Abstract
We characterized the de novo biosynthetic pathway of tetrahydrobiopterin (BH4) in the lipid-producing fungus Mortierella alpina. The BH4 cofactor is essential for various cell processes, and is probably present in every cell or tissue of higher organisms. Genes encoding two copies of GTP cyclohydrolase I (GTPCH-1 and GTPCH-2) for the conversion of GTP to dihydroneopterin triphosphate (H2-NTP), 6-pyruvoyltetrahydropterin synthase (PTPS) for the conversion of H2-NTP to 6-pyruvoyltetrahydropterin (PPH4), and sepiapterin reductase (SR) for the conversion of PPH4 to BH4, were expressed heterologously in Escherichia coli. The recombinant enzymes were produced as His-tagged fusion proteins and were purified to homogeneity to investigate their enzymic activities. Enzyme products were analysed by HPLC and electrospray ionization-MS. Kinetic parameters and other properties of GTPCH, PTPS and SR were investigated. Physiological roles of BH4 in M. alpina are discussed, and comparative analyses between GTPCH, PTPS and SR proteins and other homologous proteins were performed. The presence of two functional GTPCH enzymes has, as far as we are aware, not been reported previously, reflecting the unique ability of this fungus to synthesize both BH4 and folate, using the GTPCH product as a common substrate. To our knowledge, this study is the first to report the comprehensive characterization of a BH4 biosynthesis pathway in a fungus.
Introduction
Tetrahydrobiopterin (BH4) is a ubiquitous, multifaceted molecule found widely in nature. It has a well-known biochemical function in higher animals as an essential cofactor for various enzymes, including phenylalanine hydroxylase (PAH), tyrosine hydroxylase, tryptophan hydroxylase, nitric oxide synthase and alkylglycerol monooxygenase (Werner-Felmayer et al., 2002). The first three of these enzymes are required for the degradation of phenylalanine and for the synthesis of neurotransmitters, such as serotonin and melatonin (Nagatsu & Ichinose, 1999). Requirements for BH4 in nitric oxide synthesis and/or phenylalanine hydroxylation have also been reported in bacteria (He & Rosazza, 2003) and some fungi of specific taxa (Maier & Ninnemann, 1995).
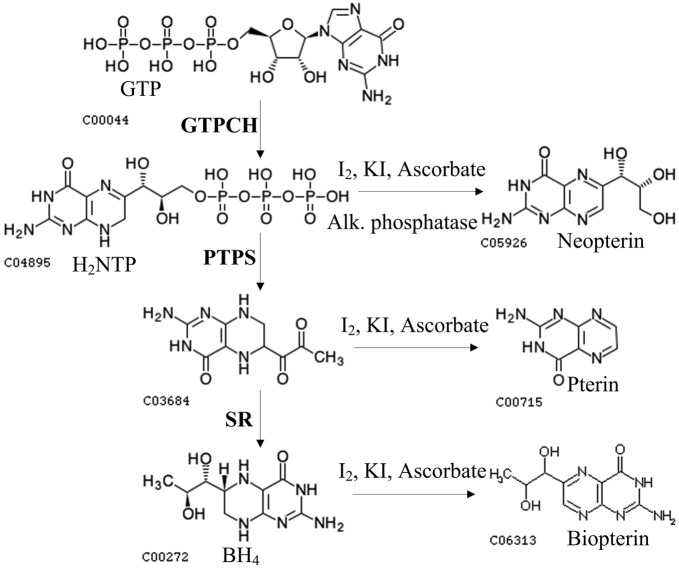
BH4 biosynthesis has been extensively characterized in higher organisms (Fig. 1). The de novo synthesis of BH4 starts with the conversion of GTP to dihydroneopterin triphosphate (H2-NTP) by GTP cyclohydrolase I (GTPCH; EC 3 . 5 . 4 . 16). Then, H2-NTP is converted to 6-pyruvoyltetrahydropterin (PPH4) by 6-pyruvoyltetrahydropterin synthase (PTPS; EC 4 . 2 . 3 . 12), before ultimately being converted to BH4 by sepiapterin reductase (SR; EC 1 . 1 . 1 . 153) (Thöny et al., 2000). The genes responsible for the synthesis of BH4 have been identified (Cha et al., 1991; Citron et al., 1990; Ohye et al., 1998; Shen et al., 1989; Thöny et al., 1992), and structural information for all pathway enzymes has been elucidated (Auerbach et al., 1997; Maita et al., 2004; Ploom et al., 1999).
Fig. 1.
Pathways for BH4 biosynthesis and the activity assays for GTPCH, PTPS and SR.
Unlike higher animals, most bacteria, fungi and plants do not synthesize BH4, but convert the first intermediate (i.e. H2-NTP) into folate (Stanger, 2002), a vitamin that serves as a coenzyme for a variety of one-carbon transfer reactions (Maden, 2000). The presence of a BH4 de novo synthesis pathway has been reported in a few bacteria based on the identification of related genes (Choi et al., 2005; Kang et al., 1998). In fungi, the presence of a BH4 de novo synthesis pathway has been reported for the zygomycete Phycomyces blakesleeanus and the ascomycete Neurospora crassa, and this was based on the isolation of related enzymes (Maier & Ninnemann, 1995). However, no putative PTPS gene involved in the de novo synthesis of BH4 has been explored in any fungi, and the molecular basis for the synthesis of BH4 in fungi has not yet been reported.
Mortierella alpina is a well-known polyunsaturated fatty acids (PUFAs)-producing oleaginous fungus commonly found in soil (Sakuradani et al., 2009). It is one of the most important industrial species for the production of PUFAs (Sakuradani et al., 2009). PUFAs play important roles as structural components of membrane phospholipids and as precursors of the eicosanoids of signalling molecules, including prostaglandins, thromboxanes and leukotrienes (Qi et al., 2004; Sakuradani et al., 2009). Recently, we have sequenced the whole genome of M. alpina (ATCC 32222) (results not shown). Analysis of this sequence has suggested the presence of a de novo pathway for the synthesis of BH4 in this fungus.
In this present study, we characterized in vitro the functions of the genes encoding GTPCH, PTPS and SR in the BH4 de novo pathway in M. alpina. Two functional GTPCH genes (GTPCH-1 and GTPCH-2) were found for the first time to our knowledge. Kinetic parameters and other properties of GTPCH, PTPS and SR were investigated. Comparative analysis between GTPCH, PTPS and SR proteins and other homologous proteins was performed, and the physiological roles of BH4 in M. alpina are discussed.
Methods
Strains and growth conditions.
M. alpina (ATCC 32222) was cultured on potato dextrose agar at 25 °C for 5–7 days. Fungal cultures were initially cultivated in 200 ml Kendrick medium (Kendrick & Ratledge, 1992) and incubated at 25 °C for 36 h. These cultures were used to provide a 10 % (v/v) seed to inoculate a 2 l (working volume) fermenter containing modified Kendrick medium, supplemented with ammonium tartrate (2 g l−1) and glucose (30 g l−1) (Wynn et al., 1999). The fermenter contents were incubated at 25 °C and stirred at 500 r.p.m. with aeration at 0.5 vol./vol. min−1. Then, the mycelia were collected by filtration through sterile cheesecloth, and frozen immediately in liquid nitrogen for RNA extraction. The ammonium concentration in the culture filtrate was determined using the indophenol test (Chaney & Marbach, 1962). Cell lipid was extracted using the method of Bligh & Dyer (1959). BH4 in M. alpina was prepared as described by Maier & Ninnemann (1995) and analysed by HPLC.
Gene searching.
Predicted proteins in the M. alpina (ATCC 32222) genome (GenBank accession no. ADAG00000000) were annotated by blast (White et al., 2005) searches with an E-value of 1E-5 against the following protein databases: NR (http://www.ncbi.nlm.nih.gov), KOGs and COGs (Tatusov et al., 2003), KEGG (Kanehisa et al., 2004), Swiss-Prot and UniRef100 (Wu et al., 2006), BRENDA (Chang et al., 2009); and by InterProScan (Quevillon et al., 2005) against protein domain databases with default parameter settings. Pathway mapping was conducted by associating EC assignment and KO assignment with KEGG metabolic pathways based on blast search results. Predicted GTPCH-1, GTPCH-2, PTPS and SR proteins in the M. alpina genome were searched against predicted proteins of sequenced fungal genomes by blast with an E-value of 1E-5.
Cloning and plasmid construction.
Total RNA extractions were performed using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. RNA was subjected to RNase-free DNase digestion and then purified using an RNeasy Mini kit (Qiagen). The quantity and quality of total RNA were evaluated using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Total RNA was reverse-transcribed with a PrimeScript RT reagent kit (Takara Bio) following the manufacturer’s instructions, before PCR amplification of GTPCH-1, GTPCH-2, PTPS and SR genes using the primer pairs shown in Table 1. The PCR conditions used were as follows: denaturation at 95 °C for 30 s, annealing at 50 °C for 45 s and extension at 72 °C for 1 min (for 25 cycles in total); the final volume in each well was 50 µl. The amplified product was cloned into pET28a+ to construct pwl39688 (containing GTPCH-1), pwl39690 (containing GTPCH-2), pwl39692 (containing PTPS) or pwl39694 (containing SR). The presence of the inserts in the plasmids was confirmed by sequencing using an ABI 3730 sequencer.
Table 1. Primers used in this study.
| Gene | Primer sequence used for cloning (5′–3′)* | Primer sequence used for qPCR (5′–3′) |
| GTPCH-1 | CGGAATTCATGGCAAGCAGCATCGAGAAC | GACGGCCTTAGCTTCCCAAG |
| GTPCH-1 | CCGCTCGAGCTAAATCCCACTGCGTCTGATAAG | CGAACTGCCTCTGCAATCCT |
| GTPCH-2 | CGGAATTCATGTCCCACACTCCCACC | TCACGAAGCAAGTCGCAATG |
| GTPCH-2 | CCGCTCGAGCTACACGCCACGTCGTCTG | TGATCAAGGACAAGAACTCCTCC |
| PTPS | CGGAATTCATGACATCCTCAACACCCGTC | TGACATCCTCAACACCCGTCAG |
| PTPS | CCGCTCGAGTTATTCTCCTCGGTAAACAACG | GCATTTGCCAAAGAGCTTCAC |
| SR | CTAGCTAGCATGTCGTCCAAGGAGCATC | CTTGGCAGGAGGCTAGAAGTTAC |
| SR | CCCAAGCTTCTACTCATCATAAAAGTCGATGTGT | ACGGCGAGCAGTGAGGATAT |
| 18S rDNA | − | CTATTGGCGGAGGTCTATTCGT |
| 18S rDNA | − | GCACGCATTCGGATAATTGGT |
Restriction sites are underlined.
Protein expression and purification.
Escherichia coli BL21 carrying pwl39688, pwl39690, pwl39692 or pwl39694 was grown overnight at 37 °C with shaking in Luria–Bertani (LB) medium containing 50 µg kanamycin ml−1. The overnight culture (10 ml) was inoculated into 500 ml fresh medium and grown until the OD600 reached 0.6. Expression of both GTPCHs was induced by the addition of 0.1 mM IPTG at 15 °C for 16 h, expression of PTPS was induced by 0.5 mM IPTG at 37 °C for 16 h, and expression of SR was induced by 0.5 mM IPTG at 37 °C for 16 h. After the IPTG induction, cells were harvested by centrifugation, washed with binding buffer (50 mM Tris/HCl, 300 mM NaCl, 10 mM imidazole, pH 8.0), resuspended in the same buffer (supplemented with 1 mM PMSF and 1 mg lysozyme ml−1) and then sonicated. Cell debris was removed by centrifugation, while the supernatant (containing the soluble proteins) was collected. The His-tagged fusion proteins were purified by nickel ion affinity chromatography using a Chelating Sepharose Fast Flow column (GE Healthcare) according to the manufacturer’s instructions. Unbound proteins were washed through with 100 ml wash buffer (50 mM Tris/HCl, 300 mM NaCl, 25 mM imidazole, pH 8.0). Fusion proteins were eluted with 3 ml elution buffer (50 mM Tris/HCl, 300 mM NaCl, 250 mM imidazole, pH 8.0) and dialysed overnight at 4 °C against 50 mM Tris/HCl buffer containing 20 % (v/v) glycerol (pH 7.4). Protein concentration was determined by the Bradford method. Purified proteins were stored at −80 °C.
Enzyme activity assays.
Enzyme activities were assayed using the method of Maier & Ninnemann (1995) with minor modifications. The reaction mixture for GTPCH contained 100 mM Tris/HCl (pH 7.5), 5 mM EDTA, 50 mM KCl, 2 mM DTT, 0.2 mM GTP (Sigma-Aldrich) and enzyme solution in a total volume of 25 µl and was incubated at 37 °C for 1 h. The PTPS assay was performed in 100 mM Tris/HCl (pH 7.5), 10 mM MgCl2, 30 µM H2-NTP, 2 mM DTT and the enzyme solution in a total volume of 25 µl at 25 °C for 1 h. The SR assay was performed in 100 mM Tris/HCl (pH 6.0), 0.4 mM NADPH (Sigma-Aldrich), 5 mM sepiapterin (Sigma-Aldrich) and enzyme solution in a total volume of 25 µl at 37 °C for 1 h. After each reaction, 10 µl PTPS or SR reaction mixture was terminated and oxidized by adding 20 µl acidic iodine solution (2 % KI and 1 % I2 in 1 M HCl) for 1 h in the dark. Excess iodine in the supernatant was reduced by adding 10 µl 5 % ascorbic acid. Samples were prepared for HPLC after centrifugation. The 10 µl GTPCH reaction mixtures were oxidized by adding 20 µl acidic iodine solution and then partially neutralized with 1 M NaOH. Neopterin, cleaved from the neopterin phosphates by alkaline phosphatase, was used as a comparison in quantification (Viveros et al., 1981). H2-NTP was freshly prepared from GTP using the recombinant M. alpina GTPCH purified in this study. The reaction mixture contained 50 mM Tris/HCl (pH 7.5), 50 mM KCl, 5 mM EDTA, 2 mM DTT, 1 mM GTP and enzyme solution, and this was incubated at 42 °C for 16 h, before being used directly in the enzyme assay in aliquots according to the concentration of the product. SR activity was also analysed using the in vivo substrate PPH4, which was provided by a mixed assay with PTPS. One unit of GTPCH was defined as the amount of enzyme that catalyses the formation of one nanomole of H2-NTP per hour at 50 °C. One unit of PTPS was defined as the amount of enzyme that catalyses the formation of one nanomole of PPH4 per hour at 25 °C. One unit of SR was defined as the amount of enzyme that catalyses the formation of 15 nanomoles of 7,8-dihydrobiopterin per hour at 25 °C.
HPLC analysis.
HPLC was performed on a Shimadzu model LC-20AT instrument equipped with a Rheodyne loop of 20 µl, an Inertsil ODS-3 C18 column (5 µm, 150×4.6 mm, GL Sciences), and a Shimadzu model RF-20A fluorescence detector (using excitation and emission wavelengths of 350 and 450 nm, respectively). Aliquots of reaction mixtures were injected into the column in 10 mM Na2HPO4 (pH 6.0) at a flow rate of 1.2 ml min−1 (Maier & Ninnemann, 1995). Quantities were calculated by comparison with external pteridine standards (Sigma-Aldrich). In order to isolate pteridine compounds, HPLC was performed under the same conditions as described above, except that 10 % methanol was used as the mobile phase and the flow rate was set at 1 ml min−1 (Son & Rosazza, 2000).
Electrospray ionization-MS (ESI-MS).
The samples were injected into a Finnigan LCQ Advantage MAX ion trap mass spectrometer (Thermo Electron) in negative mode (4.5 kV, 250 °C) for ESI-MS analysis. For the SR product only, the apparatus was used in positive mode.
Measurement of kinetic parameters.
To measure the Km and Vmax values of both GTPCHs for using GTP, reactions were carried out using various concentrations of GTP (0.4–1.5 mM), in conjunction with a fixed concentration of GTPCH (0.67 mM). To measure Km and Vmax values for PTPS acting on H2-NTP, reactions were carried out using various concentrations of H2-NTP (28–50 µM), but a fixed concentration of PTPS (4.48 mM). To measure Km and Vmax values for SR interacting with sepiapterin, reactions were carried out using various concentrations of sepiapterin (0.4–1.5 mM), and fixed concentrations of NADPH (1 mM) and SR (0.59 mM). To measure Km and Vmax values for SR using NAPDH, reactions were carried out using various concentrations of NAPDH (0.4–1.5 mM), with fixed concentrations of sepiapterin (5 mM) and SR (0.59 mM). The GTPCH reaction was performed at 45 °C for 5 min, the PTPS reaction was performed at 20 °C for 5 min, and the SR reaction was performed at 25 °C for 5 min. All reactions had final volumes of 25 µl. Conversions of GTP to H2-NTP, H2-NTP to PPH4, and sepiapterin to 7,8-dihydrobiopterin were monitored by HPLC. Kinetic parameters were obtained by fitting initial rates of reactions measured at varying concentrations of one substrate, at a fixed concentration of the other, to the Michaelis–Menten equation using non-linear regression.
Determination of temperature and pH optima, cation requirements, and the effects of a protein-reducing reagent.
To determine the temperature optima for GTPCH, PTPS and SR, reactions were carried out at 5, 15, 25, 37, 50, 65 and 80 °C for 30 min at pH 8.0. For the determination of the optimum pH, Tris/HCl was used over a range of pH 2.0 to 12.0 for 15 min at 37 °C. To test the effects of various cations and a protein-reducing reagent on enzymic activity, reactions were carried out for 15 min at 37 °C in the presence of 5 mM of the following: KCl, MgCl2, MnCl2, FeCl2, CuCl2, CoCl2 or DTT.
Real-time quantitative PCR (qPCR).
qPCR was conducted with 2 µl reverse-transcribed cDNA using a 7300 Real-Time PCR system (Applied Biosystems) and Power SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s guidelines. The primer pairs used for qPCR are shown in Table 1. The PCR cycling conditions were: 10 s at 95 °C, followed by 30 s at 60 °C for a total of 40 cycles. The 18S rDNA gene was used as the reference gene. Fold changes in gene expression were calculated using the formula 2−ΔΔCt, where ΔΔCt is ΔCt (treatment)−ΔCt (control), ΔCt is Ct (target gene)−Ct (18S), and Ct is the cycle value at which the fluorescence of the gene of interest crosses the threshold level (User’s Manual for ABI 7300 Real-Time PCR system).
Results
Gene searching
Putative GTPCH-1, GTPCH-2, PTPS and SR genes were found in M. alpina genomes (GenBank accession nos JF746874, JF746875, JF746876 and JF746877), and the presence of BH4 in M. alpina is shown in Fig. 2. These results revealed the presence of a de novo pathway for the synthesis of BH4 in this fungus. The search results for GTPCH, PTPS and SR genes in all sequenced fungal genomes are shown in Supplementary Table S1.
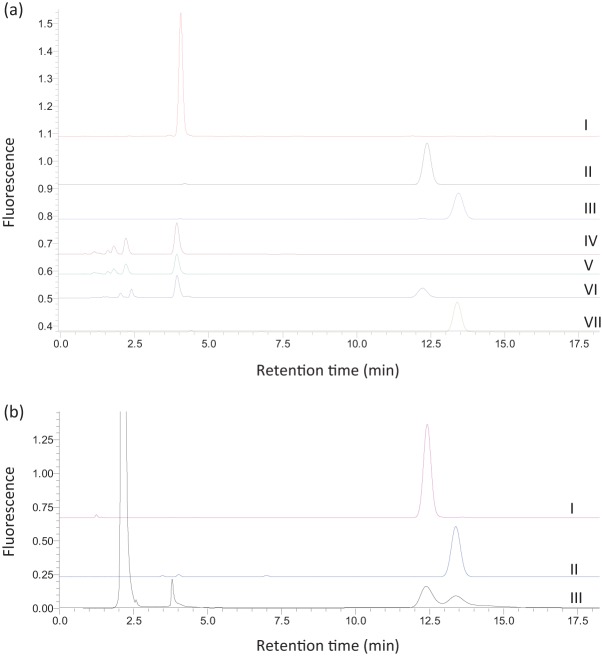
Fig. 2.
(a) HPLC chromatographs of reaction products. Shown are chromatographs of a neopterin standard (I), a pterin standard (II), a biopterin standard (III), the GTPCH-1 reaction (IV), the GTPCH-2 reaction (V), a mixed assay of GTPCH and PTPS (VI), and a mixed assay of GTPCH, PTPS and SR (VII). (b) HPLC chromatographs of a pterin standard (I), a biopterin standard (II) and the pteridines in M. alpina (III).
Expression and purification of GTPCHs, PTPS and SR
GTPCH-1, GTPCH-2, PTPS and SR were expressed as His-tagged fusion proteins in E. coli BL21 by IPTG induction, and purified to near homogeneity by nickel ion affinity chromatography (Supplementary Fig. S1). The molecular masses estimated from SDS-PAGE analysis were 34 kDa for GTPCH-1, 34 kDa for GTPCH-2, 21 kDa for PTPS and 33 kDa for SR, corresponding well to the calculated masses (excluding the His-tag, these were 30, 30, 16 and 30 kDa, respectively).
HPLC analysis of GTPCHs, PTPS and SR products
The enzymic products of the recombinant proteins were identified by HPLC as shown in Fig. 2. GTP was converted to a peak eluted at 4 min, corresponding to the position of the neopterin standard in the reaction catalysed by GTPCH-1 (Fig. 2, IV) or GTPCH-2 (Fig. 2, V). In the reaction catalysed by PTPS, the 4 min peak (the GTPCH oxidized product) was converted to a new peak eluted at 12 min, corresponding to the position of the pterin standard (Fig. 2, VI). When sepiapterin was used as a substrate for PTPS, sepiapterin side chain-releasing (SSCR) activity was not obtained (data not shown), contrary to the human, Drosophila melanogaster, Synechocystis sp. and E. coli cases (Woo et al., 2002). A new fluorescent peak at 13.5 min, which corresponded to the position of biopterin, was seen when sepiapterin was incubated with SR and NADPH and subsequently oxidized by iodine (not shown). The same result was obtained when the in vivo substrate of SR, PPH4 (provided by a mixed assay with GTPCH and PTPS), was used (Fig. 2, VII). None of these peaks was observed in the sample incubated in the absence of the enzymes, while the other minor peaks remained unchanged (data not shown). It was therefore confirmed that the recombinant proteins possess GTPCH, PTPS and SR activity, respectively.
Analysis of GTPCHs, PTPS and SR products by ESI-MS
To further confirm the identities of the GTPCH, PTPS and SR reaction products, the oxidized products from the GTPCHs (the 4 min peak), PTPS (the 12 min peak) and SR (the 13.5 min peak) were purified by HPLC and analysed by ESI-MS. In negative mode, ion peaks were obtained at m/z 252.62 and m/z 252.87 for the products of GTPCH-1 and GTPCH-2, respectively. These peaks corresponded well to the mass of neopterin (m/z 253). Ion peaks were obtained at m/z 162.88 (negative mode) for the PTPS product and m/z 238.87 (positive mode) for the SR product, and these corresponded well to the masses of pterin (m/z 163) and biopterin (m/z 237), respectively (Supplementary Fig. S2).
Kinetic parameters of the GTPCHs, PTPS and SR
Kinetic parameters of GTPCH (for GTP), PTPS (for H2-NTP) and SR (for sepiapterin and NADPH) were measured. The kinetics of the reactions catalysed by these enzymes fit well with the Michaelis–Menten model, and non-linear regression of the experimental data to the Michaelis–Menten equation yielded the kinetic parameters listed in Table 2.
Table 2. Kinetic parameters.
The data shown are the mean of three independent experiments.
| Enzyme | Substrate | Km (mM) | Vmax (µM min−1) |
| GTPCH-1 | GTP | 0.88±0.04 | 0.145±0.006 |
| GTPCH-2 | GTP | 0.37±0.02 | 0.408±0.004 |
| PTPS | H2-NTP | 0.164±0.005 | 4.61±0.21 |
| SR | Sepiapterin | 2.14±0.12 | 15.93±0.75 |
| SR | NADPH | 0.303±0.004 | 13.05±0.05 |
Effects of temperature, pH, cations and a protein-reducing reagent on the activities of the GTPCHs, PTPS and SR
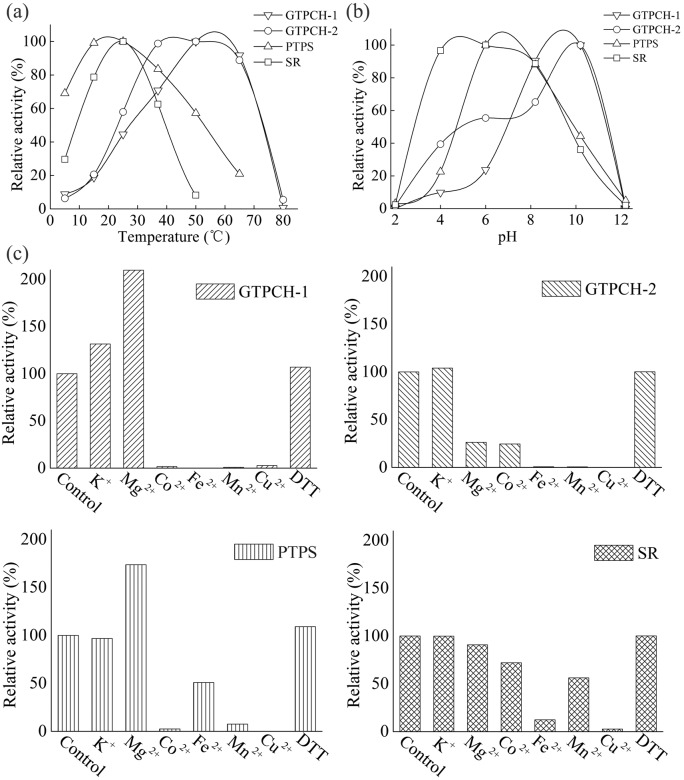
GTPCH-1, GTPCH-2, PTPS and SR were active at each of the temperatures tested (15–65 °C), with the greatest activities detected at 55, 45, 20 and 25 °C, respectively (Fig. 3a). The 100 % activities were defined as the GTPCH, PTPS and SR reaction at 50, 37 and 25 °C, respectively, under the above conditions. For GTPCH-1 and GTPCH-2, high activity (90 %) was observed at temperatures up to 65 °C (Fig. 3a), which is in accordance with the data previously obtained for D. melanogaster (Weisberg & O’Donnell, 1986), Dictyostelium discoideum (Witter et al., 1996) and Nocardia sp. (He & Rosazza, 2003). GTPCH-1 and GTPCH-2 from M. alpina were heat-stable, although this organism is thermosensitive and cannot withstand temperatures above 37 °C. This may reflect the ancient origin of GTPCH from a putative thermophilic ancestor, and this property of the enzyme may have been maintained during evolution. PTPS remained active across a broad temperature range, and high activity (70 %) could even be sustained at temperatures as low as 5 °C, which is not known to have been reported in any other species before. SR had greatest activity at 25 °C, but this decreased quickly at higher temperatures (Fig. 3a), which contrasts to data obtained for SR activity in Chlorobium tepidum (Cho et al., 1999).
Fig. 3.
Effects of temperature (a), pH (b), and cations and DTT (c) on the activity of GTPCH-1 (▽), GTPCH-2 (○) PTPS (▵) and SR (□). Each point represents the mean of three independent measurements.
GTPCH-1, GTPCH-2, PTPS and SR showed optimal activities at pH 9.0, 9.5, 7.0 and 5.0, respectively (Fig. 3b). The 100 % activities were defined as the GTPCH, PTPS and SR reactions at pH 10.2, 10.2 and 6.2, respectively, under the above conditions. The GTPCHs, PTPS and SR were all active over a broad range of pH values (4–10). GTPCH-1 and GTPCH-2 were most active in more alkaline conditions than the optimal pH ranges for human, D. melanogaster and Nocardia sp. GTPCHs (He & Rosazza, 2003; Shen et al., 1989; Weisberg & O’Donnell, 1986). PTPS had optimal activity at pH 7.0, which is similar to that for recombinant rat PTPS (optimal pH 6.5–7.0) (Inoue et al., 1991). SR had optimal activity at pH 5.0, which is consistent with the acidic pH optima observed for human and rat SRs (Sueoka & Katoh, 1982), but is lower than the optimal pH for SR derived from C. tepidum (Cho et al., 1999).
GTPCH-1 activity was enhanced significantly by Mg2+ (100 % increase), and completely inhibited by Co2+, Fe2+, Mn2+ and Cu2+. The 100 % activities were defined as the GTPCH, PTPS and SR reactions without cations under the above conditions. GTPCH-2 activity was completely inhibited by Fe2+, Mn2+ and Cu2+, and inhibited to different degrees by Mg2+ and Co2+ (Fig. 3c). The inhibitory effect of Mg2+ has also been observed for the GTPCHs of D. melanogaster and Bacillus subtilis (De Saizieu et al., 1995; Weisberg & O’Donnell, 1986). Neither K+ nor DTT was found to be necessary for the activity of the two M. alpina GTPCHs, in contrast to the reported increase in activity with other homologous enzymes in the presence of K+ and DTT (Milstien et al., 1996; Yim & Brown, 1976). PTPS activity was enhanced (75 %) by Mg2+, completely inhibited by Co2+, Mn2+ and Cu2+, slightly inhibited by Fe2+, but unaffected by DTT. A positive effect of Mg2+ on PTPS activity has been reported before (Lee et al., 1999; Park et al., 1990). SR activity was completely inhibited by Cu2+, inhibited to different degrees by Co2+, Fe2+ and Mn2+, but unaffected by K+, Mg2+ and DTT (Fig. 3c). M. alpina SR activity was not enhanced by those cations and that protein-reducing reagent, in accordance with the fact that cations and DTT are not involved in the SR reaction (Cho et al., 1999; Kim et al., 2000; Seong et al., 1998).
qPCR analysis
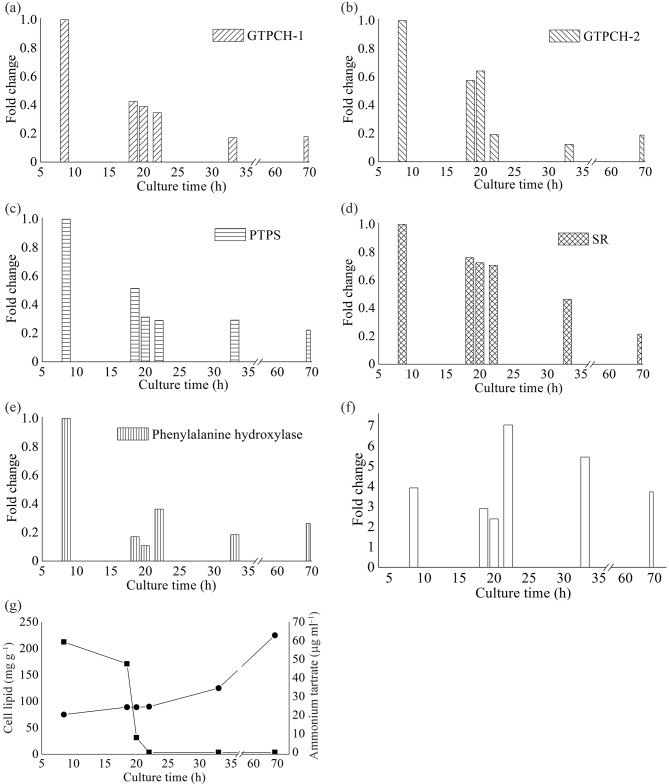
To investigate whether BH4 synthesis is related to lipid accumulation in M. alpina, qPCR was performed to analyse the changes in the expression levels of genes involved in BH4 biosynthesis during the course of the lipid accumulation which is induced by the exhaustion of nitrogen (Wynn et al., 1999). Total RNA samples were purified from mycelia collected from cultures in different growth phases. RNA samples were collected before lipid accumulation when nitrogen availability was sufficient (8.5 h), during the initial stage of lipid accumulation just before nitrogen was used up completely (18.5, 20 and 22 h), and after the lipid had accumulated for a long and sustained period of time (33 and 69 h). The relative expression levels of these genes were measured in triplicate by qPCR (Fig. 4). Ct values obtained after 8 h of incubation were chosen as the control values in order to calculate the fold change in gene expression over time for each treatment. GTPCH-1, GTPCH-2, PTPS and SR genes were transcribed in all growth phases. In comparison with the sample collected at 8 h, when nitrogen was sufficient, the expression of GTPCH-1, GTPCH-2, PTPS and SR was downregulated 0.4-, 0.6-, 0.5- and 0.8-fold, respectively, in cells collected at 18.5 h when nitrogen started to decrease; 0.4-, 0.6-, 0.3- and 0.7-fold, respectively, in cells collected at 20 h when nitrogen was almost used up; and 0.3-, 0.2-, 0.3- and 0.7-fold, respectively, in cells collected at 22 h when nitrogen was completely exhausted and lipids started to accumulate (Fig. 4). Compared with the 22 h cells, the expression levels of the GTPCH-1, GTPCH-2 and PTPS genes in cells collected from other time points (33 and 69 h), when lipids were rapidly accumulating, were not changed significantly (Fig. 4). To evaluate the GTPCH-1 transcript/GTPCH-2 transcript ratio, Ct values of GTPCH-2 obtained at each phase were chosen as the control values. The GTPCH-1/GTPCH-2 transcript ratios were 3.9, 2.9, 2.4, 7.0, 5.5 and 3.7 at the respective sampling times (Fig. 4f), which suggests that GTPCH-1 is more active than GTPCH-2 in all phases of growth.
Fig. 4.
Relative expression levels of GTPCH-1 (a), GTPCH-2 (b), PTPS (c), SR (d) and PAH (e) genes during the accumulation of lipids in M. alpina; (f) the ratio of the transcripts of GTPCH-1/GTPCH-2; (g) exhaustion of ammonium tartrate (▪) and accumulation of cell lipid (•) in M. alpina. Each point represents the mean of three independent measurements.
Discussion
GTPCH-1 and GTPCH-2 share 28–48 % identity with other characterized GTPCH proteins and are most similar to those of Gallus gallus. Multiple sequence alignment of the M. alpina GTPCHs and other characterized GTPCH proteins reveals the presence of certain conserved motifs and residues (Supplementary Fig. S3), including five conserved motifs (Lys-Thr-Pro-Xaa-Arg-Xaa-Ala, Ser-Xaa-Cys-Glu-His-His, Gly-Leu-Ser-Lys-Leu-Ala-Arg, Gln-Val-Gln-Glu-Arg-Leu-Thr and Cys-Met-Val-Met-Arg-Gly-Val) that form a GTP-binding pocket (Du et al., 2009; He et al., 2004; Maita et al., 2004); a Lys-Thr-Arg-Glu motif at the C terminus involved in 7,8-dihydrobiopterin binding, resulting in inhibition of GTPCH activity (Du et al., 2009; Maita et al., 2004); four residues (Thr108, Ser153, Ser191 and Thr255) involved in multiple phosphorylation regulation, both positively and negatively (Du et al., 2009); and six residues (Ala205, Ala207, Pro214, Gly247, Arg250 and Lys254) involved in stimulatory or inhibitory effects on the GTPCH feedback regulatory protein (GFRP) (Maita et al., 2004). Compared with the highly conserved central and C-terminal amino acids of rat GTPCH, which are responsible for substrate binding and catalysis, the N-terminal parts of GTPCH differ depending on the species examined. The N terminus exerts auto-inhibitory control over rat GTPCH and is required for GFRP binding on its own (Higgins & Gross, 2011). This indicates that the N termini of M. alpina GTPCHs may exert allosteric control over the enzymic activities on their own, or via interactions with GFRP, or even with other regulatory factors. All of these conserved motifs and residues in M. alpina GTPCHs are also present in other fungi, and these fungal GTPCHs cluster in the same branch of the phylogenetic tree (Supplementary Fig. S4). However, two GTPCH genes were found and functionally confirmed in M. alpina, and such a case has never been reported before to our knowledge. In most fungi, the GTPCH product is converted to folate and not BH4. In M. alpina, there also exists a folate synthesis pathway (as suggested by genome analysis), indicating that both folate and BH4 are synthesized by this fungus. As the reaction catalysed by GTPCH is the first and rate-limiting step in the de novo biosynthesis of BH4 (Higgins & Gross, 2011), the presence of two distinct GTPCH genes likely reflects the importance of BH4 synthesis in M. alpina. Thus, the presence of two GTPCH genes is advantageous for the fungus to produce sufficient H2-NTP to supply substrates for both the BH4 pathway and the folate pathway. The enzymic properties of GTPCH-1 and GTPCH-2 are generally the same, but the contrasting effects of Mg2+ on GTPCH-1 and GTPCH-2 may reflect different origins for the two genes. GTPCH-1 is more active than GTPCH-2 in all phases of growth, indicating that GTPCH-1 makes the major contribution to BH4 synthesis. GTPCH-1 and GTPCH-2 display different substrate binding and catalytic activity, contributing to the increased Km of GTPCH-1 compared with GTPCH-2. The physiological roles of the two GTPCH genes in the synthesis of BH4 and folate in M. alpina require further investigation.
PTPS shares 28–48 % identity with other characterized PTPS proteins and shares most homology with the human protein. Multiple sequence alignment of the PTPS proteins reveals the presence of various conserved residues characteristic of PTPSs (Supplementary Fig. S5), including four catalytic residues (Cys41, Asp87, His88 and Gly145) and two residues (Thr104 and Thr105) responsible for substrate binding (Ben et al., 2003; Woo et al., 2002). The first 20 amino acid residues at the N terminus are highly variable, but the other portions of the protein are well conserved between vertebrates and M. alpina. Putative PPTS genes were found in the genomes of the fungi Allomyces macrogynus, Batrachochytrium dendrobatidis, Rhizopus oryzae, Spizellomyces punctatus and P. blakesleeanus. The conserved residues characteristic of PTPS in M. alpina are also shown by these fungal PTPSs. When displayed on a phylogenetic tree, the fungal PTPSs were more related to animal than to microbial enzymes (Supplementary Fig. S6).
M. alpina SR shares 15–36 % identity with other characterized SR proteins, and again is most similar to the human enzyme. Multiple sequence alignment of the SR proteins reveals the presence of conserved motifs and residues described before for SRs (Supplementary Fig. S7), including a motif (Gly-Xaa-Xaa-Xaa-Gly-Xaa-Gly) that acts as an NADP-binding site at the N terminus (Cho et al., 1999); a motif (Tyr-Xaa-Xaa-Xaa-Lys) known to be a catalytic site (Cho et al., 1999); three residues (Ser176, Tyr189 and Met226) involved in the active site cavity in the NADP–sepiapterin complex (Supangat et al., 2006); a specific anchor (Asp278) for pterin substrates (Auerbach et al., 1997); and a residue (Arg42) conserved in all known members of the SR family that prefer NADPH to NADH (Auerbach et al., 1997). However, the Tyr165 of mouse SR is not conserved in the M. alpina enzyme, since this residue is proposed to interact with indoleamines via π electrons (Auerbach et al., 1997). Moreover, the Phe99 and Trp196 residues of the C. tepidum SR, which are known to play a key role in stereospecific enzyme catalysis by holding the substrate in the active site (Supangat et al., 2008), are also not conserved in M. alpina. The significantly increased Km of M. alpina SR compared with C. tepidum SR could be due to the loss of one of these residues (i.e. Phe99 or Trp196), which might consequently impair substrate binding and decrease catalytic activity. Compared with SRs from fungi which cannot synthesize BH4, M. alpina SR contains a specific anchor (Asp278) for pterin substrates that is essential in determining the substrate-binding specificity and anchors pterin derivatives for reduction at the monoketo or diketo side chains (Auerbach et al., 1997). This is why M. alpina SR maintains the ability to bind PPH4, which is not synthesized in most fungi due to the absence of PTPS. Based on the search results of fungal genomes (Supplementary Table S1), M. alpina, Batrachochytrium dendrobatidis, R. oryzae, S. punctatus and P. blakesleeanus are the fungi which contain all of the GTPCH, PTPS and SR enzymes involved in the BH4 de novo pathway. The existence of these fungi and other members of the fungal SR cluster as a distinct group in the phylogenetic tree could also explain why M. alpina SR has the ability to synthesize BH4 (Supplementary Fig. S8). This tree illustrates the clear distinction between M. alpina SR and other fungi that cannot synthesize BH4, and also shows the close relationship between M. alpina and mammalian SRs (Supplementary Fig. S8). Based on the sequence comparisons and enzymic properties, SRs from M. alpina and mammals are thought to have evolved from a common ancestor.
The functions of BH4 in fungi are not well understood, but it can act as a cofactor in nitric oxide synthesis (Maier et al., 2001). To investigate the functions of BH4 in M. alpina, the genome was searched for the presence of potential BH4-dependent genes, including PAH, tyrosine hydroxylase, tryptophan hydroxylase, nitric oxide synthase and alkylglycerol monooxygenase, but only a putative PAH (EC 1 . 14 . 16 . 1) gene was found. The expression of this enzyme was confirmed by qPCR analysis (Fig. 4e). BH4 functions as a biological cofactor for three aromatic amino acid hydroxylases, including PAH, which is required for the catabolism of phenylalanine and the synthesis of neurotransmitters in animals (Nagatsu & Ichinose, 1999). Although the same function has never been demonstrated in fungi, it is highly likely that BH4 plays the similar role in M. alpina, based on the identification of the PAH homologue. Still, this needs to be confirmed experimentally in future studies. We did not find the other expected genes based on homology searching, and it is possible that the fungus uses non-homologous genes instead.
M. alpina is noteworthy for its production of various PUFAs, especially arachidonic acid, and the accumulation of lipids, such as glycerolipids, phospholipids, sphingolipids and sterols. It has been suggested that BH4 might be essential for the desaturation or omega oxidation of long-chain fatty acids, the conversion of dihydrosphingosine into sphingosine, or in sterol metabolism, but the functional significance of BH4 and closely related compounds remains to be fully elucidated (Forrest & Van Baalen, 1970; Kaufman, 1967, 1993). The effect of BH4 on lipid metabolism has been investigated in vitro (Rudzite et al., 1998). BH4 increases the incorporation of unsaturated fatty acids into phospholipids (particularly arachidonic acid) at the expense of saturated varieties, and it also decreases the cholesterol content, which means that BH4 may act to increase membrane fluidity, stimulate the cell cycle and prevent cholesterol precipitation. In phenylketonuria patients treated with BH4, n-3 PUFA composition increases significantly (Vilaseca et al., 2010), indicating that BH4 is important for PUFA status in humans. These findings suggest that BH4 biosynthesis may exert control over lipid accumulation on its own or via interactions with other as-yet-unrecognized regulatory factors in M. alpina. In our gene regulation study, the downregulation of BH4 synthesis may simply be due to N limitation and have no close relationship with lipid accumulation (Fig. 4). In order to determine the exact relationships between them, gene knockout experiments could be performed in further work.
Apart from GTPCH, PTPS and SR genes for the de novo pathway, a putative dihydrofolate reductase (EC 1 . 5 . 1 . 3) gene responsible for the sepiapterin-dependent salvage pathway (Hasegawa et al., 2005) and genes encoding pterin-4α-carbinolamine dehydratase (EC 4 . 2 . 1 . 96) and dihydropteridine reductase (EC 1 . 6 . 99 . 7), which are required for the regeneration of BH4, which is essential in phenylalanine metabolism (Thöny et al., 2000), were found in the M. alpina genome (data not shown). This provides further evidence for the importance of BH4 in this fungus. According to KEGG (Kanehisa, 1997), the salvage pathway is rarely found in micro-organisms, and no other fungal species is known to contain the BH4 de novo synthesis and salvage pathways simultaneously with the BH4 regeneration pathway.
To the best of our knowledge, this is the first time that a BH4 synthesis pathway has been characterized at the molecular level in a fungus. Genes encoding GTPCH, PTPS and SR were functionally characterized in vitro. The roles of BH4 in M. alpina PAH interaction and lipid accumulation are deserving of further investigation. BH4 is commercially produced by pharmaceutical companies for the treatment of hyperphenylalaninaemia and endothelial dysfunction (Werner-Felmayer et al., 2002). As the method currently used for the organic synthesis of BH4 is expensive, M. alpina could be used as an alternative source of this molecule.
Acknowledgements
This study was supported by the National Science Foundation of China (NSFC) Key Program Grants 2083600, NSFC General Program Grants 30871952, and the Research Program of State Key Laboratory of Food Science and Technology (SKLF-MB-200802).
Abbreviations:
- BH4
tetrahydrobiopterin
- ESI-MS
electrospray ionization-MS
- GTPCH
GTP cyclohydrolase I
- H2-NTP
dihydroneopterin triphosphate
- PAH
phenylalanine hydroxylase
- PPH4
6-pyruvoyltetrahydropterin
- PTPS
6-pyruvoyltetrahydropterin synthase
- PUFA
polyunsaturated fatty acid
- qPCR
real-time quantitative PCR
- SR
sepiapterin reductase
Supplementary sequence information, eight supplementary figures and a supplementary table are available with the online version of this paper.
Footnotes
Edited by: R. P. Oliver
References
- Auerbach G., Herrmann A., Gütlich M., Fischer M., Jacob U., Bacher A., Huber R. (1997). The 1.25 A� crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters. EMBO J 16, 7219–7230. 10.1093/emboj/16.24.7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben J., Lim T. M., Phang V. P., Chan W. K. (2003). Cloning and tissue expression of 6-pyruvoyl tetrahydropterin synthase and xanthine dehydrogenase from Poecilia reticulata. Mar Biotechnol (NY) 5, 568–578. 10.1007/s10126-002-0121-y [DOI] [PubMed] [Google Scholar]
- Bligh E. G., Dyer W. J. (1959). A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37, 911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- Cha K. W., Jacobson K. B., Yim J. J. (1991). Isolation and characterization of GTP cyclohydrolase I from mouse liver. Comparison of normal and the hph-1 mutant. J Biol Chem 266, 12294–12300. [PubMed] [Google Scholar]
- Chaney A. L., Marbach E. P. (1962). Modified reagents for determination of urea and ammonia. Clin Chem 8, 130–132. [PubMed] [Google Scholar]
- Chang A., Scheer M., Grote A., Schomburg I., Schomburg D. (2009). brenda, amenda and frenda the enzyme information system: new content and tools in 2009. Nucleic Acids Res 37 (Database issue), D588–D592. 10.1093/nar/gkn820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. H., Na J. U., Youn H., Hwang C. S., Lee C. H., Kang S. O. (1999). Sepiapterin reductase producing l-threo-dihydrobiopterin from Chlorobium tepidum. Biochem J 340, 497–503. 10.1042/0264-6021:3400497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. K., Jun S. R., Cha E. Y., Park J. S., Park Y. S. (2005). Sepiapterin reductases from Chlorobium tepidum and Chlorobium limicola catalyze the synthesis of l-threo-tetrahydrobiopterin from 6-pyruvoyltetrahydropterin. FEMS Microbiol Lett 242, 95–99. 10.1016/j.femsle.2004.10.044 [DOI] [PubMed] [Google Scholar]
- Citron B. A., Milstien S., Gutierrez J. C., Levine R. A., Yanak B. L., Kaufman S. (1990). Isolation and expression of rat liver sepiapterin reductase cDNA. Proc Natl Acad Sci U S A 87, 6436–6440. 10.1073/pnas.87.16.6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Saizieu A., Vankan P., van Loon A. P. (1995). Enzymic characterization of Bacillus subtilis GTP cyclohydrolase I. Evidence for a chemical dephosphorylation of dihydroneopterin triphosphate. Biochem J 306, 371–377. [PMC free article] [PubMed] [Google Scholar]
- Du J., Wei N., Xu H., Ge Y., Vásquez-Vivar J., Guan T., Oldham K. T., Pritchard K. A., Jr, Shi Y. (2009). Identification and functional characterization of phosphorylation sites on GTP cyclohydrolase I. Arterioscler Thromb Vasc Biol 29, 2161–2168. 10.1161/ATVBAHA.109.194464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Mistry J., Schuster-Böckler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., et al. & other authors (2006). Pfam: clans, web tools and services. Nucleic Acids Res 34 (Database issue), D247–D251. 10.1093/nar/gkj149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest H. S., Van Baalen C. (1970). Microbiology of unconjugated pteridines. Annu Rev Microbiol 24, 91–108. 10.1146/annurev.mi.24.100170.000515 [DOI] [PubMed] [Google Scholar]
- Hasegawa H., Sawabe K., Nakanishi N., Wakasugi O. K. (2005). Delivery of exogenous tetrahydrobiopterin (BH4) to cells of target organs: role of salvage pathway and uptake of its precursor in effective elevation of tissue BH4. Mol Genet Metab 86 (Suppl. 1), S2–S10. 10.1016/j.ymgme.2005.09.002 [DOI] [PubMed] [Google Scholar]
- He A., Rosazza J. P. (2003). GTP cyclohydrolase I: purification, characterization, and effects of inhibition on nitric oxide synthase in Nocardia species. Appl Environ Microbiol 69, 7507–7513. 10.1128/AEM.69.12.7507-7513.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A., Simpson D. R., Daniels L., Rosazza J. P. (2004). Cloning, expression, purification, and characterization of Nocardia sp. GTP cyclohydrolase I. Protein Expr Purif 35, 171–180. 10.1016/j.pep.2004.02.008 [DOI] [PubMed] [Google Scholar]
- Higgins C. E., Gross S. S. (2011). The N-terminal peptide of mammalian GTP cyclohydrolase I is an autoinhibitory control element and contributes to binding the allosteric regulatory protein GFRP. J Biol Chem 286, 11919–11928. 10.1074/jbc.M110.196204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Kawasaki Y., Harada T., Hatakeyama K., Kagamiyama H. (1991). Purification and cDNA cloning of rat 6-pyruvoyl-tetrahydropterin synthase. J Biol Chem 266, 20791–20796. [PubMed] [Google Scholar]
- Kanehisa M. (1997). A database for post-genome analysis. Trends Genet 13, 375–376. 10.1016/S0168-9525(97)01223-7 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res 32 (Database issue), D277–D280. 10.1093/nar/gkh063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D., Kim S., Yim J. (1998). Biosynthetic enzymes of tetrahydrolimipterin from green sulfur bacterium Chlorobium limicola. Pteridines 9, 69–84. [Google Scholar]
- Kaufman S. (1967). Pteridine cofactors. Annu Rev Biochem 36, 171–184. 10.1146/annurev.bi.36.070167.001131 [DOI] [PubMed] [Google Scholar]
- Kaufman S. (1993). New tetrahydrobiopterin-dependent systems. Annu Rev Nutr 13, 261–286. 10.1146/annurev.nu.13.070193.001401 [DOI] [PubMed] [Google Scholar]
- Kendrick A., Ratledge C. (1992). Desaturation of polyunsaturated fatty acids in Mucor circinelloides and the involvement of a novel membrane-bound malic enzyme. Eur J Biochem 209, 667–673. 10.1111/j.1432-1033.1992.tb17334.x [DOI] [PubMed] [Google Scholar]
- Kim Y. A., Chung H. J., Kim Y. J., Choi Y. K., Hwang Y. K., Lee S. W., Park Y. S. (2000). Characterization of recombinant Dictyostelium discoideum sepiapterin reductase expressed in E. coli. Mol Cells 10, 405–410. [PubMed] [Google Scholar]
- Lee S. W., Lee H. W., Chung H. J., Kim Y. A., Kim Y. J., Hahn Y., Chung J. H., Park Y. S. (1999). Identification of the genes encoding enzymes involved in the early biosynthetic pathway of pteridines in Synechocystis sp. PCC 6803. FEMS Microbiol Lett 176, 169–176. 10.1111/j.1574-6968.1999.tb13658.x [DOI] [PubMed] [Google Scholar]
- Maden B. E. (2000). Tetrahydrofolate and tetrahydromethanopterin compared: functionally distinct carriers in C1 metabolism. Biochem J 350, 609–629. 10.1042/0264-6021:3500609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier J., Ninnemann H. (1995). Biosynthesis of pteridines in Neurospora crassa, Phycomyces blakesleeanus and Euglena gracilis: detection and characterization of biosynthetic enzymes. Photochem Photobiol 61, 43–53. 10.1111/j.1751-1097.1995.tb09241.x [DOI] [PubMed] [Google Scholar]
- Maier J., Hecker R., Rockel P., Ninnemann H. (2001). Role of nitric oxide synthase in the light-induced development of sporangiophores in Phycomyces blakesleeanus. Plant Physiol 126, 1323–1330. 10.1104/pp.126.3.1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maita N., Hatakeyama K., Okada K., Hakoshima T. (2004). Structural basis of biopterin-induced inhibition of GTP cyclohydrolase I by GFRP, its feedback regulatory protein. J Biol Chem 279, 51534–51540. 10.1074/jbc.M409440200 [DOI] [PubMed] [Google Scholar]
- Milstien S., Jaffe H., Kowlessur D., Bonner T. I. (1996). Purification and cloning of the GTP cyclohydrolase I feedback regulatory protein, GFRP. J Biol Chem 271, 19743–19751. 10.1074/jbc.271.33.19743 [DOI] [PubMed] [Google Scholar]
- Nagatsu T., Ichinose H. (1999). Regulation of pteridine-requiring enzymes by the cofactor tetrahydrobiopterin. Mol Neurobiol 19, 79–96. 10.1007/BF02741379 [DOI] [PubMed] [Google Scholar]
- Ohye T., Hori T. A., Katoh S., Nagatsu T., Ichinose H. (1998). Genomic organization and chromosomal localization of the human sepiapterin reductase gene. Biochem Biophys Res Commun 251, 597–602. 10.1006/bbrc.1998.9503 [DOI] [PubMed] [Google Scholar]
- Park Y. S., Kim J. H., Jacobson K. B., Yim J. J. (1990). Purification and characterization of 6-pyruvoyl-tetrahydropterin synthase from Drosophila melanogaster. Biochim Biophys Acta 1038, 186–194. 10.1016/0167-4838(90)90203-R [DOI] [PubMed] [Google Scholar]
- Ploom T., Thöny B., Yim J., Lee S., Nar H., Leimbacher W., Richardson J., Huber R., Auerbach G. (1999). Crystallographic and kinetic investigations on the mechanism of 6-pyruvoyl tetrahydropterin synthase. J Mol Biol 286, 851–860. 10.1006/jmbi.1998.2511 [DOI] [PubMed] [Google Scholar]
- Qi B. X., Fraser T., Mugford S., Dobson G., Sayanova O., Butler J., Napier J. A., Stobart A. K., Lazarus C. M. (2004). Production of very long chain polyunsaturated omega-3 and omega-6 fatty acids in plants. Nat Biotechnol 22, 739–745. 10.1038/nbt972 [DOI] [PubMed] [Google Scholar]
- Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R. (2005). InterProScan: protein domains identifier. Nucleic Acids Res 33 (Web Server issue), W116–W120. 10.1093/nar/gki442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudzite V., Jurika E., Baier-Bitterlich G., Widner B., Reibnegger G., Fuchs D. (1998). Pteridines and lipid metabolism. Pteridines 9, 103–112. [Google Scholar]
- Sakuradani E., Ando A., Ogawa J., Shimizu S. (2009). Improved production of various polyunsaturated fatty acids through filamentous fungus Mortierella alpina breeding. Appl Microbiol Biotechnol 84, 1–10. 10.1007/s00253-009-2076-7 [DOI] [PubMed] [Google Scholar]
- Seong C., Kim Y. A., Chung H. J., Park D., Yim J., Baek K., Park Y. S., Han K., Yoon J. (1998). Isolation and characterization of the Drosophila melanogaster cDNA encoding the sepiapterin reductase. Biochim Biophys Acta 1443, 239–244. [DOI] [PubMed] [Google Scholar]
- Shen R. S., Alam A., Zhang Y. X. (1989). Human liver GTP cyclohydrolase I: purification and some properties. Biochimie 71, 343–349. 10.1016/0300-9084(89)90006-0 [DOI] [PubMed] [Google Scholar]
- Son J. K., Rosazza J. P. N. (2000). Cyclic guanosine-3′,5′-monophosphate and biopteridine biosynthesis in Nocardia sp. J Bacteriol 182, 3644–3648. 10.1128/JB.182.13.3644-3648.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger O. (2002). Physiology of folic acid in health and disease. Curr Drug Metab 3, 211–223. 10.2174/1389200024605163 [DOI] [PubMed] [Google Scholar]
- Sueoka T., Katoh S. (1982). Purification and characterization of sepiapterin reductase from rat erythrocytes. Biochim Biophys Acta 717, 265–271. [DOI] [PubMed] [Google Scholar]
- Supangat S., Seo K. H., Choi Y. K., Park Y. S., Son D., Han C. D., Lee K. H. (2006). Structure of Chlorobium tepidum sepiapterin reductase complex reveals the novel substrate binding mode for stereospecific production of l-threo-tetrahydrobiopterin. J Biol Chem 281, 2249–2256. 10.1074/jbc.M509343200 [DOI] [PubMed] [Google Scholar]
- Supangat S., Park S. O., Seo K. H., Lee S. Y., Park Y. S., Lee K. H. (2008). Role of Phe-99 and Trp-196 of sepiapterin reductase from Chlorobium tepidum in the production of l-threo-tetrahydrobiopterin. Acta Biochim Biophys Sin (Shanghai) 40, 513–518. 10.1111/j.1745-7270.2008.00422.x [DOI] [PubMed] [Google Scholar]
- Tatusov R. L., Fedorova N. D., Jackson J. D., Jacobs A. R., Kiryutin B., Koonin E. V., Krylov D. M., Mazumder R., Mekhedov S. L., et al. & other authors (2003). The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4, 41. 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny B., Leimbacher W., Bürgisser D., Heizmann C. W. (1992). Human 6-pyruvoyltetrahydropterin synthase: cDNA cloning and heterologous expression of the recombinant enzyme. Biochem Biophys Res Commun 189, 1437–1443. 10.1016/0006-291X(92)90235-D [DOI] [PubMed] [Google Scholar]
- Thöny B., Auerbach G., Blau N. (2000). Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem J 347, 1–16. 10.1042/0264-6021:3470001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilaseca M. A., Lambruschini N., Gómez-López L., Gutiérrez A., Moreno J., Tondo M., Artuch R., Campistol J. (2010). Long-chain polyunsaturated fatty acid status in phenylketonuric patients treated with tetrahydrobiopterin. Clin Biochem 43, 411–415. 10.1016/j.clinbiochem.2009.11.013 [DOI] [PubMed] [Google Scholar]
- Viveros O. H., Lee C. L., Abou-Donia M. M., Nixon J. C., Nichol C. A. (1981). Biopterin cofactor biosynthesis: independent regulation of GTP cyclohydrolase in adrenal medulla and cortex. Science 213, 349–350. 10.1126/science.7017928 [DOI] [PubMed] [Google Scholar]
- Weisberg E. P., O’Donnell J. M. (1986). Purification and characterization of GTP cyclohydrolase I from Drosophila melanogaster. J Biol Chem 261, 1453–1458. [PubMed] [Google Scholar]
- Werner-Felmayer G., Golderer G., Werner E. R. (2002). Tetrahydrobiopterin biosynthesis, utilization and pharmacological effects. Curr Drug Metab 3, 159–173. 10.2174/1389200024605073 [DOI] [PubMed] [Google Scholar]
- White S. W., Zheng J., Zhang Y. M., Rock C. O. (2005). The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem 74, 791–831. 10.1146/annurev.biochem.74.082803.133524 [DOI] [PubMed] [Google Scholar]
- Witter K., Cahill D. J., Werner T., Ziegler I., Rödl W., Bacher A., Gütlich M. (1996). Molecular cloning of a cDNA coding for GTP cyclohydrolase I from Dictyostelium discoideum. Biochem J 319, 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H. J., Hwang Y. K., Kim Y. J., Kang J. Y., Choi Y. K., Kim C. G., Park Y. S. (2002). Escherichia coli 6-pyruvoyltetrahydropterin synthase ortholog encoded by ygcM has a new catalytic activity for conversion of sepiapterin to 7,8-dihydropterin. FEBS Lett 523, 234–238. 10.1016/S0014-5793(02)02997-6 [DOI] [PubMed] [Google Scholar]
- Wu C. H., Apweiler R., Bairoch A., Natale D. A., Barker W. C., Boeckmann B., Ferro S., Gasteiger E., Huang H., et al. & other authors (2006). The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res 34 (Database issue), D187–D191. 10.1093/nar/gkj161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn J. P., bin Abdul Hamid A., Ratledge C. (1999). The role of malic enzyme in the regulation of lipid accumulation in filamentous fungi. Microbiology 145, 1911–1917. 10.1099/13500872-145-8-1911 [DOI] [PubMed] [Google Scholar]
- Yim J. J., Brown G. M. (1976). Characteristics of guanosine triphosphate cyclohydrolase I purified from Escherichia coli. J Biol Chem 251, 5087–5094. [PubMed] [Google Scholar]