Summary
Targeting sirtuins for cancer treatment has been a topic of debate due to conflicting reports and lack of potent and specific inhibitors. We have developed a thiomyristoyl lysine compound, TM, as a potent SIRT2-specific inhibitor with broad anticancer effect in various human cancer cells and mouse models of breast cancer. Mechanistically, SIRT2 inhibition promotes c-Myc ubiquitination and degradation. The anticancer effect of TM correlates with its ability to decrease c-Myc level. TM had limited effects on non-cancerous cells and tumor-free mice, suggesting that cancer cells have an increased dependency on SIRT2 that can be exploited for therapeutic benefit. Our studies demonstrate that SIRT2-selective inhibitors are promising anticancer agents and may represent a general strategy to target certain c-Myc-driven cancers.
Introduction
Oncogenes that drive tumorigenesis have attracted extensive interest as therapeutic targets for treating cancers. MYC, and c-Myc in particular, is one such oncogene. MYC was discovered in studies of fulminant chicken tumors caused by oncogenic retroviruses, which co-opted cellular c-Myc to generate the oncogenic v-Myc (Meyer and Penn, 2008). Subsequently, mouse plasmacytomas and human Burkitt lymphomas were found to be caused by c-Myc activation due to chromosomal translocations that fused c-Myc to the immunoglobin (Ig) gene loci (Meyer and Penn, 2008). Recent genomic sequencing efforts identified c-Myc as one of the most highly amplified oncogenes in many different human cancers, further highlighting the oncogenic role of c-Myc activation (Beroukhim et al., 2010). The identification of effective therapeutic strategies targeting Myc has been challenging. Recently it was demonstrated that bromodomain inhibitors that target BRD4 could suppress c-Myc transcription and lead to tumor inhibition in vivo (Delmore et al., 2011). This finding underscores the therapeutic value of targeting Myc.
The sirtuin family of NAD-dependent protein lysine deacylases has been shown to play important roles in many physiological processes, including the regulation of transcription, metabolism, and DNA repair (Haigis and Sinclair, 2010; Imai et al., 2000; Imai and Guarente, 2010). Many of these functions are achieved by their ability to deacylate various substrate proteins, including histones, transcription factors, and metabolic enzymes (Du et al., 2011; Haigis and Sinclair, 2010; Imai et al., 2000; Imai and Guarente, 2010; Jiang et al., 2013; Peng et al., 2011; Zhu et al., 2012). Because the functionally related but structurally distinct zinc-dependent histone deacetylases (HDACs) are established cancer targets (Lee et al., 2012; Marks and Breslow, 2007), there is interest in exploring whether sirtuins can also be important targets for cancers (Fang and Nicholl, 2011; Herranz and Serrano, 2010; Stünkel and Campbell, 2011). However, there is evidence suggesting both tumor suppressor and oncogenic roles of sirtuins (Fang and Nicholl, 2011; Herranz and Serrano, 2010; Stünkel and Campbell, 2011). In the case of SIRT2, genetic studies indicated that aged Sirt2 knockout (KO) mice show increased tumor incidence as compared to wild-type (WT) (Kim et al., 2011a) controls. In contrast, SIRT2 was also observed to have tumor promoting activity in several studies (Chen et al., 2013; Liu et al., 2013; McGlynn et al., 2014; Soung et al., 2014; Yang et al., 2013; Zhao et al., 2014; Zhao et al., 2013). Moreover, several SIRT2 inhibitors have also been reported to have anticancer effects (Cheon et al., 2015; He et al., 2014; Heltweg et al., 2006; Hoffmann et al., 2014; Kim et al., 2011b; Mahajan et al., 2014; McCarthy et al., 2013; Neugebauer et al., 2008; Rotili et al., 2012; Zhang et al., 2009). However, the moderate potency and specificity of the existing sirtuin inhibitors are insufficient to draw conclusions about the anticancer potential of sirtuin inhibition. Thus, whether sirtuin inhibitors are useful anticancer agents is still an open question. Here we set out to develop sirtuin inhibitors with improved potency and selectivity to explore the potential of targeting sirtuins for treating human cancers, especially c-Myc driven cancers.
Results
Development of a highly selective and potent SIRT2 inhibitor
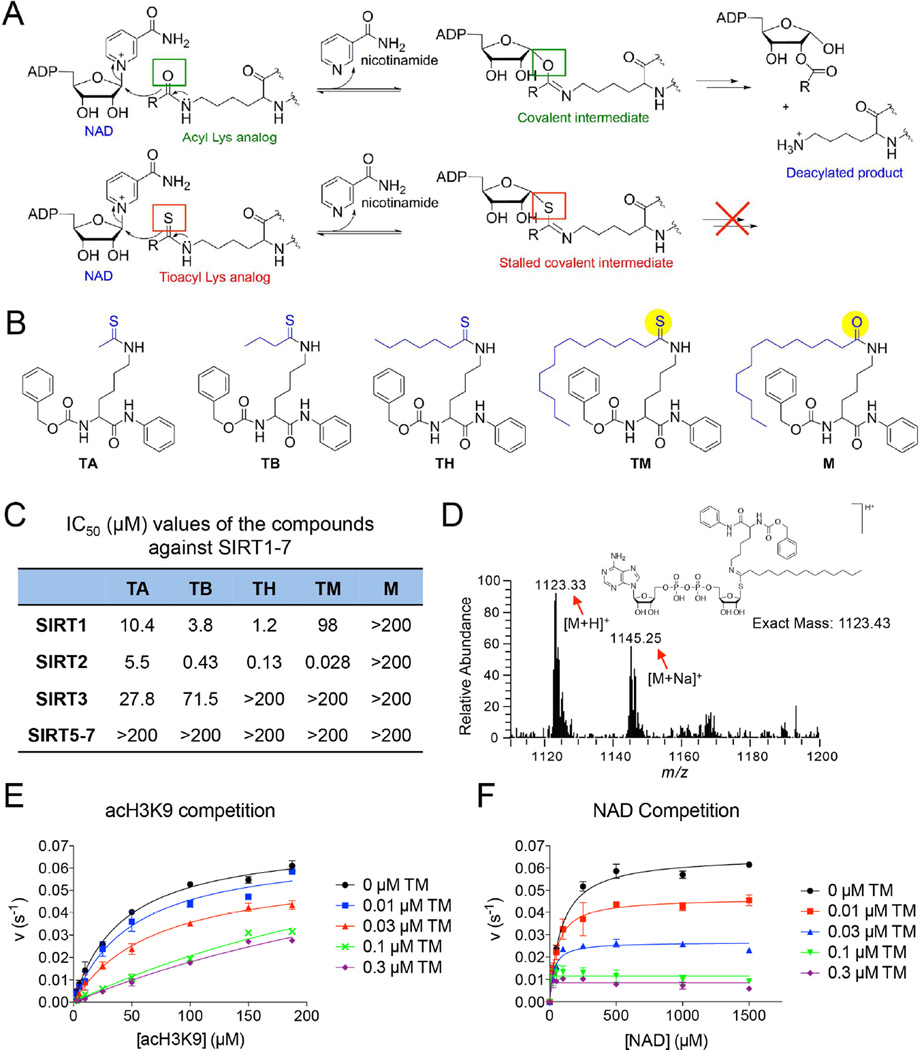
Most existing sirtuin inhibitors are either not very potent (e.g. with IC50 values in the high micromolar range) or not very selective (i.e. they inhibit several different sirtuins). More potent and more selective sirtuin inhibitors would greatly aid in evaluating the therapeutic potential of targeting sirtuins. To develop potent inhibitors specific for a particular sirtuin, we used mechanism-based thioacyl lysine compounds. Thioacyl lysine peptides can react with NAD in the sirtuin active site, forming a relatively stable intermediate that inhibits sirtuin (Figure 1A) (Fatkins et al., 2006; Hawse et al., 2008; Smith and Denu, 2007). Recent studies suggested that different sirtuins may have different acyl group specificity (Du et al., 2011; Feldman et al., 2013; et al., 2013a; Zhu et al., 2012), which can be utilized to design inhibitors specific for different sirtuins (He et al., 2012; He et al., 2014). To target the sirtuins that can recognize aliphatic acyl groups, we synthesized four thioacyl lysine compounds, TA (thioacetyl) (Suzuki et al., 2009), TB (thiobutyryl), TH (thioheptanoyl), and TM (thiomyristoyl) (Figure 1B), and then analyzed their ability to inhibit different sirtuins.
Figure 1.
Development of mechanism-based inhibitor of sirtuins. (A) The enzymatic reaction mechanism of sirtuin-catalyzed NAD-dependent deacylation (upper panel). Thioacyl lysine compounds act as suicide substrates to inhibit sirtuins (lower panel). (B) Structures of four different thioacyl lysine sirtuin inhibitors, TA, TB, TH, and TM. M, which differs from TM by just one atom (highlighted by yellow color), is an inactive control of TM. (C) IC50 (µM) values of the TA, TB, TH, TM and M against SIRT1–7. IC50 values derived from Graphpad Prism are presented as mean values from three independent experiments. (D) Mass spectrometry detection of the stable covalent intermediate formed by TM and NAD. (E, F) Henri-Michaelie-Menten plots showing acH3K9 (E) and NAD (F) competition analyses of TM-mediated SIRT2 inhibition. Error bars represent mean ± sd. See also Figure S1.
Remarkable differences in the potency and selectivity of these compounds were observed by sirtuin activity assays in vitro (Figure 1C, Figure S1A). TA could inhibit SIRT1, SIRT2 and SIRT3, but not very potently. TB was a better SIRT1/SIRT2 inhibitor than TA. The IC50 of TB for SIRT1 (3.8 µM) and SIRT2 (0.43 µM) were about 3 fold and >10-fold, respectively, better than those of TA (Figure 1C). Further increasing the size of the thioacyl group by three methylene groups lead to TH, which had even lower IC50 values for SIRT1 (1.2 µM) and SIRT2 (0.13 µM). Remarkably, TM, with a 14-carbon thioacyl group, could inhibit SIRT2 with an IC50 value of 0.028 µM, but inhibited SIRT1 with an IC50 value of 98 µM and did not inhibit SIRT3 even at 200 µM (Figure 1C). None of these compounds can efficiently inhibit SIRT5, SIRT6, or SIRT7. Thus, TM is a SIRT2-speific inhibitor in vitro. To facilitate later investigations of TM, we also synthesized the corresponding myristoyl lysine compound (M, Figure 1B) as an inactive control for TM. M differs from TM by only one atom (the S atom in TM is changed to an O atom in M). As expected, M did not show sirtuin inhibition even at 200 µM (Figure 1C).
To further confirm that TM is a mechanism-based inhibitor of SIRT2, we performed substrate competition analyses for TM-mediated SIRT2 inhibition. At saturating NAD concentration, the apparent Km value for acetyl-H3K9 peptide (acH3K9) increased with increasing TM concentrations, whereas the vmax remained relatively constant (Figure 1E). The double-reciprocal plot of 1/v versus 1/[acH3K9] revealed a series of lines that intersect at the 1/v axis (Figure S1C), suggesting that TM is competitive with acH3K9. This is consistent with our recent finding that SIRT2 possesses a large hydrophobic pocket that can accommodate the myristoyl group (Teng et al., 2015). At saturating acH3K9 concentration, both the apparent Km value for NAD and vmax decreased with increasing TM concentrations (Figure 1F), suggesting that TM is uncompetitive with NAD, which is consistent with the fact that formation of the inhibitory covalent intermediate requires NAD. We then used liquid chromatographymass spectrometry (LC-MS) to examine the formation of the stalled covalent intermediate. Ions with m/z of 1123.33 (the protonated intermediate) and 1145.25 (the sodium adduct of the intermediate) were detected only when TM was incubated with both SIRT2 and NAD (Figure 1D), but not without SIRT2 or NAD (Figure S1B). Overall, these results indicate that TM acts as a mechanism-based inhibitor of SIRT2.
TM exhibited potent anticancer activity
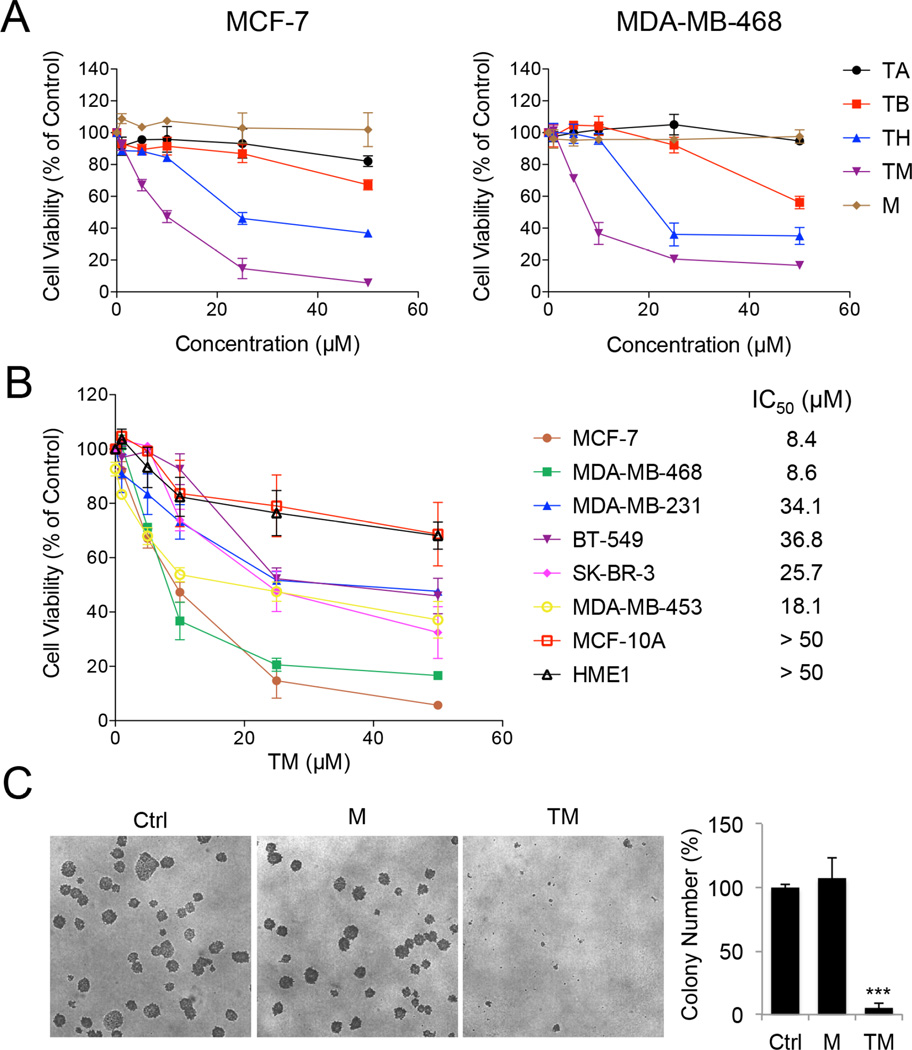
Sirtuin inhibitors have been reported to have anticancer properties. However, most of the inhibitors used are not very selective and thus, inhibiting which sirtuins can provide beneficial effects remains unclear. Having a potent and very selective SIRT2 inhibitor provided a unique opportunity to investigate whether inhibiting SIRT2 can be useful as an anticancer strategy. We initially explored this in several breast cancer cell lines because of the substantial tumor-promoting role of SIRT2 in breast cancer (McGlynn et al., 2014; Soung et al., 2014; Zhao et al., 2014) and the previous studies showing that SIRT2 inhibitors exert anti-proliferative effect against breast cancer cell lines (Di Fruscia et al., 2012; Neugebauer et al., 2008; Rotili et al., 2012; Seifert et al., 2014; Yoon et al., 2014). We assayed the ability of TA, TB, TH, and TM to inhibit three human breast cancer cell lines, MCF-7, MDA-MB-468 and MDA-MB-231. The cytotoxicity of these compounds correlated with their in vitro SIRT2 inhibitory effects (Figure 2A and Figure S2A). TA, which showed modest SIRT1, SIRT2 and SIRT3 inhibition in vitro, did not inhibit cell viability at 50 µM. TB had greater inhibitory effect on cell viability than TA, but only showed inhibition at 50 µM. TH and TM were more potent than TA and TB. Compared with TH, the SIRT2-selective inhibitor TM showed greater inhibition of cell viability. The inactive inhibitor mimic M did not affect cell viability at 50 µM. Similar result was also obtained in HeLa cells (Figure S2B). Next we treated eight different human normal and breast cancer cell lines with TM. As shown in Figure 2B, different malignant cells showed differential susceptibility to TM. And the two non-cancerous cell lines, MCF-10A and HME1, were much less sensitive to TM, suggesting that the cytotoxicity of TM is relatively selective toward cancer cells. We further evaluated the anticancer activity of TM using soft agar colony formation assay. TM significantly inhibited anchorage-independent growth of various cancer cells tested (Figure 2C and Figure S2C), while the control compound M did not (Figure 2C).
Figure 2.
TM inhibits human cancer cells. (A) Cell viability of MCF-7 and MDA-MB-468 cells treated with the indicated inhibitors for 72 hr. (B) Cell viability of the indicated human normal and breast cancer cells treated with TM for 72 hr. IC50 values were means from 3 independent experiments. (C) Soft agar colony formation of MCF-7 cells treated with ethanol, TM (25 µM in ethanol) or M (25 µM in ethanol). Representative images of colonies were shown on the left panel. Quantification of the colony numbers was shown on the right panel. The y axis represents percent colony number relative to ethanol-treated cells. Statistics, two-tailed Student’s t-test. Error bars represent mean ± sd. ***p < 0.001. See also Figure S2.
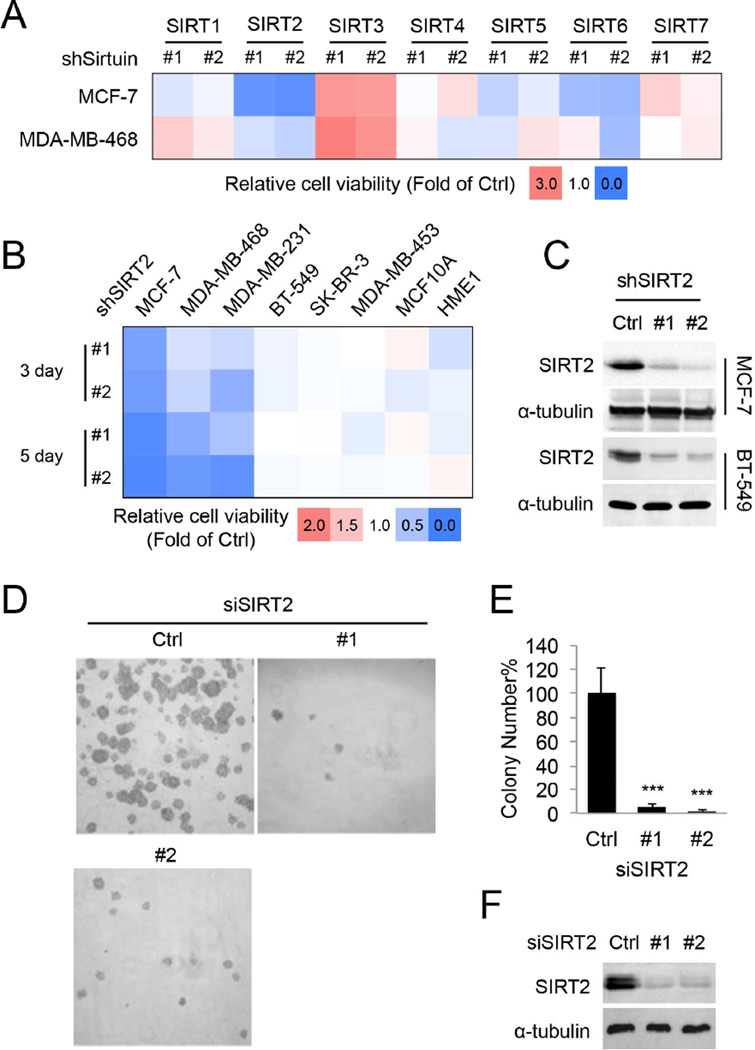
The correlation between the cytotoxic effects of TA, TB, TH, TM and M and their in vitro SIRT2 inhibitory activities suggests that SIRT2 inhibition could have anticancer effects. To further confirm this, we knocked down all seven sirtuins individually in MCF-7, MDA-MB-468, and HeLa cells, which were relatively sensitive to TM (Figure S3A). SIRT2 knockdown (KD) produced the strongest cytotoxicity in all the three cell lines tested (Figure 3A & S3B), which further supported SIRT2 inhibition as a promising anticancer strategy.
Figure 3.
SIRT2 KD decreases the viability of various cancer cell lines. (A) Cell viability of MCF-7 and MDA-MB-468 cells infected with lentivirus carrying luciferase shRNA (Ctrl) or SIRT1–7 shRNAs for 72 hr. The heat map presents average relative cell viability compared to Ctrl shRNA-infected cells from three independent experiments. (B) Cell viability of various human normal and breast cancer cells infected with lentivirus carrying luciferase (Ctrl) or SIRT2 shRNAs. (C) Representative Western blots showing the knockdown efficiency of SIRT2 in MCF-7 and BT-549 cells. (D) Soft agar colony formation of MCF-7 cells transfected with scrambled siRNA or SIRT2 siRNA. (E) Quantification of the colony numbers in (D). The y axis represents percent colony number relative to scrambled siRNA-transfected cells. Statistics, two-tailed Student’s t-test. (F) Representative Western blots showing the knockdown efficiency of SIRT2 by siRNAs in MCF-7 cells. Error bars represent mean ± sd. ***p < 0.001. See also Figure S3.
We then further examined the effect of SIRT2 KD in the same set of human breast cancer and non-tumorigenic mammary cell lines in which the cytotoxic effect of TM was tested. SIRT2 KD significantly decreased cell viability in a time-dependent manner in MCF-7, MDA-MB-468, and MDA-MB-231 cells, but did not show much cytotoxicity in BT-549, SK-BR-3, and MDA-MB-453 cells or the non-transformed MCF-10A and HME1 cells (Figure 3B). In MCF-7 and MDA-MB-468 cells, SIRT2 KD resulted in less than 1% cell viability after 10 days of lentiviral infection (Figure S3C). Moreover, colony formation in soft agar by MCF-7 cells was dramatically diminished by SIRT2 KD (Figure 3D–3F). The knockdown data are thus consistent with the small molecule data, indicating that SIRT2 inhibition can effectively suppress cancer cell proliferation and that the anticancer effect of TM is likely through SIRT2 inhibition.
TM inhibits SIRT2 in cells
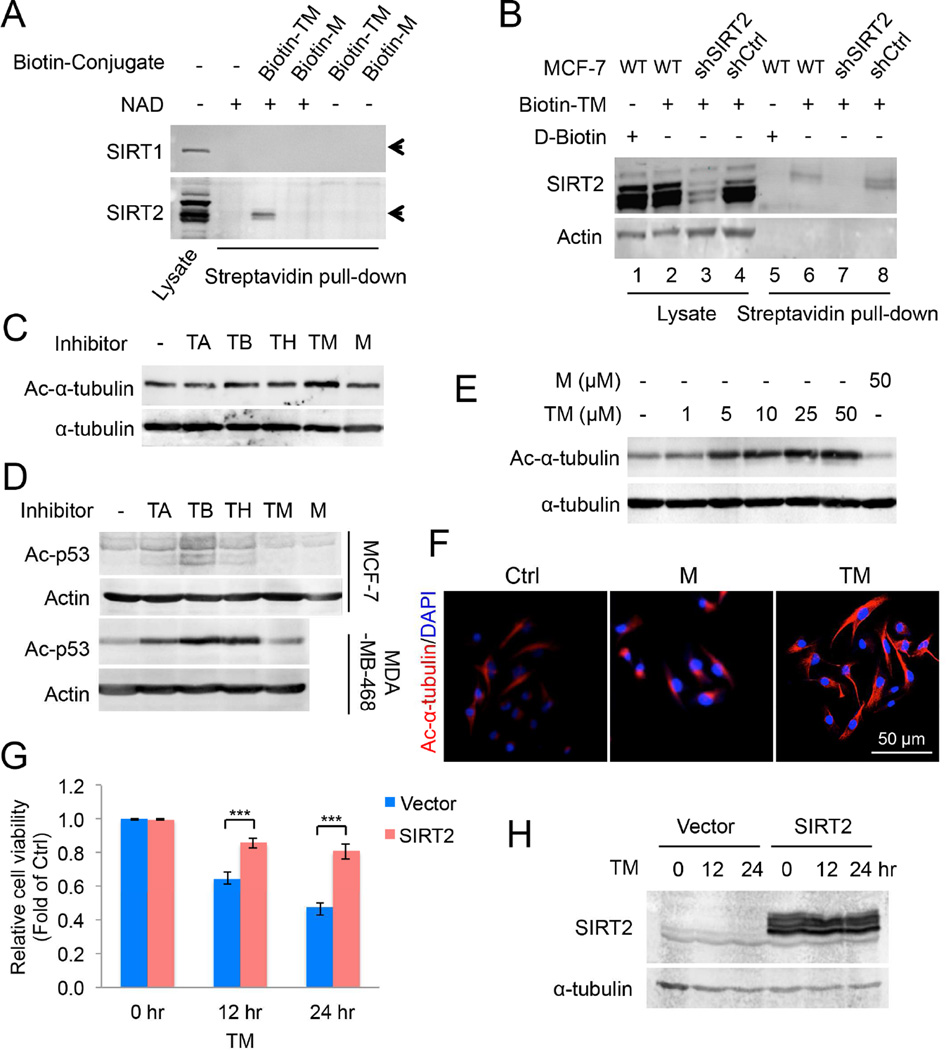
We next wanted to determine whether TM inhibits cancer cells by targeting SIRT2. We first carried out a number of experiments to validate that SIRT2 is the target of TM in cells. We conjugated biotin to TM and M to generate Biotin-TM and Biotin-M compounds (Figure S4A). We then added these compounds to either total protein extract (Figure 4A) or live cells (Figure 4B) to pull down sirtuins. Biotin-TM was able to pull down SIRT2 but not SIRT1 from the HEK293T cell extract. In contrast, Biotin-M, the inactive control compound, did not pull out SIRT2 (Figure 4A). When assayed using SIRT2 KD cells, the amount of SIRT2 pulled down by Biotin-TM was also decreased (Figure 4B). These data suggest that TM targets SIRT2 but not SIRT1 in cells.
Figure 4.
TM specifically inhibits SIRT2 in cells. (A) Pull-down assay to detect the binding of Biotin-M (10 µM) and Biotin-TM (10 µM) to SIRT1 and SIRT2 in HEK293T total cell lysate. (B) Pull-down assay to detect the binding of Biotin-TM (50 µM) to SIRT2 in MCF-7 cells. D-Biotin (50 µM) was used as a negative control. (C) Immunoblot for the acetyl-α-tubulin (K40) levels in SIRT2-overexpressing MCF-7 cells treated with indicated inhibitors (25 µM) for 6 hr. (D) Immunoblot for the acetylation of p53 (K382) in In MCF-7 or MDA-MB-468 cells treated with TSA (200 nM) and the indicated inhibitors (25 µM) for 6 hr. (E) Immunoblot for acetyl-α-tubulin (K40) levels in MCF-7 cells treated with TM or M for 6 hr. (F) Immunofluorescence detection of the acetyl-α-tubulin (K40) level in MDA-MB-231 cells treated with ethanol, M or TM (25 µM in ethanol) for 6 hr. (G) Effect of SIRT2 overexpression on the cytotoxicity effect of TM. MCF-7 cells were transfected with pCMV vector or pCMV-SIRT2 for 12 hr before being treated with 25 µM of TM for 12 or 24 hr. The y axis represents relative cell viability compared to ethanol-treated controls. Statistics, two-tailed Student’s t-test. (H) SIRT2 overexpression in (G) was confirmed by Western blot. Error bars represent mean ± sd. ***p < 0.001. See also Figure S4.
Second, we confirmed that TM inhibits SIRT2 in cells by detecting the acetylation level of known SIRT2 as well as SIRT1 targets. In MCF-7 and MDA-MB-468 cells, TA, TB, and TH, inhibited SIRT1, based on the acetylation level of a known SIRT1 deacetylation target, p53 (Figure 4D). In contrast, TM showed almost no inhibition of p53 deacetylation. By detecting the acetylation of α-tubulin, a known SIRT2 target, we monitored SIRT2 inhibition. TA or M, which did not inhibit SIRT2 well, did not affect the acetylation of α-tubulin. TB and TH, which have intermediate SIRT2 inhibition potency, slightly increased the acetylation of α-tubulin. TM, the best SIRT2 inhibitor, led to the greatest increase in α-tubulin acetylation (Figure 4C). The effect of TM on α-tubulin acetylation was dose-dependent, whereas M did not affect acetyl-α-tubulin level at 50 µM (Figure 4E). Similarly, TM, but not M, increased the level of α-tubulin acetylation in MDA-MB-231 cells based on immunofluorescence imaging (Figure 4F). SIRT2 has been reported to be not only a deacetylase, but also a defatty-acylase (He et al., 2014; Liu et al., 2014), so we further examined the effect of TM on the defatty-acylase activity of SIRT2 in cells. Metabolic labeling of fatty-acylated proteins revealed that SIRT2 KD (Figure S4B) but not TM (Figure S4C) was able to elevate the fatty-acylation levels of many proteins, suggesting that in cells TM is a potent inhibitor of SIRT2 deacetylase but not defatty-acylase.
Finally, to confirm that the anticancer effect of TM is due to SIRT2 inhibition, we tested the sensitivity of cells to TM under SIRT2 overexpression or knockdown conditions. If TM inhibits cancer cells by targeting SIRT2, overexpression of SIRT2 would decrease the sensitivity of cells to TM (the increased SIRT2 level would require more TM for inhibition), while partial and transient knockdown of SIRT2 would increase the sensitivity. Indeed, overexpression of SIRT2 (Figure 4H) significantly decreased the cytotoxicity of TM (Figure 4G), while transient and partial knockdown of SIRT2 (Figure S4E) sensitized cells to TM (Figure S4D). These results support the conclusion that the anticancer effect of TM is through SIRT2 inhibition instead of other off-target effects.
TM inhibits tumor growth in mouse models of breast cancer
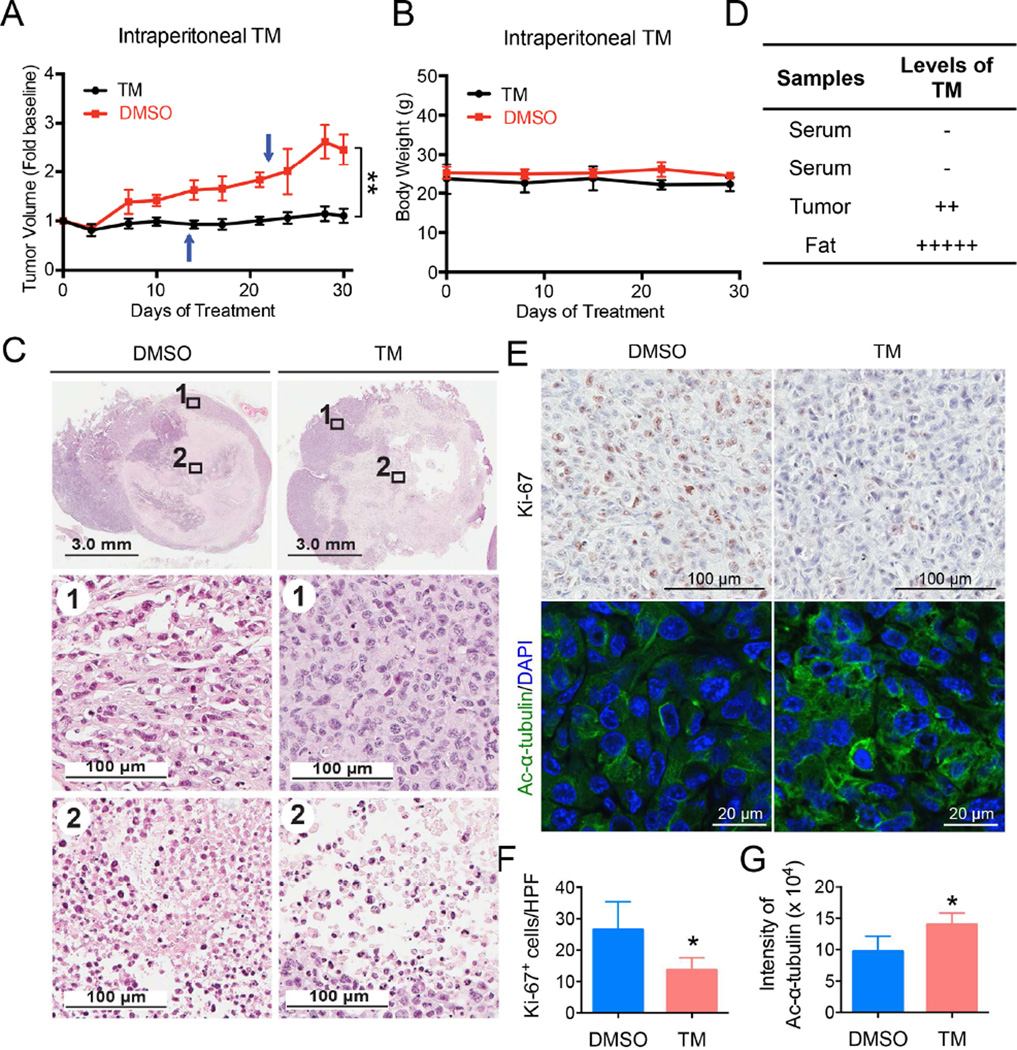
To further demonstrate that SIRT2 inhibition can be useful for treating cancers, we tested TM in two mouse models of cancer. The first was a xenograft model in which the triple-negative breast cancer cell line, MDA-MB-231, was injected subcutaneously into immunocompromised mice. When tumor size reached ~200 mm3, the mice were divided into two groups and treated by either direct intratumor (IT) (Figure S5) or intraperitoneal (IP) (Figure 5) injection of the control vehicle solvent (DMSO) or TM (1.5 mg TM in 50 µL DMSO; n = 5) daily. Tumors were collected after 30-days of treatment and analyzed. TM treatment significantly inhibited tumor growth as compared to the control (Figure S5A, S5B, and 5A). Histopathological examination revealed central areas of necrosis in tumors from both DMSO and TM treated mice, but the necrosis was more extensive and the overall tumor size was smaller in the TM treated mice (Figures S5D and 5C). IT TM injection showed a stronger effect in reducing tumor volume and increasing areas of necrosis as compared to IP TM injection. Analysis of TM content in tissue samples from TM-treated mice showed that IP-administered TM reached the tumors, even though the serum concentration of TM was low and a significant amount of TM accumulated in abdominal fat (Figure 5D). TM did not cause significant toxicity in mice (one mouse from each treatment group died, likely due to infection caused by repeated IP injection but not due to TM toxicity) and no significant weight loss was observed in TM-treated mice (Figure S5C and 5B). Immunohistochemistry staining of Ki-67 was performed to assess the effect of TM on the proliferation of tumor cells in vivo. As shown Figure 5E (upper panel) and 5F, as well as Figure S5F (upper panel) and S5G, a significant decrease in Ki-67+ cells was observed with TM treatment relative to vehicle treatment. To determine whether TM inhibits SIRT2 in vivo, we performed immunofluorescence staining of acetyl-α-tubulin in the xenograft tumors. As shown in Figure 5E (lower panel) and 5G, and Figure S5F (lower panel) and S5H, the acetyl-α-tubulin level was moderately but statistically significantly increased in tumors from TM treated mice compared with those from vehicle-treated mice, suggesting that TM indeed inhibits SIRT2 in vivo.
Figure 5.
Analysis of tumor growth and histopathological findings of xenografted mice treated by intraperitoneal TM injection. Mice bearing MDA-MB-231 human breast cancer xenograft were divided into two groups and treated by IP injection with either the vehicle (DMSO) or TM (1.5 mg TM in 50 µL DMSO; n = 5) daily. Tumors were collected after 30-day treatment. (A) Tumor growth chart. Arrows indicate time point when an animal was found dead (1 untreated, 1 treated). Statistics, paired Student’s t-test. (B) Mouse body weight chart. (C) Hematoxylin and eosin staining of tumor tissues after 30 days of treatment with DMSO or TM. (D) Detection of TM in mouse serum, fat and tumor tissues by mass spectrometry. (E) Representative images of Ki-67 immunohistochemistry staining and acetyl-α-tubulin (K40) immunofluorescence staining of tumor tissues after 30 days of treatment with DMSO or TM. (F) Quantification of Ki-67+ cells in (E). The y axis represents Ki-67+ cells per high power field (10 HPFs/tumor for all the tumors analyzed, n = 3 for DMSO, n = 4 for TM). Statistics, unpaired Student’s t-test. (F) Quantification of acetyl-α-tubulin fluorescence intensity in (E) by ImageJ. The y axis represents integrated intensity per cell. (10 HPFs/tumor for all the tumors analyzed, n = 3 for DMSO, n = 4 for TM). Statistics, unpaired Student’s t-test. Error bars represent mean ± sd. *p < 0.05, **p < 0.01. See also Figure S5.
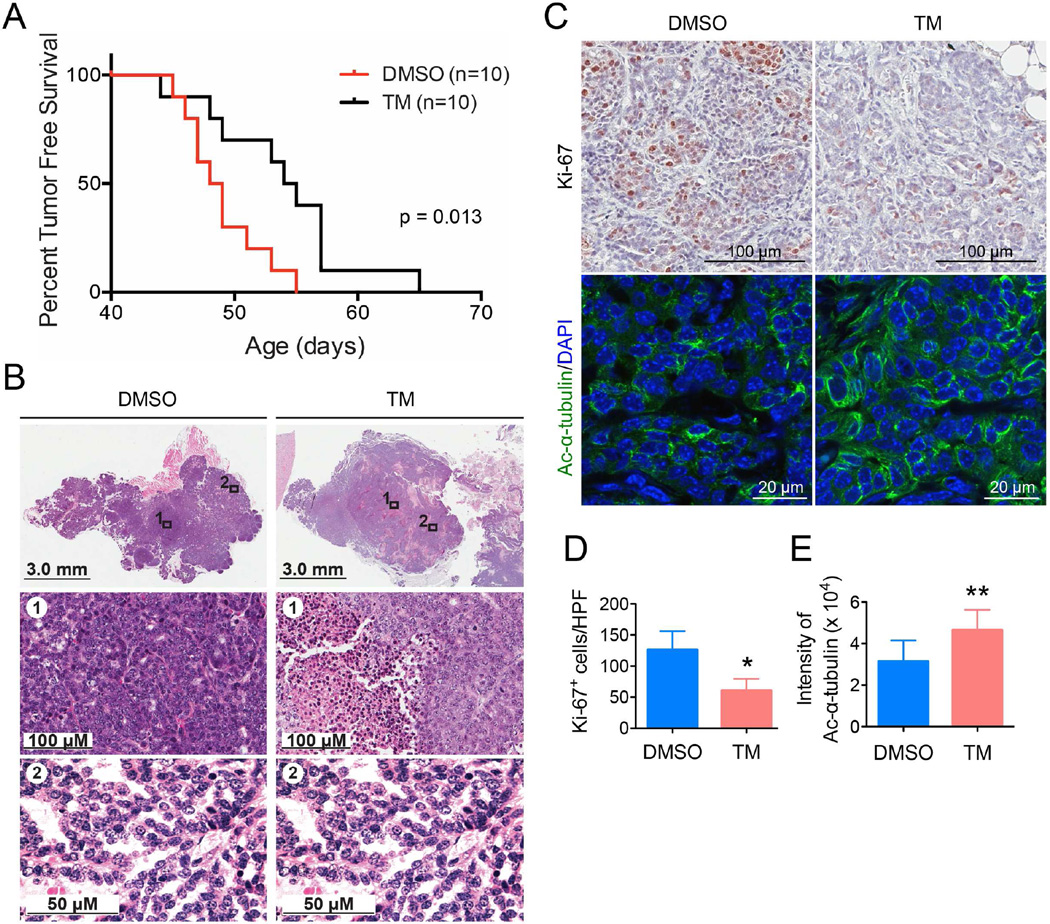
The second mouse model was the mammary tumor model driven by mammary gland-specific expression of polyoma middle T antigen under the control of mouse mammary tumor virus promoter/enhancer (MMTV-PyMT model) (Guy et al., 1992). The MMTV-PyMT mice received daily IP injections with either the control vehicle solvent (DMSO) or TM (1.5 mg TM in 50 µL DMSO; n = 10). The Kaplan-Meier tumor-free survival curve showed that TM treatment significantly prolonged the tumor-free survival of mice compared with vehicle-treated mice (Figure 6A). While the average time to tumor onset in the control group was 48 days, the mean latency for TM-treated mice was 54 days. Histopathological examination revealed more extensive areas of necrosis in the neoplasms from TM-treated mice as compared to the control group (Figure 6B). A significant decrease in proliferation of tumor cells was observed with TM treatment relative to vehicle treatment as measured by Ki-67 staining (Figure 6C, upper panel, and 6D). A modest but statistically significant increase in the acetyl-α-tubulin level was observed in tumors from TM-treated mice compared to those from vehicle-treated mice (Figure 6C, lower panel, and 6E), indicating that SIRT2 was inhibited by TM in vivo. The data demonstrate that SIRT2 inhibition with TM delays tumor onset in the MMTV-PyMT model and reduces tumor growth in vivo.
Figure 6.
Mammary tumorigenesis in MMTV-PyMT female mice following intraperitoneal TM injection. (A) Kaplan-Meier tumor-free survival curve of MMTV-PyMT mice treated by IP injection with either the vehicle (DMSO) or TM (1.5 mg TM in 50 µL DMSO; n = 10) daily. The x-axis shows mice age; the y-axis shows proportion of mice remaining tumor-free. Statistics, log-rank test. (B) Hematoxylin and eosin staining of mammary tumors after 30 days of treatment with either DMSO or TM. (C) Representative images of Ki-67 immunohistochemistry staining and acetyl-α-tubulin (K40) immunofluorescence staining of tumor tissues after 30 days of treatment with either DMSO or TM. (D) Quantification of Ki-67+ cells in (C). The y axis shows Ki-67+ cells per high power field (10 HPFs/tumor for all the tumors analyzed, n = 4 for DMSO, n = 4 for TM). Statistics, unpaired Student’s t-test. (E) Quantification of acetyl-α-tubulin fluorescence intensity in (C) by ImageJ. The y axis shows integrated intensity per cell. (10 HPFs/tumor for all the tumors analyzed, n = 8 for DMSO, n = 8 for TM). Statistics, unpaired Student’s t-test. Error bars represent mean ± sd. *p < 0.05, **p < 0.01.
COMPARE analysis with the NCI-60 cancer cell panel points to possible mechanism of action for the SIRT2 inhibitor TM
To further investigate the anticancer effects of TM, we first examined whether the level of SIRT2 in different cell lines could be used to predict which cell lines would be more sensitive to SIRT2 inhibitors. We checked the SIRT2 protein level in all the eight human normal and breast cancer cell lines above (Figure 2B & 3B) to see if the sensitivity to TM correlated with SIRT2 level in these cell lines. Compared to MCF-10A and HME1 cells, the cancer cell lines showed relatively high SIRT2 expression. However, we did not see an obvious correlation between SIRT2 level and TM sensitivity (Figure S6A and S6B) among the cancer cell lines, suggesting that other factors account for the SIRT2 inhibitor sensitivity.
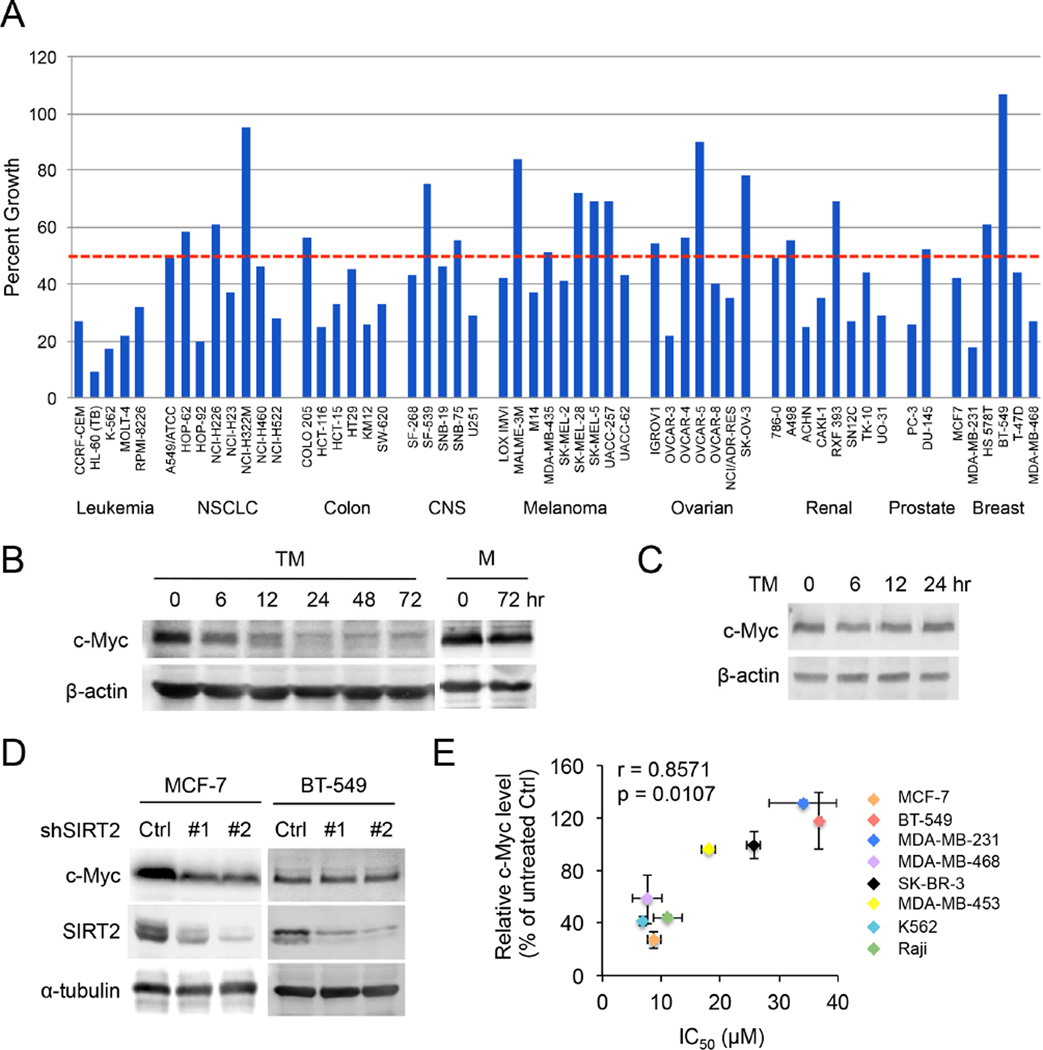
To examine the anticancer activity of TM against other malignancies and the molecular mechanisms underlying its activity, we submitted the TM compound to the Developmental Therapeutics Program of the National Cancer Institute (NCI) at the National Institutes of Health for screening against the NCI-60 panel of human cancer cell lines (Shoemaker, 2006) at a single dose of 10 µM. The screening result showed that TM inhibited 36/56 of the NCI-60 cell lines by >50% at 10 µM (Figure 7A). In particular, all the leukemia cell lines were very sensitive to TM and most of colon cancer cell lines were sensitive to TM. In contrast, melanoma and ovarian cancer cells were less sensitive to TM. Consistent with our earlier findings (Figure 2), MCF-7 and MDA-MB-468 cells were very sensitive to TM. One discrepancy was noted for MDA-MB-231 cells, which were very sensitive to TM in the NCI-60 screening, but not very sensitive to TM in our study (IC50 34 µM). This could be due to differences in the MDA-MB-231 cells or the culture conditions used in NCI-60 screening and our laboratory. To confirm our findings with MDA-MB-231 cells, we purchased a new batch of MDA-MB-231 cells and showed that the sensitivity to TM was similar to that of the cells we tested earlier (Figure S2D). Despite the discrepancy with MDA-MB-231 cells, the screening results suggest that SIRT2 inhibitors can potentially be used to treat many types of cancers.
Figure 7.
TM inhibits various types of human cancer cell lines and decreases c-Myc protein level. (A) NCI-60 cell line screening of TM. NCI-60 cell lines were cultured with and without 10 µM TM for 24 hr. The percent growth of TM-treated cells compared to the controls is shown. The horizontal dotted red line shows 50% growth. (B) The c-Myc protein levels in MCF-7 cells treated with TM (25 µM) or M (25 µM). (C) The c-Myc protein levels in BT-549 cells treated with TM (25 µM). (D) The levels of c-Myc, SIRT2 and α–tubulin in MCF-7 or BT-549 cells infected with luciferase or SIRT2 shRNAs for 72 hr. (E) The correlation between the ability of TM to inhibit cancer cell lines and its ability to decrease c-Myc level. The x axis shows IC50 values of TM in different cell lines. The y axis shows the TM-induced decreases in c-Myc level. Relative c-Myc level was obtained by comparing the c-Myc protein level in cells treated with TM for 24 hr to that in vehicle-treated control cells. Error bars represent mean ± sd. See also Figure S6.
To investigate how SIRT2 inhibition halts cancer cell proliferation, we took advantage of NCI molecular target COMPARE analysis (Zaharevitz et al., 2002). NCI has accumulated many data sets regarding the properties of the NCI-60 cell lines, including gene expression, DNA methylation, protein expression, and post-translational modifications. The molecular target COMPARE analysis serves to correlate the response of the NCI-60 panel to a small molecule (TM in this case) to known molecular patterns. From this analysis, we found that the sensitivity of NCI-60 cell lines to TM correlated best with c-Myc phosphorylation/protein levels. In other words, cell lines with higher c-Myc phosphorylation/protein levels were more sensitive to TM (Table S1). The correlation between TM sensitivity and c-Myc is intriguing as c-Myc is an oncoprotein that is up-regulated in many cancers.
TM decreases c-Myc oncoprotein level in cancer cells
The correlation between TM efficacy and c-Myc was informative, but the small correlation value (~0.5) was not sufficient to establish a mechanistic relationship. To further understand the connection, we measured c-Myc level in the cells treated with and without TM or M. TM decreased c-Myc protein level in a time-dependent manner in MCF-7 cells, whereas M treatment had no effect on the c-Myc protein level (Figure 7B). Similar effects of TM on c-Myc were also observed in K562 and MDA-MB-468 cells (Figure S6C and S6D). Consistent with the effect of TM, c-Myc abundance was also reduced by SIRT2 KD (Figure 7D), suggesting that TM works through SIRT2 inhibition to decrease c-Myc. To further establish that the reduction in c-Myc protein is important for the anticancer effect of TM, we examined whether the sensitivity of cancer cell lines to TM correlated with the decrease in c-Myc level induced by TM treatment. Among the six breast cancer cell lines in the NCI-60 panel, BT-549 did not respond to treatment with 10 µM TM. At 10 µM TM, the viability of BT-549 was close to 100%. This result was in line with our own findings (Figure 2B). Although higher concentrations of TM did decrease the viability of BT-549, the sensitivity was much lower than that of MCF-7 cells. Consistent with the reduced sensitivity to TM, SIRT2 KD in BT-549 cells did not decrease cell viability (Figure 3B and S3C). We therefore examined whether TM could affect c-Myc protein level in BT-549 cells. Consistent with the decreased TM sensitivity, TM treatment did not have a significant effect on c-Myc protein abundance in BT-549 cells (Figure 7C). SIRT2 KD also failed to decrease c-Myc level in BT-549 cells (Figure 7D). These data collectively suggest that the sensitivity of cancer cell lines to TM correlates with the ability of TM to decrease c-Myc level via SIRT2 inhibition in these cell lines. We further measured the IC50 values of TM in six different cancer cell lines and the corresponding decrease in c-Myc level in these cell lines upon TM treatment. Plotting the IC50 values against the decreases in c-Myc levels indicates that there was an excellent correlation between them (Figure 7E), supporting that the ability of TM to decrease c-Myc is important for its anticancer effect in the cell lines that are very sensitive to TM.
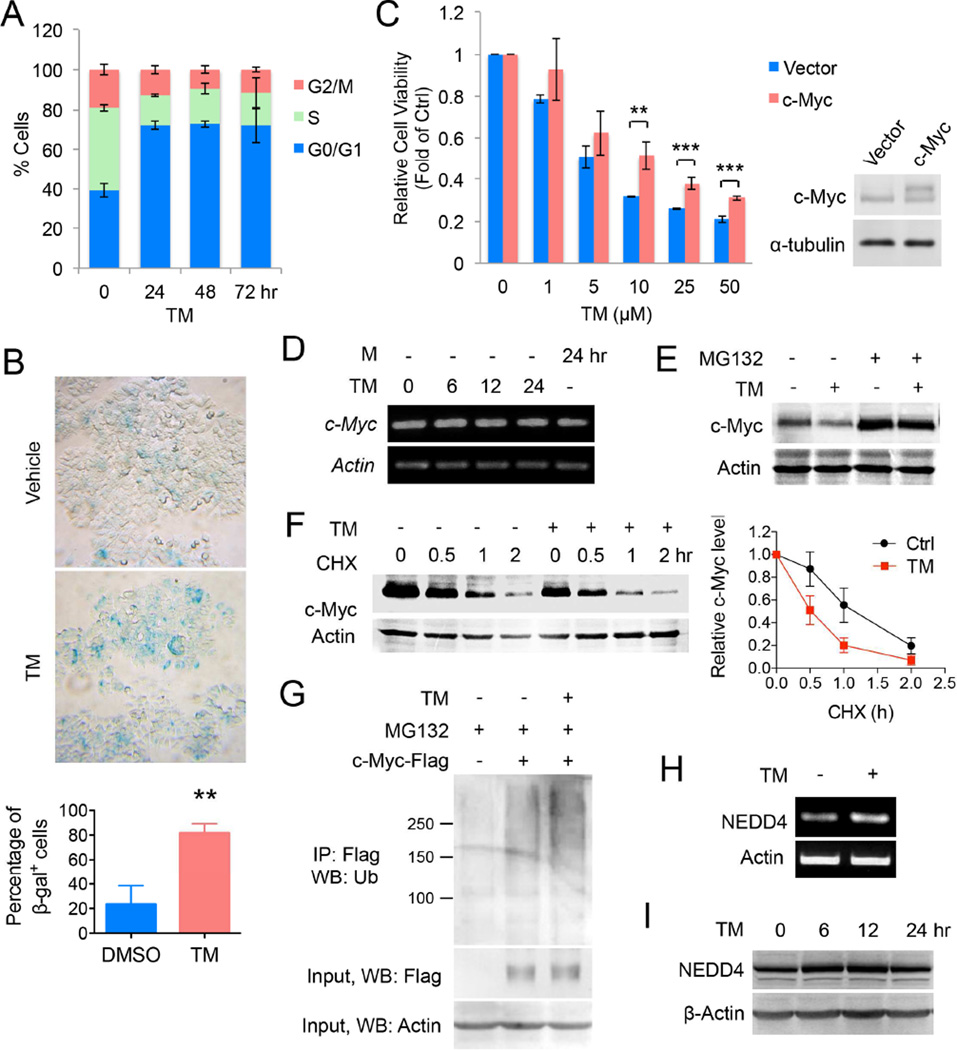
MCF-7 cells were then further analyzed for Myc-specific biological effects. Flow cytometry of TM-treated cells revealed a pronounced increase in cells arrested in G0/G1 phase, with a concomitant decrease of cells in S phase (Figure 8A). Treatment of TM resulted in significant cellular senescence by β-galactosidase staining (Figure 8B). Similar effects of TM on cell cycle progression and cellular senescence were also observed in K562 cells (Figure S6E and S6F), suggesting that the effect of TM-induced c-Myc decrease is not restricted to breast cancer cells. Overall, the phenotypes of G0/G1 cell cycle arrest and cellular senescence are consistent with the anticipated effects of inhibiting cellular c-Myc function (Wu et al., 2007).
Figure 8.
Decreasing c-Myc protein abundance contributes to the anticancer effect of TM. (A) Cell cycle distribution (assessed by propidium iodide staining-coupled flow cytometry) of MCF-7 cells treated with TM (25 µM) for 0, 24, 48 or 72 hr. (B) Acidic β-gal (β-gal) staining in MCF-7 cells treated with TM (25 µM) for 5 days. Representative images were shown in the upper panel, quantification was shown as percentage of β-gal+ cells in the lower panel. Statistics, two-tailed Student’s t-test. (C) Effect of c-Myc overexpression on the cytotoxicity effect of TM. MCF-7 cells were transfected with pCDH vector or pCDH-c-Myc for 12 hr before being treated with TM for 72 hr. Statistics, two-tailed Student’s t-test. (D) The mRNA levels of c-Myc in MCF-7 cells treated with TM (25 µM) or M (25 µM) analyzed by RT-PCR. (E) Effect of MG132 on TM-mediated decrease in c-Myc protein level in MCF-7 cells. Cells were treated with ethanol or TM (25 µM in ethanol) for 4 hr and then MG132 (10 µM) for 2 hr. (F) Effect of TM on c-Myc degradation in MCF-7 cells. Cells were incubated ethanol or TM (25 µM in ethanol) for 4 hr and then with CHX (10 µg/mL) for 0, 0.5, 1, or 2 hr. Loading was normalized based on the level of the internal control, actin. The relative c-Myc protein levels at different time point of CHX treatment were calculated by normalizing to the corresponding level without CHX treatment. The relative c-Myc levels were plotted against the time of treatment with CHX. (G) Effect of TM (25 µM) on the polyubiquitination of c-Myc in MCF-7. (H) The mRNA level of NEDD4 in MCF-7 cells treated with TM (25 µM) for 12 hr. (I) Western blot analysis of NEDD4 protein level in MCF-7 cells treated with TM (25 µM). Error bars represent mean ± sd. **p < 0.01, ***p < 0.001. See also Figure S6.
To further establish that decreasing c-Myc is important for the anticancer effect of TM, we examined whether forced overexpression of c-Myc in MCF-7 cells is able to reduce TM-mediated cytotoxicity. Cells were transfected with c-Myc for 12 hr before being treated with TM. As shown in Figure 8C and S6G, overexpression of c-Myc significantly reduced the cytotoxicity effect of TM. Together, these results demonstrate TM decreases c-Myc, which is important for the cytotoxicity of TM in tumor cell, although it is likely not the only mechanism that underlies the cytotoxicity.
The c-Myc mRNA level was not affected by TM treatment, suggesting that TM does not affect c-Myc transcription (Figure 8D, Figure S6H). Therefore, the effect of TM on c-Myc protein turnover was tested. The half-life of c-Myc was shortened by TM treatment, suggesting that TM promoted c-Myc degradation (Figure 8F). Treatment with a proteasome inhibitor, MG132, prevented the TM-induced down-regulation of c-Myc, suggesting that TM promoted the proteasomal degradation of c-Myc (Figure 8E). Increased proteasomal degradation was associated with increased c-Myc ubiquitination (Figure 8G). It was previously reported that SIRT2 can suppress the expression of NEDD4, an E3 ubiquitin ligase for c-Myc (Liu et al., 2013), which could explain why SIRT2 inhibition promotes c-Myc degradation. Indeed, NEDD4 was up-regulated by TM at the transcriptional level (Figure 8H and Figure S6H) and also modestly at the protein level (Figure 8I and Figure S6C) in both MCF-7 and K562 cells. However, this is not a universal mechanism as alteration of NEDD4 level was not detected in TM-treated MDA-MB-468 cells despite the observed reduction in c-Myc protein abundance (Figure S6D & H). As TM regulates the protein stability of c-Myc in all three cell lines, we checked the effect of TM on the transcription levels of several additional known E3 ligases that destabilize c-Myc (Choi et al., 2010; Kim et al., 2003; Liu et al., 2013; Paul et al., 2013; Welcker et al., 2004). As shown in Figure S6H, NEDD4 and TRPC4AP were increased in MCF-7 and K562 cells, but not in MDA-MB-468 cells; FBXW7 and STUB1 were up-regulated only in MDA-MB-468 cells; FBXO32 was increased in all the three cell lines. However, none of the E3 ligase genes was obviously up-regulated by TM in BT-549 cells in which neither cell viability nor c-Myc level was affected by TM. These results suggested that SIRT2 inhibition led to up-regulation of several c-Myc E3 enzymes, which may result in the destabilization of c-Myc by TM.
Discussion
Previous reports have suggested that SIRT1 or SIRT2 inhibitors can have anticancer activity. However, the potency of most of these inhibitors is modest, with IC50 values in the micromolar range at inhibiting purified sirtuins. Most of the sirtuin inhibitors tested for anticancer activity are also not very selective and can inhibit several sirtuins. The modest potency and selectivity make it hard to rule out off-target effects and pinpoint which sirtuin should be targeted for treating cancers. Our SIRT2 inhibitor TM described here has an excellent combination of potency and selectivity that allowed us to conclude that inhibiting SIRT2 produces anticancer effects in a variety of human cancer cell lines. Knocking down of all seven sirtuins also confirmed that SIRT2 is important for the viability of various cancer cell lines while knocking down other sirtuins either had no significant effect or much less effect on cancer cell viability.
C-Myc is an important oncoprotein and is up-regulated in many human tumors. Thus, it has been considered as a promising cancer target. So far, no small molecules can directly target c-Myc in vivo. Recent studies showed that bromodomain inhibitors targeting BRD4 can suppress c-Myc transcription and inhibit tumorigenesis (Delmore et al., 2011). Our studies demonstrate that inhibiting SIRT2 offers a different way to target c-Myc. We show here that our SIRT2 inhibitor TM can effectively decrease the level of c-Myc in various cancer cell lines. Our data suggest that the ability of TM to decrease c-Myc abundance in different cell lines correlates with the sensitivity of the cell lines to TM. We further demonstrate here that decreasing c-Myc protein level is an important mechanism that accounts for hypersensitivity of certain cancer cell lines to TM. However, it should be pointed out that effects on other SIRT2-regulated pathways may also contribute to the activity of TM in cancer cells. This is especially true given that even cells without TM-induced c-Myc decrease (e.g. MDA-MB-231 and BT-549 cells) can still be inhibited by TM at higher concentrations. This also likely explains why c-Myc overexpression confers some but not complete resistance to TM (Figure 8C). We found that TM promotes the proteolytic degradation of c-Myc without affecting its transcription, which serves as an important but perhaps not the only mechanism by which TM destabilizes c-Myc. Aberrant translational control of the Myc oncoprotein has been implicated in many cancers (Chappell et al., 2000; Wolfe et al., 2014) and might also be involved in TM-induced reduction in c-Myc level. Our work establishes SIRT2 inhibition as a strategy to target the oncoprotein c-Myc, which is effective in many human cancer cell lines. Future detailed mechanistic investigations of the SIRT2/c-Myc regulatory pathway could potentially lead to the identification of additional therapeutic targets.
The roles of sirtuins in cancer have been a topic of debate. Both tumor-promoting and tumor-suppressing roles of SIRT1 have been reported. For SIRT2, Kim et al. reported that SIRT2 is a tumor suppressor because Sirt2 KO mice develop tumors earlier than WT mice (Kim et al., 2011a). Serrano and co-workers did not find a cancer-prone phenotype in unchallenged Sirt2 KO mice that they generated, although they did observe that Sirt2 KO mice had increased tumorigenesis when challenged with carcinogens (Serrano et al., 2013). Contradictory to these genetic studies that pointed to a weak tumor suppressor role of SIRT2, we found that inhibiting SIRT2 with TM has broad anticancer activity in many cancer cell lines.
Different outcomes for mouse genetic studies and pharmacological studies in cancer cells are not without precedent. Similar cases have been well documented in the literature (Weiss et al., 2007). There are several possible explanations. First, there are several examples of factors that have tumor suppressor activity in normal cells but nevertheless are required for the growth and survival of transformed cells. For example, loss of function for the DNA damage checkpoint kinase ATR causes modest tumor predisposition, but greatly impairs the growth of established tumors (Bartek et al., 2012). SIRT2 has been identified as a regulator of mitotic chromosome segregation (Kim et al., 2011a), a function that could account for the weak tumor predisposition phenotype in Sirt2-deficient mice given the oncogenic consequences of genomic instability. Nevertheless, a greater dependency of transformed cells on SIRT2 due to increased mitotic and other stresses, or because of the regulation of other targets such as c-Myc by SIRT2, result in heightened sensitivity to SIRT2 inhibition in cancer cells. It also should be noted that small molecules may have off target effects, which could contribute to observed pharmacological effects. While it is difficult to completely rule out this possibility for the anticancer effect of TM, our studies using the inactive control compound (M) and the SIRT2 KD studies suggest that the anticancer effect is largely through SIRT2 inhibition.
An alternative explanation relates to the fact that in a genetic knockout, the protein is gone and thus all the enzymatic activities and protein-protein interactions involving the enzyme also are gone. In contrast, when using a small molecule to inhibit the enzyme, the protein is intact and so are the protein-protein interactions that involve the protein. In the case of SIRT2, another layer of complexity is that SIRT2 has multiple enzymatic functions. We and others recently found that sirtuins are not only deacetylases. Some sirtuins, such as SIRT5 (Du et al., 2011) and SIRT6 (Jiang et al., 2013), prefer to hydrolyze other acyl lysine modifications. Perhaps more surprising is the fact that even the well-studied deacetylases (SIRT1, SIRT2, and SIRT3) can remove long chain fatty acyl groups efficiently (He et al., 2014; Liu et al., 2014). Although the exact substrate proteins for the defatty-acylase activity of SIRT2 remain to be identified, our preliminary studies showed that the fatty-acylation levels of many proteins were elevated when SIRT2 was knocked down (Figure S4B), but not when SIRT2 inhibitor TM was used (Figure S4C). Thus, the small molecule inhibitor may selectively target one of the enzymatic functions of SIRT2, thus contributing to the fact that small molecule inhibitors may produce beneficial pharmacological effects that are different from genetic knockout.
Experimental Procedures
For more details, see Supplemental Experimental Procedures.
Synthesis of Compounds used in the study
Detailed synthetic routes are presented in Supplemental Experimental Procedures. The NMR spectra of the synthesized compounds are shown in Figure S7.
Inhibition assay for different sirtuins
The assays were carried out using an HPLC-based method with different acyl peptides. The detailed method is described in Supplemental Experimental Procedures.
Cell viability assay
Cells were seeded into 96-well plates at 3000–4000 cells per well. After 24 hr, test compounds were added to cells to final concentrations ranging from 1–50 µM. Cells were then incubated for 72 hr and cell viability was measured using the CellTiter-Blue viability assay (Promega) following the manufacturer’s instruction. Relative cell viability in the presence of test compounds was normalized to the vehicle-treated controls after background subtraction. Graphpad Prism software was used to determine the IC50 values.
Knockdown of SIRT1–7 in various cell lines was achieved by lentiviral infection. Lentiviral supernatants were generated as described previously. Cell viability was assessed after 3, 5 or 10 days of infection by using CellTiter-Blue.
Animal experiments
All animals used in this study were handled in accordance with federal and institutional guidelines, under a protocol approved by the Cornell University Institutional Animal Care and Use Committee (IACUC). For more animal experimental details, please see the Supplemental Experimental Procedures.
Statistical analysis
Quantitative data were expressed as mean ± sd (standard deviation, shown as error bar) from at least three independent experiments. Differences between two groups were examined statistically as indicated (*p < 0.05, **p < 0.01, and ***p < 0.001).
Supplementary Material
Significance.
Inhibiting oncoproteins frequently found in cancers is a common theme in targeted cancer therapy. One such oncoprotein is c-Myc, which is up-regulated in ~50% human tumors. Small molecules targeting c-Myc are highly sought as anticancer agents. Our studies identified SIRT2 as a promising target to treat c-Myc-driven cancers. We have developed a SIRT2 inhibitor with the best combination of potency and selectivity reported and showed that it has broad anticancer activity with little effects on non-cancerous cells. The small molecule inhibitor studies combined with knockdown and/or overexpression of sirtuins provide extensive evidence establishing SIRT2 as a promising anticancer target, especially for certain c-Myc-driven cancers.
Highlights.
TM, a SIRT2 inhibitor with excellent potency and specificity, has been developed
TM has broad anticancer activity
SIRT2 inhibition promotes c-Myc ubiquitination and degradation
SIRT2 is a promising target for c-Myc-driven cancers
Acknowledgments
We thank the Development Therapeutic Program at National Cancer Institute (NCI) for screening TM in the NCI60 cell lines. This work is supported in part by an intercampus seed grant from Cornell University and a transformative R01 grant from NIH (CA163255).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
HJ designed and performed all the biochemical studies except those noted below. BH and HL designed the sirtuin inhibitors. JH, B. and YLC synthesized the inhibitors and biotin-conjugated compounds. HJ, JH, and BH purified the sirtuin enzymes, performed in vitro inhibitor assay and determined the mechanism of SIRT2 inhibition. YLNA and JS performed the animal studies. KW, MC and PG carried out the Biotin-TM pull-down assay and immunofluorescence of acetyl-α-tubulin. PG suggested the NCI60 screening, which helped making the c-Myc connection. TS performed pathologic review. RSW directed the animal studies. HL directed the inhibitor development and biochemical studies, and wrote the manuscript with help from HJ, RSW, PG, JH and YLNA.
Reference
- Bartek J, Mistrik M, Bartkova J. Thresholds of replication stress signaling in cancer development and treatment. Nat. Struct Mol. Biol. 2012;19:5–7. doi: 10.1038/nsmb.2220. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell SA, LeQuesne JP, Paulin FE, deSchoolmeester ML, Stoneley M, Soutar RL, Ralston SH, Helfrich MH, Willis AE. A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: a novel mechanism of oncogene de-regulation. Oncogene. 2000;19:4437–4440. doi: 10.1038/sj.onc.1203791. [DOI] [PubMed] [Google Scholar]
- Chen J, Chan AW, To KF, Chen W, Zhang Z, Ren J, Song C, Cheung YS, Lai PB, Cheng SH, et al. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3beta/beta-catenin signaling. Hepatology. 2013;57:2287–2298. doi: 10.1002/hep.26278. [DOI] [PubMed] [Google Scholar]
- Cheon MG, Kim W, Choi M, Kim JE. AK-1, a specific SIRT2 inhibitor, induces cell cycle arrest by downregulating Snail in HCT116 human colon carcinoma cells. Cancer Lett. 2015;356:637–645. doi: 10.1016/j.canlet.2014.10.012. [DOI] [PubMed] [Google Scholar]
- Choi SH, Wright JB, Gerber SA, Cole MD. Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 2010;24:1236–1241. doi: 10.1101/gad.1920310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fruscia P, Ho KK, Laohasinnarong S, Khongkow M, Kroll SH, Islam SA, Sternberg MJ, Schmidtkunz K, Jung M, Lam EW, Fuchter MJ. The Discovery of Novel 10,11-Dihydro-5H-dibenz[b,f]azepine SIRT2 Inhibitors. Medchemcomm. 2012 doi: 10.1039/C2MD00290F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu J, Khan SH, Kim J, Woo J, Kim JH, Choi BH, et al. Sirt5 is an NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Nicholl MB. Sirtuin 1 in malignant transformation: Friend or foe? Cancer Lett. 2011;306:10–14. doi: 10.1016/j.canlet.2011.02.019. [DOI] [PubMed] [Google Scholar]
- Fatkins DG, Monnot AD, Zheng W. Nε-Thioacetyl-lysine: A multi-facet functional probe for enzymatic protein lysine Nε-deacetylation. Bioorg. Med. Chem. Lett. 2006;16:3651–3656. doi: 10.1016/j.bmcl.2006.04.075. [DOI] [PubMed] [Google Scholar]
- Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian Sirtuins: Biological Insights and Disease Relevance. Annu. Rev. Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawse WF, Hoff KG, Fatkins DG, Daines A, Zubkova OV, Schramm VL, Zheng W, Wolberger C. Structural insights into intermediate steps in the Sir2 deacetylation reaction. Structure. 2008;16:1368–1377. doi: 10.1016/j.str.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Du J, Lin H. Thiosuccinyl peptides as Sirt5-specific inhibitors. J. Am. Chem. Soc. 2012;134:1922–1925. doi: 10.1021/ja2090417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Hu J, Zhang X, Lin H. Thiomyristoyl peptides as cell-permeable Sirt6 inhibitors. Org. Biomol. Chem. 2014;12:7498–7502. doi: 10.1039/c4ob00860j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA, Bedalov A. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nat. Rev. Cancer. 2010;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann G, Breitenbucher F, Schuler M, Ehrenhofer-Murray AE. A novel sirtuin 2 (SIRT2) inhibitor with p53-dependent pro-apoptotic activity in non-small cell lung cancer. J. Biol. Chem. 2014;289:5208–5216. doi: 10.1074/jbc.M113.487736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S-i, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Imai S-i, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol. Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011a;20:487–499. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol. Cell. 2003;11:1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- Kim WJ, Lee JW, Quan C, Youn HJ, Kim HM, Bae SC. Nicotinamide inhibits growth of carcinogen induced mouse bladder tumor and human bladder tumor xenograft through up-regulation of RUNX3 and p300. J. Urol. 2011b;185:2366–2375. doi: 10.1016/j.juro.2011.02.017. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Choy ML, Marks PA. Chapter Two - Mechanisms of Resistance to Histone Deacetylase Inhibitors. In: Steven G, editor. Adv. Cancer Res. Academic Press; 2012. pp. 39–86. [DOI] [PubMed] [Google Scholar]
- Liu PY, Xu N, Malyukova A, Scarlett CJ, Sun YT, Zhang XD, Ling D, Su SP, Nelson C, Chang DK, et al. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ. 2013;20:503–514. doi: 10.1038/cdd.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yang T, Li X, Peng T, Hang HC, Li XD. Integrative Chemical Biology Approaches for Identification and Characterization of "Erasers" for Fatty-Acid-Acylated Lysine Residues within Proteins. Angew Chem. Int Ed Engl. 2014 doi: 10.1002/anie.201408763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan SS, Scian M, Sripathy S, Posakony J, Lao U, Loe TK, Leko V, Thalhofer A, Schuler AD, Bedalov A, Simon JA. Development of pyrazolone and isoxazol-5-one cambinol analogues as sirtuin inhibitors. J. Med. Chem. 2014;57:3283–3294. doi: 10.1021/jm4018064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- McCarthy AR, Sachweh MC, Higgins M, Campbell J, Drummond CJ, van Leeuwen IM, Pirrie L, Ladds MJ, Westwood NJ, Lain S. Tenovin-D3, a novel small-molecule inhibitor of sirtuin SirT2, increases p21 (CDKN1A) expression in a p53-independent manner. Mol. Cancer Ther. 2013;12:352–360. doi: 10.1158/1535-7163.MCT-12-0900. [DOI] [PubMed] [Google Scholar]
- McGlynn LM, Zino S, MacDonald AI, Curle J, Reilly JE, Mohammed ZM, McMillan DC, Mallon E, Payne AP, Edwards J, Shiels PG. SIRT2: tumour suppressor or tumour promoter in operable breast cancer? Eur. J. Cancer. 2014;50:290– 301. doi: 10.1016/j.ejca.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat. Rev. Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- Neugebauer RC, Uchiechowska U, Meier R, Hruby H, Valkov V, Verdin E, Sippl W, Jung M. Structure-activity studies on splitomicin derivatives as sirtuin inhibitors and computational prediction of binding mode. J. Med. Chem. 2008;51:1203–1213. doi: 10.1021/jm700972e. [DOI] [PubMed] [Google Scholar]
- Paul I, Ahmed SF, Bhowmik A, Deb S, Ghosh MK. The ubiquitin ligase CHIP regulates c-Myc stability and transcriptional activity. Oncogene. 2013;32:1284–1295. doi: 10.1038/onc.2012.144. [DOI] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotili D, Tarantino D, Nebbioso A, Paolini C, Huidobro C, Lara E, Mellini P, Lenoci A, Pezzi R, Botta G, et al. Discovery of salermide-related sirtuin inhibitors: binding mode studies and antiproliferative effects in cancer cells including cancer stem cells. J. Med. Chem. 2012;55:10937–10947. doi: 10.1021/jm3011614. [DOI] [PubMed] [Google Scholar]
- Seifert T, Malo M, Kokkola T, Engen K, Friden-Saxin M, Wallen EA, Lahtela-Kakkonen M, Jarho EM, Luthman K. Chroman-4-one- and chromone-based sirtuin 2 inhibitors with antiproliferative properties in cancer cells. J. Med. Chem. 2014;57:9870–9888. doi: 10.1021/jm500930h. [DOI] [PubMed] [Google Scholar]
- Serrano L, Martinez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, Beck DB, Kane-Goldsmith N, Tong Q, Rabanal RM, et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 2013;27:639–653. doi: 10.1101/gad.211342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Smith BC, Denu JM. Mechanism-based Inhibition of Sir2 deacetylases by thioacetyl-lysine peptide. Biochemistry. 2007;46:14478–14486. doi: 10.1021/bi7013294. [DOI] [PubMed] [Google Scholar]
- Soung YH, Pruitt K, Chung J. Epigenetic silencing of ARRDC3 expression in basal-like breast cancer cells. Sci. Rep. 2014;4:3846. doi: 10.1038/srep03846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stünkel W, Campbell RM. Sirtuin 1 (SIRT1): the misunderstood HDAC. J. Biomol. Screen. 2011;16:1153–1169. doi: 10.1177/1087057111422103. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Asaba T, Imai E, Tsumoto H, Nakagawa H, Miyata N. Identification of a cell-active non-peptide sirtuin inhibitor containing N-thioacetyl lysine. Bioorg. Med. Chem. Lett. 2009;19:5670–5672. doi: 10.1016/j.bmcl.2009.08.028. [DOI] [PubMed] [Google Scholar]
- Teng YB, Jing H, Aramsangtienchai P, He B, Khan S, Hu J, Lin H, Hao Q. Efficient demyristoylase activity of SIRT2 revealed by kinetic and structural studies. Sci. Rep. 2015;5:8529. doi: 10.1038/srep08529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss WA, Taylor SS, Shokat KM. Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat. Chem. Biol. 2007;3:739–744. doi: 10.1038/nchembio1207-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U S A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJM, Roderick JE, Van der Meulen J, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc. Natl. Acad. Sci. U S A. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Laurent G, Bause AS, Spang R, German N, Haigis MC, Haigis KM. HDAC6 and SIRT2 regulate the acetylation state and oncogenic activity of mutant K-RAS. Mol. Cancer Res. 2013;11:1072–1077. doi: 10.1158/1541-7786.MCR-13-0040-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YK, Ali MA, Wei AC, Shirazi AN, Parang K, Choon TS. Benzimidazoles as new scaffold of sirtuin inhibitors: green synthesis, in vitro studies, molecular docking analysis and evaluation of their anti-cancer properties. Eur. J. Med. Chem. 2014;83:448–454. doi: 10.1016/j.ejmech.2014.06.060. [DOI] [PubMed] [Google Scholar]
- Zaharevitz DW, Holbeck SL, Bowerman C, Svetlik PA. COMPARE: a web accessible tool for investigating mechanisms of cell growth inhibition. J. Mol. Graph Model. 2002;20:297–303. doi: 10.1016/s1093-3263(01)00126-7. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Au Q, Zhang M, Barber JR, Ng SC, Zhang B. Identification of a small molecule SIRT2 inhibitor with selective tumor cytotoxicity. Biochem. Biophys. Res. Commun. 2009;386:729–733. doi: 10.1016/j.bbrc.2009.06.113. [DOI] [PubMed] [Google Scholar]
- Zhao D, Mo Y, Li MT, Zou SW, Cheng ZL, Sun YP, Xiong Y, Guan KL, Lei QY. NOTCH-induced aldehyde dehydrogenase 1A1 deacetylation promotes breast cancer stem cells. J. Clin Invest. 2014;124:5453–5465. doi: 10.1172/JCI76611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Zou S-W, Liu Y, Zhou X, Mo Y, Wang P, Xu Y-H, Dong B, Xiong Y, Lei Q-Y, Guan K-L. Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer cell. 2013;23:464–476. doi: 10.1016/j.ccr.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AY, Zhou Y, Khan S, Deitsch KW, Hao Q, Lin H. Plasmodium falciparum Sir2A preferentially hydrolyzes medium and long chain fatty acyl lysine. ACS Chem. Biol. 2012;7:155–159. doi: 10.1021/cb200230x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.