Abstract
Vibrio cholerae is a neutrophilic enteric pathogen that is extremely sensitive to acid. As V. cholerae passages through the host gastrointestinal tract it is exposed to a variety of environmental stresses including low pH and volatile fatty acids. Exposure to acidic environments induces expression of the V. cholerae acid tolerance response. A key component of the acid tolerance response is the cad system, which is encoded by cadC and the cadBA operon. CadB is a lysine/cadaverine antiporter and CadA is a lysine decarboxylase and these function together to counter low intracellular and extracellular pH. CadC is a membrane-associated transcription factor that activates cadBA expression in response to acidic conditions. Herein we investigated the role of the LysR-type transcriptional regulator LeuO in the V. cholerae acid tolerance response. Transcriptional reporter assays revealed that leuO expression repressed cadC transcription, indicating that LeuO was a cadC repressor. Consistent with this, leuO expression was inversely linked to lysine decarboxylase production and leuO overexpression resulted in increased sensitivity to organic acids. Overexpression of leuO in a cadA mutant potentiated killing by organic acids, suggesting that the function of leuO in the acid tolerance response extended beyond its regulation of the cad system. Collectively, these studies have identified a new physiological role for LeuO in V. cholerae acid tolerance.
Introduction
Vibrio cholerae is a neutrophilic bacterium that is extremely sensitive to even mild acidic conditions (Wachsmuth et al., 1994). V. cholerae naturally persists in aquatic reservoirs with a neutral pH, variable nutrient availability and ambient temperatures. It is also an enteric pathogen that encounters a variety of environmental stresses while passing through the human gastrointestinal tract. Following ingestion V. cholerae encounters a dramatic change in pH from near neutral to ≤ 2 in the human stomach. Passage of V. cholerae from the stomach into the small intestine further exposes the bacterium to an environment that contains a combination of inorganic acids and organic acids (Audia et al., 2001). Exposure of V. cholerae to acidic conditions results in the induction of an acid tolerance response. The acid tolerance response can be divided into two distinct branches: an inorganic acid tolerance response and an organic acid tolerance response (Merrell & Camilli, 1999).
The V. cholerae acid tolerance response encompasses diverse genes that function together to mitigate the effects of acid stress. This includes alterations in the outer membrane, the expression of genes that function in the regulation of K+ and Na+ homeostasis, and biofilm production (Merrell et al., 2001, 2002; Zhu & Mekalanos, 2003). The acid tolerance response is probably an important factor for V. cholerae pathogenesis. For example, biofilm production has been shown to enhance acid tolerance, which contributes to pathogenesis by providing protection from acid stress during passage through the gastric acid barrier of the stomach (Tamayo et al., 2010; Zhu & Mekalanos, 2003). In addition, pre-activation of the V. cholerae acid tolerance response has been shown to impart a competitive advantage for colonization of the infant mouse intestine relative to unadapted cells (Merrell & Camilli, 1999). Taken together these results suggest that the acid tolerance response may play a crucial role in both the initial infection with V. cholerae and the subsequent development of hyper-infectivity that has been observed in human and animal-shed V. cholerae (Alam et al., 2005; Angelichio et al., 2004; Merrell et al., 2002).
An important subset of genes that are induced in both the inorganic and the organic acid tolerance response of V. cholerae is the cad system. The contribution of the cad system to acid resistance is conserved among a number of enteric bacteria (Bearson et al., 1997). The cad system includes three genes that are involved in maintaining the intracellular pH while also neutralizing the external pH. CadC is a ToxR-family transcriptional regulator that positively regulates expression of the cadBA operon (Merrell & Camilli, 2000). CadA is a lysine decarboxylase that converts lysine to cadaverine while consuming a proton and producing carbon dioxide. CadB is a lysine–cadaverine antiporter that is localized to the cytoplasmic membrane. Tight regulation of the cad system are necessary as alterations in the intracellular pH are detrimental to the cell (Booth, 1985).
In V. cholerae it has been shown that AphB, a cytoplasmic DNA-binding protein, positively regulates the cad system in response to low pH or low oxygen by directly binding to the cadC promoter (Kovacikova et al., 2010). Upregulation of the cad system contributes to the maintenance of the intracellular pH. Expression of the cad system returns to a low constitutive level upon neutralization of the external environmental. The molecular mechanisms by which V. cholerae downregulates the cad system are not known. In Escherichia coli, the cad system is repressed in two ways: the first is through feedback inhibition by cadaverine, and the second is through the transcriptional regulator LeuO which functions by repressing cadC expression (Shi & Bennett, 1995). In V. cholerae cadaverine does not repress the cad system (Merrell & Camilli, 1999), but it is unknown if LeuO influences cadC expression.
LeuO is a LysR-type transcriptional regulator that shares 50 % identity and 75 % similarity with E. coli LeuO. Our laboratory has shown that V. cholerae leuO is positively regulated by the virulence regulator ToxR, often in response to environmental signals (Ante et al., 2015; Bina et al., 2013). Expression of leuO is induced by the endogenously produced cyclic dipeptide cyclo(Phe–Pro). In response to cyclo(Phe–Pro) LeuO has was shown to repress the production of essential virulence factors by downregulating the ToxR regulon. Expression of leuO is also induced by bile salts and contributes to V. cholerae bile resistance (Ante et al., 2015). Preliminary transcriptomic profiling experiments performed in our laboratory indicated that the cad system was differentially regulated in a V. cholerae leuO mutant, suggesting that LeuO may regulate the cad system. In the present study, we expanded upon this observation and tested the hypothesis that LeuO functioned as a regulator of the V. cholerae cad system. The results showed that LeuO was a repressor of cadC expression and directly bound to the cadC promoter. LeuO was also shown to regulate the production of CadA (lysine decarboxylase) and to contribute to V. cholerae survival after exposure to organic acid. LeuO overproduction in a cadA mutant also resulted in increased acid sensitivity, suggesting that the contribution of LeuO to acid tolerance extends beyond the cad system. Taken together, our studies have identified a new physiological role for LeuO and indicate that LeuO is a component of the V. cholerae acid tolerance response.
Methods
Strains, media and growth conditions
The bacterial strains used in this study are listed in Table 1. E. coli strains EC100λpir and SM10λpir were used for cloning and plasmid mobilization, respectively. E. coli strain BW25113 was used for the two-plasmid β-galactosidase reporter assays. E. coli strain ER2566 was used for purification of LeuO-MBP and maltose binding protein (MBP). The V. cholerae strains used in this study were seventh pandemic O1 El Tor clinical isolates. V. cholerae strain JB58 (N16961ΔlacZ SmR) or strain XBV144 (C6706ΔlacZ SmR) were used as the wild-type (WT) control strains. Bacterial strains were grown at 37 °C in Luria–Bertani (LB) broth or on LB agar. AKI growth conditions which are used to induce the ToxR regulon have been described previously (Iwanaga et al., 1986). An organic acid cocktail (1 × ) consisting of 87 mM acetic acid, 25 mM butyric acid and 37 mM propionic acid was used for the organic acid challenge assays. Acid adaptation media contained 0.1 × organic acid cocktail in LB broth at pH 5.7. Bacterial stocks were maintained at (80 °C in LB broth containing 25 % glycerol. Growth media were supplemented with carbenicillin (Cb) and streptomycin (Sm) at 100 μg ml− 1, kanamycin (Km) at 50 μg ml− 1, or chloramphenicol (Cml) at 1 μg ml− 1 for V. cholerae or at 25 μg ml− 1 for E. coli as required. Arabinose was added to growth media at the indicated concentrations to induce expression from the arabinose-regulated promoter in pBAD18, pBAD18Km and pBAD33.
Table 1. Strains, plasmids and oligonucleotides used in this study.
| Strain/plasmid/oligonuncleotide | Characteristic/sequence (5′ to 3′) | Source |
|---|---|---|
| E. coli | ||
| EC100λpir | supE44 ΔlacU169 (ϕ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 (λpirR6K) | Epicenter |
| SM10λpir | thi-1 thr leu tonA lacY supE recA: :RP4-2-Tc: :Mu kmR (λpirR6K) | Lab collection |
| BW25113 | F − Δ(araD-araB)567 lacZ4787Δ: :rrnB-3 LAM − rph-1 Δ(rhaD-rhaB)568 hsdR514 | Baba et al. (2006) |
| ER2566 | fhuA2 lacZ: :T7 gene1 [lon] ompT gal sulA11 R(mcr-73: :miniTn10–TetS)2 [dcm] R(zgb-210: :Tn10–TetS) endA1 Δ(mcrCmrr)114: :IS10 | New England BioLabs |
| V. cholerae | ||
| JB58 | V. cholerae O1 El Tor strain N16961 ΔlacZ, SmR | Lab collection |
| XBV222 | JB58ΔleuO | Bina et al. (2013) |
| XBV148 | JB58ΔaphB | This study |
| JB804 | V. cholerae O1 El Tor strain C6706, SmR | Thelin & Taylor (1996) |
| XBV144 | JB804 ΔlacZ | This study |
| VA412 | XBV144 ΔleuO | This study |
| EC20568 | C6706 Tn: :VC2485 (leuO) | Cameron et al. (2008) |
| EC17926 | C6706 Tn: :VC0278 (cadA) | Cameron et al. (2008) |
| Plasmid | ||
| pTL61T | lacZ transcriptional reporter plasmid, CbR | Linn & St Pierre (1990) |
| pXB239 | pTL61T containing the cadC promoter region | This study |
| pXB203 | pTL61T containing the aphB promoter region | Bina & Bina (2010) |
| pBAD18 | Arabinose-regulated expression plasmid, CbR | Guzman et al. (1995) |
| pVA94 | pBAD18 expressing leuO | Ante et al. (2015) |
| pBAD18Km | Arabinose-regulated expression plasmid, KmR | Guzman et al. (1995) |
| pXB298 | pBAD18Km expressing leuO | Bina et al. (2013) |
| pBAD33 | Arabinose-regulated expression plasmid, CmlR | Guzman et al. (1995) |
| pVA126 | pBAD33 expressing leuO | This study |
| pWM91 | Suicide plasmid vector used for allelic exchange | Metcalf et al. (1996) |
| pDLT | pWM91 containing a fragment of lacZ harbouring an internal deletion | Fullner & Mekalanos (1999) |
| pWM91ΔleuO | pWM91 containing a fragment of leuO harbouring an internal deletion | Moorthy & Watnick (2005) |
| pWM91ΔaphB | pWM91 containing a fragment of aphB harbouring an internal deletion | This study |
| pMAL-c2 | IPTG-inducible expression vector for fusion of proteins to MBP and cytoplasmic expression, CbR | New England BioLabs |
| pVA175 | pMAL-c2 expressing leuO | This study |
| Oligonucleotide | ||
| PcadC-F | TTCTCGAGTCGGGCTATCGACTGTACGATG | |
| PcadC-R | GTTCTAGACACCACACACCGATGAAGAGCGAAATTATAA | |
| aphB-F1 | TTGGATCCGCCCCACGATGGCTCGCG | |
| aphB-F2 | CGACTGGTTGTCACAAAGATCACCAGCCGGAAAAAGTGCGCCTG | |
| aphB-R1 | GCGAGCTCCAGTGGGCGATATGGGCG | |
| aphB-R2 | GGTGATCTTTGTGACAACCAGTCGAAAGAGGTTTAGGTCATCTAG | |
| LeuO-F | CCCCCGGGTTAGATAAAAAAGACGCAATGAGTGCC | |
| LeuO-R | CCTCTAGATAGAAACGTAGAATGAACAAAGGATC | |
| cadC-EMSA-F1 | GCGGGAGTCGGCAGCGGATGGTTAAACAACCTAAGTT | |
| cadC-EMSA-R1 | GCGGGAGTCGGCAGCGGAGCGAAATTATAAGTGCAC | |
| cadC-EMSA-F2 | GCGGGAGTCGGCAGCGAATTTCGCTCTTCATCGGTG | |
| cadC-EMSA-R2 | GCGGGAGTCGGCAGCGCATAGAATAGCTCTTTGTATC | |
| 5′BIO | GCGGGAGTCGGCAGCG | |
Plasmid and mutant construction
Plasmids and oligonucleotides used in this study are listed in Table 1. The cadC-lacZ reporter plasmid pXB239 was constructed as follows. Briefly, the PcadC-F/PcadC-R PCR primer pair was used to amplify the cadC promoter region from the V. cholerae N16961 genome. The resulting PCR amplicon was then digested with XhoI and XbaI restriction endonucleases before being ligated into similarly digested pTL61T to generate pXB239. The aphB deletion plasmid, pWM91: :ΔaphB, was constructed by PCR stitching as previously described (Bina & Mekalanos, 2001; Imai et al., 1991). Briefly, the aphB-F1/aphB-R2/ and aphB-F2/aphB-R1 PCR primer pairs were used to amplify ∼1 kb regions flanking aphB. The resulting PCR amplicons were used as the template for a second round of PCR using the aphB-F1 and aphB-R1 PCR primers. The resulting ∼2 kb amplicon was digested with BamHI and SacI restriction endonucleases before being ligated into similarly digested pWM91 to generate pWM91: :ΔaphB. The leuO expression plasmid pVA126 (pBAD33: :leuO) was constructed by removing the leuO fragment from pXB298 using XbaI and SspI restriction enzymes. The resulting ∼1 kb leuO fragment was collected and ligated into pBAD33 digested with XbaI and SmaI. The LeuO-MBP purification plasmid pVA175 (pMAL-c2: :leuO) was constructed by amplifying the leuO gene from N16961 using the LeuO-F/LeuO-R PCR primers. The resulting PCR amplicon was then digested with XbaI and SmaI restriction endonucleases and ligated to pMAL-c2 which had been restricted with XbaI and XmnI endonucleases to generate pVA175. This ligation resulted in a translational fusion of leuO to the C terminus of malE (MBP). The DNA sequence of the protein purification construct was subsequently verified by sequencing.
Deletion of V. cholerae aphB (VC1049) was performed by allelic exchange as previously described (Bina et al., 2006). Briefly, E. coli SM10λpir was used to conjugate plasmid pWM91: :ΔaphB into V. cholerae JB58 and co-integrants were selected for Sm/Cb resistance. Several Sm/Cb resistant colonies were cultured on LB agar (without NaCl) containing 5 % sucrose to select for resolution of the integrated plasmid. Sucrose-resistant and Cb-sensitive colonies were then screened by PCR using the aphB-F1/aphB-R1 PCR primers to confirm aphB deletion. Deletion of lacZ (VC2338) in JB804 was accomplished in an identical manner using pDLT to generate strain XBV144. Deletion of leuO (VC2485) in XBV144 was accomplished as described by Moorthy & Watnick (2005) to generate strain VA412. The C6706 transposon insertion mutants were generously supplied by Dr John Mekalanos (Harvard Medical School).
β-Galactosidase assays
V. cholerae strains harbouring the cadC-lacZ reporter plasmid pXB239 were grown under AKI conditions and culture aliquots were taken in triplicate at various times to quantify β-galactosidase activity as described by Miller (1972). The effect of LeuO on cadC expression in V. cholerae consisted of growing strain JB58 containing the cadC-lacZ plasmid pXB239 and pBAD33-leuO plasmid pVA126 under AKI conditions in the presence or absence of 0.02 % arabinose. Culture aliquots were collected in triplicate after 5 h to quantify cadC-lacZ expression. The effect of LeuO on aphB expression in V. cholerae was determined by growing strain JB58 containing the aphB-lacZ plasmid pXB203 and the pBAD33-leuO plasmid pVA126 under AKI conditions in the presence or absence of 0.02 % arabinose. Culture aliquots were collected in triplicate after 5 h to quantify aphB-lacZ expression. LeuO repression of cadC expression in E. coli was accomplished as follows. Overnight cultures of E. coli strain BW25113 containing the cadC-lacZ plasmid pXB239 and the pBAD33-leuO plasmid pVA126 were diluted 1 : 100 in LB broth with or without 0.02 % arabinose. The cultures were incubated at 37 °C with shaking and aliquots were collected after 5 h to quantify cadC-lacZ expression using β-galactosidase activity. Expression from the reporter plasmids was calculated and displayed as Miller units (MU).
Purification of LeuO-MBP and MBP
Proteins for the gel shift assays were purified as follows. E. coli ER2566 carrying plasmid pMAL-c2 or the pMAL-c2: :leuO plasmid pVA175 were grown in LB broth overnight at 37 °C with aeration. The cultures were then diluted 100-fold into LB broth containing Cb and incubated at 37 °C with shaking to an OD600 of ∼0.5 when 0.3 mM IPTG was added and the cultures were incubated for an additional 2 h. The cells were then harvested by centrifugation and the pellet was resuspended in column buffer (20 mM Tris/HCl, 200 mM NaCl, 1 mM EDTA) plus 1 mM PMSF. The cells were then lysed with an M-11P Microfluidizer according to the manufacturer's instructions (Microfluidics). The resulting cell lysates were cleared of particulate matter by centrifugation at 15 000 g for 20 min at 4 °C. The clarified supernatant (i.e. LeuO-MBP or MBP) was then diluted 1 : 6 with column buffer and loaded onto a 0.8 × 7.0 cm chromatography column containing 1 ml amylose resin (New England Biolabs). The column was equilibrated with 12 ml column buffer before the clarified supernatant was run through. Bound proteins were eluted from the resin using elution buffer (20 mM Tris/HCl, 200 mM NaCl, 1 mM EDTA, 10 mM maltose). The purity of the eluted fusion proteins was analysed by SDS-PAGE with Coomassie brilliant blue R-250 staining. Protein concentrations were determined using the Coomassie Plus (Bradford) Assay kit according to the manufacturer's instructions (Thermo Scientific).
Electrophoretic mobility shift assays (EMSAs)
The DNA fragments designated cadC1 (the nucleotide sequence between (79 and +1 relative to the cadC transcriptional start site) and cadC2 (the nucleotide sequence between (8 and +77 relative to the cadC transcriptional start site) were PCR amplified from the N16961 genome using the cadC-EMSA-F1/cadC-EMSA-R1 and cadC-EMSA-F2/cadC-EMSA-R2 oligonucleotide primers, respectively. The PCR fragments were then gel purified and 100 ng was used as a template for a second PCR using the biotinylated 5′BIO oligonucleotide primer purchased from Integrated DNA Technologies. The resulting DNA fragments were end-labelled with biotin. The biotin-labelled probes (1.5 nM) were incubated with purified LeuO-MBP or MBP in amounts ranging from 0 to 30 μM in binding buffer containing 10 mM Tris (pH 7.4), 150 mM KCl, 0.1 mM DTT, 0.1 mM EDTA (pH 8.0) and 200 μg sheared salmon sperm ml− 1. The binding reactions were incubated at room temperature for 20 min before being subjected to electrophoresis on a non-denaturing 5 % TBE-PAGE in 0.25 × Tris/borate/EDTA (TBE) buffer at 200 V for 45 min. The DNA in the gel was transferred to a nylon membrane in 0.5 × TBE buffer at 380 mA for 1 h. The nylon membrane was then UV cross-linked at 120 mJ using a Stratalinker 1800 crosslinker (Stratagene). Biotin-labelled DNA was detected using the Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific) and visualized using a Fluorchem E Digital Darkroom imager (Protein Simple).
Lysine decarboxylase assays
Strains were grown in AKI media static at 37 °C until the cultures reached an OD600 of ∼0.1 (4 h). Strains containing pBAD18Km-leuO were grown in AKI media in the presence or absence of 0.02 % arabinose. After 4 h, culture aliquots were collected and processed for the quantification of lysine decarboxylase activity as described by Lemonnier & Lane (1998) with slight modification. Briefly, the cells were collected by centrifugation and normalized to an OD600 of 1. The cell pellet was then washed with 1 ml of cold (4 °C) Buffer A (1 M NaCl, 0.05 M potassium Pi buffer pH 6.5) before being centrifuged and resuspended in 200 μl cold (4 °C) Buffer B (20 mM potassium Pi buffer pH 5.8). CHCl3 (20 μl) was then added to each sample followed by vortexing for 15 s to disrupt the cell membrane. Quantification of lysine decarboxylase activity was then carried out in triplicate by combining 10 μl of the cell lysate with 110 μl prewarmed Buffer C (5 mM lysine, 0.1 mM pyridoxal 5′-phosphate, 16 mM potassium Pi biffer pH 5.8); a parallel mixture without lysine was also prepared to control the level of endogenous polyamines, as these react in the assay as cadaverine. The enzymic reaction was incubated at 37 °C for 15 min before adding 120 μl of Stop Solution (1 M Na2CO3) and placing on ice. Lysine and cadaverine were then derivatized by adding 120 μl of 10 mM 2,4,6-trinitrobenzene sulphonate to the mixture and incubating at 40 °C for 4 min. After incubation, samples were chilled on ice. For phase separation, 1 ml toluene was added and thoroughly vortexed for 20 s; N,N′-bistrinitrophenylcadaverine (TNP-cadaverine) is soluble in toluene and N,N′-bistrinitrophenyllysine (TNP-lysine) is toluene-insoluble. Samples were then centrifuged at 2000 r.p.m. for 5 min to allow the phases to separate. The concentration of TNP-cadaverine was measured by removing the upper aqueous phase and reading the A 340 in quartz cuvettes with a Genesys 10S UV-Vis spectrophotometer (Thermo Scientific). Lysine decarboxylase activity was determined as the difference in A 340 between the sample incubated with lysine and that incubated without. Specific activity was calculated using the equation (A 340/(time × OD600)) × 1000, and is a measure of lysine converted to cadaverine per time (min) per unit cell density.
Organic acid challenge assays
The acid challenge assays were facilitated by obtaining mutant strains from an ordered V. cholerae C6706 transposon library (Cameron et al., 2008). C6706 is highly conserved with N16961 (Reimer et al., 2011) and we have not observed differences in the LeuO regulon or acid tolerance between the two strains. Overnight cultures of each test strain were diluted 1 : 10 000 into 10 ml of AKI broth, in the presence or absence of 0.2 % arabinose, in a test tube and incubated statically at 37 °C for 4 h before the cultures were normalized to an OD600 of 0.1 before use. The analysis of unadapted cells was performed as follows. Aliquots (100 μl) of the respective normalized cultures were distributed into the wells of a 96-well microtitre plate that contained a linear range of the organic acid cocktail in LB broth. For the acid adaptation analysis, the cells were resuspended in organic acid adaptation media at pH 5.7 and incubated for 1 h at 37 °C. The cells were then collected by centrifugation and resuspended in fresh LB broth from which 100 μl aliquots were distributed into the wells of a 96-well microtitre plate that contained a linear range of the organic acid cocktail in LB broth. The inoculated microtitre plates were then incubated at 37 °C and ∼10 μl aliquots from each well were replica-plated at the indicated time points onto LB agar plates using a 96-pin replicator. The agar plates were then incubated at 37 °C for 18 h after which they were imaged using a Fluorchem E Digital Darkroom imager (Protein Simple).
Results
LeuO regulates cadC expression
Our preliminary transcriptome studies suggested that LeuO may regulate the V. cholerae cad system. The cad system is regulated by AphB, which functions as an activator of cadC. Once CadC is produced, it directly activates expression of the cadBA operon. Therefore, we tested if LeuO affected the expression of either of these two regulatory genes in V. cholerae. We first investigated cadC transcription by quantifying cadC expression levels in WT strain JB58 and an isogenic ΔleuO strain XBV222 using the cadC-lacZ transcriptional reporter pXB239. The test strains were cultured under AKI virulence-gene-inducing conditions and cadC-lacZ expression was quantified using β-galactosidase assays. The results showed that cadC expression peaked at 5 h and declined thereafter (Fig. 1a). Growth of V. cholerae under AKI conditions resulted in acidification of the culture media during static growth (i.e. the first 4 h). The reduction in pH appeared to correlate with the generation of organic acid byproducts from fermentation metabolism. After the initial 4 h of static growth, the cultures shifted to aerobic growth, which resulted in alkalinization of the media (data not shown). Thus, cadC expression appeared to correlate with the changes in the pH of the growth medium during growth under AKI conditions. Expression of cadC in the ΔleuO mutant mirrored expression in the WT strain except that the expression level was elevated in the ΔleuO mutant compared with WT. The elevated cadC expression observed in the absence of leuO supported the hypothesis that LeuO was a regulator of the cad system and was acting as a repressor of cadC.
Fig. 1.
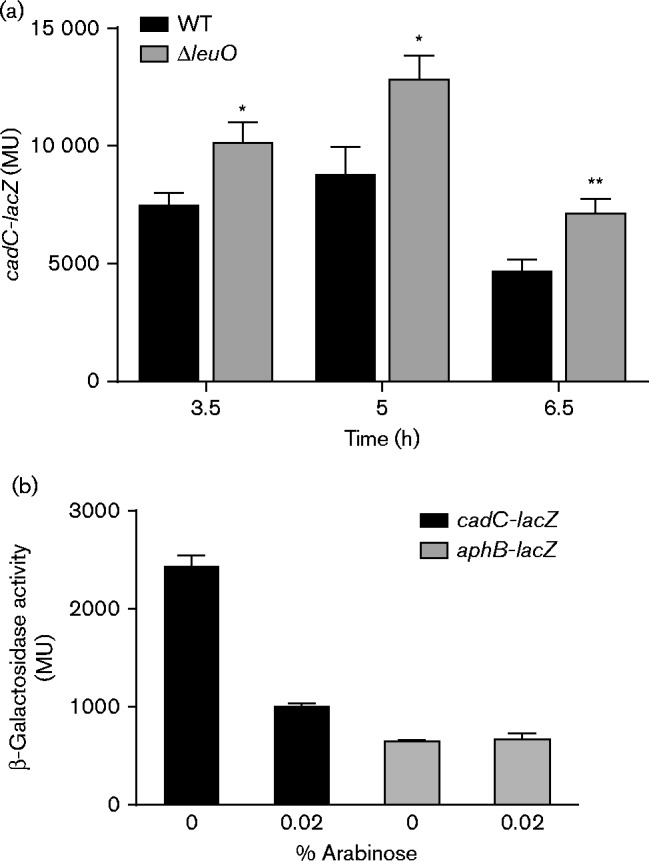
Effect of LeuO on cadC and aphB expression. (a) WT V. cholerae strain JB58 and ΔleuO strain XBV222 carrying the cadC-lacZ reporter plasmid pXB239 were grown under AKI conditions. Culture aliquots were taken at the indicated times and assayed for β-galactosidase activity as described in Methods. The presented data are the mean ± sd of three independent biological replicates. (b) WT strain JB58 bearing pBAD33-leuO plasmid pVA126 and either the cadC-lacZ transcriptional reporter pXB239 (black bars) or the aphB-lacZ transcriptional reporter pXB203 (grey bars) were grown under AKI conditions in the presence or absence of 0.02 % arabinose. Expression of the indicated reporter gene was assessed at 5 h by measuring β-galactosidase production. The presented data are the mean ± sd of three technical replicates and are representative of two independent experiments. Statistical significance was determined using a t-test comparing the sample mean with the WT control mean; *P < 0.05, **P < 0.01.
If LeuO was a cadC repressor, then we hypothesized that leuO overexpression would repress cadC transcription in V. cholerae. To test this hypothesis, V. cholerae WT strain JB58 was transformed with the expression plasmid pVA126 (pBAD33-leuO) and the cadC-lacZ reporter plasmid pXB239. The resulting strain was cultured under AKI growth conditions in AKI broth alone or AKI broth containing arabinose to induce leuO expression. Expression of cadC was then quantified at 5 h post-inoculation. The results showed that the induction of leuO expression by the addition of 0.02 % arabinose resulted in an ∼60 % reduction in cadC expression (Fig. 1b). This further supported the conclusion that LeuO was a cadC repressor in V. cholerae.
There are several potential mechanisms for LeuO to affect cadC expression. LeuO could act directly at cadC by binding to its promoter and inhibiting transcription. Alternatively, LeuO could repress cadC expression indirectly by repressing the expression of its upstream activator aphB. To differentiate between these two possibilities, we examined the effect of leuO overexpression on aphB transcription. We therefore repeated the above experiments using WT strain JB58 carrying pVA126 (pBAD33-leuO) and an aphB-lacZ transcriptional reporter plasmid (pXB203). The results showed that induction of leuO expression by the addition of 0.02 % arabinose did not alter aphB expression (Fig. 1b). This indicated that aphB is not regulated by LeuO and that the effects of LeuO on cadC transcription were probably independent of aphB.
LeuO represses cadC expression by directly binding to its promoter
The above results suggested that LeuO reduced cadC expression independently of aphB, but did not discriminate between LeuO affecting cadC expression directly or indirectly. To address this we examined whether leuO expression affected cadC-lacZ expression in a heterologous host. We introduced both the pBAD33-leuO expression plasmid pVA126 and the cadC-lacZ reporter plasmid pXB239 into E. coli and quantified cadC-lacZ expression following growth in LB broth for 5 h in the presence and absence of arabinose. The results showed an ∼65 % decrease in cadC-lacZ expression in LB broth containing 0.02 % arabinose (Fig. 2a). This result indicated that genes unique to V. cholerae were not required for LeuO repression of cadC and suggested that LeuO may act directly at the cadC promoter. Note that these results do not exclude the possibility that LeuO could be acting indirectly through an intermediate gene present in E. coli.
Fig. 2.
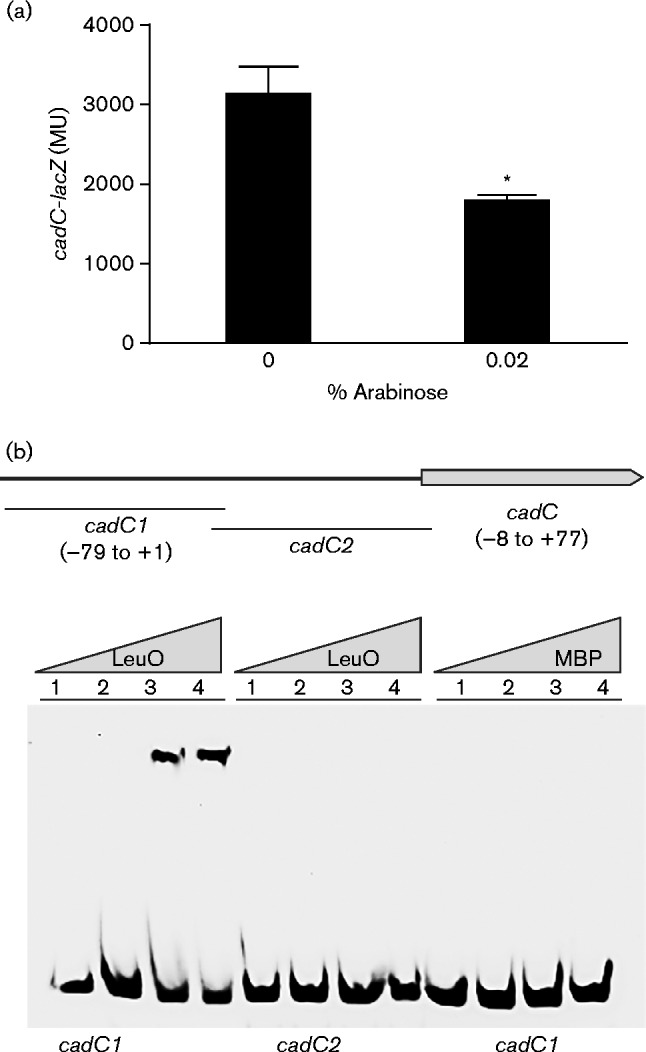
Influence of LeuO on the cadC promoter. (a) E. coli containing the cadC-lacZ reporter plasmid pXB239 and the pBAD33-leuO plasmid pVA126 was grown in LB broth in the presence or absence of 0.02 % arabinose for 5 h when β-galactosidase activity was determined. The presented data are the mean ± sd of three independent experiments. Statistical significance was determined using a t-test comparing the mean of the induced strain with the mean of 0 % arabinose control; *P < 0.005. (b) Gel shift assays were performed using purified LeuO-MBP or MBP and the two indicated DNA fragments from the cadC promoter. Nucleotide numbering listed for the cadC1 and cadC2 DNA fragments are relative to the cadC transcriptional start site. Biotin-labelled cadC1 or cadC2 DNA fragments (1.5 nM) were incubated with either purified LeuO-MBP or MBP at 0 μM (lane 1), 10 μM (lane 2), 20 μM (lane 3) or 30 μM (lane 4) prior to electrophoresis.
To confirm further that LeuO was acting directly at the cadC promoter we performed gel shift assays. For these experiments, we purified LeuO as a translational fusion to MBP and generated two biotin-labelled DNA probes from the cadC locus. The first DNA probe, named cadC1, contained the cadC promoter region from (79 to +1 relative to the cadC transcriptional start site as defined by Merrell & Camilli (2000) (Fig. 2b). This region of the cadC promoter also included the AphB binding site which was mapped to nucleotides (71 to 55 (Kovacikova et al., 2010). The second DNA probe, called cadC2, was used as a negative control and contained nucleotides (8 to +77 relative to the cadC transcriptional start site. The results of the gel shift assays showed that LeuO-MBP bound to the cadC1 DNA probe, but not to the cadC2 DNA probe (Fig. 2b). Incubation of the cadC1 DNA probe with MBP alone did not result in a shift, confirming that LeuO was responsible for the shift of the cadC1 probe by the LeuO-MBP fusion protein. Taken together these results confirmed that LeuO directly binds to a region in the cadC promoter that is present in the cadC1 probe.
Lysine decarboxylase activity is influenced by LeuO
CadC positively regulates the expression of cadBA, and thus the production of lysine decarboxylase (CadA), in response to low environmental pH (Merrell & Camilli, 2000). Based on this, we hypothesized that if LeuO repressed cadC, then deletion of leuO should result in increased cadC expression, and a corresponding increase in cadBA expression and lysine decarboxylase production. Likewise, leuO overexpression should result in decreased cadC expression and a corresponding decrease in cadBA expression and lysine decarboxylase activity. To test this hypothesis we quantified lysine decarboxylase activity in V. cholerae strains lacking leuO or aphB and in a V. cholerae leuO-negative mutant in which we ectopically expressed leuO. In contrast to E. coli (Tabor & Tabor, 1985), V. cholerae encodes only one lysine decarboxylase (i.e. CadA), which facilitates direct measurement of lysine decarboxylase production in V. cholerae cell lysates as a reporter for cadA expression (Merrell & Camilli, 1999).
We first quantified lysine decarboxylase production in WT strain JB58, ΔleuO strain XBV222 and ΔaphB strain XBV148. The results revealed a 29 % increase in lysine decarboxylase activity in the leuO mutant relative to WT (Fig. 3). Although this increase in lysine decarboxylase activity did not reach statistical significance (P = 0.16), lysine decarboxylase activity was consistently elevated in the leuO mutant in multiple independent experiments. By contrast, deletion of aphB resulted in a 79 % reduction in lysine decarboxylase activity. This was expected, as AphB is a positive regulator of cadC. To provide further evidence that LeuO negatively regulated lysine decarboxylase production we quantified the effect of leuO overexpression from pBAD18Km-leuO (pXB298) on lysine decarboxylase production in a ΔleuO mutant (XBV222). The results showed that the addition of 0.02 % arabinose to the growth media resulted in a 53 % reduction in lysine decarboxylase activity (Fig. 3). The observation that leuO deletion appeared to increase lysine decarboxylase activity, while leuO overexpression decreased it, provided additional evidence to support the conclusion that LeuO was a negative regulator of the cad system in V. cholerae.
Fig. 3.
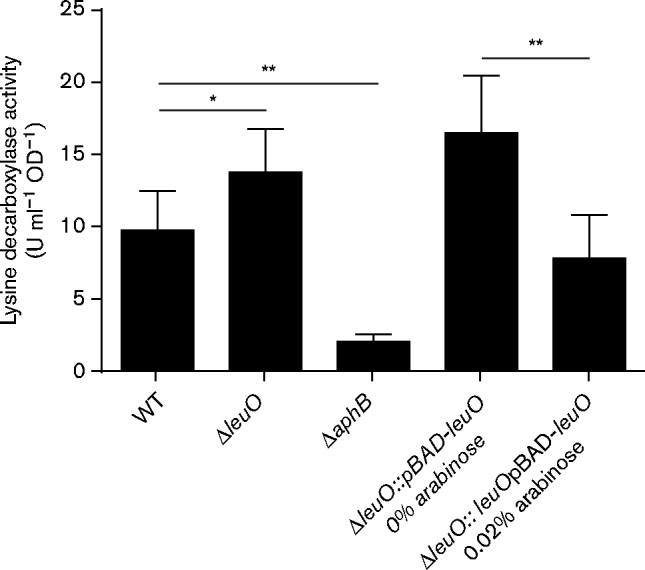
Impact of leuO on lysine decarboxylase production in V. cholerae. The WT strain JB58, ΔleuO strain XBV222, ΔaphB strain XBV148 and ΔleuO strain carrying the pBAD18Km-leuO plasmid pXB298 were grown in AKI media under AKI conditions at 37 °C for 4 h when lysine decarboxylase activity was quantified as described in Methods. Strains containing the arabinose inducible pBAD18Km-leuO were grown in the presence or absence of 0.02 % arabinose. Lysine-decarboxylase-specific activity was defined as the amount of lysine converted to cadaverine per minute divided by the optical density at 600 nm. The presented data are the mean ± sd of three independent experiments. *P = 0.16; **P>0.05.
Effect of LeuO on V. cholerae survival following organic acidic challenge
Studies have shown that the cad system contributes to an inducible acid tolerance phenotype whereby V. cholerae cells pre-adapted to mild acid conditions (i.e. pH 5.7) exhibit increased resistance to lethal acid challenge relative to unadapted cells (Merrell & Camilli, 1999). As our genetic and biochemical data suggested that LeuO repressed the cad system, we hypothesized that LeuO should also negatively affect V. cholerae acid tolerance (Merrell & Camilli, 2000). We tested this by challenging unadapted V. cholerae cells with varying concentrations of organic acids as described in Methods. As both leuO and the acid tolerance response were expressed in vivo (Bina et al., 2013; Merrell & Camilli, 2000; Merrell et al., 2002), we chose to perform these assays using cells cultured under virulence-gene-inducing conditions (i.e. AKI conditions). We cultured WT, leuO and cadA mutant strains for 4 h under AKI conditions, which is the point at which cadC expression was greatest (Fig. 1a), before exposing the cells to varying concentrations of organic acids that were present in the wells of microtitre plates. We then assessed cell viability 15 and 30 min after organic acid challenge with the unadapted cells and 45 and 60 min after challenge with the adapted cells by replica plating culture aliquots from the microtitre plates onto LB agar plates.
The results for the unadapted cells revealed that there was no significant difference in the susceptibility of WT or the leuO mutant to organic acid challenge at either time point (Fig. 4a). This was an expected result given that LeuO appeared to be a cadC repressor. In contrast, the cadA mutant exhibited an increase in susceptibility to the acid challenge, as shown by decreased survival at 30 min relative to the WT control (Fig. 4a). This confirmed previous reports that cadA contributed to the V. cholerae acid tolerance response (Merrell & Camilli, 1999). In contrast to the unadapted cells, there was no apparent difference in organic acid susceptibility among any of the acid-adapted mutant strains at either time point (Fig. 4b). This suggests that under virulence-gene-inducing conditions, other components of the acid tolerance response can compensate for the loss of cadA.
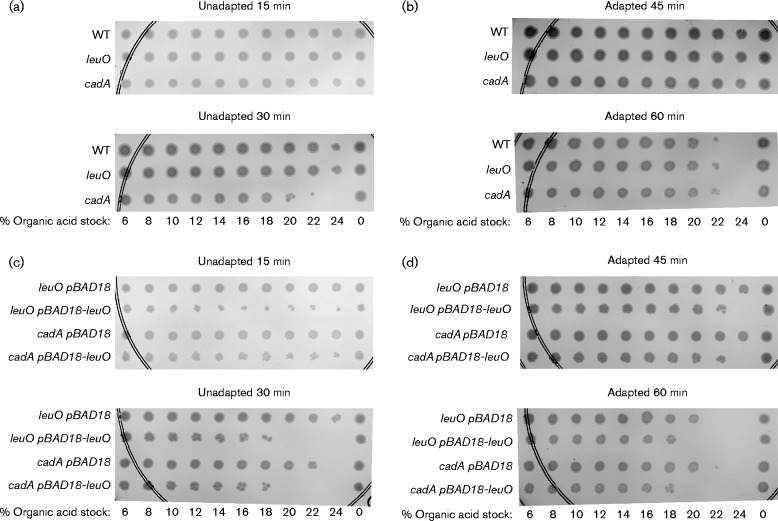
Fig. 4.
Effect of leuO on V. cholerae survival in organic acid. (a) Survival of unadapted WT (XBV144), leuO (VA412) and cadA (EC17926) mutants following organic acid challenge for 15 and 30 min. (b) Survival of adapted WT (XBV144), leuO (VA412) and cadA (EC17926) mutants following organic acid challenge for 45 and 60 min. (c) Survival of unadapted leuO (EC20568) and cadA (EC17926) mutants containing pBAD18 or pBAD-leuO following organic acid challenge for 45 and 60 min. (d) Survival of adapted leuO (EC20568) and cadA (EC17926) mutants containing pBAD18 or pBAD-leuO following organic acid challenge for 45 and 60 min. All strains were cultured for 4 h under AKI conditions before the organic acid challenge; 0.02 % arabinose was added to the broth for strains containing pBAD18 or pBAD18-leuO. Unadapted cells (a, c) and adapted cells (b, d) were inoculated into microtitre plates containing the indicated final concentrations of the organic acid stock solution. The microtitre plates were then incubated at 37 °C and cell viability was assessed over time by replica plating ∼10 μl from each well of the microtitre plates onto the surface of an LB agar plate using a 96-well pin replicator. The agar plates were incubated overnight at 37 °C before being photographed. The presented results are representative of at least three independent experiments.
LeuO is a global regulator in the Enterobacteriaceae, a phenotype that appears to be conserved in the Vibrionaceae. This suggested the that LeuO might affect the expression of other acid tolerance genes in addition to cadC. If this were true, leuO overexpression in the cadA mutant should result in increased organic acid susceptibility. To test this, we repeated the acid killing assays using leuO and cadA mutants in which we ectopically expressed leuO (Fig. 4c, d). The results showed that leuO overexpression in the leuO mutant resulted in increased susceptibility of the unadapted cells to organic acid challenge (Fig. 4c). This confirmed that leuO expression enhanced V. cholerae susceptibility to organic acids and was consistent with the conclusion that LeuO repressed the cad system. Interestingly, ectopic expression of leuO in the cadA mutant also increased V. cholerae susceptibility to organic acid challenge (Fig. 4c). This indicated that the function of LeuO in organic acid tolerance extended beyond its regulation of the cad system.
We next tested whether LeuO affected the induction of an acid tolerance response phenotype. We therefore repeated the above experiments with AKI cultures that had been pre-adapted at pH 5.7 for 1 h prior to organic acid challenge. The results showed increased organic acid resistance among the adapted cells relative to the unadapted cells with all of the tested strains (Fig. 4). This was evidenced by comparison of cell viability between the 30 min unadapted cultures and the 45 min adapted cultures. Significantly, 60 min post-challenge, there were no observable differences in survival between the WT, leuO and cadA mutant strains (Fig. 4b), indicating that V. cholerae was able to mount an acid tolerance response in the absence of leuO and cadA. By contrast, when leuO was overexpressed in either the leuO or the cadA mutants, the cells exhibited increased susceptibility to organic acid challenge relative to the empty vector control (Fig. 4d). This indicated that leuO overexpression negatively affected the ability of V. cholerae to mount an acid tolerance response. The fact that leuO overexpression in the cadA mutant resulted in increased acid susceptibility provided additional evidence to suggest that the function of leuO to acid tolerance extends beyond the cad system.
Discussion
LeuO is a LysR-family regulator that has been shown to function downstream of ToxR in V. cholerae (Ante et al., 2015; Bina et al., 2013). Several lines of evidence suggest that LeuO is a global regulator in the Vibrionaceae that functions in host adaptation and virulence. In V. cholerae LeuO has been shown to affect virulence factor production, biofilm production and bile salt resistance (Ante et al., 2015; Bina et al., 2013; Moorthy & Watnick, 2005). In Vibrio parahaemolyticus LeuO has been shown to regulate expression of the type III secretion system, and serine protease production in Vibrio vulnificus (Kim et al., 2015; Whitaker et al., 2012). Taken together these results suggest that LeuO probably functions to regulate diverse genes involved in environmental adaptation in the Vibrionaceae.
In this study, we identified a new physiological function for LeuO in V. cholerae environmental adaptation. We found that LeuO regulated the expression of the cad system, suggesting that LeuO contributes to acid tolerance. The V. cholerae cad system is constitutively expressed at a low basal level, but is upregulated under conditions of low pH or low oxygen (Kovacikova et al., 2010). Upregulation under these conditions is mediated by AphB binding to the cadC promoter. Once CadC is produced, it upregulates cadBA expression leading to the production of CadB (a lysine/cadaverine antiporter) and CadA (a lysine decarboxylase). CadA contributes to acid tolerance through its degradation of lysine to the polyamine cadaverine, a reaction that plays a key role in maintaining pH homeostasis within the cell.
While AphB positively regulates expression of the cad system, our results showed that LeuO negatively regulates expression of the cad system. This was supported by the fact that leuO deletion increased cadC expression while leuO overexpression reduced cadC expression (Fig. 1). These results suggested strongly that LeuO was a cadC repressor. The negative effects of leuO on cadC transcription were further shown to affect the production of lysine decarboxylase production, the downstream target of CadC (Fig. 3). Taken together these results indicated that LeuO negatively regulates the expression of the cad system by repressing cadC transcription.
LeuO appeared to regulate expression of the cad system by directly binding to the cadC promoter. This suggests that there may be interplay between AphB and LeuO in regulation of the cad system. Our results show that expression of the cad system increased during static growth under AKI conditions before declining upon shift of the cultures to aerated growth (which is associated with alkalinization of the media). Growth of El Tor strains under static AKI growth conditions results in low oxygen tension and low pH, conditions that have been correlated with AphB activation of cadC (Kovacikova et al., 2010). By contrast, leuO expression appears to increase with cell density until it reaches its maximum level at the late exponential phase (data not shown). This suggests that LeuO may function to fine-tune expression of the cad system by antagonizing AphB binding to the cadC promoter. The facts that both LeuO and AphB are LysR-family regulators and that LysR-family regulators bind to T-N11-A motifs (Maddocks & Oyston, 2008) are consistent with this idea. Furthermore, LeuO has been shown to regulate many of its target genes in the Enterobacteriaceae by functioning as an antagonist (Shimada et al., 2011). Whether LeuO is functioning as an AphB antagonist in V. cholerae will require additional studies.
Overexpression of leuO in a cadA mutant increased V. cholerae susceptibility to organic acid in both adapted and unadapted cells (Fig. 4). This suggested that the contribution of LeuO to organic acid tolerance extended beyond the cad system. The mechanism by which this occurred is not known. The acid tolerance response in V. cholerae is complicated and involves diverse genes including the virulence regulator ToxR (Merrell et al., 2001, 2002). ToxR was shown to be required for the organic acid tolerance response through its regulation of the OmpU and OmpT porins. The fact that ToxR positively regulates leuO expression suggests that the role of ToxR in acid tolerance extends beyond porin regulation. In addition to cadC, AphB positively regulates other genes that contribute to acid tolerance (Ding & Waldor, 2003; Kovacikova et al., 2010). While LeuO does not appear to affect production of OmpU or OmpT (Ante et al., 2015), it is possible that LeuO could affect the expression of other AphB-regulated genes that contribute to acid tolerance via a mechanism similar to that with cadC. Alternatively, given that LeuO appears to be a global regulator, LeuO could affect acid tolerance through regulation of other unknown genes.
Although our data conclusively show that LeuO represses cadC expression, the physiological relevance of LeuO repression of cadC and the acid tolerance response is not yet clear. As leuO expression is induced by bile and LeuO contributes to bile salt resistance (Ante et al., 2015), one possibility is that downregulation of the acid tolerance response may contribute to bile resistance. In Salmonella typhimurium the acid tolerance response increased cell surface hydrophobicity (Leyer & Johnson, 1993), a phenotype that could result in increased susceptibility to detergent-like molecules such as bile salts. If the V. cholerae acid tolerance response also resulted in increased cell surface hydrophobicity, leuO induction in response to bile salts may function to downregulate the acid tolerance response to decrease cell surface hydrophobicity and positively affect bile resistance. LeuO could also function in a feedback mechanism to modulate cadaverine production via cadC repression. Cadaverine is a polyamine that has two positive charges at neutral pH. Excess polyamines are growth inhibitory, which necessitates regulation of their production (He et al., 1993). Cadaverine has also been found to reduce V. cholerae auto-agglutination, probably as a result of its positively charged amine groups electrostatically disrupting the pili interactions (Goforth et al., 2013). Thus, excess cadaverine could hinder intestinal colonization.
Acknowledgements
This research was supported by the National Institutes of Allergy and Infectious Disease of the National Institutes of Health (NIH) under award number R01AI091845. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations:
- Cb
carbenicillin
- Cml
chloramphenicol
- EMSA
electrophoretic mobility shift assay
- Km
kanamycin
- MBP
maltose binding protein
- Sm
streptomycin
References
- Alam A., Larocque R. C., Harris J. B., Vanderspurt C., Ryan E. T., Qadri F., Calderwood S. B. (2005). Hyperinfectivity of human-passaged Vibrio cholerae can be modeled by growth in the infant mouse Infect Immun 73 6674–6679 10.1016/S1534-5807(03)00295-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelichio M. J., Merrell D. S., Camilli A. (2004). Spatiotemporal analysis of acid adaptation-mediated Vibrio cholerae hyperinfectivity Infect Immun 72 2405–2407 10.1128/IAI.72.4.2405-2407.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ante V. M., Bina X. R., Howard M. F., Sayeed S., Bina J. E. (2015). Vibrio cholerae leuO transcription is positively regulated by ToxR and contributes to bile resistance J Bacteriol 197 3499–3510 10.1128/JB.00419-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audia J. P., Webb C. C., Foster J. W. (2001). Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria Int J Med Microbiol 291 97–106 10.1078/1438-4221-00106 . [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection Mol Syst Biol 2 0008 10.1038/msb4100050 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson S., Bearson B., Foster J. W. (1997). Acid stress responses in enterobacteria FEMS Microbiol Lett 147 173–180 10.1111/j.1574-6968.1997.tb10238.x . [DOI] [PubMed] [Google Scholar]
- Bina X. R., Bina J. E. (2010). The cyclic dipeptide cyclo(Phe-Pro) inhibits cholera toxin and toxin-coregulated pilus production in O1 El Tor Vibrio cholerae J Bacteriol 192 3829–3832 10.1128/JB.00191-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina J. E., Mekalanos J. J. (2001). Vibrio cholerae tolC is required for bile resistance and colonization Infect Immun 69 4681–4685 10.1128/IAI.69.7.4681-4685.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina J. E., Provenzano D., Wang C., Bina X. R., Mekalanos J. J. (2006). Characterization of the Vibrio cholerae vexAB and vexCD efflux systems Arch Microbiol 186 171–181 10.1007/s00203-006-0133-5 . [DOI] [PubMed] [Google Scholar]
- Bina X. R., Taylor D. L., Vikram A., Ante V. M., Bina J. E. (2013). Vibrio cholerae ToxR downregulates virulence factor production in response to cyclo(Phe-Pro) MBio 4 e00366–e00313 10.1128/mBio.00366-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth I. R. (1985). Regulation of cytoplasmic pH in bacteria Microbiol Rev 49 359–378 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron D. E., Urbach J. M., Mekalanos J. J. (2008). A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae Proc Natl Acad Sci U S A 105 8736–8741 10.1073/pnas.0803281105 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Waldor M. K. (2003). Deletion of a Vibrio cholerae ClC channel results in acid sensitivity and enhanced intestinal colonization Infect Immun 71 4197–4200 10.1128/IAI.71.7.4197-4200.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullner K. J., Mekalanos J. J. (1999). Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae Infect Immun 67 1393–1404 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goforth J. B., Walter N. E., Karatan E. (2013). Effects of polyamines on Vibrio cholerae virulence properties PLoS One 8 e60765 10.1371/journal.pone.0060765 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter J Bacteriol 177 4121–4130 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Kashiwagi K., Fukuchi J., Terao K., Shirahata A., Igarashi K. (1993). Correlation between the inhibition of cell growth by accumulated polyamines and the decrease of magnesium and ATP Eur J Biochem 217 89–96 10.1111/j.1432-1033.1993.tb18222.x . [DOI] [PubMed] [Google Scholar]
- Imai Y., Matsushima Y., Sugimura T., Terada M. (1991). A simple and rapid method for generating a deletion by PCR Nucleic Acids Res 19 2785 10.1093/nar/19.10.2785 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga M., Yamamoto K., Higa N., Ichinose Y., Nakasone N., Tanabe M. (1986). Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor Microbiol Immunol 30 1075–1083 10.1111/j.1348-0421.1986.tb03037.x . [DOI] [PubMed] [Google Scholar]
- Kim J. A., Park J. H., Lee M. A., Lee H. J., Park S. J., Kim K. S., Choi S. H., Lee K. H. (2015). Stationary-phase induction of vvpS expression by three transcription factors: repression by LeuO and activation by SmcR and CRP Mol Microbiol 97 330–346 10.1111/mmi.13028 . [DOI] [PubMed] [Google Scholar]
- Kovacikova G., Lin W., Skorupski K. (2010). The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis J Bacteriol 192 4181–4191 10.1128/JB.00193-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemonnier M., Lane D. (1998). Expression of the second lysine decarboxylase gene of Escherichia coli Microbiology 144 751–760 10.1099/00221287-144-3-751 . [DOI] [PubMed] [Google Scholar]
- Leyer G. J., Johnson E. A. (1993). Acid adaptation induces cross-protection against environmental stresses in Salmonella typhimurium Appl Environ Microbiol 59 1842–1847 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn T., St Pierre R. (1990). Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ J Bacteriol 172 1077–1084 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks S. E., Oyston P. C. (2008). Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins Microbiology 154 3609–3623 10.1099/mic.0.2008/022772-0 . [DOI] [PubMed] [Google Scholar]
- Merrell D. S., Camilli A. (1999). The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance Mol Microbiol 34 836–849 10.1046/j.1365-2958.1999.01650.x . [DOI] [PubMed] [Google Scholar]
- Merrell D. S., Camilli A. (2000). Regulation of Vibrio cholerae genes required for acid tolerance by a member of the ToxR-like family of transcriptional regulators J Bacteriol 182 5342–5350 10.1128/JB.182.19.5342-5350.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell D. S., Bailey C., Kaper J. B., Camilli A. (2001). The ToxR-mediated organic acid tolerance response of Vibrio cholerae requires OmpU J Bacteriol 183 2746–2754 10.1128/JB.183.9.2746-2754.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell D. S., Butler M. S., Qadri F., Dolganov N. A., Alam A., Cohen M. B., Calderwood S. B., Schoolnik G. K., Camilli A. (2002). Host-induced epidemic spread of the cholera bacterium Nature 417 642–645 10.1016/S1534-5807(03)00295-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell D. S., Hava D. L., Camilli A. (2002). Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae Mol Microbiol 43 1471–1491 10.1046/j.1365-2958.2002.02857.x . [DOI] [PubMed] [Google Scholar]
- Metcalf W. W., Jiang W., Daniels L. L., Kim S. K., Haldimann A., Wanner B. L. (1996). Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria Plasmid 35 1–13 10.1006/plas.1996.0001 . [DOI] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Moorthy S., Watnick P. I. (2005). Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development Mol Microbiol 57 1623–1635 10.1111/j.1365-2958.2005.04797.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer A. R., Van Domselaar G., Stroika S., Walker M., Kent H., Tarr C., Talkington D., Rowe L., Olsen-Rasmussen M., other authors (2011). Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa Emerg Infect Dis 17 2113–2121 10.3201/eid1711.110794 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Bennett G. N. (1995). Effects of multicopy LeuO on the expression of the acid-inducible lysine decarboxylase gene in Escherichia coli J Bacteriol 177 810–814 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Bridier A., Briandet R., Ishihama A. (2011). Novel roles of LeuO in transcription regulation of E. coli genome: antagonistic interplay with the universal silencer H-NS Mol Microbiol 82 378–397 10.1111/j.1365-2958.2011.07818.x . [DOI] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. (1985). Polyamines in microorganisms Microbiol Rev 49 81–99 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R., Patimalla B., Camilli A. (2010). Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae Infect Immun 78 3560–3569 10.1128/IAI.00048-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelin K. H., Taylor R. K. (1996). Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains Infect Immun 64 2853–2856 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsmuth K., Blake P. A., Olsvik Ø. (1994). Vibrio Cholerae and Cholera: Molecular to Global Perspectives Washington, DC: 10.1128/9781555818364 American Society for Microbiology. [DOI] [Google Scholar]
- Whitaker W. B., Parent M. A., Boyd A., Richards G. P., Boyd E. F. (2012). The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model Infect Immun 80 1834–1845 10.1128/IAI.06284-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Mekalanos J. J. (2003). Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae Dev Cell 5 647–656 10.1016/S1534-5807(03)00295-8 . [DOI] [PubMed] [Google Scholar]