Abstract
Marine sponges harbour abundant and diverse bacterial communities, providing an ideal environment for bacterial cell-density-dependent cell–cell signalling, termed quorum sensing. The marine sponge symbiont Ruegeria sp. KLH11 produces mainly long chain acylhomoserine lactones (AHLs) and has been developed as a quorum sensing model for roseobacterial sponge symbionts. Two pairs of luxR/I homologues were identified by genetic screening and were designated ssaRI and ssbRI (sponge-associated symbiont locus A or B, luxR/luxI homologue). In this study, we identified a third luxI-type gene, named sscI. The sscI gene does not have a cognate luxR homologue present at an adjacent locus and thus sscI is an AHL synthase solo. The sscI gene is required for production of long-chain hydroxylated AHLs, contributes to AHL pools and modestly influences flagellar motility in KLH11. A triple mutant for all luxI-type genes cannot produce AHLs, but still synthesizes para-coumaroyl-homoserine lactone.
Introduction
Marine sponges harbour highly diverse and dense microbial communities and in some cases up to 30–40 % of the sponge biomass is from the associated bacteria (Taylor et al., 2007). These sponges provide a highly conducive environment for bacterial quorum sensing (QS), a process by which bacteria use chemical signals to communicate with each other and coordinate group behaviours, such as bioluminescence, antibiotic production and virulence at high cell density (Ahlgren et al., 2011; Fuqua & Greenberg, 2002). Acylhomoserine lactone (AHL)-based QS was first discovered in the marine squid symbiont Vibrio fisheri about 40 years ago and since then it has been reported in over 100 different bacterial species, although mainly restricted to the phylum Proteobacteria (Ahlgren et al., 2011; Fuqua & Greenberg, 2002).
Typically, each QS circuit has a luxI-type gene, responsible for AHL synthesis, and a luxR-type gene, encoding a protein that binds and provides a response to AHL(s). These two genes are often genetically linked, arranged in tandem or convergently expressed (Fuqua & Greenberg, 2002). However, in many different bacteria, luxR-type genes without a linked luxI-type gene have been discovered and these luxR-type genes are termed luxR solos. These solos occur both in bacteria that have complete LuxR-LuxI-type QS systems and bacteria that do not (Subramoni & Venturi, 2009). LuxR solos can regulate gene expression by binding to AHLs produced by other luxI genes encoded elsewhere in the same bacterial genome, such as in the cases of QscR in Pseudomonas aeruginosa (Lequette et al., 2006) and ExpR in Sinorhizobium meliloti (McIntosh et al., 2008), or by binding to AHLs produced by other bacteria, such as in the case of SdiA in Escherichia coli and Salmonella enterica (Ahmer, 2004; Yao et al., 2006). Furthermore, some LuxR-type proteins can regulate gene expression in response to non-AHL signals or independently of ligand binding (Subramoni & Venturi, 2009). Although progress has been made in understanding these luxR solos, there is little information about the less common scenario in which functional luxI genes are not linked to luxR-type genes, which we designate as luxI solos.
We have found that members of the Silicibacter-Ruegeria (SR) subgroup of the ecologically important Roseobacter clade are the primary AHL producers among cultivatable bacterial isolates from the marine sponges Mycale laxissima and Ircinia strobilina (Mohamed et al., 2008) and over 80 % of available roseobacterial genomes encode at least one luxI homologue (Zan et al., 2014). We have developed Ruegeria sp. KLH11 (hereafter referred to as KLH11) as a model to study QS in marine sponge symbionts. We previously reported detailed analyses of two luxRI systems: ssaRI and ssbRI in KLH11 (Zan et al., 2012). SsaI and SsbI direct the synthesis of long chain AHLs ranging from C12- to C16-homoserine lactone (HSL), dominated by 3-oxo and 3-hydroxy moieties at the beta-position in the acyl chain, respectively. The SsaRI system provides QS-dependent control of flagellar motility in KLH11, functioning through the CtrA master regulator (Zan et al., 2013). Analysis of whole sponge tissues revealed the presence of ssaI transcripts and AHLs (Zan et al., 2012). The function of the SsbRI system remains unclear, but it is indirectly regulated by SsaRI. We have also presented preliminary evidence for a solo luxI-type gene, sscI, in KLH11, which is not genetically linked to a luxR homologue (Zan et al., 2012). In the current study, we have analysed the functional characteristics of the sscI solo in KLH11. Furthermore, we tested whether KLH11 can produce the novel para-coumaroyl-HSL (pC-HSL) molecule that was originally discovered in Rhodopseudomonas palustris and requires the substrate para-courmarate for synthesis by the LuxI homologue RpaI (Schaefer et al., 2008).
Methods
Bacterial strains, oligonucleotides and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA, USA). Unless stated otherwise, E. coli strains were grown in Luria–Bertani (LB) broth at 37 °C with aeration, Ruegeria sp. KLH11 strains were grown in marine broth 2216 at 28 °C (MB 2216; BD) and Agrobacterium tumefaciens strains were grown in AT minimal salt medium supplemented with 0.5 % (w/v) glucose and 15 mM (NH4)2SO4 (ATGN; Tempé et al., 1977). Rhodopseudomonas palustris CGA814 was grown in LB broth. Antibiotics were used at the following final concentrations (μg ml−1): (i) E. coli (ampicillin, Ap, 100; gentamicin, Gm, 25; kanamycin, Km, 25; spectinomycin, Sp, 100; tetracycline, Tc, 5). (ii) KLH11 (Km, 100; rifampicin, Rif, 200; Gm 25; Sp, 100; Tc, 5). (iii) A. tumefaciens (Gm, 300; Sp, 200). (iv) Rh. palustris CGA814 (Km, 50).
Table 1. Bacterial strains and plasmids used in this study.
| Bacteria/plasmid | Relevant feature | Reference |
| E. coli DH5α/λpir | Strain for propagating R6K suicide plasmids | Lab collection |
| E. coli S17-1/λpir | IncP conjugal donor | Kalogeraki & Winans (1997) |
| E. coli XL-1 Blue | Standard alpha-complementation strain | Lab collection |
| E. coli MC4100 | K-12 derivative, ΔlacZ | Lab collection |
| A. tumefaciens NTL4 | Ti plasmid-less derivative, nopaline chromosomal background | Zhu et al. (1998) |
| KLH11 | WT | Mohamed et al. (2008) |
| KLH11-EC1 | RifR | Zan et al. (2012) |
| KLH11-SK01 | ΔssaI, RifR | Zan et al. (2012) |
| KLH11-SK02 | ΔssaI ΔssbI, RifR | Zan et al. (2012) |
| KLH11-OKC2 | sscI-lacZ, RifR KmR | This study |
| KLH11-OKC6 | ΔssaI ΔssbI ssc-lacZ, RifR KmR | This study |
| CGA814 | Rh. palustris; rpaI-lacZ, KmR | Schaefer et al. (2008) |
| pCR2.1-TOPO | PCR fragment cloning vector, Ap/KmR | Invitrogen |
| pBBR1-MCS5 | BHR Plac expression vector, GmR | Kovach et al. (1995) |
| pSRKTc | BHR expression vector containing lac promoter and lacIq, TcR | Khan et al. (2008) |
| pVIK112 | R6K-based lacZ transcriptional fusion, KmR | Kalogeraki & Winans (1997) |
| pRA301 | BHR lacZ translational fusion vector | Akakura & Winans (2002) |
| pEC108 | pBBR1-MCS5 derivative carrying full-length Plac-ssaI, GmR | Zan et al. (2012) |
| pEC109 | pBBR1-MCS5 derivation carrying full-length Plac-ssbI, from pEC110, GmR | Zan et al. (2012) |
| pEC112 | pBBR1-MCS5 derivative, carrying full-length Plac-ssaR, from pEC106, GmR | Zan et al. (2012) |
| pEC116 | pRA301 derivation, PssaI-lacZ, SpR | Zan et al. (2012) |
| pEC121 | pRA301 derivative, PssbI-lacZ, Sp/SmR | Zan et al. (2012) |
| pOKC1 | pCR2.1-TOPO, carrying internal fragment of sscI, KmR | This study |
| pOKC2 | pVIK112 derivative, carrying internal fragment of sscI, KmR | This study |
| pOKC3 | pCR2.1-TOPO, carrying full-length sscI, KmR | This study |
| pOKC4 | pSRKTC derivative, carrying full-length Plac-sscI, TcR | This study |
| pOKC8 | pRA301 derivation, PsscI-lacZ, SpR | This study |
Sm, Streptomycin; Tc, tetracycline.
Plasmid construction for sscI null mutation, expression and lacZ-fusion.
Several regions around the sscI gene were isolated by PCR amplification from KLH11 genomic DNA. The method used to construct the sscI Campbell insertion mutant was similar to that described by Zan et al. (2012). Briefly, an internal fragment of the sscI gene was amplified using forward primer 5′-GAATTCATGTTTCGCGATCGAGCAGAT-3′ (the EcoRI recognition site is underlined) and reverse primer 5′-GGTACCTCTTGATACTCCCGCTC-3′ (the KpnI recognition site is underlined). The PCR amplicon was gel-purified and cloned into pCR 2.1-TOPO vector (Invitrogen) to create pOKC1 and the insert was confirmed by sequencing. For recombinational mutagenesis, pOKC1 was digested with EcoRI and KpnI, and the resulting sscI fragment was ligated to a similarly digested R6K replicon, the pVIK112 suicide vector (Kalogeraki & Winans, 1997), creating pOKC2. pOKC2 was conjugated into KLH11 and Km-resistant (KmR) transconjugants were selected and confirmed by sequencing using the forward primer 5′-ATTAACCATAATCAAGCATCTCTT-3′. To construct double and triple AHL synthase gene mutants, pOCK2 was conjugated into ΔssaI and ΔssbI, and ΔssaI ΔssbI strains, respectively, and the transconjugants were selected and confirmed as described for the sscI single mutant.
A controlled expression construct of sscI was generated by PCR amplification of the coding regions using the forward primer 5′-TCTAGACTGAAACAGGAAACAGCTATGCTCCGTTATGTTTTTGCA-3′ (the XbaI recognition site is underlined, the stop codon TGA and the start codon ATG are in bold type and the E. coli lacZ ribosome-binding site is in italics) and the reverse primer 5′-CTCGAGTCAAGCGGTTCTTTGAAACTT-3′ (the stop codon is in bold type and the XhoI recognition site is underlined). The PCR products were ligated into pCR2.1-TOPO vector (Invitrogen) to create pOKC3 and confirmed by sequencing. pOKC3 was digested by XbaI and XhoI and the insert was subcloned into the vector pSRKTc (Khan et al., 2008) to create pOKC4. The insert carried by the construct was confirmed by sequencing.
In order to generate a plasmid-borne PsscI-lacZ fusion, the presumptive promoter sequences were PCR amplified. The forward primer 5′-GAATTCGCCGAGATGAACTGTTCAAAGAAC-3′ (the EcoRI recognition site is underlined) and the reverse primer 5′-GGATCCGAGCATTTTTAACCTCTTGTTCAC-3′ (the BamHI recognition site is underlined) annealing 255 bp upstream and 3 bp downstream of the sscI translational start site were used to amplify its promoter. The PCR products were cloned into pCR2.1-TOPO vector and the inserts were confirmed by DNA sequencing. The pCR2.1-TOPO derivatives were digested with EcoRI and PstI and the resulting fragments were ligated with pRA301 vector digested with the same restriction enzymes (Akakura & Winans, 2002) to create pOKC8 with the PsscI-lacZ translational fusion.
Organic extraction, TLC and MS analysis of AHLs.
As described previously, organic extraction of KLH11-derivative cultures, followed by reverse-phase TLC of organic extracts and AHL bioassay analysis with an ultrasensitive AHL bioreporter derived from A. tumefaciens was used to characterize the AHLs specified by SscI (Zan et al., 2012; Zhu et al., 2003).
Identification of SscI AHLs by LC-MS-MS was also performed. KLH11 derivatives were grown in MB 2216 with appropriate antibiotics (and 0.5 mM IPTG to induce the Plac promoter) at 28 °C to stationary phase (OD600~2.0) in the presence of 5 g Amberlite XAD 16 resin l−1 for 36 h. Cells and resin were separated by centrifugation and extracted with 50 ml methanol and dried to 2 ml. Three nanomoles of D3-C6-HSL (D3 indicates that there are three deuterium atoms at the terminal position of the acyl chain in the AHL molecule) was added to each sample as an internal standard and a volume of 0.2 ml of each extract was purified using solid phase extraction methods as described previously (Gould et al., 2006).
Extracts were dried down and resuspended in 38 µl solvent A (8.3 mM ammonium acetate, pH 5.7) and 2 µl solvent B (methanol). This solution was injected on to a 50×3.00 mm 2.6 µ C18 Kinetex (Phenomenex) column. A mobile phase gradient was generated from 5 % B to 65 % B in 5 min, then B was increased to 95 % in 15 min and held for 8 min at a flow rate of 250 µl min−1. The HPLC system was interfaced to the electrospray source of a triple quadrupole mass spectrometer (Sciex API2000, PE Sciex). Precursor ion-scanning experiments were performed in positive-ion mode with the third quadrupole set to monitor m/z 102.3 and the first quadrupole set to scan a mass range of 170 to 700 over 9 s. The collision cell and instrument parameters were as follows: ion spray voltage of 4200 V, declustering potential of 50 V and collision energy of 25 V with nitrogen as the collision gas.
Preparation of extracts and bioassay for pC-HSL.
KLH11 and derivative strains were grown to stationary phase in MB 2216 supplemented with or without 1 mM p-coumaric acid. The cultures (acidified by 0.01 % acetic acid) were extracted twice with an equal amount of acetyl acetate and the extracts were dried in a rotary evaporator under vacuum. Each extract was concentrated 1000-fold, dissolving in 50 % (v/v) methanol. Fifty microlitres of the enriched extract was added to 1 ml of Rh. palustris CGA814 culture grown in LB. β-Galactosidase assays were performed as described previously (Zan et al., 2013).
Motility assays.
Bacterial swim assays were performed using MB 2216 with 0.25 % (w/v) agar. Plates were inoculated at the centre with freshly isolated KLH11 colonies. KLH11 crude organic extract (0.5 %, v/v) was added to MB 2216 agar. Plates were placed in an airtight container with a beaker containing 15 ml K2SO4 to maintain constant humidity, and incubated for 5–7 days at 28 °C. Photos were taken by using a Nikon D90 camera.
Results and Discussion
Identification of sscI
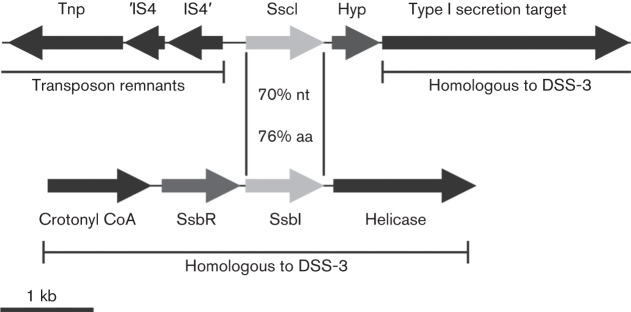
Genome sequencing of KLH11 revealed the presence of a luxI homologue, designated sscI, encoded on a large assembled sequence scaffold (>700 kb) that contains neither the ssaRI nor the ssbRI operons (Zan et al., 2011). The sscI and ssbI genes and their translation products are highly similar (70 % and 76 % nucleotide and amino acid identity, respectively) (Fig. 1), and much less similar to those of ssaI (52 % and 27 %, respectively). The sequence conservation between the ssbI and sscI genes is strikingly confined to the coding sequences, without significant similarity in their flanking regions (Fig. 1). This strongly suggests that sscI arose from a gene duplication event with ssbI. The closely related Ruegeria pomeroyi DSS-3 genome encodes ssaRI and ssbRI regions that are highly homologous and syntenic to KLH11, but does not encode an sscI gene (Moran et al., 2004). This indicates that sscI was either generated by duplication in KLH11 or, conversely, lost in DSS-3 since the time of their most recent common ancestor in the Ruegeria lineage. The region downstream of sscI in KLH11 is conserved with Ru. pomeroyi DSS-3, particularly linkage to a large gene encoding a predicted type I secretion target repeat protein (SPO2401). Upstream of sscI both genomes are chequered with several transposase and phage integrase gene remnants (Fig. 1) with a large number of frame-shift mutations, suggesting a high level of chromosomal rearrangement.
Fig. 1.
Genetic map of KLH11 sscI region and comparison with the ssbR-ssbI region. The sscI region shows homology to an analogous region of Ru. pomeroyi DSS-3 on the downstream side of the gene, and upstream of the gene there is a similar yet not identical area with several transposase and phage integrase partial gene fragments.
Solo luxR-type genes are common in bacterial genome sequences, but there are very few reported intact luxI-type solos. One exception is in another roseobacter, Dinoroseobacter shibae DFL 12T, an algal symbiont that also has two sets of luxR-luxI QS systems and one luxI solo, designated luxI3 (Wagner-Döbler et al., 2010). The genomic location of this luxI-type solo is not recognizably similar to that of sscI in KLH11 and the LuxI3 protein is not particularly similar to SsbI or SscI (~30 % identity). The luxI3 solo in D. shibae is therefore distinct from sscI in KLH11.
SscI-derived AHL production
A targeted sscI mutation using a 514 bp internal fragment of sscI and the pVIK112 suicide plasmid (Kalogeraki & Winans, 1997) was made, generating a sscI null mutant (OKC2), with a sscI-lacZ transcriptional fusion. β-Galactosidase assays of OKC2 revealed significant levels of sscI-lacZ expression that were unaffected by the addition of KLH11 whole culture extracts containing AHLs (Miller units: 189.6±20.2 and 199.8±7.9; P>0.05; unpaired Student’s t-test) that strongly activate expression of ssaI. This level of expression, although AHL-independent, was ~200-fold higher than the expression of an ssbI-lacZ fusion generated in an analogous manner with the pVIK112 plasmid (Zan et al., 2012). This difference in expression probably reflects the lack of identity in the regions immediately upstream of these coding sequences. It is worth noting that neither a Lux box nor the previously defined ssa box (Zan et al., 2012) was found in either the ssbI or sscI promoter regions (data not shown).
A triple mutant, ΔssaI ΔssbI sscI− (OKC6, with sscI disrupted using the pVIK112 derivative), was analysed for AHL production in whole-cell extracts using TLC overlaid with agar containing an A. tumefaciens AHL reporter as described by Zan et al. (2012). No AHL production was observed for this mutant (Fig. 2a). Quantitative MS, as previously described (Gould et al., 2006; Zan et al., 2012), also failed to detect AHLs in this mutant (Fig. 2b). Provision of plasmid-borne copies of each AHL synthase gene individually in this triple mutant resulted in AHL synthesis (Fig. 2a); the ssaI plasmid is weakly active and its AHLs were difficult to detect by this bioassay, but clearly detected by MS (Zan et al., 2012). MS analysis of the sscI-expressing derivative revealed high-level synthesis of several hydroxylated AHLs (Fig. 2c), consistent with our findings on its expression in E. coli (Zan et al., 2012). The high levels of AHL driven by sscI suggest that it encodes a highly active enzyme.
Fig. 2.
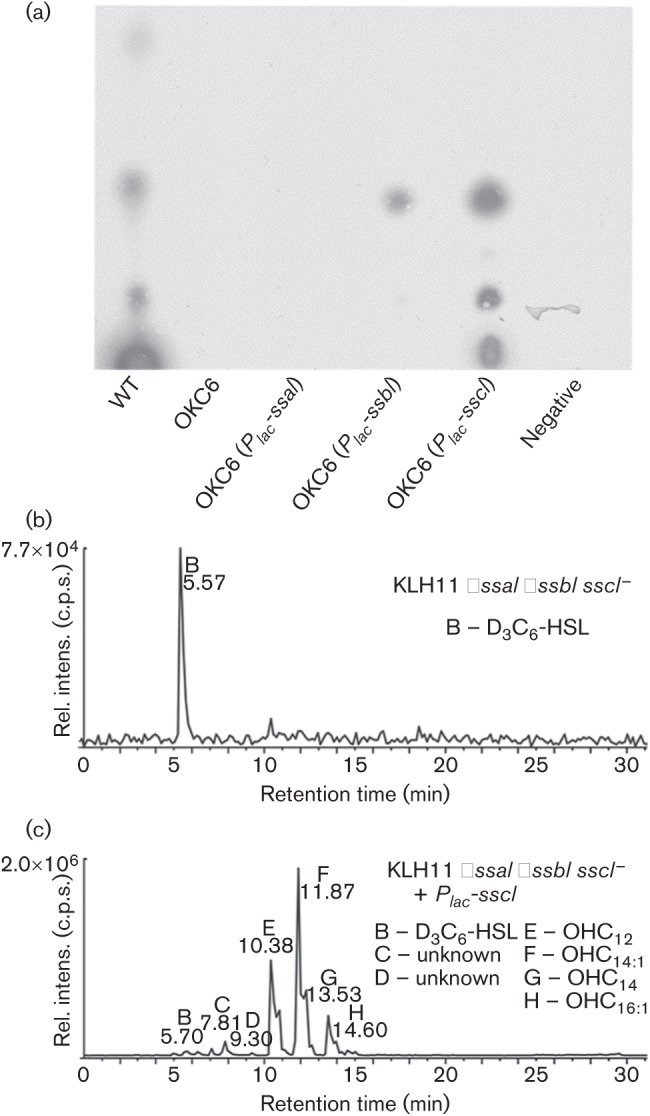
(a) Reverse-phase (RP)-TLC analysis of AHLs from KLH11 and QS mutants. TLC plate was overlaid with the A. tumefaciens ultrasensitive AHL reporter strain (Zhu et al., 2003). Organic extracts of cultures of strain OKC6 containing vector pSRKTc (Khan et al., 2008) were used as negative control. The concentration of X-Gal in the agar was 40 µg ml−1. (b, c) MS analysis of purified samples. The products of reverse-phase chromatographic separation of AHLs extracted and purified from (b) OKC6 (ΔssaI ΔssbI sscI−) and (c) OKC6 (ΔssaI ΔssbI sscI−/Plac-sscI) were examined using the precursor ion-scanning mode (transitions were monitored for precursor [M+H]+, →m/z 102.1). The peaks in the chromatograms are labelled with upper-case lettering and include the AHLs noted and as described by Zan et al. (2012).
SscI-derived AHLs are involved in ssaI expression and influence swimming motility
SsaR responds to SsaI-directed 3-oxo-HSL derivatives, and also, but more weakly, to those synthesized by SsbI (Zan et al., 2012). To test whether SsaR can also respond to SscI-derived AHLs, a plasmid-borne copy of ssaR (pEC112) or a vector control were paired with a compatible plasmid carrying the PssaI-lacZ fusion (pEC116), in an AHL−, plasmid-less derivative of A. tumefaciens NTL4. Cultures of A. tumefaciens derivatives were grown with 2.5 % (v/v) culture extracts containing SscI-derived AHLs (whole culture dichloromethane extracts from an A. tumefaciens NTL4 derivative grown with IPTG to induce expression of the Plac-sscI plasmid). Expression of ssaI increased ~fourfold in response to the extracts compared with the negative control (P<0.01) (Fig. 3a; as in prior studies, the Plac-ssaR plasmid modestly stimulates AHL-independent ssaI expression). This response of SsaR to SscI-directed AHLs adds another layer of complexity to the QS network in KLH11.
Fig. 3.
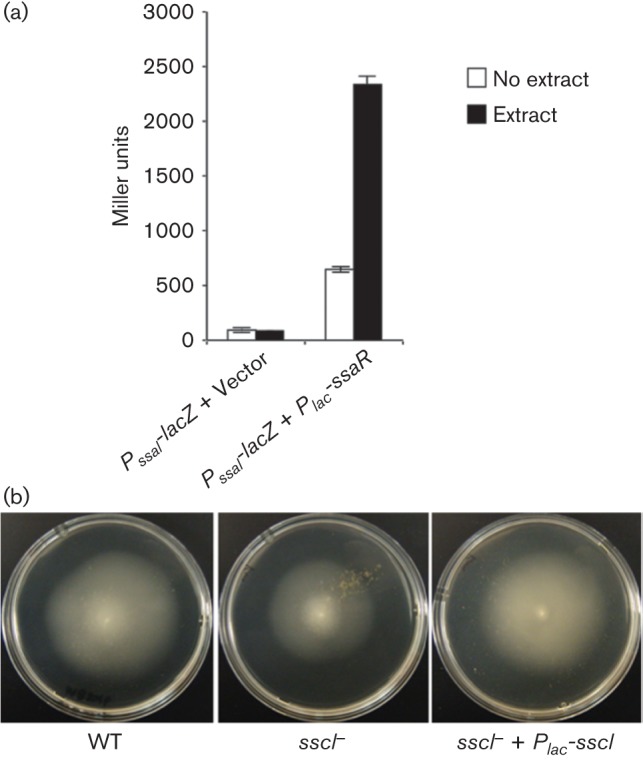
(a) SscI-derived AHLs stimulate ssaI expression and the sscI mutant is decreased for motility. Promoter fusion of ssaI with lacZ reporter (pEC116) paired with Plac-ssaR (pEC116) (Zan et al., 2012) or empty vector (pBBR1-MCS5; Kovach et al., 1995) were transformed into A. tumefaciens NTL4. β-Galactosidase assay was used to monitor the lacZ activity with and without organic extracts. Error bars represent the standard deviation from three replicates. (b) sscI is involved in motility control. WT KLH11 and derivatives were inoculated on MB 2216 (supplemented with 0.25 %, w/v, agar) swim agar plates for 1 week at 28 °C. The results shown are representatives of several independent experiments each with three biological replicates.
Flagellar motility is strictly dependent on activation by the ssaRI system through the CckA-ChpT-CtrA motility regulators (Zan et al., 2012, 2013). The sscI null mutant (OKC2) consistently showed a 20 % decrease in swim ring diameter relative to WT KLH11 (P<0.05) and plasmid-borne sscI complemented this defect (Fig. 3b). We hypothesize that this mild effect on motility is most likely due to the impact of SscI-derived AHLs on the ssaI gene expression through SsaR.
Production of pC-HSL independently from LuxI homologues
Several novel types of AHL molecules have been reported recently (Ahlgren et al., 2011; Lindemann et al., 2011; Schaefer et al., 2008). Rh. palustris produces pC-HSL, which is an arylhomoserine lactone incorporating a coumaroyl group rather than an acyl chain. Remarkably, pC-HSL is only produced in the presence of para-coumarate, a compound synthesized by plants and certain algae, and directly incorporated into the signal molecule via the LuxI-type protein RpaI. In the absence of para-coumarate, RpaI does not synthesize a product. Ru. pomeroyi DSS-3, a relative of KLH11, was also found to produce pC-HSL in cultures grown with para-coumarate (Schaefer et al., 2008). We used the pC-HSL reporter strain Rh. palustris CGA814 that cannot synthesize pC-HSL but directs RpaR-dependent expression of a target rpaI-lacZ fusion (Hirakawa et al., 2011) to examine KLH11 culture extracts. KLH11 grown in the presence of 1 mM para-coumarate can activate the expression of rpaI-lacZ to a level equivalent to 1 µM pC-HSL, suggesting the presence of pC-HSL or a structurally similar molecule. Surprisingly, a KLH11 mutant disrupted for all three luxI-type genes (ssaI, ssbI and sscI) retained this activity (Fig. 4), suggesting the existence of a novel enzyme(s) in KLH11 responsible for its synthesis.
Fig. 4.
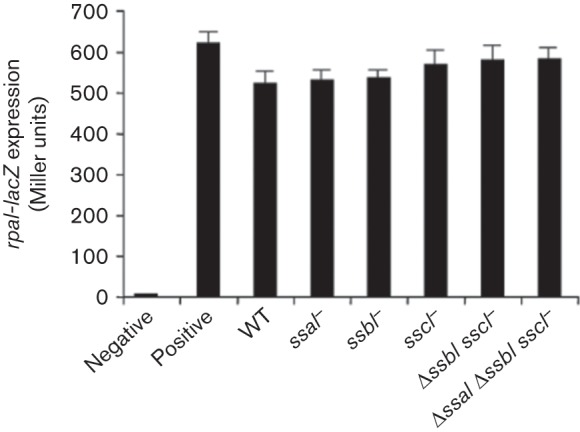
β-Galactosidase assay of the expression of an rpaI-lacZ fusion. Rh. palustris CGA814 was used as the reporter strain. Organic extracts of KLH11 strains were prepared from cultures grown in MB 2216 supplemented with 1 mM para-coumarate. An extract of MB 2216 plus para-coumarate was used as the negative control and 1 µM pC-HSL was used as the positive control. Bars represent the mean of three biological replicates and the error bars represent the sd of triplicates.
The roseobacter Phaeobacter gallaeciensis BS107 can respond to the presence of para-coumarate produced by the microalga Emiliania huxleyi potentially via pC-HSL (Seyedsayamdost et al., 2011). Novel signal molecules are synthesized by other roseobacters, including Silicibacter sp. TM1040, which does not encode luxI homologues or luxM (Cao & Meighen, 1989; Ng & Bassler, 2009), the gene encoding an alternative AHL synthase that directs the synthesis of 3-OH-C4 HSL in Vibrio harveyi and exists in several Vibrio species, but rather produces the Roseobacter Motility Inducer (RMI) that can be induced by addition of para-coumarate (Sule & Belas, 2013). Several roseobacters produce the antibiotic and novel QS molecule tropodithietic acid (TDA), which regulates its own synthesis. Our findings contribute to the emerging impression that the roseobacter group may be an underexplored and rich source of novel signalling molecules.
Acknowledgements
We are grateful to Carrie Harwood and Amy Schaefer for their assistance in analysing pC-HSL and providing the reporter strain. The MS instrumentation is supported by the University of Colorado Denver Lipid Maps Large Scale Collaborative Grant (NIH GM069338). Dr Yue Liu is thanked for her assistance in making figures. This study was supported by grants from the National Science Foundation (MCB 0703467 to C. F. and others, 0821220 to M. E. A. C. and IOS-0919728 to R. T. H). This is IMET contribution no. 14-138 and UMCES contribution no. 4972.
Abbreviations:
- AHL
acylhomoserine lactone
- Ap
ampicillin
- Gm
gentamicin
- HSL
homoserine lactone
- Km
kanamycin
- pC
para-coumaroyl
- QS
quorum sensing
- Rif
rifampicin
- Sp
spectinomycin
Edited by: M. Whiteley
References
- Ahlgren N. A., Harwood C. S., Schaefer A. L., Giraud E., Greenberg E. P. (2011). Aryl-homoserine lactone quorum sensing in stem-nodulating photosynthetic bradyrhizobia. Proc Natl Acad Sci U S A 108, 7183–7188. 10.1073/pnas.1103821108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmer B. M. (2004). Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol Microbiol 52, 933–945. 10.1111/j.1365-2958.2004.04054.x [DOI] [PubMed] [Google Scholar]
- Akakura R., Winans S. C. (2002). Mutations in the occQ operator that decrease OccR-induced DNA bending do not cause constitutive promoter activity. J Biol Chem 277, 15773–15780. 10.1074/jbc.M200109200 [DOI] [PubMed] [Google Scholar]
- Cao J. G., Meighen E. A. (1989). Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem 264, 21670–21676. [PubMed] [Google Scholar]
- Fuqua C., Greenberg E. P. (2002). Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol 3, 685–695. 10.1038/nrm907 [DOI] [PubMed] [Google Scholar]
- Gould T. A., Herman J., Krank J., Murphy R. C., Churchill M. E. (2006). Specificity of acyl-homoserine lactone synthases examined by mass spectrometry. J Bacteriol 188, 773–783. 10.1128/JB.188.2.773-783.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa H., Oda Y., Phattarasukol S., Armour C. D., Castle J. C., Raymond C. K., Lappala C. R., Schaefer A. L., Harwood C. S., Greenberg E. P. (2011). Activity of the Rhodopseudomonas palustris p-coumaroyl-homoserine lactone-responsive transcription factor RpaR. J Bacteriol 193, 2598–2607. 10.1128/JB.01479-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalogeraki V. S., Winans S. C. (1997). Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 188, 69–75. 10.1016/S0378-1119(96)00778-0 [DOI] [PubMed] [Google Scholar]
- Khan S. R., Gaines J., Roop R. M., II, Farrand S. K. (2008). Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74, 5053–5062. 10.1128/AEM.01098-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M., II, Peterson K. M. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
- Lequette Y., Lee J. H., Ledgham F., Lazdunski A., Greenberg E. P. (2006). A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J Bacteriol 188, 3365–3370. 10.1128/JB.188.9.3365-3370.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann A., Pessi G., Schaefer A. L., Mattmann M. E., Christensen Q. H., Kessler A., Hennecke H., Blackwell H. E., Greenberg E. P., Harwood C. S. (2011). Isovaleryl-homoserine lactone, an unusual branched-chain quorum-sensing signal from the soybean symbiont Bradyrhizobium japonicum. Proc Natl Acad Sci U S A 108, 16765–16770. 10.1073/pnas.1114125108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh M., Krol E., Becker A. (2008). Competitive and cooperative effects in quorum-sensing-regulated galactoglucan biosynthesis in Sinorhizobium meliloti. J Bacteriol 190, 5308–5317. 10.1128/JB.00063-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed N. M., Cicirelli E. M., Kan J., Chen F., Fuqua C., Hill R. T. (2008). Diversity and quorum-sensing signal production of Proteobacteria associated with marine sponges. Environ Microbiol 10, 75–86. [DOI] [PubMed] [Google Scholar]
- Moran M. A., Buchan A., González J. M., Heidelberg J. F., Whitman W. B., Kiene R. P., Henriksen J. R., King G. M., Belas R. & other authors (2004). Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432, 910–913. 10.1038/nature03170 [DOI] [PubMed] [Google Scholar]
- Ng W. L., Bassler B. L. (2009). Bacterial quorum-sensing network architectures. Annu Rev Genet 43, 197–222. 10.1146/annurev-genet-102108-134304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A. L., Greenberg E. P., Oliver C. M., Oda Y., Huang J. J., Bittan-Banin G., Peres C. M., Schmidt S., Juhaszova K. & other authors (2008). A new class of homoserine lactone quorum-sensing signals. Nature 454, 595–599. 10.1038/nature07088 [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost M. R., Case R. J., Kolter R., Clardy J. (2011). The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem 3, 331–335. 10.1038/nchem.1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramoni S., Venturi V. (2009). LuxR-family ‘solos’: bachelor sensors/regulators of signalling molecules. Microbiology 155, 1377–1385. 10.1099/mic.0.026849-0 [DOI] [PubMed] [Google Scholar]
- Sule P., Belas R. (2013). A novel inducer of Roseobacter motility is also a disruptor of algal symbiosis. J Bacteriol 195, 637–646. 10.1128/JB.01777-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. W., Radax R., Steger D., Wagner M. (2007). Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71, 295–347. 10.1128/MMBR.00040-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempé J., Petit A., Holsters M., Montagu M., Schell J. (1977). Thermosensitive step associated with transfer of the Ti plasmid during conjugation: possible relation to transformation in crown gall. Proc Natl Acad Sci U S A 74, 2848–2849. 10.1073/pnas.74.7.2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Döbler I., Ballhausen B., Berger M., Brinkhoff T., Buchholz I., Bunk B., Cypionka H., Daniel R., Drepper T. & other authors (2010). The complete genome sequence of the algal symbiont Dinoroseobacter shibae: a hitchhiker’s guide to life in the sea. ISME J 4, 61–77. 10.1038/ismej.2009.94 [DOI] [PubMed] [Google Scholar]
- Yao Y., Martinez-Yamout M. A., Dickerson T. J., Brogan A. P., Wright P. E., Dyson H. J. (2006). Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J Mol Biol 355, 262–273. 10.1016/j.jmb.2005.10.041 [DOI] [PubMed] [Google Scholar]
- Zan J., Fricke W. F., Fuqua C., Ravel J., Hill R. T. (2011). Genome sequence of Ruegeria sp. strain KLH11, an N-acylhomoserine lactone-producing bacterium isolated from the marine sponge Mycale laxissima. J Bacteriol 193, 5011–5012. 10.1128/JB.05556-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan J., Cicirelli E. M., Mohamed N. M., Sibhatu H., Kroll S., Choi O., Uhlson C. L., Wysoczynski C. L., Murphy R. C. & other authors (2012). A complex LuxR-LuxI type quorum sensing network in a roseobacterial marine sponge symbiont activates flagellar motility and inhibits biofilm formation. Mol Microbiol 85, 916–933. 10.1111/j.1365-2958.2012.08149.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan J., Heindl J. E., Liu Y., Fuqua C., Hill R. T. (2013). The CckA-ChpT-CtrA phosphorelay system is regulated by quorum sensing and controls flagellar motility in the marine sponge symbiont Ruegeria sp. KLH11. PLoS ONE 8, e66346. 10.1371/journal.pone.0066346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan J., Liu Y., Fuqua C., Hill R. T. (2014). Acyl-homoserine lactone quorum sensing in the Roseobacter clade. Int J Mol Sci 15, 654–669. 10.3390/ijms15010654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Beaber J. W., More M. I., Fuqua C., Eberhard A., Winans S. C. (1998). Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J Bacteriol 180, 5398–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Chai Y., Zhong Z., Li S., Winans S. C. (2003). Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl Environ Microbiol 69, 6949–6953. 10.1128/AEM.69.11.6949-6953.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]