP2, as we previously found that HCoV-NL63 PLP2 can antagonize type I IFN induction which is independent of catalytic activity (Clementz et al., 2010). The underlying mechanism(s) that lead to the difference of IFN antagonism profile of PLP2 from different CoVs was not completely clear. One possibility is that the transmembrane (TM) domain downstream of CoV PLP2 has an essential effect on PLP2 DUB and IFN antagonistic activity. In fact, our previous studies showed that HCoV-NL63 PLP2 has DUB activity (Clementz et al., 2010) and IFN antagonistic activity (Sun et al., 2010a) which are dependent on the protease activity. In contrast, both the wt and the catalytic mutants of TM-containing PLP2 (PLP2-TM) have DUB activity and IFN antagonistic activity, although the DUB and IFN antagonism of PLP2-TM catalytic mutants are less than that of wt PLP-TM (Sun et al., 2012). While many reports have demonstrated that cellular and viral DUBs play important roles in negative regulation of host innate immunity, as discussed as below, future work will be needed to determine the exact functions of PLP2 protease/DUB activity in coronavirus interaction with host innate immune response.
Ubiquitination and deubiquitination are critically involved in regulation of virus-induced type I IFN signalling pathways (Bhoj & Chen, 2009; Bibeau-Poirier & Servant, 2008; Isaacson & Ploegh, 2009; Zhong et al., 2010). The activation of related receptors (such as RIG-I) and the transduction of the cell signal pathway in the innate immune response require ubiquitination (Zeng et al., 2010). Many cellular DUBs were identified as negative regulators of innate immunity. For example, a central gatekeeper in inflammation and immunity, A20, has DUB activity and removes K-63 linked polyubiquitin lines from RIP1, TRAF6, RIP2 and NEMO, which results in negative regulation of the innate immune response (Coornaert et al., 2008). Cellular proteins DUBA and CYLD also negatively regulate the innate immune response (Kayagaki et al., 2007; Sun, 2008; Yoshida et al., 2005). We have reported that human CoV NL63 and SARS PLpro reduce the ubiquitinated forms of STING, RIG-I, TBK1 and IRF3 in cells (Sun et al., 2012). It is interesting that coronaviruses, including human CoV NL63, SARS-CoV, transmissible gastroenteritis virus, murine hepatitis virus and PEDV reported here, have evolved to encode DUBs (Barretto et al., 2005; Clementz et al., 2010; Lindner et al., 2005; Wang et al., 2011b; Wojdyla et al., 2010; Zheng et al., 2008), likely to modulate the innate immune response. Indeed, DUBs have now been reported in a variety of viruses such as foot-and-mouth disease virus Lpro (Wang et al., 2011a), human cytomegalovirus UL48 (Kim et al., 2009), herpes simplex virus type 1 UL36 (Kattenhorn et al., 2005), porcine reproductive and respiratory syndrome virus nsp2 (Chen et al., 2010b; Sun et al., 2010b).
In recent years, many important regulators in the IFN pathway have been discovered and studied intensively, such as STING, MAVS, ZAPS and TRAF (Belgnaoui et al., 2011; Hayakawa et al., 2011; Ishikawa & Barber, 2008; Oganesyan et al., 2006). In our recent study, human CoV NL63 and SARS PLPs were found to be co-immunoprecipitated with STING, block STING dimer formation and negatively regulate assembly of STING-MAVS-TBK1/IKKe complexes, and remove polyubiquitin chains from STING, RIG-I, TBK1 and IRF3 (Sun et al., 2012). We report here that PEDV PLP2 interferes with and significantly inhibits ubiquitination of RIG-I and STING, which are essential regulators for activation of type I IFN signaling. Three catalytically inactive mutants of PEDV PLP2 (C1729A, H1888A and D1901A), which are defective for DUB activity and the capability of reducing ubiquitinated RIG-I and STING, failed to inhibit virus-induced IFN-β activation, indicating that the DUB activity of PEDV PLP2 is directly involved in the inhibition of type I IFN induction.
Taken together, our research suggests that PEDV PLP2 is not only a classical papain-like protease encoded by a CoV genome, but also a multifunctional protein which plays important roles in regulation of the interactions of PEDV–host antiviral innate immune response: (i) PEDV PLP2 is an IFN antagonist and its IFN antagonistic activity depends on the intact catalytic sites of C1729, H1888 and D1901; (ii) PEDV PLP2 is a viral DUB that cleaves ubiquitin chains from RIG-I and STING, thereby inhibiting the activation of type I IFN signaling. These characteristics of PEDV PLP2 as a viral IFN antagonist and DUB reveal the multilayered counteracting of host defence by PEDV and suggest the possibility of developing effective new strategies that target PLP for control of PEDV infections.
Methods
Cells and virus.
Vero E6 and HEK293T cells were cultured using Dulbecco’s modified Eagle’s medium (Invitrogen) containing 10 % (v/v) FCS, supplemented with penicillin (100 U ml−1) and streptomycin (100 µg ml−1). PEDV strain CV777 was propagated in Vero cells kept in the Harbin Veterinary Research Institute, Harbin, China, and as previously described (Chen et al., 2008, 2010a). Sendai virus was kindly provided by Dr Shaobo Xiao (Huazhong Agricultural University, China).
Plasmid DNAs.
The plasmids of IFN-β-Luc, PRD(III-I)4-Luc and HA-tagged Ub were previously described (Clementz et al., 2010; Devaraj et al., 2007). pNF-κB-Luc and pAP-1-Luc were kindly donated by Dr Shaobo Xiao (Huazhong Agricultural University, China). Flag-hRIG-I, Flag-RIG-IN, the N-terminal helicase domain of RIG-I as its constitutively active mutant, and pCMV14-Flag-ERIS were previously described (Sun et al., 2012). HCoV NL63 PLP2, a positive inhibitor of IFN-β expression, was described previously (Clementz et al., 2010; Sun et al., 2012). DNA constructs containing wt and catalytic mutants of PEDV PLP1 and PLP2 were generated as described previously (Chen et al., 2007). The details of the optimized DNA sequences of PEDV PLP1 and PLP2 and the specific primers used in mutagenesis steps are available from the authors on request. All constructs were confirmed by DNA sequencing.
Luciferase activity assay.
HEK293T cells were transfected with the reporter plasmid DNA (pRL-TK, IFN-β-Luc, PRD(III-I)4-Luc, pNF-κB-Luc or pAP-1-Luc) and PEDV PLP1 or PLP2 using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Luciferase activity was assayed as previously described (Clementz et al., 2010; Sun et al., 2012). Data were shown as mean relative luciferase (firefly luciferase activity divided by Renilla luciferase activity) with standard deviation from repeated experiments that were carried out in triplicate. For statistical analysis, the data between Vector and PLPs were subjected to unpaired, two-tailed Student’s t-test using Microsoft SPSS 12.0 software, and P-values of <0.05 were considered to indicate statistical significance (Vaux et al., 2012).
Assay of deubiquitinase activity in cultured cells.
HEK293T cells were co-transfected with pcDNA3.1-HA-Ub or pcDNA3.1-HA-Ub K48, pcDNA3.1-HA-Ub K63 plus indicated amounts of constructs containing PEDV PLP contructs or the corresponding catalytic mutants. The effect of PLP1 and PLP2 of PEDV on ubiquitinated proteins in cultured cells was then assessed as described previously (Clementz et al., 2010; Evans et al., 2004).
Western blotting assay.
HEK293T cells were transfected with PEDV PLP1, PEDV PLP2 and specific mutations in the PLP2 catalytic residues (C1729, H1888 and D1901), and were lysed in buffer containing 0.5 % Triton X-100, 150 mM NaCl, 12.5 mM β-glycerolphosphate, 1.5 mM MgCl2, 2 mM EGTA, 10 mM NaF, 1 mM Na3VO4, 2 mM DTT plus protease inhibitor cocktail (Sigma). Western blotting assay was performed as previously described (Clementz et al., 2010; Sun et al., 2012).
Co-immunoprecipitation analysis.
HEK293T cells were transfected with the indicated expression plasmids and then co-immunoprecipitation analysis was performed as previously described (Clementz et al., 2010; Sun et al., 2012).
Assessing ubiquitination of RIG-I and STING in cultured cells.
Flag-tagged RIG-I and STING were co-transfected into HEK293T cells together with pcDNA3.1-HA-Ub-K63, plus wt or catalytic mutant PEDV PLP2 DNA. Then the effect of PEDV PLP2 on ubiquitinated proteins in cultured cells was assessed as described previously (Friedman et al., 2008; Kayagaki et al., 2007; Sun et al., 2012).
IRF3 nuclear translocation assay.
Vero E6 cells were transfeced with 2.0 µg IRF3 expression construct, and then were mock-infected or infected with PEDV at an m.o.i. of 0.1. Twelve hours post-infection, cells were transfected with 2.0 µg of poly(I : C) or left untransfected. IRF3 nuclear translocation assay was performed as described previously (Fitzgerald et al., 2003).
Acknowledgments
This work was supported by grants from the State Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, China (no. SKLVBF201101 to Z. C. and L. F.) and the National Natural Science Foundation of China (nos 81172799 and 81273231 to Z. C.; no. 30901081 to J. C.; no. 81102478 to Y. X.). All authors declare that there are no financial or other relationships that might lead to a conflict of interest. All authors have seen and approved the manuscript and have contributed significantly to the work.
References
- Albina E., Carrat C., Charley B. (1998). Interferon-alpha response to swine arterivirus (PoAV), the porcine reproductive and respiratory syndrome virus. J Interferon Cytokine Res 18, 485–490. 10.1089/jir.1998.18.485 [DOI] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A. D., Baker S. C. (2005). The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol 79, 15189–15198. 10.1128/JVI.79.24.15189-15198.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgnaoui S. M., Paz S., Hiscott J. (2011). Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol 23, 564–572. 10.1016/j.coi.2011.08.001 [DOI] [PubMed] [Google Scholar]
- Bhoj V. G., Chen Z. J. (2009). Ubiquitylation in innate and adaptive immunity. Nature 458, 430–437. 10.1038/nature07959 [DOI] [PubMed] [Google Scholar]
- Bibeau-Poirier A., Servant M. J. (2008). Roles of ubiquitination in pattern-recognition receptors and type I interferon receptor signaling. Cytokine 43, 359–367. 10.1016/j.cyto.2008.07.012 [DOI] [PubMed] [Google Scholar]
- Bovolenta C., Lou J., Kanno Y., Park B. K., Thornton A. M., Coligan J. E., Schubert M., Ozato K. (1995). Vesicular stomatitis virus infection induces a nuclear DNA-binding factor specific for the interferon-stimulated response element. J Virol 69, 4173–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charley B., Riffault S., Van Reeth K. (2006). Porcine innate and adaptative immune responses to influenza and coronavirus infections. Ann N Y Acad Sci 1081, 130–136. 10.1196/annals.1373.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Wang Y., Ratia K., Mesecar A. D., Wilkinson K. D., Baker S. C. (2007). Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J Virol 81, 6007–6018. 10.1128/JVI.02747-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. F., Sun D. B., Wang C. B., Shi H. Y., Cui X. C., Liu S. W., Qiu H. J., Feng L. (2008). Molecular characterization and phylogenetic analysis of membrane protein genes of porcine epidemic diarrhea virus isolates in China. Virus Genes 36, 355–364. 10.1007/s11262-007-0196-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang C., Shi H., Qiu H., Liu S., Chen X., Zhang Z., Feng L. (2010a). Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch Virol 155, 1471–1476. 10.1007/s00705-010-0720-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhou X., Lunney J. K., Lawson S., Sun Z., Brown E., Christopher-Hennings J., Knudsen D., Nelson E., Fang Y. (2010b). Immunodominant epitopes in nsp2 of porcine reproductive and respiratory syndrome virus are dispensable for replication, but play an important role in modulation of the host immune response. J Gen Virol 91, 1047–1057. 10.1099/vir.0.016212-0 [DOI] [PubMed] [Google Scholar]
- Clementz M. A., Chen Z., Banach B. S., Wang Y., Sun L., Ratia K., Baez-Santos Y. M., Wang J., Takayama J. & other authors (2010). Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol 84, 4619–4629. 10.1128/JVI.02406-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert B., Carpentier I., Beyaert R. (2008). A20: central gatekeeper in inflammation and immunity. J Biol Chem 284, 8217–8221. 10.1074/jbc.R800032200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S. G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C. J., Tseng C. T. & other authors (2007). Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem 282, 32208–32221. 10.1074/jbc.M704870200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. C., Ovaa H., Hamon M., Kilshaw P. J., Hamm S., Bauer S., Ploegh H. L., Smith T. S. (2004). Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem J 378, 727–734. 10.1042/BJ20031377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003). IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4, 491–496. 10.1038/ni921 [DOI] [PubMed] [Google Scholar]
- Friedman C. S., O’Donnell M. A., Legarda-Addison D., Ng A., Cárdenas W. B., Yount J. S., Moran T. M., Basler C. F., Komuro A. & other authors (2008). The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep 9, 930–936. 10.1038/embor.2008.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O., Weber F. (2007). Pathogenic viruses: smart manipulators of the interferon system. Curr Top Microbiol Immunol 316, 315–334. 10.1007/978-3-540-71329-6_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa S., Shiratori S., Yamato H., Kameyama T., Kitatsuji C., Kashigi F., Goto S., Kameoka S., Fujikura D. & other authors (2011). ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat Immunol 12, 37–44. 10.1038/ni.1963 [DOI] [PubMed] [Google Scholar]
- Isaacson M. K., Ploegh H. L. (2009). Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5, 559–570. 10.1016/j.chom.2009.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Barber G. N. (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678. 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattenhorn L. M., Korbel G. A., Kessler B. M., Spooner E., Ploegh H. L. (2005). A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol Cell 19, 547–557. 10.1016/j.molcel.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Phung Q., Chan S., Chaudhari R., Quan C., O’Rourke K. M., Eby M., Pietras E., Cheng G. & other authors (2007). DUBA: a deubiquitinase that regulates type I interferon production. Science 318, 1628–1632. 10.1126/science.1145918 [DOI] [PubMed] [Google Scholar]
- Kim E. T., Oh S. E., Lee Y. O., Gibson W., Ahn J. H. (2009). Cleavage specificity of the UL48 deubiquitinating protease activity of human cytomegalovirus and the growth of an active-site mutant virus in cultured cells. J Virol 83, 12046–12056. 10.1128/JVI.00411-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S. A., Martínez-Sobrido L., Frieman M., Baric R. A., Palese P. (2007). Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81, 548–557. 10.1128/JVI.01782-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Van Reeth K., Pensaert M. (1993). Porcine respiratory coronavirus: molecular features and virus-host interactions. Vet Res 24, 125–150. [PubMed] [Google Scholar]
- Lindner H. A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. (2005). The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol 79, 15199–15208. 10.1128/JVI.79.24.15199-15208.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Pan J., Tao J., Guo D. (2011). SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes 42, 37–45. 10.1007/s11262-010-0544-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. C., Laegreid W. W., Bono J. L., Chitko-McKown C. G., Fox J. M. (2004). Interferon type I response in porcine reproductive and respiratory syndrome virus-infected MARC-145 cells. Arch Virol 149, 2453–2463. 10.1007/s00705-004-0377-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C. T., Makino S. (2008). Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol 82, 4471–4479. 10.1128/JVI.02472-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill L. A., Bowie A. G. (2010). Sensing and signaling in antiviral innate immunity. Curr Biol 20, R328–R333. 10.1016/j.cub.2010.01.044 [DOI] [PubMed] [Google Scholar]
- Oganesyan G., Saha S. K., Guo B., He J. Q., Shahangian A., Zarnegar B., Perry A., Cheng G. (2006). Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439, 208–211. 10.1038/nature04374 [DOI] [PubMed] [Google Scholar]
- Pensaert M. B., de Bouck P. (1978). A new coronavirus-like particle associated with diarrhea in swine. Arch Virol 58, 243–247. 10.1007/BF01317606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M., Fushman D. (2004). Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 8, 610–616. 10.1016/j.cbpa.2004.09.009 [DOI] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L., Siddell S. G. (2007). A contemporary view of coronavirus transcription. J Virol 81, 20–29. 10.1128/JVI.01358-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C., Levy D. E., Decker T. (2007). JAK-STAT signaling: from interferons to cytokines. J Biol Chem 282, 20059–20063. 10.1074/jbc.R700016200 [DOI] [PubMed] [Google Scholar]
- Siu K. L., Kok K. H., Ng M. H., Poon V. K., Yuen K. Y., Zheng B. J., Jin D. Y. (2009). Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3·TANK·TBK1/IKKϵ complex. J Biol Chem 284, 16202–16209. 10.1074/jbc.M109.008227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S. C. (2008). Deubiquitylation and regulation of the immune response. Nat Rev Immunol 8, 501–511. 10.1038/nri2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Yang Y. D., Liu D. B., Xing Y. L., Chen X. J., Chen Z. B. (2010a). Deubiquitinase activity and regulation of antiviral innate immune responses by papain-like proteases of human coronavirus NL63. Prog Biochem Biophys 37, 871–880. 10.3724/SP.J.1206.2010.00111 [DOI] [Google Scholar]
- Sun Z., Chen Z., Lawson S. R., Fang Y. (2010b). The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J Virol 84, 7832–7846. 10.1128/JVI.00217-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Xing Y., Chen X., Zheng Y., Yang Y., Nichols D. B., Clementz M. A., Banach B. S., Li K. & other authors (2012). Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS ONE 7, e30802. 10.1371/journal.pone.0030802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos D., Maniatis T. (1995). Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83, 1091–1100. 10.1016/0092-8674(95)90136-1 [DOI] [PubMed] [Google Scholar]
- van der Hoek L., Sure K., Ihorst G., Stang A., Pyrc K., Jebbink M. F., Petersen G., Forster J., Berkhout B., Uberla K. (2005). Croup is associated with the novel coronavirus NL63. PLoS Med 2, e240. 10.1371/journal.pmed.0020240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren P. B., Beugeling C., Ninaber D. K., Frias-Staheli N., van Boheemen S., García-Sastre A., Snijder E. J., Kikkert M. (2012). Arterivirus and nairovirus ovarian tumor domain-containing deubiquitinases target activated RIG-I to control innate immune signaling. J Virol 86, 773–785. 10.1128/JVI.06277-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux D. L., Fidler F., Cumming G. (2012). Replicates and repeats–what is the difference and is it significant? A brief discussion of statistics and experimental design. EMBO Rep 13, 291–296. 10.1038/embor.2012.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Li P., Sun L., Fan J., Zhang Q., Luo R., Liu X., Li K. & other authors (2011a). The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J Virol 85, 3758–3766. 10.1128/JVI.02589-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Chen G., Zheng D., Cheng G., Tang H. (2011b). PLP2 of mouse hepatitis virus A59 (MHV-A59) targets TBK1 to negatively regulate cellular type I interferon signaling pathway. PLoS ONE 6, e17192. 10.1371/journal.pone.0017192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins C., Gale M., Jr (2010). Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol 22, 41–47. 10.1016/j.coi.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojdyla J. A., Manolaridis I., van Kasteren P. B., Kikkert M., Snijder E. J., Gorbalenya A. E., Tucker P. A. (2010). Papain-like protease 1 from transmissible gastroenteritis virus: crystal structure and enzymatic activity toward viral and cellular substrates. J Virol 84, 10063–10073. 10.1128/JVI.00898-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Jono H., Kai H., Li J. D. (2005). The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative cross-talk with TRAF6 and TRAF7. J Biol Chem 280, 41111–41121. 10.1074/jbc.M509526200 [DOI] [PubMed] [Google Scholar]
- Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., Chen Z. J. (2010). Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141, 315–330. 10.1016/j.cell.2010.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Chen G., Guo B., Cheng G., Tang H. (2008). PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res 18, 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B., Zhang Y., Tan B., Liu T. T., Wang Y. Y., Shu H. B. (2010). The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J Immunol 184, 6249–6255. 10.4049/jimmunol.0903748 [DOI] [PubMed] [Google Scholar]