Abstract
Klebsiella pneumoniae causes a range of clinical disease in paediatric patients and is of increasing concern due to growing antibiotic resistance, yet little is known about the relative distribution of commensal and pathogens throughout the population structure of K. pneumoniae. We conducted a prospective, observational study of 92 isolates from Seattle Children’s Hospital, including 49 disease isolates from blood and urine (13 and 36 isolates, respectively) and 43 colonization isolates from stool. Susceptibility to 20 antimicrobials was evaluated using disc diffusion, VITEK 2 and Etest. Strain relatedness was investigated using multilocus sequence typing (MLST). Demographic and clinical characteristics were largely similar between disease and colonization cohorts, with 85.7 and 74.4 % of disease and colonization cohort patients, respectively, having an underlying medical condition; the sole exception was a relative abundance of patients with urologic or renal abnormalities in the disease cohort, consistent with the predominance of urine specimens among the disease isolates. With regard to antibiotic susceptibility properties, no significant differences were noted between the disease and colonization cohorts. Using molecular analysis, 71 unique sequence types (STs) were distinguished, with novel MLST findings evident in both cohorts; 43 (46.7 %) isolates represented novel STs, including 22 with a novel allele sequence. Thirteen STs contained multiple isolates and all seven isolates with resistance to three or more antibiotic classes were within one of four multirepresentative STs. This study demonstrates that nearly half of paediatric Klebsiella isolates represent novel STs, with clustering of multidrug resistance within specific STs. These findings expand our understanding of the intersection of bacterial population structure, human colonization ecology and multidrug resistance in K. pneumoniae.
Introduction
With the emergence and spread of bacterial strains resistant to multiple classes of antibiotics, Klebsiella pneumoniae has captured worldwide attention as a human pathogen of great concern (Nordmann et al., 2009). Specifically, resistant strains such as those with KPC- and NDM-type carbapenemases have spread rapidly and are associated with poor outcomes in vulnerable patients (Ben-David et al., 2012; Bratu et al., 2005; Kitchel et al., 2009; Moellering, 2010). In paediatric populations, K. pneumoniae is identified in a wide range of diseases, ranging from bacteraemia in hospitalized patients in industrialized countries to neonatal sepsis in community settings in the developing world (Mandell et al., 2010).
Currently, relatively little is known about the co-distribution of fitness/virulence and antibiotic resistance properties throughout the K. pneumoniae species. Within the closely related species Escherichia coli, molecular analysis has demonstrated virulence factor carriage and extraintestinal disease to be associated with particular phylogroups (e.g. phylogroups B2 and D) and sequence types (STs) (Johnson et al., 2012a, b; Pitout, 2012). With the advent of multidrug resistance, predominant STs within both E. coli and K. pneumoniae have been found to be responsible for the dissemination of resistant strains into new clinical settings (Bratu et al., 2005; Johnson et al., 2010; Kitchel et al., 2009; Kumarasamy et al., 2010; Oteo et al., 2009; Sánchez-Romero et al., 2012; Woerther et al., 2011). Colonization of the gastrointestinal tract is recognized as a risk factor for development of infection with resistant bacteria (Salyers et al., 2004; Schjørring et al., 2008; Sidjabat et al., 2009; Snyder et al., 2011), and analysis of K. pneumoniae isolated from stool has demonstrated that specific STs are associated with multidrug resistance and with subsequent clinical infection (Borer et al., 2012; Oteo et al., 2009; Sánchez-Romero et al., 2012; Viau et al., 2012; Woerther et al., 2011). However, significant gaps exist in the understanding of K. pneumoniae molecular epidemiology as it relates to antimicrobial resistance, colonization and disease in the human host.
Investigations of the clinical and molecular characteristics of K. pneumoniae in a non-outbreak setting are limited. In paediatrics, in particular, studies of K. pneumoniae have focused primarily on outbreak descriptions of multidrug-resistant pathogens, with limited attention to population structure or to the relationship between colonizing and disease-causing strains. In turn, the international K. pneumoniae multilocus sequence typing (MLST) database relies heavily on samples from human disease, with limited sampling from human intestinal ecology. The aim of this study was to compare the characteristics of paediatric patients with K. pneumoniae isolated from sterile sites with those with K. pneumoniae isolated from stools, and to describe the resistance phenotypes and clonal properties of isolates from these cohorts.
Methods
Study design and setting.
We conducted a prospective, observational study of K. pneumoniae isolated from clinical samples submitted to the clinical microbiology laboratory of Seattle Children’s Hospital (SCH; Seattle, WA, USA) between December 2009 and December 2010. SCH is a free-standing, tertiary care hospital with 250 paediatric beds, over 30 000 emergency room visits and over 260 000 outpatient clinic visits per year. The Institutional Review Board at SCH approved the study.
This study included a convenience sample of 92 isolates obtained from specimens submitted for clinical purposes. The disease cohort was derived from 49 sequential isolates from blood and urine cultures. The colonization cohort of 43 isolates was obtained from stool specimens submitted for evaluation of bacterial pathogens. All patients were eligible for study inclusion regardless of demographics, hospital service or clinical indication for specimen collection. Patients were allowed to contribute only a single isolate, regardless of specimen type. Information about patient demographics and clinical comorbidities was collected from the medical record at SCH.
Laboratory methods.
K. pneumoniae from blood and urine specimens were identified clinically by standard biochemical methods and the VITEK 2 GN card (bioMérieux). For K. pneumoniae from faecal specimens, mucoid, lactose-fermenting colonies were screened for a negative reaction to indole; definitive species identification was then accomplished using the VITEK 2 GN card. Antibiotic susceptibility testing for all study isolates was conducted using a combination of the VITEK GN30 card, disc diffusion and Etest methodology. Results were interpreted according to the Clinical and Laboratory Standards Institute M100 Guidelines (CLSI, 2011). Standard agents tested included ampicillin, ampicillin/sulbactam, piperacillin/tazobactam, cefazolin, ceftazidime, ceftriaxone, cefepime, meropenem, amikacin, gentamicin, tobramycin, ciprofloxacin, levofloxacin, nitrofurantoin and trimethoprim/sulfamethoxazole. In addition, we tested agents that were not tested routinely for this organism, including cefoxitin, imipenem, nalidixic acid, sulfamethoxazole and fosfomycin. Among isolates with third-generation cephalosporin resistance, extended-spectrum β-lactamase phenotypes were confirmed using disc approximation by previously described methods (Jacoby & Han, 1996).
Molecular epidemiology.
MLST was used to evaluate the genetic relatedness of study isolates. PCR amplification and sequencing of seven housekeeping gene loci were completed according to the Institut Pasteur scheme (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html) by previously described methodology (Diancourt et al., 2005). ST assignment and phylogenetic analysis were performed using BioNumerics software (Applied Maths). The most recent Institut Pasteur database query was performed on 14 May 2012. Novel allele sequences and profiles were submitted to the Institut Pasteur on 24 May 2012 for allele ST assignments and inclusion in the international public access database.
Statistical analysis.
Chi-squared tests were used to evaluate dichotomous variables; if assumptions were not met, Fisher’s exact test was used. Continuously scaled independent variables were tested using Students’ t-test or non-parametrically by the Mann–Whitney U test.
Results
Patients and clinical characteristics
This study included 92 unique K. pneumoniae isolates from 92 paediatric patients. The disease cohort included 49 total isolates, including 36 (73.5 %) from urine cultures and 13 (26.5 %) from blood cultures. Among the entire cohort, median patient age was 2.3 years old (interquartile range: 0.7, 14.9) and 53.3 % of isolates were from female patients (Table 1). Clinical and demographic characteristics, including history of prior transplantation, history of prior hospitalization and previous antibiotic use, were similar between cohorts. Whilst the proportions of patients with a pre-existing condition were also similar between disease and colonizing cohorts (85.7 and 74.4 %, respectively; P = 0.173), patients from the disease cohort were far more likely to have a renal or urologic comorbidity (42.9 %) when compared with patients from the colonization cohort (3.1 %; P<0.001).
Table 1. Demographics and clinical characteristics of 92 patients with K. pneumoniae isolated from colonization or disease cohorts.
| Colonization | Disease | P-value | Total | |
| Isolates [n (%)] | 43 (46.7) | 49 (53.3) | 92 (100) | |
| Blood | 13 (14.1) | |||
| Urine | 36 (39.1) | |||
| Age [years (median)] | 2.3 | 2.2 | 0.85 | 2.3 |
| Range | 0.06–22.84 | 0.01–20.94 | 0.01–22.84 | |
| Interquartile range | 0.72, 18.64 | 0.53, 15.3 | 0.7, 14.9 | |
| Female gender [n (%)] | 26 (60.5) | 23 (46.9) | 0.194 | 49 (53.3) |
| Caucasian race [n (%)] | 29 (67.4) | 32 (65.3) | 0.83 | 61 (66.3) |
| Hispanic ethnicity [n (%)] | 9 (20.9) | 11 (22.5) | 0.97 | 20 (21.7) |
| Clinical comorbidity [n (%)] | 32 (74.4) | 42 (85.7) | 0.173 | 74 (80.4) |
| Oncologic | 13 (40.6) | 10 (23.8) | 23 (25.0) | |
| Urologic/renal | 1 (3.1) | 18 (42.9) | <0.001 | 19 (25.6) |
| Gastrointestinal | 10 (31.3) | 8 (19) | 18 (24.3) | |
| Other | 8 (25) | 6 (14.3) | 14 (19) | |
| Post-transplantation [n (%)] | 12 (27.9) | 8 (16.3) | 0.179 | 20 (21.7) |
| Hospitalized in the previous year [n (%)] | 28 (65.1) | 32 (65.3) | 0.985 | 60 (65.2) |
| Mean number of hospitalizations (sd)* | 4.5 (3.6) | 3.8 (2.9) | 4.13 (3.22) | |
| Antibiotics in the previous year [n (%)] | 27 (62.8) | 37 (75.5) | 0.186 | 64 (69.5) |
| Prophylaxis only | 1 (3.7) | 3 (8.1) | 4 (6.2) | |
| One to two courses (intravenous or per os) | 6 (22.2) | 6 (16.2) | 2 (3.1) | |
| More than two courses (intravenous or per os) | 20 (74.1) | 28 (75.7) | 48 (75.0) |
As a result of rounding, percentages may not add up to 100%.
Only includes patients who were hospitalized in the previous year.
Antibiotic susceptibility
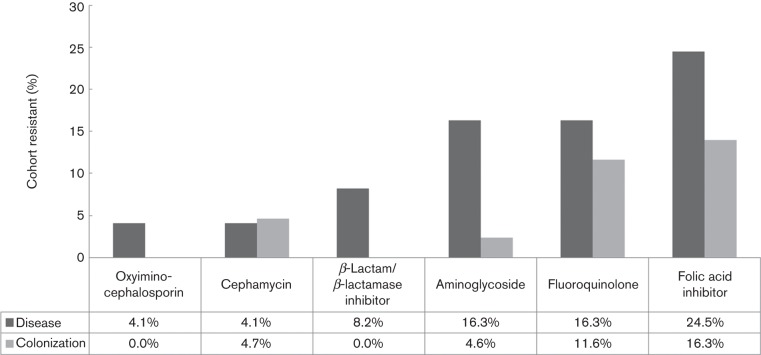
No evidence of antimicrobial resistance to carbapenem agents or the fourth-generation cephalosporin, cefepime, was detected in this collection. There were non-significant trends indicating more frequent resistance to β-lactam/β-lactamase inhibitor combinations (P = 0.12) and aminoglycosides (P = 0.1) among isolates from the disease cohort relative to those from the colonizing cohort (Fig. 1). Eleven isolates were resistant to one clinically relevant antibiotic class (β-lactam/β-lactamase inhibitor combination, oxyimino-cephalosporins, cephamycin, folic acid inhibitors, aminoglycosides or fluoroquinolones); five isolates were resistant to two classes and seven isolates were resistant to three or more classes. Six of the seven isolates with three or more classes of resistance were found in the disease cohort. There were no statistical differences in prevalence of resistance to one, two, or three or more antibiotic classes between cohorts.
Fig. 1.
Resistance to antimicrobial classes among study isolates. Specific agents with resistance, by class: oxyimino-cephalosporin: ceftriaxone; β-lactam/β-lactamase inhibitor: piperacillin/tazobactam; aminoglycoside: gentamicin, amikacin, tobramycin; fluoroquinolone: ciprofloxacin, levofloxacin; folic acid inhibitor: trimethoprim/sulfamethoxazole.
Molecular epidemiology
We identified 71 distinct ST profiles among the 92 K. pneumoniae isolates. Most ST profiles were represented by a single strain; however, 13 STs contained multiple representatives. Among these 13 STs, nine were described previously in the international K. pneumoniae MLST database: ST11, ST20, ST34, ST37, ST45, ST252, ST461, ST629 and ST661 (Table 2). However, four STs with multiple representatives were novel to the K. pneumoniae database (ST938, ST939, ST950 and ST952, each with two isolates); two of the four (ST938 and ST939) contained at least one novel allele sequence (Table 2). In all, 43 isolates (46.7 %) represented novel STs, of which 22 (23.9 % of all isolates) demonstrated at least one novel allele sequence (Table 3); isolates with novel STs were no more likely in either the colonizing or disease-causing cohort.
Table 2. ST profiles with multiple representatives identified among 92 K. pneumoniae isolates.
| ST | Resistance phenotype | Isolate type | Comorbidity |
| ST11 | AG, FQ, T/S | Urine | Urologic |
| AG, FQ, T/S | Blood | Urologic | |
| ST20 | – | Urine | Urologic |
| – | Urine | Other | |
| – | Blood | Oncologic | |
| – | Stool | Other | |
| – | Urine | Other | |
| ST34 | – | Blood | Urologic |
| FOX | Stool | Oncologic | |
| ST37 | AG, FQ, T/S | Stool | Oncologic (bone marrow transplant) |
| – | Stool | Other | |
| – | Stool | Other | |
| ST45 | T/S | Urine | Previously healthy |
| T/S | Stool | Oncologic (bone marrow transplant) | |
| – | Stool | Gastrointestinal | |
| ST252 | – | Blood | Gastrointestinal |
| – | Urine | Urologic | |
| – | Urine | Previously healthy | |
| – | Urine | Urologic | |
| ST461 | – | Blood | Oncologic (bone marrow transplant) |
| AG, T/S | Urine | Urologic | |
| ST629 | CRO, FQ, AG, T/S | Urine | Oncologic (bone marrow transplant) |
| CRO, FQ, AG, T/S | Urine | Oncologic | |
| ST661 | – | Urine | Urologic |
| – | Stool | Oncologic | |
| ST938*,† | FOX | Stool | Gastrointestinal (liver transplant) |
| FOX | Blood | Other (heart transplant) | |
| ST939*,† | – | Stool | Previously healthy |
| – | Stool | Gastrointestinal | |
| ST950* | – | Urine | Previously healthy |
| – | Stool | Urologic (renal transplant) | |
| ST952* | AG, FQ, T/S | Urine | Urologic (renal transplant) |
| AG, FQ, T/S | Urine | Urologic | |
| FQ, T/S | Stool | Oncologic (bone marrow transplant) |
BL/BLI, β-lactam/β-lactamase inhibitor (i.e. piperacillin/tazobactam); AG, aminoglycoside, any; CRO, ceftriaxone; FOX, cefoxitin; FQ, fluoroquinolone, any; T/S, trimethoprim/sulfamethoxazole.
Novel ST.
Novel allele sequence.
Table 3. Molecular typing features of 92 K. pneumoniae isolates.
| Colonization [n (%)] | Disease [n (%)] | P-value | Total [n (%)] | |
| Isolates | 43 | 49 | 92 | |
| STs | 39 | 39 | 71* | |
| Isolates in novel STs | 24 (55.8) | 19 (38.8) | 0.102 | 43 (46.7) |
| Isolates in novel STs with novel allele(s) | 13 (30.2) | 9 (18.4) | 0.183 | 22 (23.9) |
Includes seven STs containing both colonizing and disease-associated isolates.
Discussion
This study is, to the best of our knowledge, the first to compare colonizing and disease-causing strains of K. pneumoniae at a tertiary care children’s hospital. Within this high-risk clinical population, we found a significant proportion of strains demonstrating novel allele sequences or profiles. Additionally, while multidrug resistance and extended-spectrum β-lactam resistance were relatively low, we found that multidrug resistance phenotypes clustered within multirepresentative STs and within the disease cohort.
K. pneumoniae is known to cause opportunistic infections in vulnerable patients. Thus, it would be expected that patients in our disease cohort, but not necessarily those in the colonization cohort, would exhibit clinical characteristics reflecting this vulnerability (e.g. underlying comorbidities, multiple hospitalizations and courses of broad-spectrum antibiotics in the preceding year). Indeed, based on clinical features, the study cohort as a whole appeared to be at risk for invasive infection; 80 % overall had a documented comorbidity and 65 % had been hospitalized in the preceding year, with a mean of over four hospitalizations among those hospitalized. Our study found that clinical and demographic characteristics of patients with colonizing K. pneumoniae were similar to those with K. pneumoniae disease, with a single exception: urologic or renal abnormalities were more frequent in the disease cohort. However, given that 36/49 (73.5 %) of disease-associated isolates were recovered from urine, this finding is not unexpected, given the propensity of Klebsiella to produce urinary tract disease where anatomy or function is abnormal. Nonetheless, our findings suggest that both Klebsiella-infected and Klebsiella-colonized individuals have had extensive exposure to the hospital environment and selective antibiotic pressure. Additionally, persistent intestinal colonization may promote emergence in these hosts of increasingly resistant pathogens through porin alteration and/or plasmid sharing under frequent selection pressure from antibiotic therapy. The gastrointestinal tract is an essential reservoir to investigate further; more studies of colonization ecology and dynamics are warranted, as these factors likely impact patterns of disease and resistance in high-risk populations.
MSLT analysis revealed novel allele sequences or profiles in almost half (46.7 %) of study isolates, with novel STs detected with comparable frequency in disease and colonization isolates. The MLST scheme for K. pneumoniae was described in 2005 and the international K. pneumoniae MLST database remains dominated by isolates associated with human disease; as of May 2012, <10 % of human K. pneumoniae isolates with documented source came from the gastrointestinal tract (data not shown). The results presented here contribute to our understanding of the population structure of K. pneumoniae and demonstrate that a greater diversity of STs than captured to date may be present in human hosts. This expanded sampling contributes to our understanding of K. pneumoniae with human-adapted fitness traits, yet also indicates that the human ecology of K. pneumoniae remains to be characterized fully. Further delineation of the population structure is important to elucidate the molecular determinants of K. pneumoniae virulence, as well as the determinants contributing to successful colonization and to the molecular dynamics of antibiotic resistance.
Prior investigations of Enterobacteriaceae have demonstrated the epidemiological success of specific STs in association with multidrug resistance, e.g. E. coli ST131 and CTX-M-15, and K. pneumoniae ST258 and KPC. Our convenience sample, drawn from a community with low endemic rates of multidrug resistance, demonstrated 13 STs with multiple representatives, including those previously described in the K. pneumoniae MLST database, as well as novel STs described by this study. Four previously known STs have been associated with multidrug resistance in K. pneumoniae: ST11, ST20, ST37 and ST252 (Coelho et al., 2012; Orsi et al., 2011; Oteo et al., 2009; Shin et al., 2012). Phenotypic analysis of isolates in this study reflects antimicrobial resistance rates at an academic paediatric hospital in the USA with limited multidrug resistance. However, all seven isolates with resistance to three or more antibiotic classes were found in STs with multiple representatives. In addition, both ceftriaxone-resistant isolates and three of four cefoxitin-resistant isolates were found in STs with multiple representatives. The majority of isolates with worrisome resistance patterns were from the disease cohort. The finding that resistant isolates were found primarily in STs within the disease cohort supports the concept that certain STs may have fitness advantages (potentially related to accessory genetic features that are distributed differentially throughout the species), and that antibiotic resistance may play a role in the persistence and spread of pathogens throughout populations with healthcare exposure.
This study was limited by small sample size and the use of convenience sampling for the colonization cohort. The lack of Klebsiella colonization-negative controls, for example, precluded us from determining whether healthy children are less frequently colonized by K. pneumoniae than children with comorbidities. In addition, this study was not designed to evaluate outbreak dynamics, namely whether some same-ST isolates had indistinguishable pulsotypes and may have represented transmission within the hospital [e.g. two multidrug-resistant isolates from ST629 recovered on successive days (data not shown) from ‘Oncology' patients] or in the community. A larger study with higher discriminatory power would be helpful in distinguishing local (institutional or community) dynamics from the national or global dissemination of successful, emerging clones. Finally, the study was not designed to determine the impact of specific antibiotic exposures on the susceptibility patterns of colonizing or disease isolates; we cannot, for example, demonstrate differential exposure to aminoglycoside between the groups to explain the relative prominence of gentamicin resistance in the disease isolates. The strengths of this study include attention to population structure in an environment with low endemic resistance rates, focus on paediatric hosts, and inclusion of isolates from both gastrointestinal colonization and sterile sites.
In summary, this study highlights important avenues for further investigation of K. pneumoniae, including the significance of colonization, clinical risk factors for development of infection, and further understanding of population structure and virulence characteristics. Our investigation in an environment of limited multidrug and extended-spectrum β-lactam resistance found a significant proportion of novel STs, and observed multidrug resistance phenotypes in association with a limited number of ST profiles. Further extension of these findings may help to address the challenge of the global spread of virulent and multidrug-resistant K. pneumoniae.
Abbreviations:
- MLST
multilocus sequence typing
- SCH
Seattle Children’s Hospital
- ST
sequence type
References
- Ben-David D., Kordevani R., Keller N., Tal I., Marzel A., Gal-Mor O., Maor Y., Rahav G. (2012). Outcome of carbapenem resistant Klebsiella pneumoniae bloodstream infections. Clin Microbiol Infect 18, 54–60. 10.1111/j.1469-0691.2011.03478.x [DOI] [PubMed] [Google Scholar]
- Borer A., Saidel-Odes L., Eskira S., Nativ R., Riesenberg K., Livshiz-Riven I., Schlaeffer F., Sherf M., Peled N. (2012). Risk factors for developing clinical infection with carbapenem-resistant Klebsiella pneumoniae in hospital patients initially only colonized with carbapenem-resistant K. pneumoniae. Am J Infect Control 40, 421–425. 10.1016/j.ajic.2011.05.022 [DOI] [PubMed] [Google Scholar]
- Bratu S., Landman D., Haag R., Recco R., Eramo A., Alam M., Quale J. (2005). Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch Intern Med 165, 1430–1435. 10.1001/archinte.165.12.1430 [DOI] [PubMed] [Google Scholar]
- CLSI (2011). Performance Standards for Antimicrobial Susceptibility Testing; Approved Standard, 24th Informational Supplement M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Coelho A., Piedra-Carrasco N., Bartolomé R., Quintero-Zarate J. N., Larrosa N., Cornejo-Sánchez T., Prats G., Garcillán-Barcia M. P., de la Cruz F., González-Lopéz J. J. (2012). Role of IncHI2 plasmids harbouring blaVIM-1, blaCTX-M-9, aac(6′)-Ib and qnrA genes in the spread of multiresistant Enterobacter cloacae and Klebsiella pneumoniae strains in different units at Hospital Vall d’Hebron, Barcelona, Spain. Int J Antimicrob Agents 39, 514–517. 10.1016/j.ijantimicag.2012.01.006 [DOI] [PubMed] [Google Scholar]
- Diancourt L., Passet V., Verhoef J., Grimont P. A., Brisse S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43, 4178–4182. 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Han P. (1996). Detection of extended-spectrum beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol 34, 908–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Johnston B., Clabots C., Kuskowski M. A., Castanheira M. (2010). Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 51, 286–294. 10.1086/653932 [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Porter S. B., Zhanel G., Kuskowski M. A., Denamur E. (2012a). Virulence of Escherichia coli clinical isolates in a murine sepsis model in relation to sequence type ST131 status, fluoroquinolone resistance, and virulence genotype. Infect Immun 80, 1554–1562. 10.1128/IAI.06388-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Urban C., Weissman S. J., Jorgensen J. H., Lewis J. S., II, Hansen G., Edelstein P. H., Robicsek A., Cleary T. & other authors (2012b). Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4) and blaCTX-M-15 among extended-spectrum-β-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob Agents Chemother 56, 2364–2370. 10.1128/AAC.05824-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchel B., Rasheed J. K., Patel J. B., Srinivasan A., Navon-Venezia S., Carmeli Y., Brolund A., Giske C. G. (2009). Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53, 3365–3370. 10.1128/AAC.00126-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarasamy K. K., Toleman M. A., Walsh T. R., Bagaria J., Butt F., Balakrishnan R., Chaudhary U., Doumith M., Giske C. G. & other authors (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10, 597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell G. L., Bennett J. E., Dolin R. (2010). Principles and Practice of Infectious Diseases, 7th edn Philadelphia, PA: Churchill Livingstone. [Google Scholar]
- Moellering R. C., Jr (2010). NDM-1 – a cause for worldwide concern. N Engl J Med 363, 2377–2379. 10.1056/NEJMp1011715 [DOI] [PubMed] [Google Scholar]
- Nordmann P., Cuzon G., Naas T. (2009). The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9, 228–236. 10.1016/S1473-3099(09)70054-4 [DOI] [PubMed] [Google Scholar]
- Orsi G. B., García-Fernández A., Giordano A., Venditti C., Bencardino A., Gianfreda R., Falcone M., Carattoli A., Venditti M. (2011). Risk factors and clinical significance of ertapenem-resistant Klebsiella pneumoniae in hospitalised patients. J Hosp Infect 78, 54–58. 10.1016/j.jhin.2011.01.014 [DOI] [PubMed] [Google Scholar]
- Oteo J., Cuevas O., López-Rodríguez I., Banderas-Florido A., Vindel A., Pérez-Vázquez M., Bautista V., Arroyo M., García-Caballero J. & other authors (2009). Emergence of CTX-M-15-producing Klebsiella pneumoniae of multilocus sequence types 1, 11, 14, 17, 20, 35 and 36 as pathogens and colonizers in newborns and adults. J Antimicrob Chemother 64, 524–528. 10.1093/jac/dkp211 [DOI] [PubMed] [Google Scholar]
- Pitout J. D. (2012). Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol 3, 9. 10.3389/fmicb.2012.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers A. A., Gupta A., Wang Y. (2004). Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol 12, 412–416. 10.1016/j.tim.2004.07.004 [DOI] [PubMed] [Google Scholar]
- Sánchez-Romero I., Asensio A., Oteo J., Muñoz-Algarra M., Isidoro B., Vindel A., Alvarez-Avello J., Balandín-Moreno B., Cuevas O. & other authors (2012). Nosocomial outbreak of VIM-1-producing Klebsiella pneumoniae isolates of multilocus sequence type 15: molecular basis, clinical risk factors, and outcome. Antimicrob Agents Chemother 56, 420–427. 10.1128/AAC.05036-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjørring S., Struve C., Krogfelt K. A. (2008). Transfer of antimicrobial resistance plasmids from Klebsiella pneumoniae to Escherichia coli in the mouse intestine. J Antimicrob Chemother 62, 1086–1093. 10.1093/jac/dkn323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S. Y., Bae I. K., Kim J., Jeong S. H., Yong D., Kim J. M., Lee K. (2012). Resistance to carbapenems in sequence type 11 Klebsiella pneumoniae is related to DHA-1 and loss of OmpK35 and/or OmpK36. J Med Microbiol 61, 239–245. 10.1099/jmm.0.037036-0 [DOI] [PubMed] [Google Scholar]
- Sidjabat H. E., Silveira F. P., Potoski B. A., Abu-Elmagd K. M., Adams-Haduch J. M., Paterson D. L., Doi Y. (2009). Interspecies spread of Klebsiella pneumoniae carbapenemase gene in a single patient. Clin Infect Dis 49, 1736–1738. 10.1086/648077 [DOI] [PubMed] [Google Scholar]
- Snyder G. M., O’Fallon E., D’Agata E. M. (2011). Co-colonization with multiple different species of multidrug-resistant gram-negative bacteria. Am J Infect Control 39, 506–510. 10.1016/j.ajic.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viau R. A., Hujer A. M., Marshall S. H., Perez F., Hujer K. M., Briceño D. F., Dul M., Jacobs M. R., Grossberg R. & other authors (2012). “Silent” dissemination of Klebsiella pneumoniae isolates bearing K. pneumoniae carbapenemase in a long-term care facility for children and young adults in Northeast Ohio. Clin Infect Dis 54, 1314–1321. 10.1093/cid/cis036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerther P. L., Angebault C., Jacquier H., Hugede H. C., Janssens A. C., Sayadi S., El Mniai A., Armand-Lefèvre L., Ruppé E. & other authors (2011). Massive increase, spread, and exchange of extended spectrum β-lactamase-encoding genes among intestinal Enterobacteriaceae in hospitalized children with severe acute malnutrition in Niger. Clin Infect Dis 53, 677–685. 10.1093/cid/cir522 [DOI] [PubMed] [Google Scholar]