Abstract
Rats are known as reservoirs and vectors for several zoonotic pathogens. However, information on the viruses shed by urban wild rats that could pose a zoonotic risk to human health is scare. Here, intestinal contents from 20 wild Norway rats (Rattus norvegicus) collected in the city of Berlin, Germany, were subjected to metagenomic analysis of viral nucleic acids. The determined faecal viromes of rats consisted of a variety of known and unknown viruses, and were highly variable among the individuals. Members of the families Parvoviridae and Picobirnaviridae represented the most abundant species. Novel picornaviruses, bocaviruses, sapoviruses and stool-associated circular ssDNA viruses were identified, which showed only low sequence identity to known representatives of the corresponding taxa. In addition, noroviruses and rotaviruses were detected as potential zoonotic gastroenteritis viruses. However, partial-genome sequence analyses indicated that the norovirus was closely related to the recently identified rat norovirus and the rotavirus B was closely related to the rat rotavirus strain IDIR; both viruses clustered separately from respective human virus strains in phylogenetic trees. In contrast, the rotavirus A sequences showed high identity to human and animal strains. Analysis of the nearly complete genome of this virus revealed the known genotypes G3, P[3] and N2 for three of the genome segments, whereas the remaining eight genome segments represented the novel genotypes I20–R11–C11–M10–A22–T14–E18–H13. Our results indicated a high heterogeneity of enteric viruses present in urban wild rats; their ability to be transmitted to humans remains to be assessed in the future.
Introduction
The Norway or Brown rat (Rattus norvegicus Berkenhout, 1769) is a cosmopolitan species broadly distributed throughout Europe (Amori & Cristaldi, 1999). The original distribution of this rodent is assumed to be south-east Siberia, north China and the Honod region of Japan, with spread to Europe during the 18th century (reviewed by Musser & Carleton, 2005). Where the habitat is synanthropic, it is linked to the waste and refuse water produced by humans, but Norway rats are also found in crop fields near water (Amori & Cristaldi, 1999). Due to the close proximity of rat habitats to human dwellings, humans are at risk of infection by rat-borne zoonotic pathogens. These pathogens are represented by different bacteria, e.g. Yersinia pestis, Leptospira spp. and Coxiella burnetii, and viruses, e.g. Seoul hantavirus and cowpox virus (Meerburg et al., 2009; Himsworth et al., 2013). Additional rat viruses with so far unclear zoonotic potential include herpesviruses (Ehlers et al., 2007), papillomaviruses (Schulz et al., 2012) and rat hepatitis E virus (HEV) (Johne et al., 2010). Human pathogenic agents may also be spread passively by rats after ingestion of human excretions and their passage through the intestinal tract (Wolf et al., 2013). Transmission of pathogens to humans may occur via direct contact with rats, virus spread by arthropod vectors, or inhalation and ingestion of virus-containing aerosols or excretions. Depending on the efficiency of pest control measures, contamination of human food with rat faeces may occur, which represents a risk of foodborne transmission of zoonotic pathogens.
Enteric viruses of rats were first studied as pathogens infecting breeding colonies of laboratory rats (Baker, 1998). Among them, parvoviruses, picornaviruses, reoviruses and rotaviruses were detected, which can also cause disease in laboratory rats (Baker, 1998; Jacoby et al., 1996; Doi, 2011). In contrast, the presence of viruses in the faeces of wild rats has been analysed only scarcely. Recently, next-generation sequencing (NGS) techniques have been used to analyse the viruses present in the faeces of different wild-living rodent species (Phan et al., 2011). Several members of the families Circoviridae, Picobirnaviridae, Picornaviridae, Astroviridae, Parvoviridae, Papillomaviridae, Adenoviridae and Coronaviridae have been detected by NGS, some of them representing so far unknown virus genera and species. Phan et al. (2011) focused on wild neotomine rodents, voles and house mice collected within the USA; however, studies focusing on urban wild Norway rats are so far missing.
It is conceivable that some enteric human viruses may have originally been zoonotically transmitted from animals (Brugere-Picoux & Tessier, 2010; Machnowska et al., 2014). Noroviruses, sapoviruses and rotaviruses are among the most important human gastroenteritis viruses (Hansman et al., 2007; Wiegering et al., 2011; Eckhart & Baumgart, 2011; Lee et al., 2013), and the enterically transmitted HEV is a causative agent of acute hepatitis in humans (Kamar et al., 2014). Although human noroviruses are not considered as zoonotic infections, a genogroup I norovirus closely related to human strains has been detected recently in a Norway rat (Wolf et al., 2013). Another norovirus closely related to murine noroviruses of genogroup V has been identified recently in Norway rats from Hong Kong (Tse et al., 2012). Detection of sapoviruses in rats has not been reported. A rotavirus has been identified as the causative agent of an outbreak of gastroenteritis in suckling Norway rats of a laboratory animal breeding colony (Vonderfecht et al., 1984). The virus strain [subsequently designated IDIR (infectious diarrhoea of infant rats) agent] was thereafter identified as a rotavirus B strain (Huber et al., 1989). Very recently, genome fragments of a rotavirus A have been detected simultaneously in wild Norway rats and pigs from a pig farm in Brazil, suggesting cross-species transmission of the virus (Tonietti et al., 2013). A HEV-related agent has been identified in wild Norway rats in Germany and designated rat HEV, which is only distantly related to the human HEV genotypes (Johne et al., 2010). Although the presence of rat HEV-specific antibodies has been described for a few forestry workers in eastern Germany, the zoonotic potential of rat HEV seems to be low (Dremsek et al., 2012).
The aim of the current study was to assess the presence of viruses in the faeces of wild Norway rats living in an urban environment. A total of 20 intestinal contents were derived from wild rats collected in the city of Berlin, Germany, and analysed by NGS. The identified viruses should give a first general overview on the composition of the faecal virome of Norway rats and assess their risk of transmission to humans. Therefore, a special focus of the study was the identification of viruses with a close phylogenetic relationship to human viruses.
Results
Metagenomic analysis of viruses present in urban wild rat intestinal contents
Intestinal contents of 20 rats trapped between 2010 and 2011 at two locations in the city of Berlin, Germany, were analysed by NGS (Table 1). A total of 5 762 165 reads were generated from the virus-enriched faecal samples. Out of these, 179 257 reads showed significant similarities (E-values ≤10−2) to known virus sequences present in the RefSeq virus genome database of GenBank. Sequences of bacteriophages were not studied further. The total number of reads per sample ranged from 29 923 to 910 647 and the proportion of virus-specific reads (without bacteriophages) compared with all generated reads in a sample ranged from 0.04 to 15.95 %. The viral reads were assigned to 34 known virus families and 75 virus genera. As the significance of sequence assignments to specific viruses is lower if only a few sequence reads are detected in a sample, further analysis was performed mainly with viruses that represented at least 1 % of the virus reads in a sample. The abundance of these viruses in the respective samples is shown in Table 2. In this comparison, most of the sequences could be assigned to 21 virus genera infecting mammalian hosts, whereas other virus genera infecting birds, invertebrates, insects, amoeba, fungi and plants were found at a markedly lower frequency.
Table 1. Samples of wild Norway rats (R. norvegicus) collected in the city of Berlin, Germany.
nd, Not determined.
| Sample designation | Sex | Weight (g) | Trapping location | Trapping time point (month/year) |
| Mu/10/1772 | Male | 391 | A | 05/2010 |
| Mu/10/1773 | Male | 309 | A | 05/2010 |
| Mu/10/1775 | Male | 272 | A | 05/2010 |
| Mu/10/1781 | Male | 257 | B | 04/2010 |
| Mu/10/1786 | nd | 193 | A | 04/2010 |
| Mu/10/1787 | Female | 241 | A | 04/2010 |
| Mu/10/1789 | Female | 211 | A | 04/2010 |
| Mu/10/1796 | Male | 125 | A | 04/2010 |
| Mu/10/1799 | Female | 48 | B | 06/2010 |
| Mu/10/1805 | Male | 201 | A | 04/2010 |
| KS/11/0572 | Female | 86 | B | 01/2011 |
| KS/11/0573 | Male | 128 | B | 12/2010 |
| KS/11/0576 | Male | 221 | B | 02/2011 |
| KS/11/0577 | Male | 77 | B | 01/2011 |
| KS/11/0578 | Male | 75 | B | 01/2011 |
| KS/11/0579 | Female | 75 | B | 01/2011 |
| KS/11/0580 | Female | 132 | B | 01/2011 |
| KS/11/0581 | Female | 78 | B | 02/2011 |
| KS/11/0582 | Male | 87 | B | 02/2011 |
| KS/11/0586 | Female | 211 | B | 01/2011 |
Table 2. Read numbers derived from the rat faecal samples, listed according to their similarities to virus groups or genera.
Only virus genera or groups are shown that were detected in at least 1 % of the reads of at least one sample; bacteriophages have been excluded.
| Host | Virus genus / group | Sample | Total | |||||||||||||||||||
| Mu/10/1772 | Mu/10/1773 | Mu/10/1775 | Mu/10/1781 | Mu/10/1786 | Mu/10/1787 | Mu/10/1789 | Mu/10/1796 | Mu/10/1799 | Mu/10/1805 | KS/11/0572 | KS/11/0573 | KS/11/0576 | KS/11/0577 | KS/11/0578 | KS/11/0579 | KS/11/0580 | KS/11/0581 | KS/11/0582 | KS/11/0586 | |||
| Mammalian | Protoparvovirus | 151 | 117 | 22 777 | 98 475 | 436 | 528 | 28 | 1256 | 6093 | 721 | 25 | 33 | 813 | 139 | 39 | 67 | 185 | 56 | 134 | 8946 | 14 1019 |
| Picobirnavirus | 4244 | 64 | 9 | 51 | 126 | 12 | 136 | 14 051 | 49 | 75 | 21 | 10 | 2 | 8 | 21 | 14 | 18 893 | |||||
| Bocaparvovirus | 46 | 84 | 6 | 137 | 14 | 576 | 8 | 456 | 991 | 4850 | 16 | 2080 | 11 | 5 | 2 | 5 | 4 | 140 | 9431 | |||
| Rotavirus | 1 | 9 | 1 | 32 | 8 | 2 | 61 | 5 | 2086 | 7 | 5 | 1 | 2218 | |||||||||
| SCV | 87 | 5 | 2 | 2 | 527 | 11 | 2 | 891 | 15 | 274 | 1816 | |||||||||||
| Dependoparvovirus | 42 | 13 | 93 | 26 | 1349 | 83 | 6 | 10 | 31 | 4 | 26 | 1683 | ||||||||||
| Mastadenovirus | 4 | 3 | 4 | 2 | 2 | 34 | 2 | 54 | 22 | 1 | 3 | 271 | 2 | 120 | 72 | 2 | 598 | |||||
| Circovirus-like genome | 31 | 32 | 26 | 26 | 9 | 115 | 2 | 124 | 2 | 7 | 9 | 5 | 10 | 77 | 5 | 10 | 490 | |||||
| Cardiovirus | 132 | 2 | 139 | 1 | 3 | 277 | ||||||||||||||||
| Unclassified picornavirus | 2 | 149 | 2 | 2 | 2 | 1 | 158 | |||||||||||||||
| Enterovirus | 111 | 1 | 4 | 1 | 117 | |||||||||||||||||
| Hepevirus | 8 | 3 | 65 | 76 | ||||||||||||||||||
| Mouse_rosavirus | 29 | 33 | 6 | 2 | 70 | |||||||||||||||||
| Kobuvirus | 54 | 6 | 60 | |||||||||||||||||||
| Circovirus | 3 | 2 | 21 | 2 | 3 | 2 | 23 | 1 | 2 | 59 | ||||||||||||
| Sapelovirus | 50 | 1 | 51 | |||||||||||||||||||
| Astrovirus | 3 | 8 | 2 | 13 | 2 | 2 | 9 | 39 | ||||||||||||||
| Sapovirus | 38 | 38 | ||||||||||||||||||||
| Hungarovirus | 24 | 2 | 26 | |||||||||||||||||||
| Simplexvirus | 1 | 5 | 1 | 2 | 2 | 2 | 13 | |||||||||||||||
| Norovirus | 2 | 7 | 9 | |||||||||||||||||||
| Aves | Mardivirus | 5 | 2 | 5 | 1 | 6 | 19 | |||||||||||||||
| Invertebrate | Cripavirus | 57 | 57 | |||||||||||||||||||
| Insect | Iridovirus | 4 | 16 | 1 | 1 | 1 | 3 | 5 | 6 | 3 | 2 | 1 | 1 | 1 | 45 | |||||||
| Chloriridovirus | 5 | 1 | 5 | 1 | 1 | 1 | 14 | |||||||||||||||
| Alphabaculovirus | 1 | 1 | 2 | 4 | ||||||||||||||||||
| Amoeba | Mimivirus | 40 | 86 | 1 | 4 | 38 | 12 | 11 | 35 | 17 | 2 | 10 | 10 | 23 | 1 | 38 | 328 | |||||
| Marseillevirus | 7 | 9 | 5 | 12 | 1 | 7 | 18 | 37 | 2 | 14 | 6 | 4 | 16 | 138 | ||||||||
| Fungi | Partitivirus | 45 | 2 | 5 | 9 | 61 | ||||||||||||||||
| Victorivirus | 15 | 19 | 8 | 42 | ||||||||||||||||||
| Plant | Chlorovirus | 12 | 37 | 3 | 21 | 26 | 5 | 9 | 6 | 4 | 2 | 15 | 6 | 9 | 34 | 4 | 1 | 7 | 1 | 202 | ||
| Tobamovirus | 20 | 45 | 2 | 67 | ||||||||||||||||||
| Prasinovirus | 1 | 4 | 3 | 6 | 1 | 5 | 5 | 4 | 2 | 3 | 1 | 2 | 3 | 40 | ||||||||
| Cheravirus | 40 | 40 | ||||||||||||||||||||
| Begomovirus | 2 | 2 | 4 | |||||||||||||||||||
| Phaeovirus | 1 | 2 | 3 | |||||||||||||||||||
| No. of virus reads | 4728 | 515 | 22 903 | 98 859 | 981 | 1381 | 338 | 1902 | 9226 | 20 146 | 879 | 2419 | 3263 | 1159 | 131 | 339 | 348 | 110 | 647 | 8983 | ||
Although the relative abundance of the reads of the different viruses varied between rat samples, some of the virus genera were generally more abundant overall. Overall, 98.8 % of all non-bacteriophage virus reads belonged to mammalian viruses. Within this group, the most abundant genera were Protoparvovirus, Picobirnavirus and Bocaparvovirus, followed with lower abundance by Rotavirus, stool-associated circular ssDNA viruses (SCVs), Dependoparvovirus, Mastadenovirus, circovirus-like viruses, Cardiovirus and Enterovirus (in descending order). Reads with similarities to parvoviruses represented >20 % of all detected virus reads in 14 of 20 samples and the genus Protoparvovirus was the only genus that was detected in all 20 samples. Bocavirus reads were detected in 90 % of the rat samples, whereas reads of picobirnaviruses, mastadenoviruses and circovirus-like viruses were present in 80 % of all samples. Reads of rotaviruses, SCVs and dependoviruses were detected in 50–60 % of the samples.
In order to characterize the detected viruses in more detail, similarity searches were performed with the reads. Divergent viruses showing low sequence identity with known viruses of rats as well as viruses closely related to known human enteric pathogens were selected for further studies. The procedure resulted in the analysis of picornaviruses, bocaviruses, SCVs, sapoviruses, noroviruses and rotaviruses. For these viruses, the respective reads were assembled into contigs, which were used to generate primers for [reverse transcription (RT)]-PCR amplification of genome fragments followed by Sanger sequencing.
Picornavirus
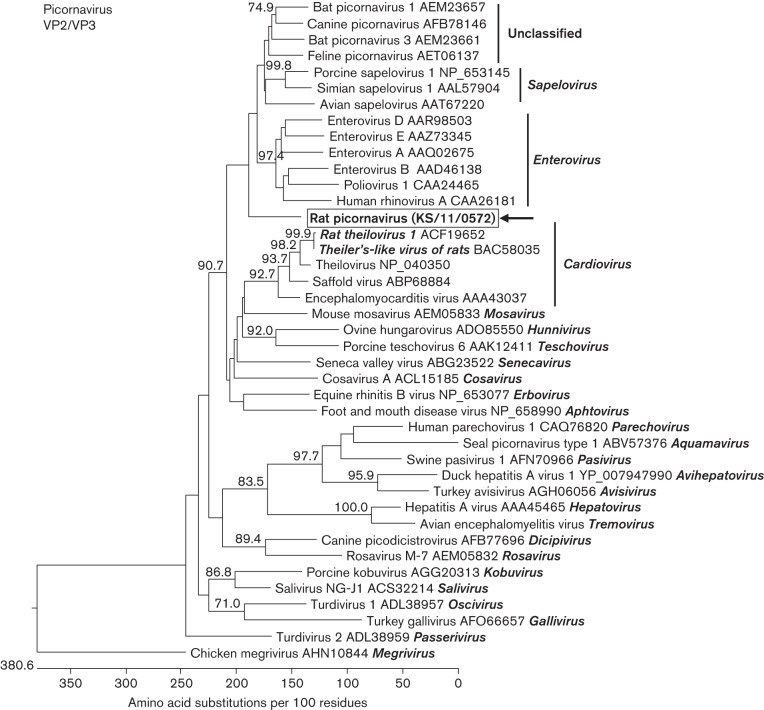
Three different contigs with low sequence similarities to the VP2, VP3 and 3B/3C genes of picornavirus genomes were identified and used for PCR primer design. Using primers with binding sites in the VP2/VP3 gene region, an RT-PCR product of 786 bp could be generated from five of the 20 samples. In contrast, primers specific for the 3B/C region amplified a 412 bp product in 12 of the 20 samples. Some of the samples were positive for both genes. A combination of the VP3-specific forward primer with the 3B/C-specific reverse primer did not amplify a 5 kb PCR product (data not shown). In a blastp search, the deduced amino acid sequence of the VP2/VP3 gene fragment of sample KS/11/0572 showed the highest similarity (46.8 %) to canine picornavirus strain 325F, an unclassified picornavirus. The deduced amino acid sequence of the 3B/C sequence of strain KS/11/0576 had the highest similarity (41.2 %) to bat picornavirus 3 strain TLC5F, also an unclassified picornavirus. A phylogenetic tree reconstrucuted using 40 picornaviruses from different genera showed clustering of the VP2/VP3 gene fragment in a separate branch close to the common root of the genera Enterovirus and Sapelovirus, and a group of unclassified picornaviruses (Fig. 1). The 3B/C fragment of strain KS/11/0576 clustered between avian and mammalian sapeloviruses, in close proximity to the roots of enteroviruses and the unclassified picornaviruses (Fig. S1, available in the online Supplementary Material). However, in both trees, the bootstrap support for the grouping was low and thus a precise phylogenetic placement of the virus was not possible. The previously described picornaviruses of rats clustered in other branches of the trees within the genus Cardiovirus, suggesting that the sequences represented a novel rat picornavirus.
Fig. 1.
Phylogenetic relationship of a picornavirus detected in the faeces of rat KS/11/0572 with other picornaviruses. The tree is based on a deduced 248 aa sequence corresponding to a fragment of the VP2/VP3-encoding region of picornaviruses. At least one representative of the different genera of the family Picornaviridae is included in the analysis (genus designation in italics); more virus strains are included for the related enteroviruses and sapeloviruses as well as for the genus Cardiovirus, which contains several viruses from rodents (rat viruses are shown in italics). GenBank accession numbers are indicated at the branches of the tree. The rat picornavirus is boxed and marked with an arrow. The tree was reconstructed using a neighbour-joining method implemented in the megalign module of dnastar with 1000 bootstrap simulations. Bootstrap values >70 % are shown.
Bocavirus
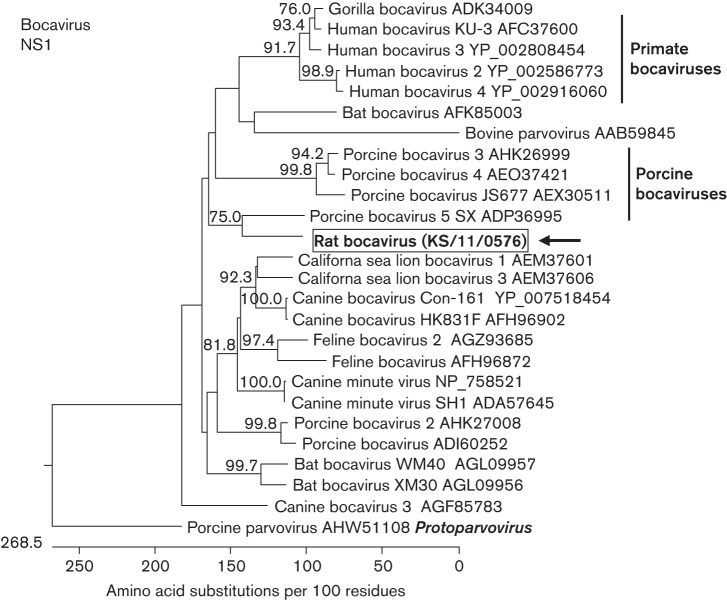
A divergent bocavirus was identified in sample KS/11/0576. Deduced amino acid sequence analysis of a 483 bp RT-PCR product of the NS1 gene showed the highest identity (49.7 %) with porcine bocavirus isolate Buk8_1 by blastp search. The presence of this rat bocavirus was confirmed by RT-PCR in 17 out of the 20 rat samples. In a phylogenetic analysis with 25 bocavirus species from different hosts, the rat bocavirus clustered together with porcine bocavirus 5 prototype SX strain (bootstrap value 75.0 %; Fig. 2).
Fig. 2.
Phylogenetic relationship of a bocavirus detected in the faeces of rat KS/11/0576 with other members of the genus Bocaparvovirus. The tree is based on a deduced 161 aa sequence corresponding to a fragment of the NS1-encoding region of bocaviruses. The host species of the bocaviruses and the GenBank accession numbers are indicated at the branches of the tree. The rat bocavirus is boxed and marked with an arrow. Porcine parvovirus from the genus Protoparvovirus is used as an outgroup. The tree was reconstructed using a neighbour-joining method implemented in the megalign module of dnastar with 1000 bootstrap simulations. Bootstrap values >70 % are shown.
SCV
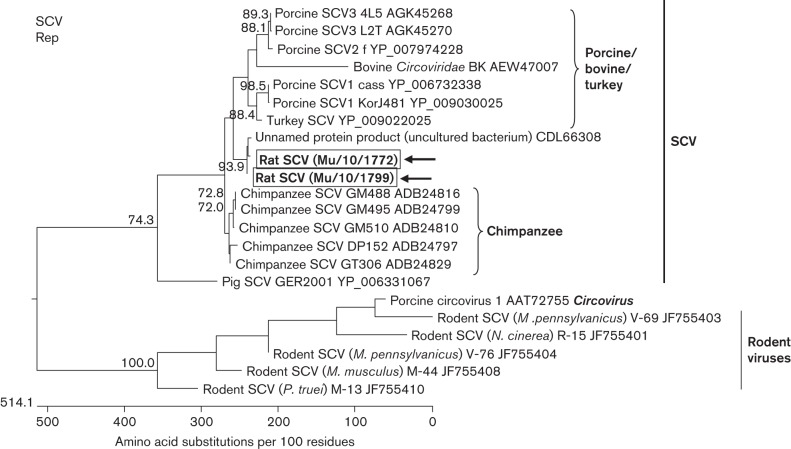
A new SCV (designated rat SCV) was identified by a contig sharing ~70 % nucleotide identity with other previously reported SCVs from different animals. Fifteen out of 20 samples were positive for the rat SCV in a specific PCR targeting 450 bp of the rep gene. Sequencing of the PCR products revealed the presence of two groups of sequences (represented by the samples Mu/10/1772 and Mu/10/1799), which differed by an insertion/deletion polymorphism of 28 nt and showed 93.6 % nucleotide sequence identity to each other. In a blastn search, the highest percentages of nucleotide sequence identity (87.4–88.1 %) showed a sequence annotated as ‘uncultured bacterium extrachromosomal DNA’ derived from rat faeces DNA amplified by rolling circle amplification (GenBank accession no. HG796401). The rat SCV also shared 65.6–68.7 % nucleotide sequence identity with chimpanzee SCVs. The deduced amino acid sequences of the PCR products showed 91.9–92.3 % identity to sequence HG796401 and 61.6–67.7 % to the Rep protein of chimpanzee SCVs. Phylogenetic analysis of related Rep sequences showed that rat SCV clustered very closely with sequence HG796401, forming a branch between chimpanzee SCVs and a group of porcine, bovine and turkey SCVs, which was supported by high bootstrap values (Fig. 3). The other known rodent Rep sequences clustered as a very diverse group in a different branch that also contained the porcine circovirus-1 (a representative of the genus Circovirus), with highest amino acid sequence identity of 46.6 % between rodent SCV V-69 and porcine circovirus-1.
Fig. 3.
Phylogenetic relationship of a SCV detected in the faeces of rats Mu/10/1772 and Mu/10/1799 with other SCVs. The tree is based on a deduced sequence of ~150 aa corresponding to a fragment of the rep-encoding region of SCVs. The host species of the SCVs and the GenBank accession numbers are indicated at the branches of the tree. The rat SCV strains are boxed and marked with arrows. The closely related sequence of an ‘uncultured bacterium’ (for details, see text), rodent circovirus-like viruses and porcine circovirus-1 from the genus Circovirus are included for comparison. The tree was reconstructed using a neighbour-joining method implemented in the megalign module of dnastar with 1000 bootstrap simulations. Bootstrap values >70 % are shown.
Sapovirus
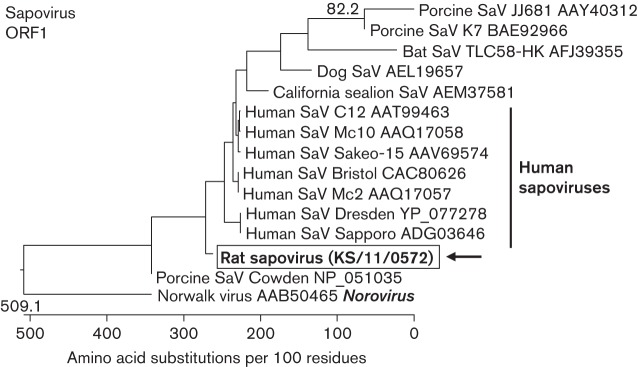
A small contig showing low sequence similarity to sapovirus genomes by blastx was used to design a specific primer pair. RT-PCR using these primers amplified a 313 bp fragment of the sapovirus ORF1 in two of the 20 rat faecal samples. The deduced amino acid sequence of the PCR product of sample KS/11/0572 showed the highest identity (53.8 %) to the human sapovirus strain SaKaeo-15 in a blastp search. As evident from the phylogenetic tree reconstructed using 14 sapoviruses of human and animal origin, the rat sapovirus branched between the porcine sapovirus strain Cowden and the group of human sapoviruses (Fig. 4). However, the tree generally has a low bootstrap support, which may have been caused by the short sequence used, and thus a precise phylogenetic placement of the new virus was not possible. Attempts to amplify larger sequence fragments of the rat sapovirus from the samples were not successful.
Fig. 4.
Phylogenetic relationship of a sapovirus detected in the faeces of rat KS/11/0572 with other members of the genus Sapovirus. The tree is based on a deduced 104 aa sequence corresponding to a fragment of the ORF1 of sapoviruses. The host species of the sapoviruses and the GenBank accession numbers are indicated at the branches of the tree. The rat sapovirus is boxed and marked with an arrow. The Norwalk virus from the genus Norovirus was used as an outgroup. The tree was constructed using a neighbour-joining method implemented in the megalign module of dnastar with 1000 bootstrap simulations. Bootstrap values >70 % are shown.
Norovirus
A total of nine sequence reads with sequence similarities to noroviruses were identified in two of the rat samples. However, using a primer pair designed on the basis of the NGS data, no norovirus-specific RT-PCR product could be amplified from the 20 rat faecal samples. From the NGS sequences of sample KS/11/0576, a 144 bp contig was assembled, which showed 79.2 % nucleotide sequence identity to the rat norovirus strain Rn/GV/HKU_KT/HKG/2012 ORF1 gene by blastn search. In a phylogenetic tree reconstructed using 14 noroviruses from humans and animals, the sequence clustered with high bootstrap support closely together with rat norovirus strains, which were most closely related to murine genotype V strains (Fig. S2).
Rotavirus B
A contig with sequence similarities to the rotavirus B VP1 gene was used to design specific PCR primers, and a 340 bp fragment could be amplified from six of the 20 rat faecal samples. The fragment derived from sample Mu/10/1805 was sequenced, which showed the highest nucleotide sequence identity of 78.6 % to the rat rotavirus B strain IDIR. A phylogenetic tree reconstructed with human, bovine and rat rotavirus B strains as well as reference strains of other rotavirus species confirmed with high bootstrap support the close relationship between the novel rat rotavirus B strain and the IDIR strain (Fig. S3).
Rotavirus A
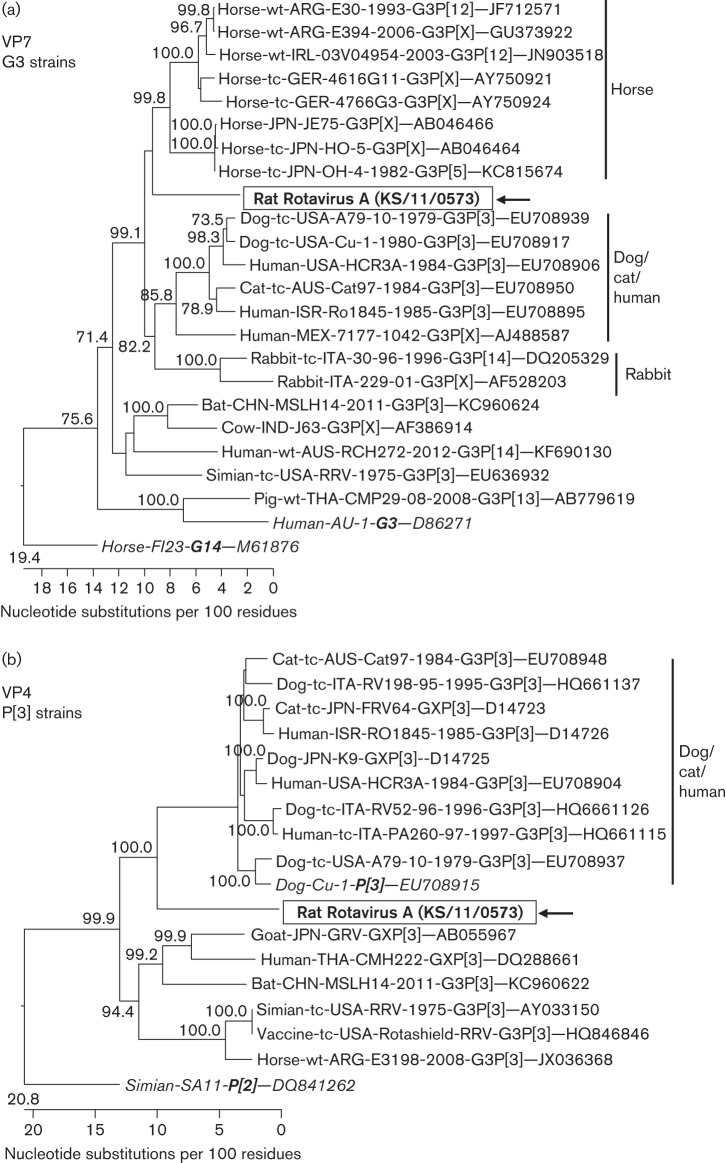
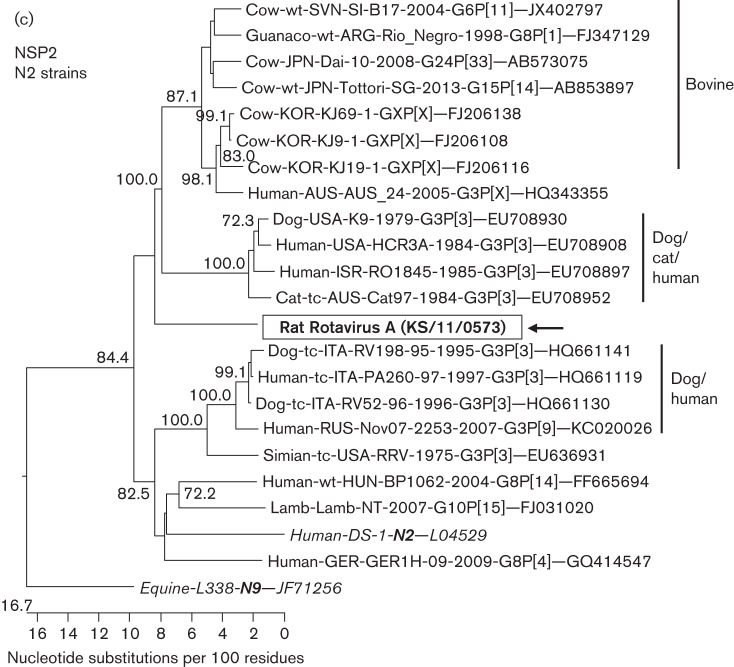
Approximately 2000 sequence reads with identities to rotavirus A were identified using blastx in rat sample KS/11/0573 and additional single rotavirus A sequences were detected in other rat samples. Using a real-time RT-PCR for detection of rotavirus A, four of the 20 rat samples turned out to be positive, whilst sample KS/11/0573 showed the lowest Ct value (not shown). As a preliminary blastn search of sequences from the VP4 and NSP2 genes derived by NGS from sample KS/11/0573 indicated a close relationship with human rotavirus A, analysis of the nearly whole genome of this rotavirus strain was attempted. Therefore, the sequences of the complete ORFs of all 11 genome segments of this strain were derived by amplification of 22 overlapping RT-PCR fragments and subsequent Sanger sequencing. A blastn search of the nucleotide sequences revealed identities between 71.2 % (NSP1 gene) and 89.6 % (VP7 gene) with known rotavirus A strains (Table 3). Whereas six of the genome segments showed the highest sequence identity with human rotaviruses, the remaining five segments were most closely related to rotaviruses from monkey, dog, horse and bat. For assignment of genotypes, nucleotide sequence identities with closely related strains of known genotypes were calculated as recommended for rotavirus A classification (Matthijnssens et al., 2008). Using the established cut-off values for definition of rotavirus A genotypes (Table 3), only the VP7, VP4 and NSP2 genes could be assigned to the known genotypes G3, P[3] and N2, respectively. The other genes showed sequence identities below the cut-offs and new genotypes were assigned in agreement with the Rotavirus Classification Working Group (RCWG). The resulting genotype constellation of the rotavirus A strain KS/11/0573 was G3–P[3]–I20–R11–C11–M10–A22–N2–T14–E18–H13. Phylogenetic trees reconstructed for the nucleotide sequences of the genome segments using all reference strains for the distinct genotypes confirmed the grouping and the assigned genotypes (Fig. S4A–K).
Table 3. Nucleotide sequence identities of rat rotavirus A strain KS/11/0573 compared with the most closely related rotavirus A strains and assignment of genotypes.
| Gene | GenBank accession no. | Most closely related genotype (strain) | Nucleotide sequence identity (%) | Cut-off for genotypes (%)* | Assigned genotype |
| VP7 | KJ879456 | G3 (RVA/Horse-wt/ARG/E30/1993/G3P[12]) | 89.6 | 80 | G3 |
| VP4 | KJ879451 | P[3] (RVA/Dog-tc/USA/CU-1/1980/G3P[3]) | 84.7 | 80 | P[3] |
| VP6 | KJ879453 | I2 (RVA/Human-wt/SEN/MRC_DPRU2053/2009/G8P[6]) | 80.6 | 85 | I20 |
| VP1 | KJ879448 | R3 (RVA/Human-wt/PRY/74IPN/2003/G12P[8]) | 81.1 | 83 | R11 |
| VP2 | KJ879449 | C3 (RVA/Human-tc/ITA/PA260-97/1997/G3P[3]) | 80.6 | 84 | C11 |
| VP3 | KJ879450 | M5 (RVA/Simian-tc/ZAF/SA11-N2/1958/G3P[2]) | 77.2 | 81 | M10 |
| NSP1 | KJ879452 | A9 (RVA/Bat-tc/CHN/MSLH14/2012/G3P[3]) | 71.2 | 79 | A22 |
| NSP2 | KJ879455 | N2 (RVA/Human-wt/AUS/AUS_24/2005/G3P[X]) | 89.4 | 85 | N2 |
| NSP3 | KJ879454 | T3 (RVA/Horse-wt/ARG/E4040/2008/G14P[12]) | 81.2 | 85 | T14 |
| NSP4 | KJ879457 | E3 (RVA/Human-wt/THA/CMH079/2005/G3P[10]) | 75.9 | 85 | E18 |
| NSP5 | KJ879458 | H1 (RVA/Human-wt/PRY/1809SR/2009/G4P[6]) | 86.9 | 91 | H13 |
According to Matthijnssens et al. (2008).
In order to identify a probable origin of the genome segments with already known genotypes, the sequences were compared with other rotavirus strains of the same genotype derived from other animal species and humans. For the VP7 genotype G3, a branching of the rat strain between a cluster of strains from horses and another cluster of strains from dogs, cats, humans and rabbits was evident in the phylogenetic tree, although with low bootstrap support (Fig. 5a). In a tree based on VP4 genotype P[3] strains, the rat strain also branched separately, between a cluster of strains from dogs, cats and humans and another cluster of strains from several mammalian hosts (high bootstrap support, Fig. 5b). In an NSP2 genotype N2-based tree, the rat strain branched out very close to the root of a cluster containing strains from dogs, cats, humans and cattle (low bootstrap support, Fig. 5c).
Fig. 5.
Phylogenetic relationship of a rotavirus A strain detected in faeces of rat KS/11/0573 with closely related rotavirus strains from other hosts. The trees are based on the complete nucleotide sequences of the ORFs encoding the VP7 gene (a), the VP4 gene (b) and the NSP2 gene (c). The host species, isolation in tissue culture (tc) or not (wt), geographical origin, strain designation, year of detection, G- and P-type, and GenBank accession numbers are indicated at the branches of the tree if available. The rat rotavirus strain is boxed and marked with an arrow. The reference strains for the respective genotypes are shown in italics. The trees were constructed using a neighbour-joining method implemented in the MEGALIGN module of DNASTAR with 1000 bootstrap simulations. Bootstrap values .70% are shown.
Discussion
Norway rats are well-known reservoirs and vectors of several human pathogenic agents (Meerburg et al., 2009; Himsworth et al., 2013). The presence of viruses in the faeces of wild rats has been only investigated scarcely so far and only by pathogen-specific (RT)-PCR amplification or virus isolation. Here, we analysed the faecal viromes of 20 wild-living Norway rats of different age and sex, captured in Berlin, Germany. Due to the high human densities in large cities, and the close proximity between rat collection sites and human facilities, direct and indirect contacts of humans with rat excretions might represent a reasonable scenario.
A large variety of viruses was found with wide differences in their prevalence. Also, the general proportion of detected viral sequences out of all sequences varied remarkably between 0.4 and 15.9 %. However, it is not known whether these observed differences are due to different virus contents in the samples or simply reflect method-associated drawbacks. NGS has been shown to be a powerful technique for analysis of the virus content in faeces (Belák et al., 2013; Xie et al., 2013; Sachsenröder et al., 2014), but the procedures are still prone to experimental variation. The efficiency of enrichment of virus particles from the faeces and their content of RT-PCR inhibitors may vary widely between samples (Sachsenröder et al., 2012; Schrader et al., 2012). More reproducible methods have been developed for comparative analyses of viruses in pig faeces (Sachsenröder et al., 2012, 2014) and should be applied in future also for faecal samples of other mammals.
The virome analyses shed light on the different virus species present in faeces of urban wild rats. The main representatives of viral reads were members of the families Parvoviridae and Picobirnaviridae, which both contain well-known viruses of rats (Jacoby et al., 1996; Fregolente et al., 2009). A number of sequences from other known and unknown viruses were also identified. Although our study focused on potentially zoonotic viruses closely related to known human viruses, some of the more divergent viruses were also analysed here. In order to exclude contig assembly errors of the NGS sequence reads, we further confirmed most of the sequences using (RT)-PCR. Problems with contig assembly from NGS data are widely known and are the subject of intensive optimization (Ji et al., 2011; Cahais et al., 2012). Another advantage of PCR analysis is the opportunity to use the assays for assessing the distribution of specific viruses among rat samples. However, both techniques cannot determine whether the detected viruses are rat viruses that replicated in the gastrointestinal tract or represent passively transmitted viruses that only passed the gastrointestinal tract. Demonstration of the viruses within rat cells or experimental infection trials with laboratory rats should be done in the future to answer this question.
Two contigs of a novel picornavirus could be detected from approximately half of the samples, both showing only low identity with known picornaviruses including rat cardioviruses (Blinkova et al., 2009), and rodent mosaviruses and rosaviruses (Phan et al., 2011, 2013b). Rat picornavirus is phylogenetically close to the root of the genera Enterovirus and Sapelovirus, suggesting it potentially represents a novel genus of the family Picornaviridae. However, characterization of the whole genome of the novel rat picornavirus is necessary in order to verify this grouping. Using whole-genome sequencing, it could also be determined whether the two generated PCR products belonged to the same virus or to two different viruses. Also, a novel sequence of a rat bocavirus could be detected in most of the samples analysed here. Bocaviruses are members of the genus Bocaparvovirus of the family Parvoviridae (Cotmore et al., 2014). They have been described to be involved in respiratory and gastrointestinal diseases of humans and animals (Manteufel & Truyen, 2008; Chow & Esper, 2009), but have not been detected in rats so far. Although the exact relationship of the virus with other bocaviruses remains to be determined by whole-genome analysis, the data presented here show only low sequence similarities with human viruses, thus indicating a low probability of zoonotic transmission to humans.
SCVs represent a lineage within a rapidly expanding group of highly divergent viruses with small rep-encoding circular DNA genomes. Most of them were detected only recently by application of rolling circle amplification methods and/or NGS. Both techniques are highly sensitive for the detection of small circular DNAs irrespective of their origin from viruses or bacteria (Johne et al., 2009; Rosario et al., 2009, 2012; Blinkova et al., 2010). The novel rat sequences clustered together with other animal SCVs; however, they were also very closely related to a sequence annotated as a plasmid of an ‘uncultured bacterium’ from rat caecum content (Jørgensen et al., 2014). A closer inspection of this sequence shows that it represents a circular DNA of 2502 nt, which contains an ORF encoding a 276 aa protein with sequence similarities to the Rep protein of SCVs and another ORF encoding a 385 aa protein with sequence similarities to the capsid protein of SCVs. It is therefore highly probable that the sequence was originally misclassified as a bacterial plasmid, but represents a complete rat SCV genome. However, a final virus classification of the currently known SCV sequences is generally pending as, to the best of our knowledge, neither virus particles nor cell-culture-isolated virus have been demonstrated for this group so far.
Several relatives of viruses that are known to regularly induce diseases of the digestive tract in humans were detected in the rats. Among them, rat HEV was detected. However, this virus was not further analysed here as the samples were already included in a detailed RT-PCR-based phylogenetic study showing that the rat HEV strains are only distantly related to the human pathogenic viruses (Johne et al., 2012). Two of the rats contained sequences with identities to sapoviruses. Although the deduced amino acid sequences showed the closest relationship with human sapoviruses, the low percentage of identity and the phylogenetic branching between human and porcine sapoviruses may indicate that the rat virus has a low potential for zoonotic transmission to humans. However, human sapoviruses comprise a rather diverse group of viruses with high sequence variability (Hansman et al., 2007). Therefore, further characterization of the rat sapovirus, which was not possible here due to its low amount within the samples, should be done in the future in order to assess the zoonotic potential of this virus. Although the very low number of virus sequences in the samples was also problematic for the detected norovirus, a close relationship to the recently discovered rat norovirus was evident (Tse et al., 2012). The rat noroviruses grouped with murine norovirus genotype V, which has not been detected in humans. Also, the rotavirus B sequence detected in six of the rats was most closely related to the rat rotavirus B strain IDIR and clustered separately from the human rotavirus B sequences. Interestingly, this is only the second description of a rotavirus B in rats since its first detection in laboratory rats in 1984 (Vonderfecht et al., 1984). Our identification of a related strain in wild urban rats argues for the continued circulation of rotavirus B strains in rats.
Sequences of a rotavirus A were detected in four of the rats, with one of them shedding a significant amount of rotavirus (80.6 % of all virus reads). The high amount of virus in this sample may indicate a replication of the rotavirus in the intestinal tract of the rat. However, inert passage of the virus through the rat’s intestinal tract after ingestion of a high amount of human or animal faeces cannot be excluded totally. The full ORF analysis of the strain identified a G3P[3] genotype for the outer capsid protein VP7 and VP4 genes. Until now, G3P[3] strains have been mainly detected in dogs and cats, but also in rabbits, humans and a rhesus monkey (Martella et al., 2010). Recently, a G3P[3] strain derived from a bat has been characterized (He et al., 2013). Therefore, a high zoonotic potential of G3P[3] strains has to be considered. However, a more detailed phylogenetic analysis of the VP7 and VP4 genes revealed a branching of the rat rotavirus in separate branches between groups of strains from dogs, cats and humans and those from other animal species. The other genome segments represent mostly novel genotypes that have never been detected in other animal species and therefore might indicate adaptation of this strain to rats. Interestingly, the strain is not closely related to rotaviruses detected in mice, which show a G16P[16] genotype (Matthijnssens et al., 2011; Tsugawa et al., 2014). Therefore, the distinct origin of the detected rat rotavirus A strain remains unclear. The only other rotavirus A strain described in rats so far had a genotype H1 for the NSP5 gene highly similar to that found in pigs of the same location, which might indicate a spillover infection in this case (Tonietti et al., 2013). Further biological characterization of rotavirus A strains from wild rats is necessary in order to assess their host specificity and risk of transmission to humans.
In summary, the faeces of wild rats in an urban environment have been shown to contain a variety of viruses. Some of these viruses, especially the detected rotavirus A strain, are closely related to human pathogenic viruses. Future investigations should focus on an increased surveillance of zoonotic viruses present in rats with close contact to humans, domestic animals or food in urban and rural habitats, and on the genetic and biological characterization of the detected viruses.
Methods
Rat samples.
Between April 2010 and February 2011, 20 wild Norway rats (R. norvegicus) were collected during pest control from two different locations separated by ~3 km in Berlin, Germany (Guenther et al., 2012; Johne et al., 2012; unpublished data). Eleven male and eight female rats were identified, whilst sex could not be determined for one animal. Further information on the animals is provided in Table 1. The whole intestine was prepared and then stored at −20 °C. The intestinal contents were derived immediately before application of the virus particle enrichment procedure from the colon and rectum after thawing the intestines.
Enrichment of virus particles.
The intestinal contents (0.5–1.0 g) were resuspended in 1 ml sterile SM buffer (50 mM Tris/HCl, 10 mM MgSO4, 0.1 M NaCl, pH 7.5). Virus particles were enriched from the homogenates essentially as described previously (Breitbart & Rohwer, 2005; Ng et al., 2012). Briefly, the resuspended faecal samples were clarified by centrifugation at 10 000 g for 20 min and filtered through 0.45 µm syringe filters (Millipore) to remove larger debris and bacteria. The resulting viral particle-enriched filtrates were then incubated with a mixture containing 0.5 U Turbo DNase I μl–1 (Ambion/Life Technologies), 0.25 U Baseline-ZERO DNase I μl–1 (Epicentre), 0.5 U Benzonase nuclease μl–1 (Novagen/Merck) and 0.25 U RNase A μl–1 (Fermentas/Fisher Scientific) at 20 °C for 90 min for degradation of non-encapsidated nucleic acids (Phan et al., 2011, 2013a).
NGS and data analysis.
DNA and RNA were extracted simultaneously from the nuclease-treated preparations using NucliSENS magnetic extraction (bioMérieux) and eluted in 50 µl volumes. RT of 5 µl nucleic acid was performed using a SuperScript III kit (Life Technologies) and primers containing a fixed sequence followed by a randomized octamer sequence at the 3′ end. Thereafter, the cDNA was denatured at 95 °C for 5 min and then cooled to 4 °C to enable reannealing of the primers. A single round of DNA synthesis used Klenow fragment DNA polymerase (New England Biolabs). To increase the amount of nucleic acids, a PCR amplification with 10 cycles was performed using a TaqMan Gold PCR kit (Life Technologies) and primers containing the fixed sequences of the primers used in RT (Victoria et al., 2009; Ng et al., 2012). Finally, the products were purified using a QIAquick Purification kit (Qiagen). The amount of DNA in each sample was quantified using KAPA Library Quantification (Kapa Biosystems) and Nextera XT libraries were prepared according to the manufacturer’s protocol for tagged samples (Illumina). The library samples were subjected to NGS using the MiSeq platform (Illumina) with 250 bp paired ends and a unique molecular tag for each sample.
The raw sequence data have been submitted to the Sequence Read Archive (SRA) at GenBank as BioProject PRJNA242707 with accession number SRP043481. Paired-end reads were generated and debarcoded using vendor software (Illumina). In-house analysis pipelines were used for adaptor trimming of the generated reads. Reads with lengths >100 bp were compared with the GenBank virus RefSeq protein database using blastx (Altschul et al., 1990, 1997), and E-values <10−2 were considered as significant. Thereafter, reads with significant similarities to viral sequences were compared with an in-house NVNR (non-virus-non-redundant) universal protein database (Phan et al., 2013a) using blastx. Reads showing lower E-values to NVNR than to the original virus sequence were excluded from further data analysis. In addition, all reads showing highest similarities to bacteriophage sequences were not analysed further. Using these results, the abundances of species were calculated using Excel 2003. The software geneious R6.1 (http://www.geneious.com/) was used for assessing reads and contigs, as well as further genome analysis of particular virus species.
(RT)-PCR detection of specific viruses.
Nucleic acids were isolated from 10 % (w/v) suspensions of the intestinal contents in PBS using the NucliSens easyMAG system (bioMérieux) according to the manufacturer’s instructions. Genome fragments of picornavirus, bocavirus, SCV, sapovirus, norovirus and rotavirus B were amplified by conventional (RT)-PCR assays using a OneStep RT-PCR kit (Qiagen). The primers used were designed based on sequence data obtained from NGS data and are listed in Table S1. The presence of rotavirus A was analysed by real-time RT-PCR according to Pang et al. (2004) using a QuantiTect Probe RT-PCR kit (Qiagen).
Sequence analysis of specific viruses.
For sequencing of the entire genome of the detected rotavirus A strain, RT-PCRs were performed with a LongRange 2Step RT-PCR kit (Qiagen) using segment-specific primers as described by Afrad et al. (2013) in a first step. Thereafter, additional primers were designed based on the NGS data or based on alignments of known rotavirus A sequences. For sequence analysis of picornavirus, bocavirus, SCV, sapovirus, norovirus and rotavirus B, the primers mentioned above were used. All successfully used primers are listed in Table S1. The obtained PCR products were analysed by gel electrophoresis and bands of the respective size were extracted using a QIAquick Gel Extraction kit (Qiagen). The purified products were either sequenced directly in an ABI3730 DNA analyser (Applied Biosystems) or first cloned using a TOPO TA Cloning kit for sequencing (Life Technologies). The accession numbers of the additional sequences used for phylogenetic analyses are indicated on the branches of the phylogenetic trees. The trees were constructed using a neighbour-joining method implemented in the megalign module of the dnastar software package (Lasergene), and bootstrap analysis with 1000 trials and 111 random seeds was performed. The sequences of the rotavirus A strain were submitted to the RCWG for genotype assignment of the respective genome segments.
Acknowledgements
The support of Matthias Stange, Mario Heising and Daniel Krämer in the trapping of rats and Kathrin Baumann, Ute Wessels, Kerstin Tauscher, Paul Dremsek and André Schütté in the dissection of rats is kindly acknowledged. The investigations in the laboratory of R. G. U. were supported by the German Center for Infection Research (DZIF). E. D., T. F. F. and X. D. and the DNA sequencing and bioinformatics analyses were supported by the Blood Systems Research Institute. This study was funded by the Deutsche Forschungsgemeinschaft through grant SFB852/1.
References
- Afrad M. H., Matthijnssens J., Moni S., Kabir F., Ashrafi A., Rahman M. Z., Faruque A. S., Azim T., Rahman M. (2013). Genetic characterization of a rare bovine-like human VP4 mono-reassortant G6P[8] rotavirus strain detected from an infant in Bangladesh. Infect Genet Evol 19, 120–126. 10.1016/j.meegid.2013.06.030 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amori G., Cristaldi M. (1999). Rattus norvegicus. In The Atlas of European Mammals, pp. 278–279. Edited by Mitchell-Jones A. J., Amori G., Bogdanowicz W., Kryštufek B., Reijnder P. J. H., Spitzenberger F., Stubbe M., Thissen J. B. M., Vohraík V., Zima J. London: T. & A. D. Poyser. [Google Scholar]
- Baker D. G. (1998). Natural pathogens of laboratory mice, rats, and rabbits and their effects on research. Clin Microbiol Rev 11, 231–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belák S., Karlsson O. E., Blomström A. L., Berg M., Granberg F. (2013). New viruses in veterinary medicine, detected by metagenomic approaches. Vet Microbiol 165, 95–101. 10.1016/j.vetmic.2013.01.022 [DOI] [PubMed] [Google Scholar]
- Blinkova O., Kapoor A., Victoria J., Jones M., Wolfe N., Naeem A., Shaukat S., Sharif S., Alam M. M. & other authors (2009). Cardioviruses are genetically diverse and cause common enteric infections in South Asian children. J Virol 83, 4631–4641. 10.1128/JVI.02085-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinkova O., Victoria J., Li Y., Keele B. F., Sanz C., Ndjango J. B., Peeters M., Travis D., Lonsdorf E. V. & other authors (2010). Novel circular DNA viruses in stool samples of wild-living chimpanzees. J Gen Virol 91, 74–86. 10.1099/vir.0.015446-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M., Rohwer F. (2005). Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques 39, 729–736. 10.2144/000112019 [DOI] [PubMed] [Google Scholar]
- Brugere-Picoux J., Tessier P. (2010). [Viral gastroenteritis in domestic animals and zoonoses]. Bull Acad Natl Med 194, 1439–1449 (in French). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahais V., Gayral P., Tsagkogeorga G., Melo-Ferreira J., Ballenghien M., Weinert L., Chiari Y., Belkhir K., Ranwez V., Galtier N. (2012). Reference-free transcriptome assembly in non-model animals from next-generation sequencing data. Mol Ecol Resour 12, 834–845. 10.1111/j.1755-0998.2012.03148.x [DOI] [PubMed] [Google Scholar]
- Chow B. D., Esper F. P. (2009). The human bocaviruses: a review and discussion of their role in infection. Clin Lab Med 29, 695–713. 10.1016/j.cll.2009.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore S. F., Agbandje-McKenna M., Chiorini J. A., Mukha D. V., Pintel D. J., Qiu J., Soderlund-Venermo M., Tattersall P., Tijssen P. & other authors (2014). The family Parvoviridae. Arch Virol 159, 1239–1247. 10.1007/s00705-013-1914-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K. (2011). Experimental encephalomyocarditis virus infection in small laboratory rodents. J Comp Pathol 144, 25–40. 10.1016/j.jcpa.2010.05.001 [DOI] [PubMed] [Google Scholar]
- Dremsek P., Wenzel J. J., Johne R., Ziller M., Hofmann J., Groschup M. H., Werdermann S., Mohn U., Dorn S. & other authors (2012). Seroprevalence study in forestry workers from eastern Germany using novel genotype 3- and rat hepatitis E virus-specific immunoglobulin G ELISAs. Med Microbiol Immunol (Berl) 201, 189–200. 10.1007/s00430-011-0221-2 [DOI] [PubMed] [Google Scholar]
- Eckardt A. J., Baumgart D. C. (2011). Viral gastroenteritis in adults. Recent Pat Antiinfect Drug Discov 6, 54–63. 10.2174/157489111794407877 [DOI] [PubMed] [Google Scholar]
- Ehlers B., Küchler J., Yasmum N., Dural G., Voigt S., Schmidt-Chanasit J., Jäkel T., Matuschka F. R., Richter D. & other authors (2007). Identification of novel rodent herpesviruses, including the first gammaherpesvirus of Mus musculus. J Virol 81, 8091–8100. 10.1128/JVI.00255-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregolente M. C., de Castro-Dias E., Martins S. S., Spilki F. R., Allegretti S. M., Gatti M. S. (2009). Molecular characterization of picobirnaviruses from new hosts. Virus Res 143, 134–136. 10.1016/j.virusres.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Guenther S., Bethe A., Fruth A., Semmler T., Ulrich R. G., Wieler L. H., Ewers C. (2012). Frequent combination of antimicrobial multiresistance and extraintestinal pathogenicity in Escherichia coli isolates from urban rats (Rattus norvegicus) in Berlin, Germany. PLoS ONE 7, e50331. 10.1371/journal.pone.0050331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman G. S., Oka T., Katayama K., Takeda N. (2007). Human sapoviruses: genetic diversity, recombination, and classification. Rev Med Virol 17, 133–141. 10.1002/rmv.533 [DOI] [PubMed] [Google Scholar]
- He B., Yang F., Yang W., Zhang Y., Feng Y., Zhou J., Xie J., Feng Y., Bao X. & other authors (2013). Characterization of a novel G3P[3] rotavirus isolated from a lesser horseshoe bat: a distant relative of feline/canine rotaviruses. J Virol 87, 12357–12366. 10.1128/JVI.02013-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himsworth C. G., Parsons K. L., Jardine C., Patrick D. M. (2013). Rats, cities, people, and pathogens: a systematic review and narrative synthesis of literature regarding the ecology of rat-associated zoonoses in urban centers. Vector Borne Zoonotic Dis 13, 349–359. 10.1089/vbz.2012.1195 [DOI] [PubMed] [Google Scholar]
- Huber A. C., Yolken R. H., Mader L. C., Strandberg J. D., Vonderfecht S. L. (1989). Pathology of infectious diarrhea of infant rats (IDIR) induced by an antigenically distinct rotavirus. Vet Pathol 26, 376–385. [DOI] [PubMed] [Google Scholar]
- Jacoby R. O., Ball-Goodrich L. J., Besselsen D. G., McKisic M. D., Riley L. K., Smith A. L. (1996). Rodent parvovirus infections. Lab Anim Sci 46, 370–380. [PubMed] [Google Scholar]
- Ji Y., Shi Y., Ding G., Li Y. (2011). A new strategy for better genome assembly from very short reads. BMC Bioinformatics 12, 493. 10.1186/1471-2105-12-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R., Müller H., Rector A., van Ranst M., Stevens H. (2009). Rolling-circle amplification of viral DNA genomes using phi29 polymerase. Trends Microbiol 17, 205–211. 10.1016/j.tim.2009.02.004 [DOI] [PubMed] [Google Scholar]
- Johne R., Heckel G., Plenge-Bönig A., Kindler E., Maresch C., Reetz J., Schielke A., Ulrich R. G. (2010). Novel hepatitis E virus genotype in Norway rats, Germany. Emerg Infect Dis 16, 1452–1455. 10.3201/eid1609.100444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R., Dremsek P., Kindler E., Schielke A., Plenge-Bönig A., Gregersen H., Wessels U., Schmidt K., Rietschel W. & other authors (2012). Rat hepatitis E virus: geographical clustering within Germany and serological detection in wild Norway rats (Rattus norvegicus). Infect Genet Evol 12, 947–956. 10.1016/j.meegid.2012.02.021 [DOI] [PubMed] [Google Scholar]
- Jørgensen T. S., Xu Z., Hansen M. A., Sørensen S. J., Hansen L. H. (2014). Hundreds of circular novel plasmids and DNA elements identified in a rat cecum metamobilome. PLoS ONE 9, e87924. 10.1371/journal.pone.0087924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamar N., Dalton H. R., Abravanel F., Izopet J. (2014). Hepatitis E virus infection. Clin Microbiol Rev 27, 116–138. 10.1128/CMR.00057-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. M., Lessler J., Lee R. A., Rudolph K. E., Reich N. G., Perl T. M., Cummings D. A. (2013). Incubation periods of viral gastroenteritis: a systematic review. BMC Infect Dis 13, 446. 10.1186/1471-2334-13-446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnowska P., Ellerbroek L., Johne R. (2014). Detection and characterization of potentially zoonotic viruses in faeces of pigs at slaughter in Germany. Vet Microbiol 168, 60–68. 10.1016/j.vetmic.2013.10.018 [DOI] [PubMed] [Google Scholar]
- Manteufel J., Truyen U. (2008). Animal bocaviruses: a brief review. Intervirology 51, 328–334. 10.1159/000173734 [DOI] [PubMed] [Google Scholar]
- Martella V., Bányai K., Matthijnssens J., Buonavoglia C., Ciarlet M. (2010). Zoonotic aspects of rotaviruses. Vet Microbiol 140, 246–255. 10.1016/j.vetmic.2009.08.028 [DOI] [PubMed] [Google Scholar]
- Matthijnssens J., Ciarlet M., Rahman M., Attoui H., Banyai K., Estes M. K., Gentsch J. R., Iturriza-Gomara M., Kirkwood C. D. & other authors (2008). Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol 153, 1621–1629. 10.1007/s00705-008-0155-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., Ciarlet M., McDonald S. M., Attoui H., Bányai K., Brister J. R., Buesa J., Esona M. D., Estes M. K. & other authors (2011). Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol 156, 1397–1413. 10.1007/s00705-011-1006-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerburg B. G., Singleton G. R., Kijlstra A. (2009). Rodent-borne diseases and their risks for public health. Crit Rev Microbiol 35, 221–270. 10.1080/10408410902989837 [DOI] [PubMed] [Google Scholar]
- Musser G. G., Carleton M. D. (2005). Rodentia: Myomorpha: Muroidea: Muridae: Murinae. In Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd edn, vol. 1, pp. 1478–1480. Edited by Wilson D. E., Reeder D. M. Baltimore, MD: The Johns Hopkins University Press. [Google Scholar]
- Ng T. F., Marine R., Wang C., Simmonds P., Kapusinszky B., Bodhidatta L., Oderinde B. S., Wommack K. E., Delwart E. (2012). High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J Virol 86, 12161–12175. 10.1128/JVI.00869-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X. L., Lee B., Boroumand N., Leblanc B., Preiksaitis J. K., Yu Ip C. C. (2004). Increased detection of rotavirus using a real time reverse transcription-polymerase chain reaction (RT-PCR) assay in stool specimens from children with diarrhea. J Med Virol 72, 496–501. 10.1002/jmv.20009 [DOI] [PubMed] [Google Scholar]
- Phan T. G., Kapusinszky B., Wang C., Rose R. K., Lipton H. L., Delwart E. L. (2011). The fecal viral flora of wild rodents. PLoS Pathog 7, e1002218. 10.1371/journal.ppat.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. G., Vo N. P., Boros Á., Pankovics P., Reuter G., Li O. T., Wang C., Deng X., Poon L. L., Delwart E. (2013a). The viruses of wild pigeon droppings. PLoS ONE 8, e72787. 10.1371/journal.pone.0072787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. G., Vo N. P., Simmonds P., Samayoa E., Naccache S., Chiu C. Y., Delwart E. (2013b). Rosavirus: the prototype of a proposed new genus of the Picornaviridae family. Virus Genes 47, 556–558. 10.1007/s11262-013-0968-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K., Duffy S., Breitbart M. (2009). Diverse circovirus-like genome architectures revealed by environmental metagenomics. J Gen Virol 90, 2418–2424. 10.1099/vir.0.012955-0 [DOI] [PubMed] [Google Scholar]
- Rosario K., Duffy S., Breitbart M. (2012). A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch Virol 157, 1851–1871. 10.1007/s00705-012-1391-y [DOI] [PubMed] [Google Scholar]
- Sachsenröder J., Twardziok S., Hammerl J. A., Janczyk P., Wrede P., Hertwig S., Johne R. (2012). Simultaneous identification of DNA and RNA viruses present in pig faeces using process-controlled deep sequencing. PLoS ONE 7, e34631. 10.1371/journal.pone.0034631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachsenröder J., Twardziok S. O., Scheuch M., Johne R. (2014). The general composition of the faecal virome of pigs depends on age, but not on feeding with a probiotic bacterium. PLoS ONE 9, e88888. 10.1371/journal.pone.0088888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C., Schielke A., Ellerbroek L., Johne R. (2012). PCR inhibitors – occurrence, properties and removal. J Appl Microbiol 113, 1014–1026. 10.1111/j.1365-2672.2012.05384.x [DOI] [PubMed] [Google Scholar]
- Schulz E., Gottschling M., Ulrich R. G., Richter D., Stockfleth E., Nindl I. (2012). Isolation of three novel rat and mouse papillomaviruses and their genomic characterization. PLoS ONE 7, e47164. 10.1371/journal.pone.0047164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonietti P. O., da Hora A. S., Silva F. D., Ferrari K. L., Brandão P. E., Richtzenhain L. J., Gregori F. (2013). Simultaneous detection of group a rotavirus in Swine and rat on a pig farm in Brazil. ScientificWorldJournal 2013, 648406. 10.1155/2013/648406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse H., Chan W. M., Lam C. S., Lau S. K., Woo P. C., Yuen K. Y. (2012). Complete genome sequences of novel rat noroviruses in Hong Kong. J Virol 86, 12435–12436. 10.1128/JVI.01976-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa T., Tatsumi M., Tsutsumi H. (2014). Virulence-associated genome mutations of murine rotavirus identified by alternating serial passages in mice and cell cultures. J Virol 88, 5543–5558. 10.1128/JVI.00041-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria J. G., Kapoor A., Li L., Blinkova O., Slikas B., Wang C., Naeem A., Zaidi S., Delwart E. (2009). Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol 83, 4642–4651. 10.1128/JVI.02301-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderfecht S. L., Huber A. C., Eiden J., Mader L. C., Yolken R. H. (1984). Infectious diarrhea of infant rats produced by a rotavirus-like agent. J Virol 52, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegering V., Kaiser J., Tappe D., Weissbrich B., Morbach H., Girschick H. J. (2011). Gastroenteritis in childhood: a retrospective study of 650 hospitalized pediatric patients. Int J Infect Dis 15, e401–e407. 10.1016/j.ijid.2011.02.006 [DOI] [PubMed] [Google Scholar]
- Wolf S., Reetz J., Johne R., Heiberg A. C., Petri S., Kanig H., Ulrich R. G. (2013). The simultaneous occurrence of human norovirus and hepatitis E virus in a Norway rat (Rattus norvegicus). Arch Virol 158, 1575–1578. 10.1007/s00705-013-1646-2 [DOI] [PubMed] [Google Scholar]
- Xie G., Yu J., Duan Z. (2013). New strategy for virus discovery: viruses identified in human feces in the last decade. Sci China Life Sci 56, 688–696. 10.1007/s11427-013-4516-y [DOI] [PMC free article] [PubMed] [Google Scholar]