Abstract
Soil bacteria are generally capable of growth on a wide range of organic chemicals, and pseudomonads are particularly adept at utilizing aromatic compounds. Pseudomonads are motile bacteria that are capable of sensing a wide range of chemicals, using both energy taxis and chemotaxis. Whilst the identification of specific chemicals detected by the ≥26 chemoreceptors encoded in Pseudomonas genomes is ongoing, the functions of only a limited number of Pseudomonas chemoreceptors have been revealed to date. We report here that McpC, a methyl-accepting chemotaxis protein in Pseudomonas putida F1 that was previously shown to function as a receptor for cytosine, was also responsible for the chemotactic response to the carboxylated pyridine nicotinic acid.
Introduction
N-heterocyclic aromatic compounds are abundant in nature, and provide good sources of carbon, nitrogen and energy for bacterial strains capable of their degradation (Fetzner, 1998; Kaiser et al., 1996). Naturally occurring N-heteroaromatic compounds include key structural components of the nucleotides in DNA and RNA, the electron-carrying coenzymes NAD+ and NADP+, the energy currency molecules ATP and GTP, and vitamin B6. In addition to natural N-heteroaromatic compounds, many structurally similar chemicals are produced industrially as solvents, pesticides and pharmaceuticals (Fetzner, 1998). Whilst there has been significant progress in elucidating the bacterial pathways by which many of these molecules are degraded (Fetzner, 1998; Kaiser et al., 1996), little is known about whether these compounds are sensed as chemoattractants by motile bacteria.
One abundant N-heteroaromatic structure in nature is the pyridine ring, and the carboxylated pyridine nicotinic acid (also known as niacin and vitamin B3) has served as a model for N-heteroaromatic compound degradation. Nicotinic acid is an intermediate in the biosynthesis and salvage pathways for the pyridine nucleotides NAD+ and NADP+, which are essential coenzymes in redox reactions in all domains of life. Additional roles for NAD in intracellular signalling, regulation of cell life span and circadian rhythms have been described in studies that highlight the importance and ubiquity of pyridine nucleotides in natural systems (Di Stefano & Conforti, 2013). The complete pathway for aerobic nicotinic acid degradation has been characterized in Pseudomonas putida KT2440 (Jiménez et al., 2008, 2011). Members of the genus Pseudomonas are generally considered to be catabolically versatile and are particularly adept at degrading aromatic compounds; however, whilst the ability to utilize nicotinic acid seems to be characteristic of P. putida, it is rarely found in other Pseudomonas species (Jiménez et al., 2010).
Consistent with their catabolic versatility, pseudomonads have complex chemotaxis systems that allow them to sense and respond to a wide range of chemicals in the environment (Kato et al., 2008; Parales et al., 2004; Sampedro et al., 2014). The currently available sequenced Pseudomonas genomes have ≥26 genes predicted to encode methyl-accepting chemotaxis proteins (MCPs), which serve as chemoreceptors and signal transducers in the chemotaxis pathway (Parales et al., 2004; Sampedro et al., 2014). In contrast, Escherichia coli has four MCPs and Aer, an MCP-like energy taxis receptor (Hazelbauer et al., 2008). Recent analyses of bacterial and archaeal genomes have shown that the number of MCPs encoded in a given genome is generally related to the metabolic diversity of the organism rather than the size of the genome (Lacal et al., 2010). We have been using P. putida F1 as a model organism in which to study the range of attractants detected and the complexity of chemotactic signalling (Ditty et al., 2013; Liu et al., 2009; Luu et al., 2013; Parales et al., 2000, 2013). Previous work demonstrated that McpC, one of the 27 predicted MCPs/MCP-like proteins in P. putida F1, mediated the chemotactic response to the N-heteroaromatic pyrimidine base cytosine (Liu et al., 2009). In this study, we confirmed the function of the predicted nicotinic acid degradation pathway in P. putida F1, demonstrated that nicotinic acid serves as a chemoattractant for strain F1 and revealed that McpC mediates the chemotactic response to nicotinic acid.
Methods
Bacterial strains and plasmids.
The strains and plasmids used in this study are shown in Table 1. E. coli strains DH5α and DH5α(λpir) were used as hosts for cloned genes. E. coli HB101(pRK2013) was used as a helper strain for mobilizing plasmids in triparental matings, which were carried out on lysogeny broth (LB) plates (Davis et al., 1980) at 30 °C for 24 h. E. coli strains were cultured in LB medium at 37 °C. P. putida strains were grown at 30 °C in minimal salts basal medium (MSB) (Stanier et al., 1966) containing 10 mM succinate, 40 mM pyruvate or 5 mM nicotinic acid. MSB plates were solidified with 1.8 % Noble agar (BD Biosciences). Kanamycin, gentamicin and tetracycline were used at 100, 15 and 20 µg ml−1, respectively, for E. coli strains, and at 50, 15 and 20 µg ml−1, respectively, for P. putida strains.
Table 1. Bacterial strains and plasmids.
| Strain or plasmid | Relevant characteristics | Source or reference |
| Strains | ||
| Escherichia coli | ||
| DH5α | Cloning host | Life Technologies |
| DH5α(λpir) | Cloning host | William W. Metcalf University of Illinois |
| HB101 | Host for mobilization plasmid pRK2013 | Sambrook et al. (1989) |
| Pseudomonas putida | ||
| F1 | WT | Finette et al. (1984); Gibson et al. (1970) |
| VNF001 | F1 nicB : : Km | This study |
| XLF004 | F1 ΔmcpC | Liu et al. (2009) |
| XLF019 | F1 Δaer2 | Luu et al. (2013) |
| XLF119 | F1 Δaer2ΔmcpC | This study |
| Pseudomonas aeruginosa | ||
| PAO1 | WT | Stover et al. (2000) |
| Plasmids | ||
| pEX18Gm | Cloning vector, sacB, Gmr | Hoang et al. (1998) |
| pRK415 | Broad-host-range cloning vector, Tcr | Keen et al. (1988) |
| pRK2013 | ColE1 ori, RP4 mobilization function, Kmr | Figurski & Helinski (1979) |
| pVNF10 | nicB (locus tag Pput_1889) from strain F1 interrupted with a kanamycin resistance gene cloned in pEX18Gm, Gmr | This study |
| pXLF019 | aer2 deletion construct | Luu et al. (2013) |
| pXLF204 | mcpC (locus tag Pput_0623) from strain F1 cloned into XbaHI/EcoRI sites of pRK415, constitutively expressed from lac promoter of plasmid, Tcr | Liu et al. (2009) |
Chemicals.
Nicotinic acid (99.5 %), picolinic acid (99 %) and isonicotinic acid (99 %) were purchased from Acros Organics, and nicotinamide was from K & K Laboratories.
DNA methods.
Genomic DNA from strain F1 was purified using a Puregene DNA Isolation kit (Gentra Systems). Plasmids were purified using a QIAprep Miniprep kit (Qiagen), and DNA fragments and PCR products were purified with a QIAquick Gel Extraction kit (Qiagen). Standard methods were used for the manipulation of plasmids (Ausubel et al., 1993). Fluorescent automated DNA sequencing was carried out at the University of California, Davis sequencing facility using an Applied Biosystems 3730 automated sequencer.
Construction of a nicotinic acid catabolic mutant.
The nicB gene in P. putida F1 (locus tag Pput_1889) was insertionally inactivated with the kanamycin resistance gene from pRK415Km (Luu et al., 2013) using an Infusion-HD cloning kit (Clontech), and the suicide vector pEX18, which carried a gentamicin resistance gene and the sacB gene, conferring sucrose sensitivity (White & Metcalf, 2004). The primers used were pEX18_1889delfor (5′-CGACGGCCAGTGCCAAGCTTCACTCACAACAGGTGCCCAG-3′)/pEX18_1889delrev (5′-GCTATGACCATGATTACGAATTCCATCATTACGTCGATAGCTGGCA-3′), pput_1889_intfor (5′-TTGAATGGGCCCTACATGGTGTGGTCAGGTACGCAGAAC-3′)/pput_1889intrev (5′-GAGTTCGGTCCGATCAAGGTACCTGACCACACGCGGAT-3′) and pETKm_RsrI-for/pETKm_ApaI-rev (Luu et al., 2013). To generate the mutant construct, 1 kb regions at the beginning and centre of the nicB gene were amplified by PCR, and the resulting PCR fragments were fused to the amplified kanamycin resistance gene and pEX18 using the complementary overhanging ends. The resulting plasmid (pVNF10) was introduced into E. coli DH5α(λpir), verified by restriction digestion and introduced into P. putida F1 by conjugation in the presence of E. coli HB101(pRK2013) (Simon et al., 1983). Gentamicin- and kanamycin-resistant P. putida exoconjugants were subjected to counterselection in MSB containing 10 mM succinate and 20 % sucrose. Deletions in kanamycin-resistant, gentamicin-sensitive strains were verified by PCR.
Construction of a P. putida F1 Δaer2ΔmcpC double mutant.
The aer2 gene was deleted from the ΔmcpC mutant XLF004 (Liu et al., 2009) by homologous recombination using the aer2 deletion construct pXLF019 (Luu et al., 2013), generating strain XLF119.
Chemotaxis assays.
Qualitative capillary assays were carried out as described previously (Grimm & Harwood, 1997; Parales et al., 2013). Briefly, bacterial cells in mid-exponential phase (OD660 0.3–0.4) were harvested by centrifugation at 5000 x g for 5 min, washed once with chemotaxis buffer (CB; 50 mM potassium phosphate buffer, pH 7.0, 10 µM disodium EDTA and 0.05 % glycerol) (Parales et al., 2000) and gently resuspended in CB to OD660~0.10. Microcapillaries (1 µl) containing attractants in 2 % low-melting-temperature agarose (NuSieve GTG; Lonza) dissolved in CB were inserted into the suspension of bacterial cells. The response to nicotinic acid was tested at 50 mM. Negative (CB) and positive (2 % Difco Casamino acids; BD Biosciences) controls were included in all experiments. Responses were visualized at ×40 magnification on a Nikon Eclipse TE2000-S microscope, and photographed using an Evolution MicroPublisher 3.3 real-time viewing camera and Evolution MP/QImaging software (Media Cybernetics).
Quantitative capillary assays were carried out as described previously (Liu et al., 2009). For these assays, cells were grown to OD660~0.4 in MSB containing 5 mM nicotinic acid, harvested by centrifugation and resuspended in CB to a final OD660 0.15. Responses to 5, 10, 50 and 100 mM nicotinic acid were tested. The response to 0.2 % Casamino acids was tested as a positive control and the response to 50 mM cytosine (the peak attractant concentration; Liu et al., 2009) was tested for comparison. Competition assays were carried out with the competing attractant cytosine (50 mM) present in both the cell suspension and the capillary. Responses to 50 mM nicotinic acid, 0.2 % Casamino acids and buffer in the capillary were tested.
Minimal medium-soft agar swim plates (Harwood et al., 1994) contained 2 mM nicotinic acid. For these assays, P. putida strains were grown overnight in 3 ml MSB medium containing 5 mM nicotinic acid at 30 °C with shaking. Cultures were harvested by centrifugation and the pellets were washed with 5 ml MSB, resuspended in MSB to OD660~0.4, and 2 µl of each suspension was used to inoculate semisolid (0.3 % Noble agar) minimal medium in 15 mm diameter Petri plates. For complementation experiments, 20 mg tetracycline ml−1 was included in the overnight growth medium as well as the soft agar plates. Cultures were incubated at 30 °C for 26–30 h. Photographs were taken using backlighting (Parkinson, 2007). For each experiment, the measured diameters of all strains were normalized to the mean diameter of WT P. putida F1 colonies (which was set to 1). All statistical analyses were conducted using JMP Pro version 10.0.
Results
P. putida F1 is chemotactic to nicotinic acid and the response does not require nicotinic acid metabolism
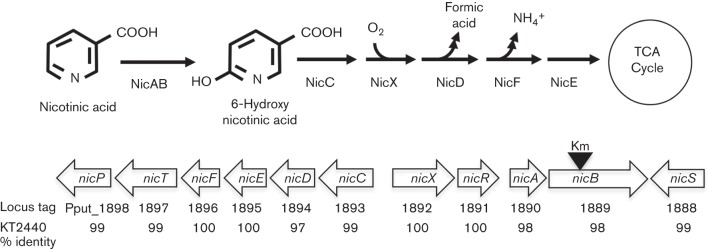
The nicotinic acid degradation pathway has been characterized in P. putida KT2440 (Jiménez et al., 2008, 2011) and a very similar gene cluster was identified in P. putida F1 (Fig. 1). Genomic analyses have suggested that key enzymes in the nicotinic acid pathway are conserved in several P. putida strains (Jiménez et al., 2010; Tang et al., 2012). As predicted by the genome sequence P. putida F1 was capable of growth on nicotinic acid as the sole carbon and energy source (doubling time 85±5 min; 5 mM nicotinic acid). The chemotactic response of P. putida F1 to 50 mM nicotinic acid was tested in qualitative capillary assays. Both pyruvate-grown (uninduced) and nicotinic acid-induced cells responded (Fig. 2). The response was specific to nicotinic acid as neither nicotinamide nor the other pyridine carboxylic acid isomers (isonicotinic acid and picolinic acid) elicited a response, and none of these N-heteroaromatic compounds served as growth substrates for strain F1 (data not shown).
Fig. 1.
Nicotinic acid degradation pathway and the nic gene cluster encoding pathway enzymes in P. putida F1. The nic genes are represented by open arrows. Locus tags for the P. putida F1 nic genes, together with the percent identities of the P. putida F1 and KT2440-deduced amino acid sequences, are indicated below the genes. The black triangle indicates the approximate location of the kanamycin resistance gene inserted into the nicB gene in strain VNF001.
Fig. 2.
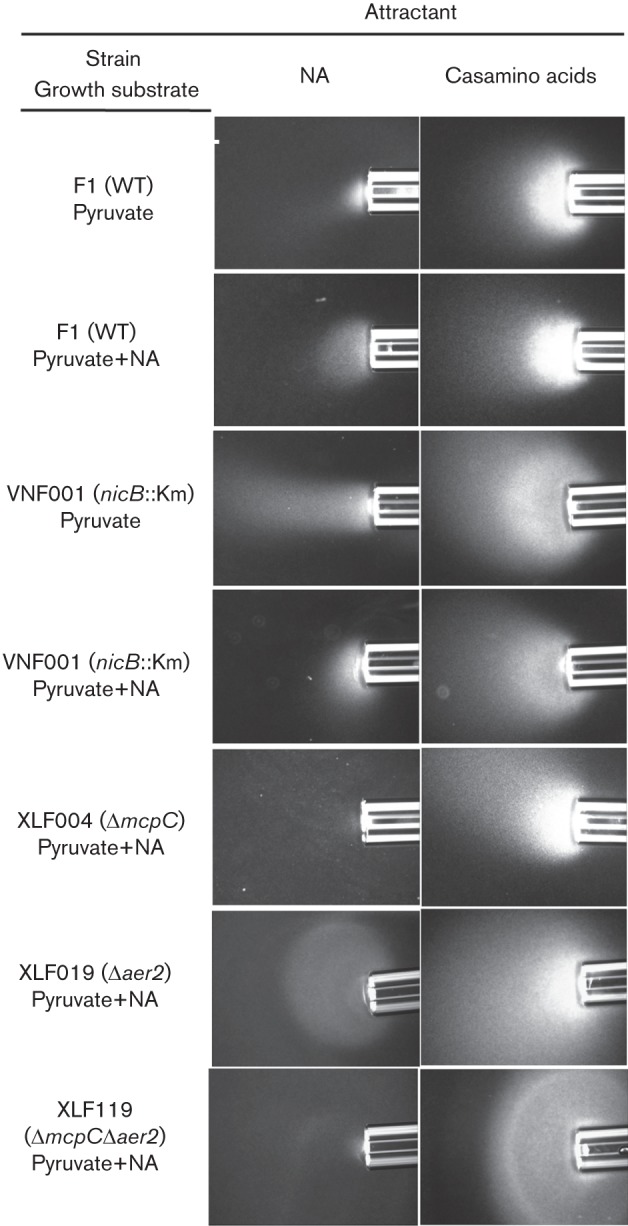
Chemotactic response of WT and mutant P. putida strains to nicotinic acid in qualitative capillary assays. P. putida F1 and the nicB (VNF001), ΔmcpC (XLF004), Δaer2 (XLF019) and Δaer2ΔmcpC (XLF119) mutants were grown either with 40 mM pyruvate (uninduced) or 40 mM pyruvate plus 5 mM nicotinic acid (NA; induced) as indicated. Nicotinic acid was provided as the attractant at 50 mM. Also shown are positive control responses of each strain to 2 % Casamino acids. No response was detected when only chemotaxis buffer was present in the capillary (not shown). Assays were repeated at least three times and representative photographs are shown. Photographs were taken after 7 min.
In order to determine whether nicotinic acid was detected directly, we insertionally inactivated the nicB gene (Fig. 1), which encoded the catalytic component of nicotinic acid hydroxylase (Jiménez et al., 2008). The resulting mutant (strain VNF001) was unable to grow on nicotinic acid (data not shown), but it had a WT response to nicotinic acid in qualitative capillary assays (Fig. 2), indicating that metabolism of nicotinic acid was not required for the chemotactic response and ruling out a role for energy taxis in the response.
McpC mediates the chemotactic response to nicotinic acid
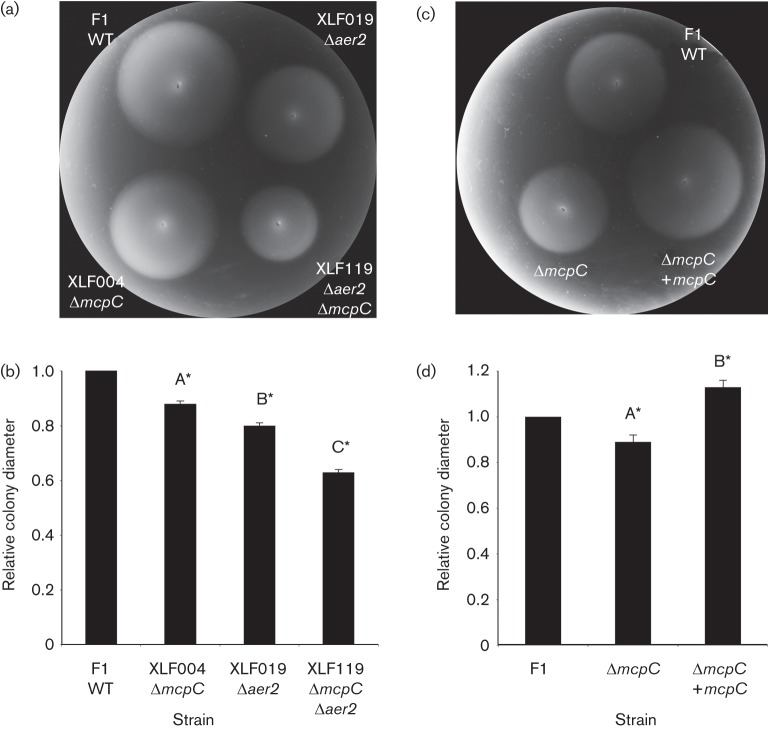
The role of McpC in the detection of nicotinic acid was evaluated using qualitative capillary assays, which showed that the response to nicotinic acid by the ΔmcpC mutant strain XLF004 was severely reduced relative to that of WT P. putida F1 (Fig. 2). The chemotaxis defect was specific, as the response to Casamino acids was as strong as that of the WT (Fig. 2). The response was not completely eliminated in XLF004, but none of the other 26 single-deletion mutants (17 mutants each lacking one of the MCP-encoding genes and nine mutants each lacking one of the genes encoding MCP-like proteins; Liu, 2009; Parales et al., 2013) had an obviously reduced response in qualitative capillary assays (data not shown). The swim-plate assay is more quantitative and can be used to detect subtle defects in chemotaxis; however, previous studies have shown that aerotaxis can mask defects in chemotaxis to specific chemicals in swim plates (Alvarez-Ortega & Harwood, 2007; Parales et al., 2013). We therefore used quantitative swim-plate assays to examine 18 double mutants of P. putida F1 (each lacking the energy taxis receptor gene aer2 and one of the 18 canonical MCP-encoding genes; Liu, 2009; Parales et al., 2013) for defects in nicotinic acid chemotaxis. However, the only strain that demonstrated an obvious mutant phenotype in response to nicotinic acid lacked aer2 and mcpC (Fig. 3a and data not shown). A slight but significant difference in colony size for WT strain F1 and the ΔmcpC single-mutant XLF004 was also detected (Fig. 3b). Growth studies demonstrated that all strains had similar growth rates in MSB medium containing 5 mM nicotinic acid (data not shown), indicating that the reduced colony size in the swim-plate assay was solely due to a chemotaxis defect. The response of the Δaer2 mutant was similar to that of the WT in qualitative capillary assays, and responses of the ΔmcpC mutant and ΔmcpCΔaer2 double mutant were comparable in this assay (Fig. 2), providing further evidence that energy taxis does not play a major role in the response to nicotinic acid. Introduction of a broad-host range plasmid carrying mcpC into strain XLF004 restored the response to nicotinic acid in both swim-plate assays (Fig. 3c, d) and qualitative capillary assays (Fig. 4).
Fig. 3.
Chemotactic response of WT, mutant and complemented P. putida strains to nicotinic acid in soft agar swim plates. (a) Representative swim plate showing responses of P. putida strains F1 (WT), XLF004 (ΔmcpC), XLF019 (Δaer2) and XLF119 (Δaer2ΔmcpC) to 2 mM nicotinic acid. (b) Quantitative analysis of swim-plate assay results in (a) (n = 3). Mean colony diameters were normalized to the diameter of WT F1. (c) Representative swim plate showing complementation of the mcpC deletion. Shown are responses of P. putida strains F1 (pRK415) (WT), XLF004 (pRK415) (ΔmcpC) and XLF004 (pXLF204) (ΔmcpC containing mcpC) to 2 mM nicotinic acid. (d) Quantitative analysis of swim-plate assay results in (c) (n = 3). Mean colony diameters were normalized to the diameter of F1 (pRK415). Bars, sd. Means with different letters are significantly different. *P<0.05, one-way ANOVA interaction, Tukey’s multiple comparison test. Ninety-five per cent confidence intervals (indicated by asterisks) are used to describe significant differences from the normalized WT controls.
Fig. 4.
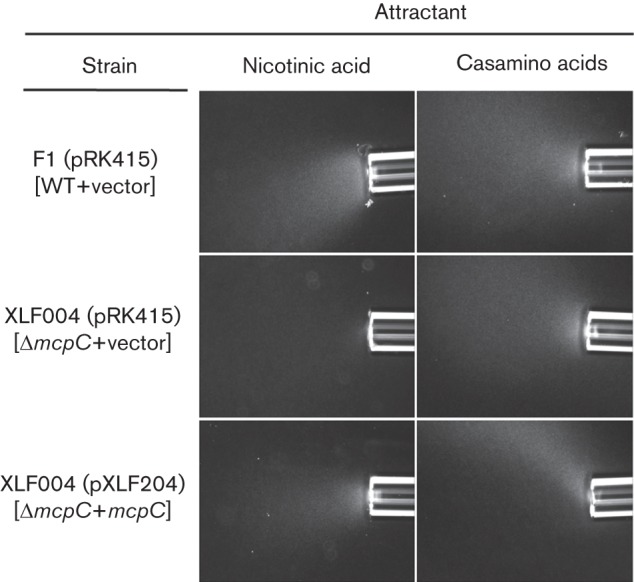
Qualitative capillary assays demonstrating complementation of the mcpC deletion. Responses of P. putida F1 (pRK415) (WT), XLF004 (pRK415) (ΔmcpC) and XLF004(pXLF204) (ΔmcpC containing mcpC) are shown. Nicotinic acid was provided at 50 mM. Also shown are positive control responses of each strain to 2 % Casamino acids. No response was detected when only chemotaxis buffer was present in the capillary (not shown). Assays were repeated at least three times and representative photographs are shown. Photographs were taken after 7 min.
Sensitivity of McpC for nicotinic acid and cytosine
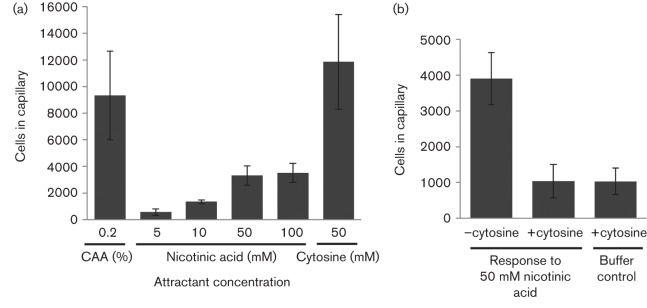
We used quantitative capillary assays to determine the sensitivity of McpC for nicotinic acid by testing the response of WT P. putida F1 to a range of nicotinic acid concentrations (5–100 mM). The strongest response to nicotinic acid was at the highest tested concentration and this was much weaker than the response to 50 mM cytosine (Fig. 5a). The response to cytosine in this experiment in which cells were grown with 5 mM nicotinic acid was comparable to the previously reported responses of succinate- and succinate plus cytosine-grown P. putida F1 (13 900±1600 and 14 200±1700 cells, respectively; Liu et al., 2009). The presence of 50 mM cytosine as a competing attractant was able to eliminate the response to nicotinic acid (Fig. 5b). These results showed that cytosine was a stronger attractant than nicotinic acid and provided evidence that both attractants were detected by the same binding site on McpC.
Fig. 5.
Sensitivity of P. putida F1 to nicotinic acid and cytosine measured by quantitative capillary assays. (a) Chemotactic responses to various concentrations of nicotinic acid compared to the response to 50 mM cytosine (the previously determined peak attractant concentration; Liu et al., 2009). Response to the positive control attractant 0.2 % Casamino acids (CAA) is also shown. The mean number of cells that accumulated in capillaries containing buffer only was subtracted from each dataset (590±140 cells). (b) Response to 50 mM nicotinic acid (attractant in the capillary) in competition capillary assays in which 50 mM cytosine (competitor) was present or absent in both the capillary and cell suspension as indicated. Cultures were grown in MSB containing 5 mM nicotinic acid and results represent the mean±sem of at least three independent experiments (nine or more capillaries in total), except the cytosine data in (a) (two independent experiments, six capillaries in total).
Heterologous expression of mcpC confers the ability to sense nicotinic acid in Pseudomonas aeruginosa
Previous studies demonstrated that P. aeruginosa PAO1 does not encode a McpC orthologue and is not chemotactic to cytosine (Liu et al., 2009). A search of the P. aeruginosa PAO1 genome sequence (Jiménez et al., 2010; Stover et al., 2000) did not identify genes encoding a nicotinic acid degradation pathway. We confirmed that P. aeruginosa PAO1 was unable to grow in minimal medium containing 5 mM nicotinic acid (data not shown) and did not respond to nicotinic acid when tested using the qualitative capillary assay (Fig. 6). However, when mcpC from P. putida F1 was expressed in P. aeruginosa PAO1, the strain acquired the ability to sense nicotinic acid (Fig. 6).
Fig. 6.
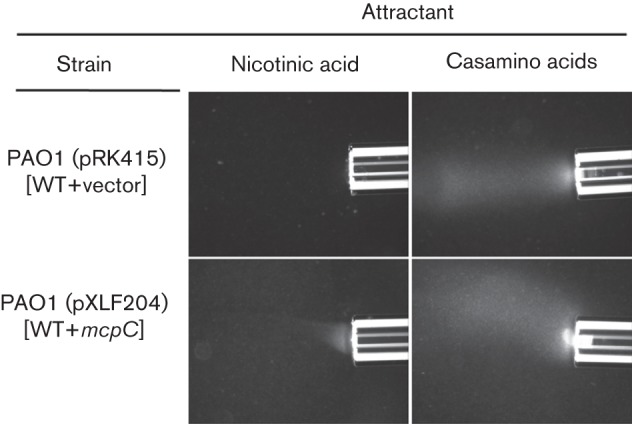
Chemotactic responses of P. aeruginosa PAO1 (pRK415) and PAO1 (pXLF204) (carrying mcpC) to 50 mM nicotinic acid in qualitative capillary assays. Cells were grown overnight in MSB containing 27.5 mM glucose and 60 µg tetracycline ml−1. As cultures grown in the presence of tetracycline were poorly motile, cultures were reinoculated in the same medium lacking tetracycline and were grown for two to three doublings prior to assays. Also shown are positive control responses of each strain to 2 % Casamino acids. No response was detected when only chemotaxis buffer was present in the capillary (not shown). Assays were repeated at least three times and representative photographs are shown. Photographs were taken after 7 min.
Discussion
The mechanism of aromatic compound sensing in Pseudomonas strains seems to depend on the particular aromatic compound under study. Responses to (methyl)phenols and phenylacetic acid are mediated through energy taxis via Aer2 (Luu et al., 2013; Sarand et al., 2008), whilst responses to naphthalene, 2-nitrobenzoate, toluene, 4-chloroaniline, catechol and aromatic amino acids are sensed by chemotaxis via specific MCPs (Grimm & Harwood, 1999; Iwaki et al., 2007; Lacal et al., 2011; Oku et al., 2012; Parales et al., 2000; Taguchi et al., 1997; Vangnai et al., 2013).
Based on previous results (Liu et al., 2009) and results reported here, we now have evidence that McpC is responsible for chemotaxis to the N-heteroaromatic compounds cytosine and nicotinic acid. The response to these two chemicals is quite specific, as the nicotinic acid isomers and nicotinamide, and the pyrimidine bases thymine and uracil (Liu et al., 2009), are not detected. Whilst a nicotinic acid chemotaxis defect was seen in the Δaer2 mutant in swim-plate assays, we interpret this as an aerotaxis defect. This conclusion is consistent with previous results in P. putida and P. aeruginosa, which have shown that responses in swim-plate assays can result from a combination of both aerotaxis and chemotaxis to specific chemicals; in some cases, aerotaxis masks chemotaxis phenotypes (Alvarez-Ortega & Harwood, 2007; Parales et al., 2013). In addition, the WT response of the nicB mutant demonstrates that metabolism of nicotinic acid by P. putida F1 is not required for the chemotactic response, as would be expected if the signal were processed primarily via energy taxis (Alexandre, 2010). In E. coli, MCPs serve as the primary chemoreceptors for some attractants, whilst other attractants are detected by a specific periplasmic binding protein; the complex then interacts with a specific MCP to transmit the signal (Wadhams & Armitage, 2004). It seems likely that McpC binds both cytosine and nicotinic acid directly without the participation of a periplasmic binding protein, as heterologous expression of mcpC alone was sufficient to confer the ability to respond to both compounds in P. aeruginosa PAO1 (Fig. 6) (Liu et al., 2009).
As reported previously, McpC has a length of 647 aa and exhibits the canonical MCP structure, with a periplasmic ligand-binding region (LBR) flanked by two hydrophobic transmembrane helices, a HAMP (histidine kinase, adenylyl cyclase, methyl-accepting chemotaxis protein, and phosphatase) domain, and a cytoplasmic signal transduction region (Liu et al., 2009). The cytoplasmic region is in class 40H (comprised of 40 heptads; Alexander & Zhulin, 2007) and has two potential methylation sites, but does not contain any C-terminal pentapeptide tether regions, which are known to be binding sites for CheB and CheR in E. coli (Li & Hazelbauer, 2006). The LBR falls into cluster II (Lacal et al., 2010), spanning 262 aa, and automated annotation identified the presence of a conserved Cache (Ca2+ channel and chemotaxis receptor) domain, which is predicted to sense small molecules (Anantharaman & Aravind, 2000; Finn et al., 2014). A closer examination revealed that the predicted Cache domain is one of two inserted PAS (Per, ARNT, Sim)-like PDC (PhoQ, DcuS, CitA) domains, in an architecture seen in the sensor domains of many bacterial histidine kinases, including chemoreceptors (Cheung & Hendrickson, 2010; Zhang & Hendrickson, 2010).
A homology search comparing the McpC periplasmic LBR to known structures in the Protein Data Bank returned an uncharacterized MCP from Vibrio cholerae (ID: 3C8C; Y. Patskovsky and others, unpublished) as the top hit. The LBRs of McpC and the Vibrio MCP are 30 % identical in amino acid sequence. Webb et al. (2014) recently reported a homologue of the Vibrio MCP (which binds alanine) in Sinorhizobium meliloti called McpU, which senses proline. The LBRs of the Vibrio MCP and McpU are 26 % identical, and have conserved aspartate residues at positions 172 and 201 (Vibrio MCP) and 155 and 182 (McpU) that coordinate their amino acid ligands via hydrogen bonds with the amino group (Webb et al., 2014). The aspartate at one of the corresponding residues in McpC, position 191, is conserved. However, position 163 contains a tryptophan, which may reflect the hydrophobic nature of the ligands nicotinic acid and cytosine.
The response to nicotinic acid was not completely abolished in the ΔmcpC mutant; however, two different screens to identify additional receptors that sense nicotinic acid did not identify any mutants with obvious phenotypes. It is possible that multiple additional MCPs participate in the response to nicotinic acid and phenotypes of mutant strains lacking any single MCP were too subtle to detect in these assays. These results suggest that McpC may not be the only chemoreceptor that detects nicotinic acid in P. putida F1.
Expression of the nic genes in P. putida KT2440 is tightly controlled by a regulatory circuit involving two repressors that respond to different effectors (Jiménez et al., 2011). Although regulation of the nic genes has not been investigated in P. putida F1, based on the conserved gene order and deduced amino acid sequences of the nine structural and two regulatory genes in P. putida KT2440, it is expected that nic genes in both strains are regulated in a similar fashion. The chemotactic response of P. putida F1 to nicotinic acid appeared to be constitutive, as was the response of P. putida F1 to cytosine (Liu et al., 2009). It seems unlikely that mcpC would be specifically regulated with genes for nicotinic acid degradation as the receptor also mediates the response to cytosine, which serves as a nitrogen source for P. putida F1 that is metabolized by a completely different pathway. The mcpC gene (locus tag Pput_0623) is not co-localized with the nic genes for nicotinic acid degradation (locus tags Pput_1888–Pput_1898), nor does it appear to be co-regulated based on the constitutive chemotaxis phenotype.
Many human, animal and plant pathogens lack pathways for de novo biosynthesis of NAD, and are nicotinic acid or nicotinamide auxotrophs; a few examples include Shigella species, enteroinvasive E. coli strains, Salmonella enterica (serovar Dublin), group A streptococci and Erwinia amylovora (Bergthorsson & Roth, 2005; Paternoster et al., 2010; Prunier et al., 2007; Sorci et al., 2013). The use of nicotinic acid-degrading strains of Pseudomonas rhizosphaerae and Pseudomonas fluorescens as biocontrol agents to protect plants from the plant pathogen Erwinia amylovora, a nicotinic acid auxotroph, was reported recently (Paternoster et al., 2010). It is unknown whether the tested Pseudomonas strains are chemotactic to nicotinic acid, but it is possible, as the genome of P. fluorescens Pf0-1 has a predicted orthologue of McpC that is >79 % identical in amino acid sequence. P. fluorescens Pf0-1, however, lacks genes for nicotinic acid degradation. It is expected that a strain with the ability not only to degrade but also to sense and respond to nicotinic acid as a specific chemoattractant could be a more efficient biocontrol strain. It would therefore be of interest to investigate the chemotactic abilities of bacterial strains that are proposed for use as biocontrol agents.
Acknowledgements
This work was supported by a grant from the National Science Foundation (MCB0919930) to R. E. P and J. L. D. JGH was supported by a NIH fellowship in Molecular and Cellular Biology (TM32 GM070377). Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors, and do not necessarily reflect the views of the National Science Foundation or the National Institutes of Health.
Abbreviations:
- LBR
ligand-binding region
- MCP
methyl-accepting chemotaxis protein
Edited by: K. Ottemann
References
- Alexander R. P., Zhulin I. B. (2007). Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci U S A 104, 2885–2890. 10.1073/pnas.0609359104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre G. (2010). Coupling metabolism and chemotaxis-dependent behaviours by energy taxis receptors. Microbiology 156, 2283–2293. 10.1099/mic.0.039214-0 [DOI] [PubMed] [Google Scholar]
- Alvarez-Ortega C., Harwood C. S. (2007). Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis-deficient mutant. Appl Environ Microbiol 73, 7793–7795. 10.1128/AEM.01898-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V., Aravind L. (2000). Cache – a signaling domain common to animal Ca2+-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem Sci 25, 535–537. 10.1016/S0968-0004(00)01672-8 [DOI] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (editors) (1993). Current Protocols in Molecular Biology. New York: Wiley. [Google Scholar]
- Bergthorsson U., Roth J. R. (2005). Natural isolates of Salmonella enterica serovar Dublin carry a single nadA missense mutation. J Bacteriol 187, 400–403. 10.1128/JB.187.1.400-403.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J., Hendrickson W. A. (2010). Sensor domains of two-component regulatory systems. Curr Opin Microbiol 13, 116–123. 10.1016/j.mib.2010.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. W., Botstein D., Roth J. R. (1980). Advanced Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Di Stefano M., Conforti L. (2013). Diversification of NAD biological role: the importance of location. FEBS J 280, 4711–4728. 10.1111/febs.12433 [DOI] [PubMed] [Google Scholar]
- Ditty J. L., Williams K. M., Keller M. M., Chen G. Y., Liu X., Parales R. E. (2013). Integrating grant-funded research into the undergraduate biology curriculum using IMG-ACT. Biochem Mol Biol Educ 41, 16–23. 10.1002/bmb.20662 [DOI] [PubMed] [Google Scholar]
- Fetzner S. (1998). Bacterial degradatoin of pyridine, indole, quinoline, and their derivatives under different redox conditions. Appl Microbiol Biotechnol 49, 237–250. 10.1007/s002530051164 [DOI] [Google Scholar]
- Figurski D. H., Helinski D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76, 1648–1652. 10.1073/pnas.76.4.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finette B. A., Subramanian V., Gibson D. T. (1984). Isolation and characterization of Pseudomonas putida PpF1 mutants defective in the toluene dioxygenase enzyme system. J Bacteriol 160, 1003–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., Heger A., Hetherington K., Holm L. & other authors (2014). Pfam: the protein families database. Nucleic Acids Res 42 (Database issue), D222–D230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. T., Hensley M., Yoshioka H., Mabry T. J. (1970). Oxidative degradation of aromatic hydrocarbons by microorganisms. III. Formation of (+)-cis-2,3-dihydroxy-1-methylcyclohexa-4,6-diene from toluene by Pseudomonas putida. Biochemistry 9, 1626–1630. 10.1021/bi00809a023 [DOI] [PubMed] [Google Scholar]
- Grimm A. C., Harwood C. S. (1997). Chemotaxis of Pseudomonas spp. to the polyaromatic hydrocarbon naphthalene. Appl Environ Microbiol 63, 4111–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm A. C., Harwood C. S. (1999). NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J Bacteriol 181, 3310–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. S., Nichols N. N., Kim M.-K., Ditty J. L., Parales R. E. (1994). Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J Bacteriol 176, 6479–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L., Falke J. J., Parkinson J. S. (2008). Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci 33, 9–19. 10.1016/j.tibs.2007.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Iwaki H., Muraki T., Ishihara S., Hasegawa Y., Rankin K. N., Sulea T., Boyd J., Lau P. C. K. (2007). Characterization of a pseudomonad 2-nitrobenzoate nitroreductase and its catabolic pathway-associated 2-hydroxylaminobenzoate mutase and a chemoreceptor involved in 2-nitrobenzoate chemotaxis. J Bacteriol 189, 3502–3514. 10.1128/JB.01098-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez J. I., Canales A., Jiménez-Barbero J., Ginalski K., Rychlewski L., García J. L., Díaz E. (2008). Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440. Proc Natl Acad Sci U S A 105, 11329–11334. 10.1073/pnas.0802273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez J. I., Nogales J., García J. L., Díaz E. (2010). A genomic view of the catabolism of aromatic compounds in Pseudomonas. In Handbook of Hydrocarbon and Lipid Microbiology, pp. 1297–1554. Edited by Timmis K. N. Berlin: Springer; 10.1007/978-3-540-77587-4_91 [DOI] [Google Scholar]
- Jiménez J. I., Juárez J. F., García J. L., Díaz E. (2011). A finely tuned regulatory circuit of the nicotinic acid degradation pathway in Pseudomonas putida. Environ Microbiol 13, 1718–1732. 10.1111/j.1462-2920.2011.02471.x [DOI] [PubMed] [Google Scholar]
- Kaiser J. P., Feng Y., Bollag J. M. (1996). Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol Rev 60, 483–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J., Kim H.-E., Takiguchi N., Kuroda A., Ohtake H. (2008). Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J Biosci Bioeng 106, 1–7. 10.1263/jbb.106.1 [DOI] [PubMed] [Google Scholar]
- Keen N. T., Tamaki S., Kobayashi D., Trollinger D. (1988). Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70, 191–197. 10.1016/0378-1119(88)90117-5 [DOI] [PubMed] [Google Scholar]
- Lacal J., García-Fontana C., Muñoz-Martínez F., Ramos J. L., Krell T. (2010). Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ Microbiol 12, 2873–2884. 10.1111/j.1462-2920.2010.02325.x [DOI] [PubMed] [Google Scholar]
- Lacal J., Muñoz-Martínez F., Reyes-Darías J. A., Duque E., Matilla M., Segura A., Calvo J. J., Jímenez-Sánchez C., Krell T., Ramos J. L. (2011). Bacterial chemotaxis towards aromatic hydrocarbons in Pseudomonas. Environ Microbiol 13, 1733–1744. 10.1111/j.1462-2920.2011.02493.x [DOI] [PubMed] [Google Scholar]
- Li M., Hazelbauer G. L. (2006). The carboxyl-terminal linker is important for chemoreceptor function. Mol Microbiol 60, 469–479. 10.1111/j.1365-2958.2006.05108.x [DOI] [PubMed] [Google Scholar]
- Liu, X. (2009). Chemotaxis to pyrimidines and s-triazines in Pseudomonas and Escherichia coli. PhD dissertation, University of California, Davis, CA, USA. [Google Scholar]
- Liu X., Wood P. L., Parales J. V., Parales R. E. (2009). Chemotaxis to pyrimidines and identification of a cytosine chemoreceptor in Pseudomonas putida. J Bacteriol 191, 2909–2916. 10.1128/JB.01708-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu R. A., Schneider B. J., Ho C. C., Nesteryuk V., Ngwesse S. E., Liu X., Parales J. V., Ditty J. L., Parales R. E. (2013). Taxis of Pseudomonas putida F1 toward phenylacetic acid is by mediated by the energy taxis receptor Aer2. Appl Environ Microbiol 79, 2416–2423. 10.1128/AEM.03895-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku S., Komatsu A., Tajima T., Nakashimada Y., Kato J. (2012). Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microbes Environ 27, 462–469. 10.1264/jsme2.ME12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales R. E., Ditty J. L., Harwood C. S. (2000). Toluene-degrading bacteria are chemotactic towards the environmental pollutants benzene, toluene, and trichloroethylene. Appl Environ Microbiol 66, 4098–4104. 10.1128/AEM.66.9.4098-4104.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales R. E., Ferrandez A., Harwood C. S. (2004). Chemotaxis in Pseudomonads. In Pseudomonas. Volume I: Genomics, Life Style and Molecular Architecture, pp. 793–815. Edited by Ramos J.-L. New York: Kluwer; 10.1007/978-1-4419-9086-0_26 [DOI] [Google Scholar]
- Parales R. E., Luu R. A., Chen G. Y., Liu X., Wu V., Lin P., Hughes J. G., Nesteryuk V., Parales J. V., Ditty J. L. (2013). Pseudomonas putida F1 has multiple chemoreceptors with overlapping specificity for organic acids. Microbiology 159, 1086–1096. 10.1099/mic.0.065698-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. (2007). A “bucket of light” for viewing bacterial colonies in soft agar. Methods Enzymol 423, 432–435. 10.1016/S0076-6879(07)23020-4 [DOI] [PubMed] [Google Scholar]
- Paternoster T., Défago G., Duffy B., Gessler C., Pertot I. (2010). Selection of a biocontrol agent based on a potential mechanism of action: degradation of nicotinic acid, a growth factor essential for Erwinia amylovora. Int Microbiol 13, 195–206. [DOI] [PubMed] [Google Scholar]
- Prunier A.-L., Schuch R., Fernández R. E., Maurelli A. T. (2007). Genetic structure of the nadA and nadB antivirulence loci in Shigella spp. J Bacteriol 189, 6482–6486. 10.1128/JB.00525-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Sampedro I., Parales R. E., Krell T., Hill J. E. (2014). Pseudomonas chemotaxis. FEMS Microbiol Rev 10.1111/1574-6976.12081 [Epub ahead of print]. 10.1111/1574-6976.12081 [DOI] [PubMed] [Google Scholar]
- Sarand I., Osterberg S., Holmqvist S., Holmfeldt P., Skärfstad E., Parales R. E., Shingler V. (2008). Metabolism-dependent taxis towards (methyl)phenols is coupled through the most abundant of three polar localized Aer-like proteins of Pseudomonas putida. Environ Microbiol 10, 1320–1334. 10.1111/j.1462-2920.2007.01546.x [DOI] [PubMed] [Google Scholar]
- Simon R., Priefer U., Pühler A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology (N Y) 1, 784–789. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- Sorci L., Blaby I. K., Rodionova I. A., De Ingeniis J., Tkachenko S., de Crécy-Lagard V., Osterman A. L. (2013). Quinolinate salvage and insights for targeting NAD biosynthesis in group A streptococci. J Bacteriol 195, 726–732. 10.1128/JB.02002-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. (1966). The aerobic pseudomonads: a taxonomic study. J Gen Microbiol 43, 159–271. 10.1099/00221287-43-2-159 [DOI] [PubMed] [Google Scholar]
- Stover C. K., Pham X. Q., Erwin A. L., Mizoguchi S. D., Warrener P., Hickey M. J., Brinkman F. S., Hufnagle W. O., Kowalik D. J. & other authors (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. 10.1038/35023079 [DOI] [PubMed] [Google Scholar]
- Taguchi K., Fukutomi H., Kuroda A., Kato J., Ohtake H. (1997). Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology 143, 3223–3229. 10.1099/00221287-143-10-3223 [DOI] [PubMed] [Google Scholar]
- Tang H., Yao Y., Wang L., Yu H., Ren Y., Wu G., Xu P. (2012). Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci Rep 2, 377. 10.1038/srep00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangnai A. S., Takeuchi K., Oku S., Kataoka N., Nitisakulkan T., Tajima T., Kato J. (2013). Identification of CtpL as a chromosomally encoded chemoreceptor for 4-chloroaniline and catechol in Pseudomonas aeruginosa PAO1. Appl Environ Microbiol 79, 7241–7248. 10.1128/AEM.02428-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams G. H., Armitage J. P. (2004). Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5, 1024–1037. 10.1038/nrm1524 [DOI] [PubMed] [Google Scholar]
- Webb B. A., Hildreth S., Helm R. F., Scharf B. E. (2014). Sinorhizobium meliloti chemoreceptor McpU mediates chemotaxis toward host plant exudates through direct proline sensing. Appl Environ Microbiol 80, 3404–3415. 10.1128/AEM.00115-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A. K., Metcalf W. W. (2004). The htx and ptx operons of Pseudomonas stutzeri WM88 are new members of the Phoregulon. J Bacteriol 186, 5876–5882. 10.1128/JB.186.17.5876-5882.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Hendrickson W. A. (2010). Structural characterization of the predominant family of histidine kinase sensor domains. J Mol Biol 400, 335–353. 10.1016/j.jmb.2010.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]