Abstract
During a succession of phocine morbillivirus outbreaks spanning the past 25 years, Bordetella bronchiseptica was identified as a frequent secondary invader and cause of death. The goal of this study was to evaluate genetic diversity and the molecular basis for host specificity among seal isolates from these outbreaks. MLST and PvuII ribotyping of 54 isolates from Scottish, English or Danish coasts of the Atlantic or North Sea revealed a single, host-restricted genotype. A single, novel genotype, unique from that of the Atlantic and North Sea isolates, was found in isolates from an outbreak in the Caspian Sea. Phylogenetic analysis based either on MLST sequence, ribotype patterns or genome-wide SNPs consistently placed both seal-specific genotypes within the same major clade but indicates a distinct evolutionary history for each. An additional isolate from the intestinal tract of a seal on the south-west coast of England has a genotype otherwise found in rabbit, guinea pig and pig isolates. To investigate the molecular basis for host specificity, DNA and predicted protein sequences of virulence genes that mediate host interactions were used in comparisons between a North Sea isolate, a Caspian Sea isolate and each of their closest relatives as inferred from genome-wide SNP analysis. Despite their phylogenetic divergence, fewer nucleotide and amino acid substitutions were found in comparisons of the two seal isolates than in comparisons with closely related strains. These data indicate isolates of B. bronchiseptica associated with respiratory disease in seals comprise unique, host-adapted and highly clonal populations.
Introducton
Bordetella bronchiseptica is a widespread respiratory pathogen in a variety of laboratory, companion and farm animals and also causes disease in terrestrial and aquatic wildlife, including sea mammals. During a succession of phocine morbillivirus outbreaks with mass mortality occurring over the past 25 years, the bacterium was identified as a frequent secondary pathogen, often believed to be the cause of death, in common or harbour seals (Phoca vitulina), grey seals (Halichoerus grypus) and Caspian seals (Phoca caspica; Heje et al., 1991; Baker & Ross, 1992; Munro et al., 1992; Kuiken et al., 2006; Rijks et al., 2008).
Expression of most virulence genes of B. bronchiseptica is regulated by a two-component sensory transduction system encoded by the bvgAS locus (Beier & Gross, 2008). In response to environmental signals, the transmembrane sensor/kinase BvgS reversibly modulates gene expression through interaction with the transcriptional activator BvgA, effecting a transition between the virulent (Bvg+) phase and the avirulent (Bvg−) phase. Growth at temperatures approximating 37 °C triggers a BvgS-dependent phosphorelay cascade that phosphorylates BvgA and activates the Bvg+ phase, characterized by expression of toxins, adhesins and numerous other virulence factors. These include an adenylate cyclase-haemolysin toxin (ACT), a dermonecrotic toxin (DNT) and several adhesins, such as filamentous haemagglutinin (FHA), pertactin and fimbriae. Also among the BvgAS-inducible genes is bvgR, located immediately downstream of bvgAS, whose product represses transcription from an alternative set of BvgAS-repressed genes. During growth at ~26 °C or below, or in the presence of millimolar concentrations of MgSO4 or nicotinic acid, the phosphorelay cascade is inactive, expression of BvgAS is greatly diminished and transcription of BvgAS-induced genes ceases. A state intermediate between Bvg+ and Bvg−, referred to as Bvgi, is elicited by growth in the presence of MgSO4 or nicotinic acid at levels insufficient to fully establish the Bvg− phase (Cotter & Miller, 1997). The Bvgi phase is characterized by expression of a subset of Bvg+ genes in addition to Bvgi-specific genes such as bipA, a surface-exposed protein with similarity to intimin in Escherichia coli and invasin of Yersinia species (Stockbauer et al., 2001).
Various typing methods have been applied to B. bronchiseptica in an effort to elucidate the population structure of the bacterium. Approaches found to be highly discriminatory include MLST, based on comparison of DNA sequences from seven housekeeping genes (Diavatopoulos et al., 2005), and PvuII ribotyping (Register et al., 1997; Register & Magyar, 1999). These methods target genes whose metabolic and structural functions are critical and, therefore, under stabilizing selection, provide a sound basis from which to infer evolutionary lineages. An additional level of discrimination among bacteria of a single species can sometimes be achieved by comparing the sequences of rapidly evolving genes subject to diversifying selection, such as those encoding antigenic proteins. The resulting data may not be optimal for establishing ancestral relationships but may better reveal transmission patterns and likely sources of exposure. In B. bronchiseptica, comparison among isolates of two repeat regions of the surface-exposed adhesin and immunogen pertactin can be informative in epidemiological investigations as polymorphisms occur at a relatively high frequency (Boursaux-Eude & Guiso, 2000; Register, 2001, 2004; Parkhill et al., 2003).
A prior study in which PvuII ribotyping and restriction endonuclease analysis were used to characterize 35 B. bronchiseptica seal isolates, most obtained from the east or west Scottish coast during the 1988 phocine distemper epidemic, suggests a single, unique clone of the bacterium was circulating among affected seals (Register et al., 2000). Isolates evaluated in that study were not typed by MLST because a scheme for Bordetella had yet to be developed. Additional outbreaks of phocine distemper virus complicated by secondary infection with B. bronchiseptica occurred in the Caspian Sea in 2000 (Kuiken et al., 2006) and in Europe in 2002 (Rijks et al., 2008). The goal of the present study was to more fully capture the degree of genetic diversity among seal isolates of B. bronchiseptica representing multiple, geographically distant outbreaks over the past 25 years based on MLST, PvuII ribotyping and pertactin gene repeat region sequence. Additionally, we undertook a comparative analysis of the newly available genome sequences from one North Sea and one Caspian Sea seal isolate, together with those of six previously sequenced isolates of B. bronchiseptica from terrestrial hosts, in an effort to reveal aspects of the molecular basis for host specificity.
Methods
Bacterial isolates, growth conditions and DNA preparation.
Fifty-nine phocine isolates of B. bronchiseptica were analysed, representing the Atlantic coast of Scotland and England and the Scottish and Danish coasts of the North Sea as well as the land-locked Caspian Sea (Table 1). Identification as B. bronchiseptica was based on characteristic colony morphology and biochemical testing (Quinn et al., 2011), including catalase and oxidase tests, and the profile obtained from the API 20NE test system (bioMérieux). All isolates were confirmed as B. bronchiseptica on the basis of a positive result with a species-specific PCR that targets a region immediately upstream of the flagellin gene (Register & DeJong, 2006). Additional information related to the provenance of the isolates can be found in the Bordetella Isolates MLST database, accessible from the home page of the Bordetella PubMLST website (http://pubmlst.org/bordetella; Jolley & Maiden, 2010). Corresponding database ID numbers are 92, 93 and 326–382.
Table 1. B. bronchiseptica seal isolates included in this study.
| Year of isolation | Geographical origin | Tissue | RT | ST | prn repeat region allele | n |
| 1988–89 | Eastern Scotland | Lung | 19 | 11 | 1-4a2/2-6a1 | 17 |
| 1988–89 | Western Scotland | Lung | 19 | 11 | 1-4a2/2-6a1 | 4 |
| 1989 | Scotland, Outer Hebrides | Lung | 19 | 11 | 1-4a2/2-6a1 | 1 |
| 1988–89 | Scotland (shore unknown) | Lung | 19 | 11 | 1-4a2/2-6a1 | 13 |
| 1993 | Eastern Scotland | Lung | 19 | 11 | 1-4a2/2-6a1 | 1 |
| 1993 | Scotland (shore unknown) | Lung | 19 | 11 | 1-4a2/2-6a1 | 2 |
| 1997 | Eastern Scotland | Lung | 19 | 11 | 1-4a2/2-6a1 | 1 |
| 1999 | Scotland (shore unknown) | Lung | 19 | 11 | 1-4a2/2-6a1 | 1 |
| 2002 | Eastern Scotland | Lung | 19 | 11 | 1-4a2/2-6a1 | 2 |
| 2002 | Northern Scotland | Lung | 19 | 11 | 1-4a2/2-6a1 | 1 |
| 2002 | Western Scotland | Lung | 19 | 11 | 1-4a2/2-6a1 | 1 |
| 2003 | Eastern Scotland | Lung | 19 | 11 | 1-4a2/2-6a1 | 3 |
| 2011 | Eastern Scotland | Lung | 19 | 11 | 1-4a2/2-6a1 | 1 |
| 2012 | Western Scotland | Lung | 19 | 11 | 1-4a2/2-6a1 | 1 |
| 2002 | South-west England | Lung | 19 | 11 | 1-4a2/2-6a1 | 1 |
| 2004 | South-west England | Lung | 19 | 11 | 1-4a2/2-6a1 | 1 |
| 2006 | South-west England | Lung | 19 | 11 | 1-4a2/2-6a1 | 2 |
| 2009 | South-west England | Intestine | 1 | 14 | 1-3a1/2-6a1 | 1 |
| 1994 or prior | Denmark (shore unknown) | Unknown | 19 | 11 | 1-4a2/2-6a1 | 1 |
| 2000 | Caspian Sea (shore unknown) | Lung | 22 | 33 | 1-4a1/2-6a1 | 4 |
Bacteria were cultured at 37 °C for 18–36 h on Bordet–Gengou agar supplemented with 10 % sterile, defibrinated sheep’s blood. Colonies harvested from plates and resuspended in PBS were the source of DNA used for ribotyping and PCR. DNA was purified using a commercially available kit (Promega). DNA used for genome sequencing was purified with a High Pure PCR Template Preparation kit (Roche Diagnostics) from cultures of Stainer–Scholte broth inoculated with a single colony and incubated for ~36 h at 37 °C, with shaking (250 r.p.m.). Purified DNA was quantified with PicoGreen reagent (Life Technologies) and evaluated on 1 % agarose gels to confirm the absence of low molecular mass fragments.
Ribotyping.
PvuII ribotyping was carried out as previously described (Register & Magyar, 1999). Photographs of ribotyping gels were scanned using a flatbed scanner and fragment patterns were compared using GelCompar II software (Applied Maths). Similarity between all possible pairs of fingerprint profiles using the coefficient of Dice (Sneath & Sokal, 1973) was calculated by the cluster analysis module of the software. Dendrograms were derived from a matrix of similarity values by UPGMA (unweighted pair group method with arithmetic mean). The PvuII ribotypes (RTs) of 35 isolates included in this study were previously reported (Register et al., 2000). At the time this study was undertaken, 21 RTs had been identified for B. bronchiseptica. These include RT1–RT19, documented in prior reports (Register & Magyar, 1999; Register et al., 2000), as well as RT20 and RT21, each represented by a single isolate from a turkey, identified in an otherwise unrelated study (K. B. Register, T. L. Nicholson and B. W. Brunelle, unpublished results). The novel pattern identified here was assigned to a newly defined RT.
Sequence-based typing.
PCR amplicons used for sequencing were purified with ExoSAP-IT (USB Corporation) and sequenced at the National Animal Disease Center Genomics unit using Applied Biosystems Big Dye Terminator version 3.1 on an Applied Biosystems 3130 XL Genetic Analyzer sequencer. Vector NTI Advance (Life Technologies) was used for sequence editing and analysis. Final consensus sequences represent a minimum of threefold coverage, with at least one read from each strand.
The scheme of Diavatopoulos et al. (2005) was used for MLST analysis. MLST allele sequences and profiles were evaluated using the PubMLST Bordetella Sequence and Profile Definitions Database, available via a link on the home page of the Bordetella PubMLST website (http://pubmlst.org/bordetella/). For each isolate, amplicon sequences from the seven target genes were concatenated and used to construct a neighbour-joining tree utilizing the maximum-composite-likelihood distance estimation model with 10 000 bootstrap replicates. All positions containing gaps were eliminated from the dataset (complete deletion option). Branches supported by <50 % of bootstrap replicates were collapsed to produce polytomies. The final dataset included a total of 2924 nt positions. Phylogenetic and molecular evolutionary analyses were conducted using mega version 6.05 (Tamura et al., 2013).
Pertactin gene repeat regions were amplified by PCR, sequenced and analysed as previously reported (Register, 2004), using the nomenclature of Register (2001) to classify predicted amino acid variants. Briefly, the epithet 1-Nx, where N is an Arabic numeral indicating the number of GGXXP repeats and x is a lower-case letter used to distinguish additional unique amino acid insertions, substitutions or deletions, is used to describe region 1 variants. Similarly, region 2 variants are categorized using the epithet 2-Nx, with N representing the number of PQP repeats. An additional Arabic numeral is used to discriminate between different DNA sequence alleles with conservative substitutions that result in identical predicted amino acid sequences.
Genome sequencing and analysis.
Genomic DNA was prepared from B. bronchiseptica isolates M435/02/3 (obtained in 2002 from the lungs of a grey seal in the North Sea) and M85/00/2 (obtained in 2000 from the lungs of a Caspian Seal), following growth overnight at 37 °C in Stainer–Scholte broth, using a High Pure PCR template preparation kit (Roche Applied Science). DNA was sequenced using a combination of 3 or 5 kb mate pair and 100 bp Illumina paired-end reads with an Illumina HiSeq 2000 system. After quality trimming, all reads were used to generate assemblies with Celera assembler 7.0 (Miller et al., 2008). Underlying consensus sequences and gaps were improved using custom scripts. After improvement, there were 182 (M435/02/3) or 159 (M85/00/2) contigs with ~79× average coverage. The sequencing of these isolates was part of a larger effort to sequence the genomes of ~100 Bordetella isolates representing several species, and involving scientists at the USDA/ARS/National Animal Disease Center, The University of Pennsylvania, the University of Bristol, Bristol, UK, and the J. Craig Venter Institute. Additional details related to genome sequencing, annotation and analysis of M435/02/3 and M85/00/2 will be presented elsewhere (manuscripts in preparation).
A tree based on genome-wide SNPs was constructed as described by Park et al. (2012). Briefly, genome assemblies of B. bronchiseptica 253 (GenBank ID: HE965806.1), B. bronchiseptica MO149 (also known as D444, GenBank ID: HE965807.1), B. bronchiseptica 1289 (GenBank ID: HE983626.1), B. bronchiseptica MO211 (also known as D445, GenBank ID: HE983627.1), B. bronchiseptica R77 (also known as Bbr77 and MBORD675, GenBank: HE983628.1), M435/02/3 and M85/00/2 were each processed into 54 bp DNA reads and subsequently mapped against the genome sequence of the B. bronchiseptica reference strain RB50 (GenBank ID: BX470250.1), using SSAHA2 version 2.5.4 (Ning et al., 2001). The resulting alignment served as an input file for RAxML version 7.0.4 software (Stamatakis, 2006) to construct a genome-wide SNP-based tree as follows. The raxmlHPC performed rapid bootstrapping (10 000 bootstrap replicates) and estimated a maximum-likelihood tree using the general time-reversible model for nucleotide substitution with gamma-distributed rate heterogeneity, or GTRGAMMA model. The final genome-wide SNP-based tree was visualized in FigTree version 1.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Gene-specific comparisons among isolates included the following genes, with the function of the proteins they encode in parentheses: bvgA (two-component system transcriptional regulator), bvgS (two-component system sensor protein), bvgR (two-component system regulatory protein), cyaA (ACT), cyaB (cyclolysin secretion ATP-binding protein), cyaC (cyclolysin-activating lysine-acyltransferase), cyaD (cyclolysin secretion protein), cyaE (cyclolysin secretion protein), prn (autotransporter, adhesin; pertactin), dnt (DNT), fimA (fimbrial protein), fimB (fimbrial chaperone protein), fimC (fimbrial usher protein), fimD (fimbrial tip protein), fim2 (serotype 2 fimbrial structural protein), fim3 (serotype 3 fimbrial structural protein), fimX (fimbrial structural protein), BB3424 (fimbrial structural protein) and bipA (putative outer membrane ligand binding protein).
Results
Ribotype analysis
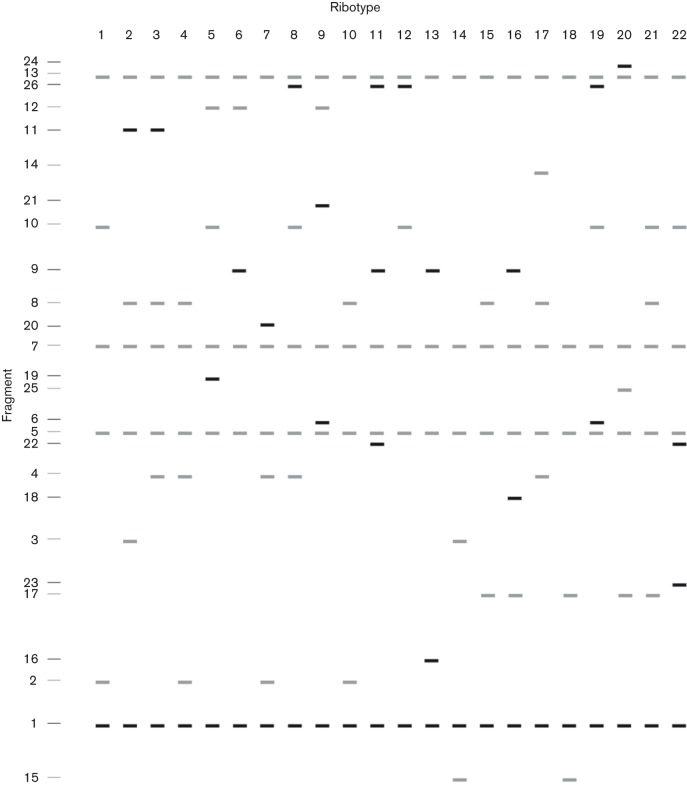
In a prior study, a single, novel RT (RT19) was found among 35 B. bronchiseptica isolates obtained between 1988 and 1997 from the lungs of seals inhabiting the shores of Scotland and Denmark (Register et al., 2000). In the current study, 19/24 newly evaluated isolates, acquired between 1988 and 2012 from pneumonic lungs of seals on the shores of south-west England or various shores of Scotland, were also found to be RT19 (Table 1). Four lung isolates from Caspian seals affected by a phocine distemper outbreak in 2000 displayed an identical but unique pattern of fragments not otherwise encountered in B. bronchiseptica, which we have designated RT22 (Fig. 1a). The remaining isolate (22/M63/4/09), acquired from the intestinal tract of a seal on the Atlantic coast of south-west England, is RT1 (Fig. 1b). A comparison of the fragment patterns from all currently known RTs is depicted in Fig. 2.
Fig. 1.
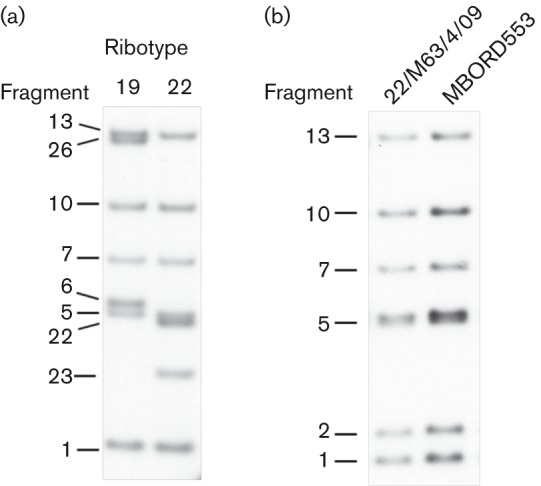
Southern blot demonstrating the PvuII RT patterns associated with phocine B. bronchiseptica isolates. (a) RTs identified from lung isolates of seals in the North Sea (RT19) and Caspian Sea (RT22). (b) RT pattern of an intestinal isolate from a North Sea seal (22/M63/4/09) and from a previously characterized swine RT1 isolate (MBORD553).
Fig. 2.
Schematic representation of currently known B. bronchiseptica PvuII RT patterns.
Analysis of the fingerprint profiles from the 22 currently defined B. bronchiseptica PvuII RTs identified five major clusters, each sharing ≥69 % similarity to other RTs within the cluster (Fig. 3a). The RTs specific for phocine lung isolates, RT19 and RT22, fall within a single cluster whose members have a minimum of 73.5 % similarity to other RTs within the group (Cluster II, Fig. 3a). However, RT22 appears to have evolved independently from RT19 and the other members of the cluster. RT1, represented by the intestinal isolate 22/M63/4/09, is also found in Cluster II and appears to have evolved from a relatively recent ancestor shared with the Atlantic and North Sea seal isolates but not with isolates from the Caspian Sea. B. bronchiseptica isolates with Cluster II RTs are relatively rare; excluding isolates RT19 and RT22, only 14/294 additional isolates evaluated, representing 11 different host species, fall within Cluster II (Register et al., 1997; Register & Magyar, 1999; Register, 2004). Others hosts from which Cluster II isolates have been acquired include rabbit, guinea pig, dog, horse and pig.
Fig. 3.
Phylogeny of B. bronchiseptica genotypes based on PvuII ribotyping (a), MLST (b) or genome-wide SNP analysis (c). (a) Dendrogram and cluster analysis of the 22 B. bronchiseptica PvuII RTs performed according to UPGMA (Tol. 1/0 %). (b) Unrooted neighbour-joining tree derived from B. bronchiseptica MLST sequences, representing 60 STs. Branches corresponding to partitions reproduced in less than 50 % of bootstrap replicates were collapsed. The percentages of replicate trees in which the associated taxa cluster together in the bootstrap test (10 000 replicates) are indicated at each node. STs identified in seal isolates are indicated by arrows. (c) Maximum-likelihood tree derived from genome-wide SNP sites of B. bronchiseptica, as compared with the reference genome of RB50. As indicated at each node, the associated taxa clustered together in the bootstrap test (10 000 replicates) in 100 % of replicate trees.
MLST analysis
Two sequence types (STs) were identified among the 58 lung isolates evaluated and there is a direct correlation between ST and RT. All lung isolates originating from seals in the Atlantic or North Sea (RT19) are ST11 while those from the Caspian Sea (RT22) are ST33. These STs appear to be associated exclusively with phocine isolates. The Bordetella pubMLST database includes no other ST11 isolates and only a single ST33 isolate, M85/00/1 (Diavatopoulos et al., 2005), which is one of the four Caspian Sea isolates also analysed in this report. The intestinal isolate 22/M63/4/09 is ST14, which has so far been found in only six other B. bronchiseptica isolates: three from rabbits, two from guinea pigs and one of unknown origin (http://pubmlst.org/bordetella/).
An MLST-based phylogeny that includes the 60 STs currently associated with B. bronchiseptica is depicted in Fig. 3(b). ST11, ST14 and ST33 are members of the same sublineage but appear no more closely related to one another than to other members of the group. Excluding seal isolates, the majority (68.1 %) of the 66 isolates representing STs most closely related to those of seal isolates, and for which the host of origin is known, were acquired from either humans, rabbits or rodents.
Comparison of host-specific diversity
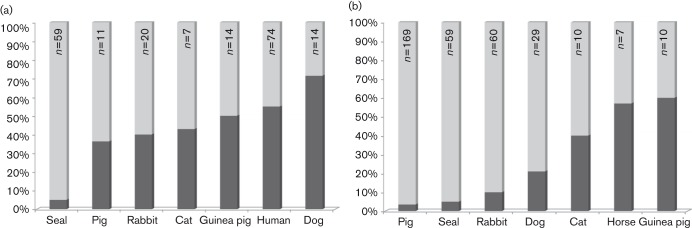
Ribotyping and MLST analyses suggest little diversity exists among B. bronchiseptica isolates from seals. To compare host-specific diversity quantitatively, we compiled the STs and RTs reported previously for other B. bronchiseptica isolates (Register et al., 1997, 2000, 2012; Register & Magyar, 1999; Diavatopoulos et al., 2005; Buboltz et al., 2008; Rath et al., 2008) or documented in the pubMLST database (http://pubmlst.org/bordetella) for all hosts represented by at least seven isolates. Most isolates included in these comparisons were acquired between 1980 and 2005, a period of time similar to that during which the seal isolates evaluated were obtained. Each host included in the ST-based comparisons is represented by isolates from at least three continents, with the exception of cats, which are represented by isolates from two continents. Hosts included in RT-based comparisons are represented by isolates originating from at least two continents each, except for pig isolates, which represent three continents. Only 5 % of seal isolates constitute unique STs, as compared with a minimum of 36.3 % for all other hosts (Fig. 4a). Diversity based on RTs is lowest for isolates from pigs, but comparable to that of seal isolates (Fig. 4b). Considering both STs and RTs, less diversity is apparent in isolates from seals than from any other host.
Fig. 4.
Host-specific diversity of B. bronchiseptica. The proportion of isolates constituting unique STs (a) or RTs (b) for the hosts indicated is represented by the darkest shaded portion of the corresponding column, with the total number of isolates available for each host indicated at the top.
Pertactin repeat region sequencing
All seal lung isolates have an identical repeat region 1 predicted amino acid sequence with four GGXXP repeats, variant 1-4a (Register, 2001), although the DNA sequences from Caspian Sea isolates have two conservative base substitutions as compared with the sequences from Atlantic and North Sea isolates. Only three predicted GGXXP repeats were found in the intestinal isolate, corresponding to the previously recognized amino acid variant 1-3a (Register, 2001), encoded by allele 1-3a1 (Table 1). The region 2 PQP predicted amino acid repeat variant 2-6a was found in all isolates and the corresponding DNA sequences are also identical to one another, representing allele 2-6a1. Table 1 lists the pertactin repeat region alleles for all isolates included in this study.
SNP analysis
Using the genome sequence of the B. bronchiseptica rabbit isolate RB50 as a reference, SNP sites were mapped to the genomes of the North Sea (M435/02/3) and Caspian Sea (M85/00/2) seal isolates as well as five additional isolates for which genome sequences were publicly available at the time we undertook this analysis, three obtained from humans and one each from a monkey and a dog (Table 2). Although the number of isolates compared is small, they collectively represent several divergent lineages of the MLST tree. The total number of SNPs per genome ranges from 8724 to 56 917, depending on the isolate. An SNP-based phylogeny suggests M435/02/3 and M85/00/2 evolved divergently, although both are found in the same major lineage as other non-human isolates (Fig. 3c). Among the strains evaluated, RB50 and M85/02/3 are predicted to share a common immediate ancestor unique from the last common ancestor of M435/00/2 and 1289.
Table 2. B. bronchiseptica isolates used for SNP analysis.
| Isolate | MLST | Host | Geographical origin | No. of SNPs vs. RB50 | Reference(s) |
| RB50 | 12 | Rabbit | USA | − | Parkhill et al. (2003) |
| M85/00/2 | 33 | Seal | Caspian Sea | 8724 | This study |
| M435/02/3 | 11 | Seal | North Sea | 9608 | This study |
| 1289 | 32 | Monkey | South America | 10 034 | Buboltz et al. (2009), Park et al. (2012) |
| 253 | 27 | Dog | USA | 34 575 | Buboltz et al. (2008), Park et al. (2012) |
| Bbr77 | 18 | Human | Germany | 52 804 | Diavatopoulos et al. (2005), Park et al. (2012) |
| MO211 | 17 | Human | USA | 53 469 | Diavatopoulos et al. (2005), Park et al. (2012) |
| MO149 | 15 | Human | USA | 56 917 | Diavatopoulos et al. (2005), Park et al. (2012) |
B. bronchiseptica has been isolated from a wide variety of animals, but many genotypes appear generally limited to particular subsets of hosts. The basis for host specificity is not well understood, although adhesins and other virulence genes whose products have a function in host interactions have been postulated to play a role. The seemingly unique, host-restricted genotypes of B. bronchiseptica associated with lung isolates from seals prompted a search for potentially host-specific virulence gene alleles based on individual comparisons of DNA sequence identity between M85/00/2, M435/02/3 and each of their closest relatives, as inferred by the genome-wide SNP tree. The 19 genes selected for analysis and the function of the proteins they encode are detailed in Methods.
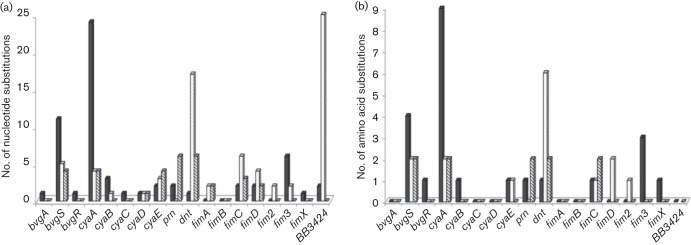
The genome-wide SNP analysis indicates that, in general, genes of M85/00/2 would be expected to share a higher level of DNA sequence identity with genes of RB50 than with genes of M435/02/3. Similarly, fewer SNPs per gene would generally be expected in a comparison of M435/02/3 and 1289, versus a comparison of the two seal isolates. However, for the majority of genes examined, the number of SNPs found when comparing the two seal isolates was either less than or no greater than the number found when comparing each seal isolate with its closest relative (Fig. 5a). Collectively, only 32 nucleotide substitutions were found in gene comparisons between M85/00/2 and M435/02/3, as opposed to 60 substitutions for M85/00/2 versus RB50 and 72 substitutions for M435/02/3 versus 1289. A similar trend was observed in a comparison of predicted protein sequences (Fig. 5b). Ten amino acid substitutions were noted between M85/00/2 and M435/02/3, compared with 23 for M85/00/2 versus RB50 and 15 for M435/02/3 versus 1289. The ORF for BB3424 from isolate 1289 is a pseudogene so was omitted from the protein comparisons.
Fig. 5.
Comparison of virulence gene polymorphisms among B. bronchiseptica seal isolates and closely related isolates from terrestrial hosts. The number of nucleotide (a) or predicted amino acid (b) substitutions in the genes evaluated is indicated for comparisons between M85/00/2 and RB50 (black), M435/02/3 and 1289 (dotted) and M85/00/2 and M435/02/3 (cross-hatched).
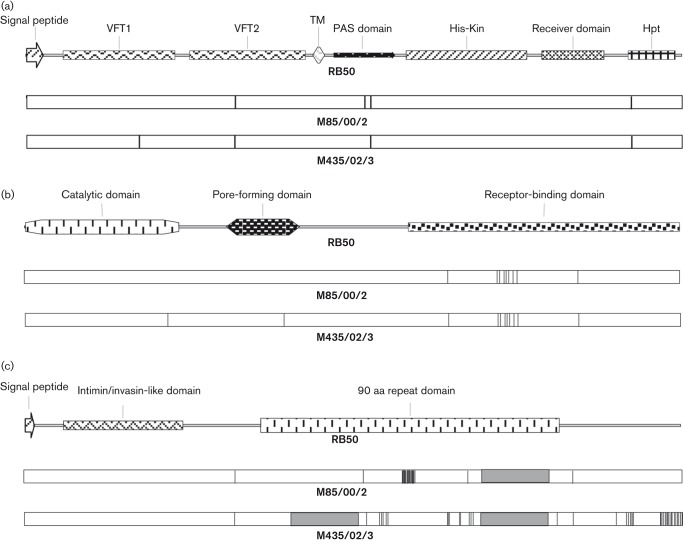
For the products of the virulence genes we evaluated, blastp queries of the NCBI non-redundant protein database revealed seal isolate-specific variants for only BvgS, ACT and DNT, with different variants of each found in M85/00/2 and M435/02/3. The five amino acid substitutions relative to RB50 predicted for the BvgS protein from seal isolates fall within functional domains involved in signal transduction or the response to virulence-modulating signals (Fig. 6a), including two in the cytoplasmic Per-ARNT-Sim (PAS) domain (Taylor & Zhulin, 1999) and one each in the VFT1 and VFT2 Venus Flytrap domains (Felder et al., 1999) and the His phosphotransfer (Hpt) domain (Uhl & Miller, 1996). For ACT, 9/11 total substitutions are clustered in the C-terminal portion of the protein involved in binding the toxin to its receptor on host cells (Vojtova et al., 2006; Fig. 6b). Only three substitutions are predicted in DNT, scattered throughout the length of the protein.
Fig. 6.
Schematic illustration comparing B. bronchiseptica virulence genes from RB50, M85/00/2 and M435/02/3. ORFs encoding BvgS (a), ACT (b) and BipA (c) are represented, with functional regions and other domains mapped to RB50, as indicated. Positions of amino acid substitutions in M85/00/2 and M435/02/3, relative to RB50, are designated with vertical lines. Grey boxes in (c) indicate the regions of RB50 bipA deleted in each seal isolate.
Of the virulence genes we examined, greatest sequence divergence was found in bipA. A tally of nucleotide and predicted amino acid substitutions is not included in Fig. 5 because of considerable variation among isolates in the size of bipA ORFs, precluding reliable comparisons based on absolute numbers. Relative to RB50, the seal isolates have either one (M85/00/2) or two (M435/02/3) in-frame, 540 bp deletions predicted to result in proteins with 180 or 360 fewer amino acids than BipA of RB50 (Fig. 6c). Both deletions fall within a repetitive region containing eight 90 aa repeats in RB50. Hence, the predicted BipA proteins of M85/00/2 and M435/02/3 are expected to contain only six and four repeats, respectively. Neither deletion is present in bipA from 1289. Amino acid identity for collinear segments of the ORF is 98.3 % for M85/00/2 versus RB50 (24 substitutions), 97.0 % for M435/02/3 versus 1289 (36 substitutions) and 95.5 % for M85/00/2 versus M435/02/3 (63 substitutions). The BipA proteins predicted for both seal isolates are unique in the NCBI non-redundant protein database.
Discussion
Results obtained in prior studies using a variety of methods suggest a considerable degree of genetic diversity exists among B. bronchiseptica isolates, including those obtained from a single host or from the same geographical region (Khattak & Matthews, 1993; Gueirard et al., 1995; Register et al., 1997; van der Zee et al., 1997; Binns et al., 1998; Keil & Fenwick, 1999; Register & Magyar, 1999; Shina et al., 2002). In contrast, ribotyping, MLST and pertactin repeat region sequence analysis reported here confirm and extend the findings of a prior investigation suggesting a highly clonal population of B. bronchiseptica is found in seals of the North Sea (Register et al., 2000). The 35 isolates previously ribotyped were obtained from 1988 to 1997. MLST analysis and pertactin repeat region sequencing of that group provide further support for a single genotype. The same apparent genotype was found among 15 additional Atlantic and North Sea lung isolates acquired between 1997 and 2012. This seems remarkable, given the nearly 25 year time span and the geographical range of the seals of origin. Isolations made from seals on the coasts of Scotland alone represent over 11 000 miles of mainland and island shoreline while the Danish isolate arose from more than 400 miles away, across open sea. A single, unique genotype was also found among isolates from the Caspian Sea, although a limited number were available for this study. The two seal-specific genotypes are distinguished not only by unique STs and RTs, but also by unique DNA sequences comprising the GGXXP region 1 repeat of pertactin, despite identical predicted amino acid sequences. The distinction between lung isolates from the Atlantic and North Sea versus those from the Caspian Sea suggests different sources of exposure for the corresponding populations of seals. However, additional isolates from the Caspian Sea should be evaluated to better determine the extent of heterogeneity within the population.
The pertactin repeat region DNA sequence representing allele 1-4a2/2-6a1, found in all Atlantic and North Sea seal lung isolates, has otherwise been reported in 22 B. bronchiseptica isolates from pigs (Register, 2004; Okada et al., 2014) and a South American monkey isolate, 1289 (Park et al., 2012). Neither the region 1 nor the region 2 sequences have been identified in any other species of Bordetella. The repeat region allele found in Caspian Sea seal isolates, 1-4a1/2-6a1, is also specific to B. bronchiseptica but the 15 isolates in which it has so far been found are of more diverse origin, including rabbits (nine isolates), humans (three isolates), guinea pig (one isolate) and two for which the source is unknown. An additional human isolate, 97-219, was reported, paradoxically, as having both the 1-4a1/2-6a1 and the 1-3a/2-6a1 alleles (Diavatopoulos et al., 2005). The region 1 variant 1-4a1 has additionally been reported for 12 human isolates of Bordetella parapertussis, but accompanied only by region 2 sequences comprising eight to ten PQP repeats (Li et al., 1991; Mäkinen et al., 2003; Diavatopoulos et al., 2005; Bouchez et al., 2011). In B. bronchiseptica, 1-4a1 has been found in association with only the six-repeat region 2 variant 2-6a1 (Boursaux-Eude & Guiso, 2000; Diavatopoulos et al., 2005; GenBank accession nos. HQ259258.1, AF411171.1, AF411172.1, AF298589.1 and AF298590.1).
Isolate 22/M63/4/09, the sole seal isolate in our study with a genotype found in other hosts (RT1/ST14), was cultured from the intestine of a dead seal recovered on the Atlantic coast of south-west England. The seal was initially found injured, subsequently treated at a rehabilitation centre and then released roughly 3 weeks before being found dead. Geographical origin seems unlikely to account for the distinct genotype of 22/M63/4/09 as four additional isolates arising from this locale are RT19/ST11. Rather, an atypical source of transmission and/or an adaptation required for survival in the digestive tract might explain this finding. RT1 is rare among the 294 B. bronchiseptica isolates so far ribotyped. Only three other RT1 isolates have been identified, one from a pig in the UK and two from guinea pigs, one each from Ireland and Switzerland (Register et al., 1997; Register & Magyar, 1999); none has been typed by MLST. Aside from 22/M63/4/09, six additional B. bronchiseptica ST14 isolates have been reported, three from rabbits, two from guinea pigs and one of unknown origin, collectively representing the USA, Ireland, Australia and South Africa (Diavatopoulos et al., 2005; Buboltz et al., 2008). The DNA sequence of the pertactin repeat region from 22/M63/4/09, allele 1-3a1/2-6a1, is similarly uncommon. With the possible exception of human isolate 97-219, discussed above, it has been identified in only two other B. bronchiseptica isolates, one from a leopard in the USA (RT6, ST unknown; Register et al., 1997) and one from an unknown host residing in South Africa (Diavatopoulos et al., 2005). The seal from which 22/M63/4/09 was cultured is not known to have had contact with any other host species while housed at the rehabilitation centre, although wild rabbits were known to be on site during that time. The possibility that the seal was infected with B. bronchiseptica from an unknown source during its stay at the rehabilitation centre cannot be ruled out.
Disease-related B. bronchiseptica infections in seals were first documented relatively recently (Heje et al., 1991). Because surveillance and, in particular, bacteriological studies of seals were largely non-existent prior to 1990, it is unknown to what extent B. bronchiseptica was present in seals before this period. Moreover, the limited data available to date were obtained from the study of diseased animals (Munro et al., 1992; Rijks et al., 2008) and thus are insufficient to determine whether the bacterium may be a constituent of the normal respiratory flora. Clinically inapparent B. bronchiseptica infection of the upper respiratory tract is known to occur in a variety of other hosts (Goodnow, 1980; Hoskins et al., 1998; Mattoo & Cherry, 2005) but additional investigation utilizing sensitive and specific methods for detection of the bacterium is necessary to establish whether it may similarly reside in healthy seals. Phylogenetic analyses presented here suggest that seal isolates are related most closely to isolates from rabbits, rodents and humans. Nevertheless, the data available are insufficient to establish whether one or more of these hosts, or perhaps some other, is the point of origin from which B. bronchiseptica was introduced into seals. Studies focused on detection of B. bronchiseptica in different seal populations and in hosts closely related to seals or in those sharing similar environments may help to more fully elucidate the population structure of the bacterium and better reveal likely sources of transmission to seals. Once present, spread of the bacterium among seal populations is generally accepted to occur via airborne transmission, with nose-to-nose transmission also occurring at haul outs, particularly at times of breeding/pupping and moulting.
The data reported here suggest there is considerably less genetic diversity among B. bronchiseptica isolates from seals than among isolates from nearly all other hosts examined, including several for which relatively few isolates are available for comparison. The geographical origins of isolates from the land-dwelling hosts used in these comparisons represent a minimum of two continents per host. Although seal isolates from the Atlantic and North Sea represent more than 15 000 miles of shoreline, it could be argued that their apparent lack of diversity is explained by the relatively limited region of the world they represent. However, both ST- and RT-based genetic variation among host-specific isolates from a limited geographical region is readily demonstrated for other hosts. For example, 12 STs are found among 22 human isolates from the USA, 14 STs are found among 23 human isolates from France and five STs are found among seven rabbit isolates from the USA (http://pubmlst.org/bordetella). Considering RTs, four RTs each are found among 12 dog isolates from the USA (Register et al., 1997) and 43 rabbit isolates from Hungary (Register & Magyar, 1999) and three RTs are found among nine rabbit isolates from Switzerland (Register et al., 1997). Rather limited variability was also found among a group of 176 swine isolates representing countries from nearly all regions of the world, including North America, Europe, Africa, Asia and Australia, isolated primarily between 1983 and 2004 (Register et al., 1997; Register & Magyar, 1999; Register, 2004; Diavatopoulos et al., 2005; http://pubmlst.org/bordetella). Multilocus electrophoresis further supports the existence in pigs of a somewhat divergent and relatively clonal population of B. bronchiseptica (Musser et al., 1987). Nonetheless, seals are the only animals for which there is convincing evidence of host-restricted genotypes of B. bronchiseptica. RTs and STs found in pigs are also frequently isolated from several other hosts (Register et al., 1997, 2012; Register & Magyar, 1999; Register, 2004; Diavatopoulos et al., 2005; Rath et al., 2008; http://pubmlst.org/bordetella) while neither RT19, RT22, ST11 nor ST33 isolates have been found in any host other than seals. Evidence at odds with the concept of seal-specific genotypes was presented by Diavatopoulos et al. (2005), who reported two North Sea seal isolates, M2936/97/3 and M861/99/1, to be ST6, a sequence type found in several other hosts. Our independent analysis of M2936/97/3 reported here indicates it is ST11 and this is consistent with the MSLT data from other North Sea seal isolates. ST6 and ST11 differ by only a single nucleotide substitution in the glyA allele. In light of our discrepant results, we repeated the MLST analysis of M2936/97/3 using a freshly grown culture and newly prepared DNA, with the same outcome. Diavatopoulos et al. (2005) indicated the remaining purported ST6 isolate, M861/99/1, was obtained from the collection of one of the authors of this report (G. F.). Because no seal isolate with that identifier exists it was not possible for us to further investigate their claim.
As expected, the phylogenies derived from MLST data and genome-wide SNP analysis are largely congruent. Both indicate that North Sea seal lung isolates (ST11) and Caspian Sea isolates (ST33) evolved divergently but share an immediate common ancestor with RB50 (ST12) and 1289 (ST32). Both additionally suggest the same ancestor is also shared with MO149 (ST15) and MO211 (ST17), although those isolates appear to have further diverged from the group in which the others are found. In general, an SNP-based phylogeny should provide higher resolution than one based on MLST given the substantially larger number of genes included in the analysis. An intriguing finding suggested by only the genome-wide SNP phylogeny is that M435/02/3 and 1289, isolated from a seal on the Scottish coast of the North Sea and a South American monkey, respectively, share an ancestor not shared by the other isolates evaluated. Whether the monkey was in captivity or had contact with other animals from which it may have acquired B. bronchiseptica is not known. Given the unlikely possibility of a common source of transmission, it could be postulated that host and environmental selective pressures drove these isolates along a somewhat convergent evolutionary path, at least from a whole-genome perspective. Yet, as discussed below, the relationship between M435/02/3 and 1289 suggested by genome-wide comparison differs from that found when comparing only virulence genes. It should further be noted that the genomes of only a few other isolates were available for comparison at the time this study was undertaken and, despite representing several divergent lineages of the MLST tree, they do not fully approximate the collective genetic diversity known to exist within the species nor the degree of diversity represented in the MSLT tree. Genome sequences from ~50 additional B. bronchiseptica isolates that have since become available provide a more robust basis for future comparisons and will probably change the topology of a related phylogeny, although M435/02/3 and 1289 may continue to share, perhaps more distantly, a common ancestor not shared by some or all other seal isolates.
Evaluation of nucleotide and predicted amino acid sequences of virulence genes from M85/00/2 and M435/02/3 revealed several homologues that share higher sequence identity with one another than with other more closely related strains, perhaps reflecting host-specific adaptations. Little is currently known about the basis for host specificity in B. bronchiseptica, although the adhesin FHA may play a role (Inatsuka et al., 2005). Due to poor sequence quality, the genes encoding FHA from M85/00/2 and M435/02/3 could not be included in our study. Among those included, unique alleles and predicted protein variants were identified for bvgS, cyaA, dnt and bipA. Nearly all deletions and substitutions relative to the RB50 reference strain occur in functional domains. The virulence gene found to be most divergent in seal isolates as compared with other closely related isolates is bipA. The function of BipA is unknown, but it is postulated to be an adhesin on the basis of sequence similarity with the bacterial adhesins intimin and invasin (Stockbauer et al., 2001). A study comparing bipA genes of B. bronchiseptica isolates from several different hosts, including the rabbit isolate RB50, found all isolates have the same allele, with eight 90 aa repeats and no nucleotide substitutions over the remaining 3′ end of the gene (Fuchslocher et al., 2003). Homologues in Bordetella pertussis and B. parapertussis also had species-specific alleles containing one to six copies of the 90 aa repeat. Because the C-terminal region of intimin and invasin interacts directly with receptors on host cells (Leong et al., 1990; Frankel et al., 1994), and the same region of BipA from RB50 is exposed on the bacterial cell surface (Stockbauer et al., 2001), it was postulated that eight 90 aa repeats may be required for BipA of B. bronchiseptica to achieve a conformation in which the C terminus is fully exposed. The novel B. bronchiseptica bipA homologues reported here, with only four to six 90 aa repeats, suggests that shorter repeat regions may not preclude exposure of the putative receptor-binding domain on the cell surface. However, Fuchslocher et al. (2003) also demonstrated that some bipA alleles are not expressed, including those with six 90 aa repeats. We have not determined whether bipA expression is detectable in either M85/00/02 or M435/02/3. A comparison of bipA expression among different alleles of B. bronchiseptica and BipA cell localization studies would help to reveal the possible effects of sequence alterations on protein synthesis and structure. Also unclear from our studies is whether the many amino acid substitutions in the C terminus predicted for BipA of M435/02/3 affects attachment or other possible functions of the protein.
In conclusion, this study reveals remarkably little genetic diversity among B. bronchiseptica isolates from seals and details some of their unique and potentially host-specific features, providing a framework for further investigation. On the basis of genome-wide comparisons, we found relatively limited divergence between isolates acquired from seals of the Atlantic or North Sea versus those from the Caspian Sea, although each has a distinct evolutionary history. The anticipated availability of genome sequence data from ~50 additional B. bronchiseptica isolates will more accurately reveal the population structure of the bacterium and the position within it of the seal genotypes described here, in addition to the possible influence of host-specific pressures on evolution and virulence-associated diversity.
Acknowledgements
Sequencing of M85/00/2 and M435/02/3 was funded, in part, by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (contract HHSN272200900007C). We gratefully acknowledge the excellent technical assistance of William Boatwright and Gwen Nordholm and the contributions of James Barnett, who conducted post-mortem evaluation and provided the clinical history of the seal that was the source of isolate 22/M63/4/09. Harry Ross, Tony Patterson, Jason Barley and Thijs Kuiken are acknowledged for provision of material from seal carcasses from which isolates were recovered. We thank Lea Ann Hobbs and David Alt at the NADC Genomics Unit for DNA sequence data derived from PCR amplicons. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
Abbreviations:
- ACT
adenylate cyclase-haemolysin toxin
- DNT
dermonecrotic toxin
- FHA
filamentous haemagglutinin
- RT
ribotype
- ST
sequence type
Edited by: P. Langford
References
- Baker J. R., Ross H. M. (1992). The role of bacteria in phocine distemper. Sci Total Environ 115, 9–14. 10.1016/0048-9697(92)90028-Q [DOI] [PubMed] [Google Scholar]
- Beier D., Gross R. (2008). The BvgS/BvgA phosphorelay system of pathogenic Bordetellae: structure, function and evolution. Adv Exp Med Biol 631, 149–160. 10.1007/978-0-387-78885-2_10 [DOI] [PubMed] [Google Scholar]
- Binns S. H., Speakman A. J., Dawson S., Bennett M., Gaskell R. M., Hart C. A. (1998). The use of pulsed-field gel electrophoresis to examine the epidemiology of Bordetella bronchiseptica isolated from cats and other species. Epidemiol Infect 120, 201–208. 10.1017/S0950268897008613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez V., Brun D., Dore G., Njamkepo E., Guiso N. (2011). Bordetella parapertussis isolates not expressing pertactin circulating in France. Clin Microbiol Infect 17, 675–682. 10.1111/j.1469-0691.2010.03303.x [DOI] [PubMed] [Google Scholar]
- Boursaux-Eude C., Guiso N. (2000). Polymorphism of repeated regions of pertactin in Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica. Infect Immun 68, 4815–4817. 10.1128/IAI.68.8.4815-4817.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buboltz A. M., Nicholson T. L., Parette M. R., Hester S. E., Parkhill J., Harvill E. T. (2008). Replacement of adenylate cyclase toxin in a lineage of Bordetella bronchiseptica. J Bacteriol 190, 5502–5511. 10.1128/JB.00226-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buboltz A. M., Nicholson T. L., Weyrich L. S., Harvill E. T. (2009). Role of the type III secretion system in a hypervirulent lineage of Bordetella bronchiseptica. Infect Immun 77, 3969–3977. 10.1128/IAI.01362-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P. A., Miller J. F. (1997). A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol Microbiol 24, 671–685. 10.1046/j.1365-2958.1997.3821741.x [DOI] [PubMed] [Google Scholar]
- Diavatopoulos D. A., Cummings C. A., Schouls L. M., Brinig M. M., Relman D. A., Mooi F. R. (2005). Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog 1, e45. 10.1371/journal.ppat.0010045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder C. B., Graul R. C., Lee A. Y., Merkle H. P., Sadee W. (1999). The Venus flytrap of periplasmic binding proteins: an ancient protein module present in multiple drug receptors. AAPS PharmSci 1, 7–26. 10.1208/ps010202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel G., Candy D. C., Everest P., Dougan G. (1994). Characterization of the C-terminal domains of intimin-like proteins of enteropathogenic and enterohemorrhagic Escherichia coli, Citrobacter freundii, and Hafnia alvei. Infect Immun 62, 1835–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchslocher B., Millar L. L., Cotter P. A. (2003). Comparison of bipA alleles within and across Bordetella species. Infect Immun 71, 3043–3052. 10.1128/IAI.71.6.3043-3052.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnow R. A. (1980). Biology of Bordetella bronchiseptica. Microbiol Rev 44, 722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueirard P., Weber C., Le Coustumier A., Guiso N. (1995). Human Bordetella bronchiseptica infection related to contact with infected animals: persistence of bacteria in host. J Clin Microbiol 33, 2002–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heje N. I., Henriksen P., Aalbaek B. (1991). The seal death in Danish waters 1988. 1. Pathological and bacteriological studies. Acta Vet Scand 32, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins J. D., Williams J., Roy A. F., Peters J. C., McDonough P. (1998). Isolation and characterization of Bordetella bronchiseptica from cats in southern Louisiana. Vet Immunol Immunopathol 65, 173–176. 10.1016/S0165-2427(98)00152-4 [DOI] [PubMed] [Google Scholar]
- Inatsuka C. S., Julio S. M., Cotter P. A. (2005). Bordetella filamentous hemagglutinin plays a critical role in immunomodulation, suggesting a mechanism for host specificity. Proc Natl Acad Sci U S A 102, 18578–18583. 10.1073/pnas.0507910102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley K. A., Maiden M. C. (2010). BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil D. J., Fenwick B. (1999). Evaluation of canine Bordetella bronchiseptica isolates using randomly amplified polymorphic DNA fingerprinting and ribotyping. Vet Microbiol 66, 41–51. 10.1016/S0378-1135(98)00306-X [DOI] [PubMed] [Google Scholar]
- Khattak M. N., Matthews R. C. (1993). Genetic relatedness of Bordetella species as determined by macrorestriction digests resolved by pulsed-field gel electrophoresis. Int J Syst Bacteriol 43, 659–664. 10.1099/00207713-43-4-659 [DOI] [PubMed] [Google Scholar]
- Kuiken T., Kennedy S., Barrett T., Van de Bildt M. W., Borgsteede F. H., Brew S. D., Codd G. A., Duck C., Deaville R. & other authors (2006). The 2000 canine distemper epidemic in Caspian seals (Phoca caspica): pathology and analysis of contributory factors. Vet Pathol 43, 321–338. 10.1354/vp.43-3-321 [DOI] [PubMed] [Google Scholar]
- Leong J. M., Fournier R. S., Isberg R. R. (1990). Identification of the integrin binding domain of the Yersinia pseudotuberculosis invasin protein. EMBO J 9, 1979–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. J., Dougan G., Novotny P., Charles I. G. (1991). P.70 pertactin, an outer-membrane protein from Bordetella parapertussis: cloning, nucleotide sequence and surface expression in Escherichia coli. Mol Microbiol 5, 409–417. 10.1111/j.1365-2958.1991.tb02123.x [DOI] [PubMed] [Google Scholar]
- Mäkinen J., Mertsola J., Soini H., Arvilommi H., Viljanen M. K., Guiso N., He Q. (2003). PFGE and pertactin gene sequencing suggest limited genetic variability within the Finnish Bordetella parapertussis population. J Med Microbiol 52, 1059–1063. 10.1099/jmm.0.05434-0 [DOI] [PubMed] [Google Scholar]
- Mattoo S., Cherry J. D. (2005). Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 18, 326–382. 10.1128/CMR.18.2.326-382.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. R., Delcher A. L., Koren S., Venter E., Walenz B. P., Brownley A., Johnson J., Li K., Mobarry C., Sutton G. (2008). Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 24, 2818–2824. 10.1093/bioinformatics/btn548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro R., Ross H., Cornwell C., Gilmour J. (1992). Disease conditions affecting common seals (Phoca vitulina) around the Scottish mainland, September–November 1988. Sci Total Environ 115, 67–82. 10.1016/0048-9697(92)90033-O [DOI] [PubMed] [Google Scholar]
- Musser J. M., Bemis D. A., Ishikawa H., Selander R. K. (1987). Clonal diversity and host distribution in Bordetella bronchiseptica. J Bacteriol 169, 2793–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z., Cox A. J., Mullikin J. C. (2001). SSAHA: a fast search method for large DNA databases. Genome Res 11, 1725–1729. 10.1101/gr.194201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Ogura Y., Hayashi T., Abe A., Kuwae A., Horiguchi Y., Abe H. (2014). Complete genome sequence of Bordetella bronchiseptica S798, an isolate from a pig with atrophic rhinitis. Genome Announc 2, e00436-14. 10.1128/genomeA.00436-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Zhang Y., Buboltz A. M., Zhang X., Schuster S. C., Ahuja U., Liu M., Miller J. F., Sebaihia M. & other authors (2012). Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics 13, 545. 10.1186/1471-2164-13-545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J., Sebaihia M., Preston A., Murphy L. D., Thomson N., Harris D. E., Holden M. T., Churcher C. M., Bentley S. D. & other authors (2003). Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 35, 32–40. 10.1038/ng1227 [DOI] [PubMed] [Google Scholar]
- Quinn P., Markey B., Leonard F., FitzPatrick E., Fanning S., Hartigan P. (2011). Bordetella species. In Veterinary Microbiology and Microbial Disease, pp. 325–329. Chichester: Wiley-Blackwell. [Google Scholar]
- Rath B. A., Register K. B., Wall J., Sokol D. M., Van Dyke R. B. (2008). Persistent Bordetella bronchiseptica pneumonia in an immunocompetent infant and genetic comparison of clinical isolates with kennel cough vaccine strains. Clin Infect Dis 46, 905–908. 10.1086/528858 [DOI] [PubMed] [Google Scholar]
- Register K. B. (2001). Novel genetic and phenotypic heterogeneity in Bordetella bronchiseptica pertactin. Infect Immun 69, 1917–1921. 10.1128/IAI.69.3.1917-1921.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Register K. B. (2004). Comparative sequence analysis of Bordetella bronchiseptica pertactin gene (prn) repeat region variants in swine vaccines and field isolates. Vaccine 23, 48–57. 10.1016/j.vaccine.2004.07.020 [DOI] [PubMed] [Google Scholar]
- Register K. B., DeJong K. D. (2006). Analytical verification of a multiplex PCR for identification of Bordetella bronchiseptica and Pasteurella multocida from swine. Vet Microbiol 117, 201–210. 10.1016/j.vetmic.2006.05.003 [DOI] [PubMed] [Google Scholar]
- Register K. B., Magyar T. (1999). Optimized ribotyping protocol applied to Hungarian Bordetella bronchiseptica isolates: identification of two novel ribotypes. Vet Microbiol 69, 277–285. 10.1016/S0378-1135(99)00118-2 [DOI] [PubMed] [Google Scholar]
- Register K. B., Boisvert A., Ackermann M. R. (1997). Use of ribotyping to distinguish Bordetella bronchiseptica isolates. Int J Syst Bacteriol 47, 678–683. 10.1099/00207713-47-3-678 [DOI] [PubMed] [Google Scholar]
- Register K. B., Sacco R. E., Foster G. (2000). Ribotyping and restriction endonuclease analysis reveal a novel clone of Bordetella bronchiseptica in seals. J Vet Diagn Invest 12, 535–540. 10.1177/104063870001200607 [DOI] [PubMed] [Google Scholar]
- Register K. B., Sukumar N., Palavecino E. L., Rubin B. K., Deora R. (2012). Bordetella bronchiseptica in a paediatric cystic fibrosis patient: possible transmission from a household cat. Zoonoses Public Health 59, 246–250. 10.1111/j.1863-2378.2011.01446.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijks J. M., Read F. L., van de Bildt M. W., van Bolhuis H. G., Martina B. E., Wagenaar J. A., van der Meulen K., Osterhaus A. D., Kuiken T. (2008). Quantitative analysis of the 2002 phocine distemper epidemic in the Netherlands. Vet Pathol 45, 516–530. 10.1354/vp.45-4-516 [DOI] [PubMed] [Google Scholar]
- Shina A., Hart C. A., Stenton M. D., Dawson S., McCracken C. M., Binns S. H., Gaskell R. M., Winstanley C. (2002). Distribution of fim3 and flaA TTGE sequence types amongst isolates of Bordetella bronchiseptica from different host animals. J Med Microbiol 51, 557–563. [DOI] [PubMed] [Google Scholar]
- Sneath P., Sokal R. (1973). The Principles and Practice of Numerical Classification. San Francisco: W. H. Freeman. [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stockbauer K. E., Fuchslocher B., Miller J. F., Cotter P. A. (2001). Identification and characterization of BipA, a Bordetella Bvg-intermediate phase protein. Mol Microbiol 39, 65–78. 10.1046/j.1365-2958.2001.02191.x [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. L., Zhulin I. B. (1999). PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63, 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl M. A., Miller J. F. (1996). Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component phosphorelay. J Biol Chem 271, 33176–33180. 10.1074/jbc.271.52.33176 [DOI] [PubMed] [Google Scholar]
- van der Zee A., Mooi F., Van Embden J., Musser J. (1997). Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J Bacteriol 179, 6609–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtova J., Kamanova J., Sebo P. (2006). Bordetella adenylate cyclase toxin: a swift saboteur of host defense. Curr Opin Microbiol 9, 69–75. 10.1016/j.mib.2005.12.011 [DOI] [PubMed] [Google Scholar]