Abstract
Two exons of the human haptoglobin (HP) gene exhibit copy number variation that affects HP multimerization and underlies one of the first protein polymorphisms identified in humans. The evolutionary origins and medical significance of this polymorphism have been uncertain. Here we show that this variation has likely arisen from the recurring reversion of an ancient hominin-specific duplication of these exons. Though this polymorphism has been largely invisible to genome-wide genetic studies to date, we describe a way to analyze it by imputation from SNP haplotypes and find among 22,288 individuals that these HP exonic deletions associate with reduced LDL and total cholesterol levels. We show that these deletions, and a SNP that affects HP expression, are the likely drivers of the strong but complex association of cholesterol levels to SNPs near HP. Recurring exonic deletions in the haptoglobin gene likely enhance human health by lowering cholesterol levels in the blood.
Introduction
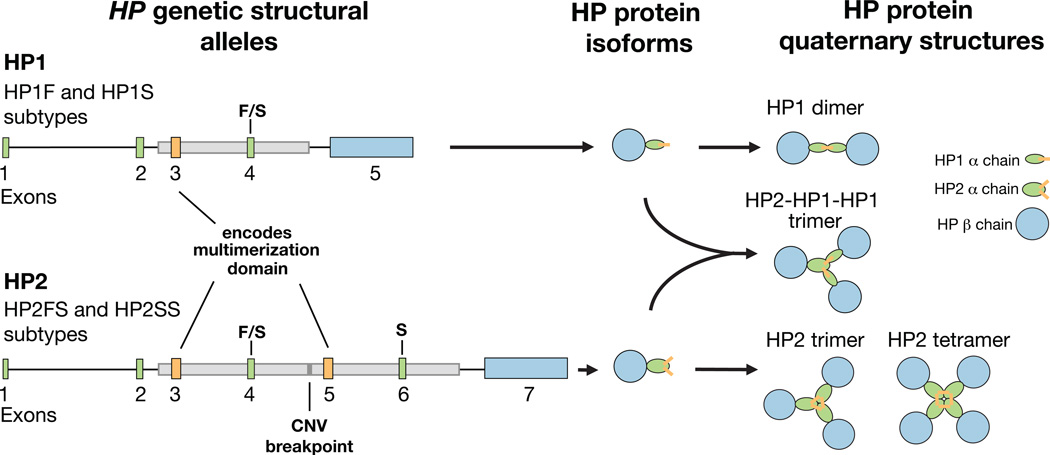
The HP protein binds free hemoglobin and facilitates its removal from the bloodstream1,2. A common 1.7 kb copy number variant (CNV) inside the HP gene determines the copy number (generally 1 or 2) of a tandem two-exon segment, including sequence that encodes a multimerization domain. This CNV is responsible for a striking protein phenotype: HP circulates as a dimer in individuals who are homozygous for the HP1 allele (encoding a single copy of the multimerization domain), but it forms multimers in individuals with the two-copy HP2 allele3–5 (Fig. 1). HP2 is also a less efficient antioxidant than HP16, and HP2 is required to make the tight-junction modulator protein, zonulin, which is the pre-processed product of HP27. Whether such functional variation contributes to human phenotypes is not well understood.
Figure 1. A common CNV in the HP gene is responsible for distinct molecular phenotypes.
The HP2 allele contains two additional exons compared to the HP1 allele: exons 3 and 4 are analogous to exons 5 and 6 respectively. The boundaries of the CNV are shown with gray boxs on the gene diagrams. The HP1 allele contains one copy of sequence in exon 3 (orange), which encodes the protein multimerization domain, allowing dimers to be formed. HP2 has two copies of this multimerization domain, which results in the formation of multimers. Exons 4 and 6 (green) contain the F/S mutations responsible for the protein running “Faster” or “Slower” on a gel. The long final exon of HP1 and HP2 encodes the beta subunit of the protein (blue), whereas the earlier exons encode the alpha subunit (green and orange). The alpha and beta subunits are cleaved apart by proteolytic processing after translation but are held together by disulfide bonds5. The protein isoform diagrams shown here were modeled after those in an earlier publication5.
The alleles of HP are further divided into subtypes by nucleotide polymorphisms that cause HP to run “Faster” or “Slower” on a protein gel8, hereafter called the “F” and “S” alleles. Both F and S segregate on the HP1 background, creating the subtypes HP1F and HP1S. The most common form of HP2 contains both alleles (as paralogous sequence variants) and is called HP2FS, but a low frequency HP2SS form also exists9. There are no known functional differences between the F and S alleles.
Despite the functional importance of haptoglobin – one of the five most abundant proteins in blood10 – and the potential functional importance of the common CNV that affects its structure, analyzing the association of this CNV to human phenotypes has proven challenging, and the CNV’s relationship to GWAS signals near HP has been unclear11. The CNV is not in strong linkage disequilibrium (LD) with any individual SNP11, and it has not been successfully genotyped with array-based copy number analysis12 or low coverage sequencing13. Instead, the polymorphism is generally typed with protein polyacrylamide gel electrophoresis14, PCR15, or quantitative PCR16, which has practically restricted the size of most association studies. While the HP polymorphism – one of the earliest polymorphisms to be discovered in humans – has been analyzed in hundreds of studies for associations to many human phenotypes, the limited sample sizes of these studies have provided insufficient power to determine whether the common HP CNV, or other nearby genetic variation, contributes to genetically complex phenotypes.
Blood cholesterol levels are one of the most important known biomarkers for future health and mortality17. A GWAS for cholesterol levels found a definitive signal (p = 3×10−24) at markers near HP18, but as at most GWAS-implicated loci, the causal variant(s) explaining this association are not known. While the HP protein’s most familiar role is to bind hemoglobin, HP also binds cholesterol molecules19–22. We hypothesized that the HP CNV might be responsible for the genetic association of cholesterol levels to this locus. To investigate this relationship, we had to develop ways to understand a surprisingly complex form of structural variation and its relationships to SNPs and haplotypes.
Results
A revised structural history of the haptoglobin gene
The alleles and mutational history of a locus provide a context for understanding whether and how the locus generates phenotypic variation. Standard genomics approaches, such as LD-based and array-based CNV analyses, have not yet successfully captured structural variation in HP11; we sought to determine why standard methods have failed, and to develop a new approach.
The long-accepted model of HP structural evolution23 proposed that HP2 arose through non-homologous recombination between HP1F and HP1S to produce HP2FS. The assumption that HP2 was formed by the fusion of human HP1 alleles arose from the observation that non-human great apes lack HP224 and that the left and right copies of the sequence in HP2FS share sequence similarities with HP1F and HP1S respectively23. However, the low LD between the HP CNV and surrounding SNPs potentially suggests a more complex structural history, as has been noted previously25. We first sought to distinguish between the two forces that reduce LD between nearby loci: (1) recombination and (2) recurrent mutation. If the low LD (of the CNV with flanking SNPs) were caused by frequent homologous recombination near the HP CNV region, then SNPs on the left and right sides of the structural variation would have low LD to one another. Conversely, if HP structure were affected by recurring intra-chromosomal structural mutations (or by non-allelic recombination between identical sister chromatids), then low LD between SNPs and the CNV might still be accompanied by high LD between SNPs on either side of the CNV.
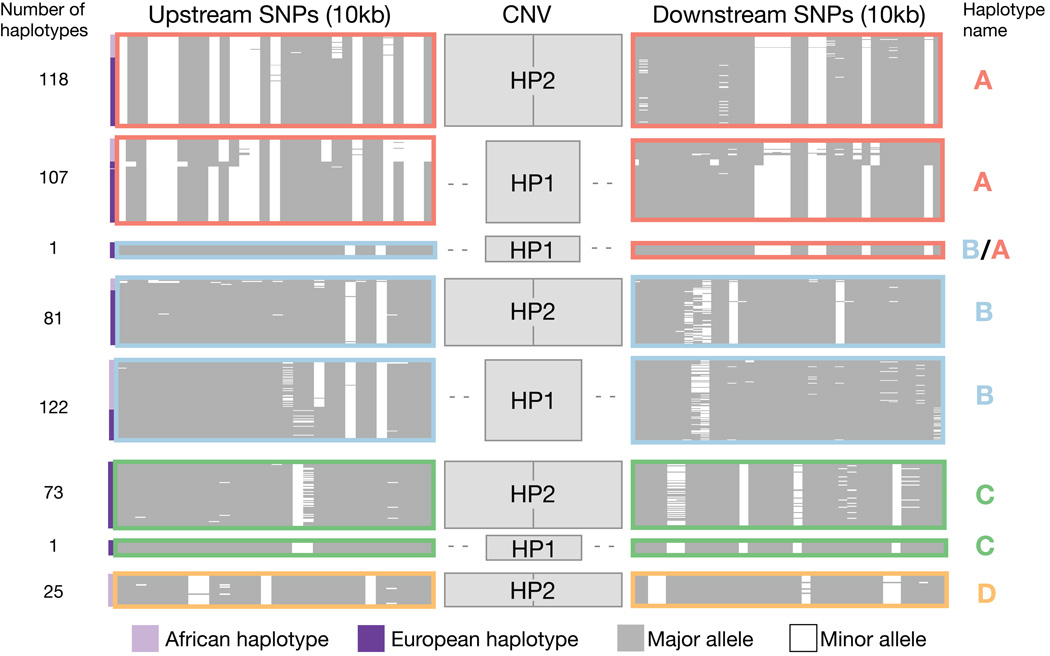
We used droplet digital PCR (ddPCR)26 to genotype the HP CNV in 264 unrelated individuals sampled by the 1000 Genomes Project13, phased the structural alleles onto SNP haplotypes using low-coverage sequence data27, and clustered similar SNP haplotypes (Methods). We observed that although many pairs of SNPs on opposite sides of the CNV were in high LD with each other (r2 >0.95) (Supplementary Fig. 1), copy number of the HP exons was not strongly correlated with any SNP on either side (maximum r2 = 0.44 in Europeans from 1000 Genomes). Three common SNP haplotypes (denoted A, B and C in Figure 2) persisted through the CNV region, yet segregated with both the HP1 and HP2 forms, a pattern that appears consistent with recurring structural mutations at HP (Fig. 2).
Figure 2. SNP haplotypes surrounding HP persist through the CNV region, yet segregate with both structural forms of HP.
This plot displays the SNP haplotypes (10 kb on each side of the HP CNV) segregating with HP1 and HP2 based on an analysis of 264 samples (528 haplotypes). The upstream SNPs are proximal to the centromere, while the downstream SNPs are distal to the centromere. Each thin horizontal line represents an individual SNP haplotype; similar or identical haplotypes are organized into clusters outlined by colored boxes. Note that the size of small clusters has been increased for visibility purposes and the number of haplotypes contained in each cluster is indicated at the left of the plot. White represents the minor allele and grey indicates the major allele across all populations in the analysis (CEU, IBS, TSI, YRI). Haplotypes ascertained from West African (HapMap YRI) individuals are indicated with lavender bars to the left of the plot, while haplotypes ascertained in European populations (CEU, IBS, TSI) are indicated with dark purple bars to the left of the plot. Haplotypes were clustered with the k-means method using upstream SNP haplotypes. Similar SNP haplotypes carrying different structures are indicated with colored outlines (dark pink, light blue, green, gold) and are designated haplotypes A–D. This figure was based on analysis of 1,000 Genomes Project samples and data (Methods).
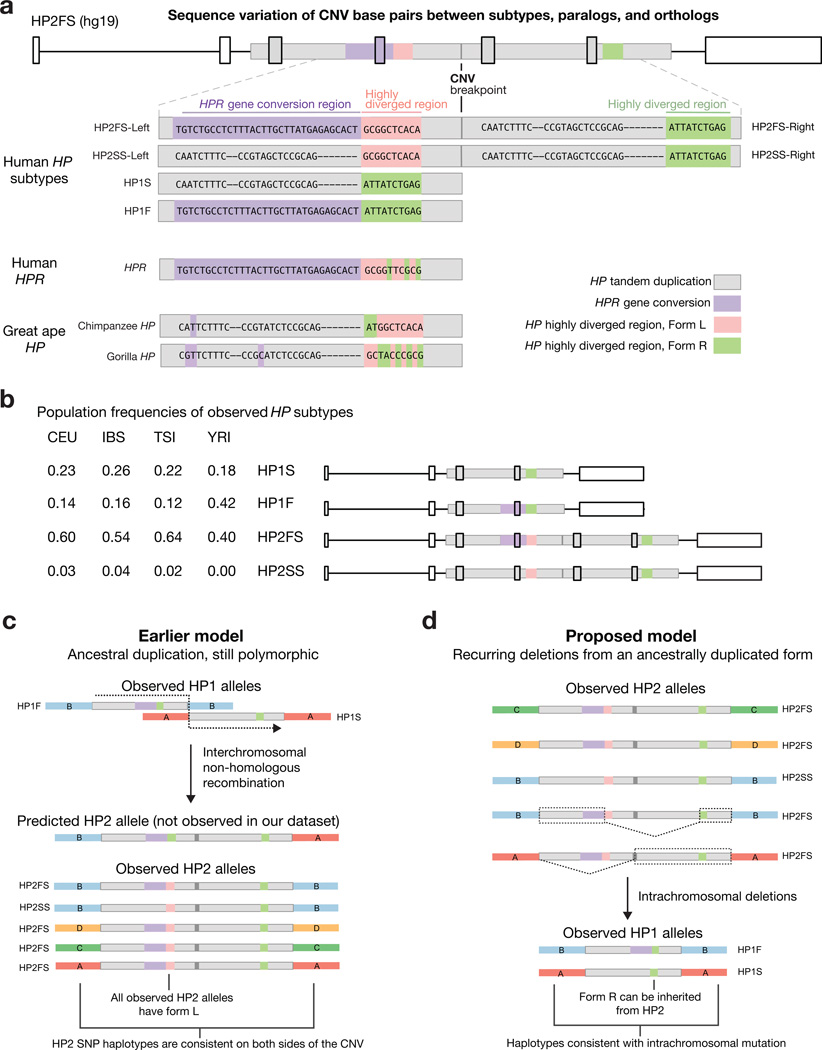
We next sought to determine whether structural mutations at HP involved deletions or duplications, by analyzing the nucleotide variation in the CNV region. We classified 27 haplotypes as one of four conventional subtypes – HP1S, HP1F, HP2FS, and HP2SS – based on the known sequence differences23. For HP2 haplotypes, we refer to the left copy of the CNV as HP2-Left (which is proximal to the centromere and 5’ on the transcribed RNA) and the right copy as HP2-Right (distal to the centromere and 3’ on the transcribed RNA) (Fig. 3a). Some 42 nucleotide polymorphisms differed among the subtypes of HP (e.g., between HP2FS and HP1S) but were consistent for any given subtype (Fig. 3a, Supplementary Fig. 2). In order to identify ancestral and derived alleles, we compared the human variants of each polymorphism to great ape versions of the HP gene, great ape paralogs of HP, and the human haptoglobin-related gene (HPR), which lies 2.2 kb downstream and shares 90% sequence identity with HP (Fig. 3a, Supplementary Fig. 3, Methods).
Figure 3. SNP haplotypes and sequence differences between HP subtypes inform structural history.
(a) This alignment shows base pair differences between HP structural forms analyzed from 27 haplotypes. Only the polymorphic bases are depicted. The HP2FS haplotype contains a 300-bp segment with derived paralogous gene conversion from HPR (lavender) and a 250-bp region that is highly diverged between subtypes (green/pink). Each allele of the highly diverged region contains a mix of ancestral and derived alleles. The dashes reflect a 2-bp and a 7-bp indel; the other sites shown are individual SNPs. The sequence data used to create this alignment are available online (GenBank: KT923758–KT923784). (b) The frequency of each HP haplotype in four populations. (c) The earlier model of HP structural evolution (interchromosomal non-homologous recombination) would predict the HP1F SNP haplotype background (haplotype B) upstream of HP2 and the HP1S SNP haplotype (haplotype A) downstream of HP2. Additionally, it would predict Form R of the highly diverged region in HP2-Left. However, neither of these predictions was observed in any of the HP2 alleles in this study. (d) Both HP1F and HP1S can be created through simple deletions in HP2FS. The dashed lines indicate deleted sequence, while the dashed boxes indicate the sequence required to create each HP1 haplotype. The deletion model is also consistent with the observed SNP haplotype backgrounds surrounding the CNV.
This analysis revealed that HP1F and HP2FS-Left (the left copy of the CNV segment on HP2FS) share a 300-bp segment containing 30 derived variants that is nearly identical to a portion of the human HPR gene. This segment is likely the result of paralogous gene conversion, through which a segment of HPR sequence was transferred into the HP gene (Fig. 3a, Supplementary Figs. 2–3). This gene conversion is responsible for the “F” mutations in HP1F and HP2FS. We believe that this gene conversion event has complicated detection of the CNV in genomic studies, since the copy-number-variable sequence can appear to arise partly from HP and partly from HPR, and likely explains why CNV data resources12,13 have lacked genotypes for this CNV. Our analysis also identified a highly diverged 250-bp region that has 10 fixed differences (between subtypes) including a mix of derived and ancestral alleles in each segment (Fig. 3a, Supplementary Figs. 2–3). We refer to this sequence as the “highly diverged region” and call the allele present in HP1S, HP1F and HP2-Right “Form R”, and the allele in HP2-Left “Form L”. We confirmed that these sequence differences are consistent at the population level by genotyping the boundaries of each variable region using ddPCR in DNA from 590 individuals sampled by HapMap28 and the 1000 Genomes Project27 (Fig. 3b, Supplementary Fig. 4, Methods).
The sequence differences between the HP subtypes shown in Figure 3 indicate that neither modern HP2 subtype (HP2FS nor HP2SS) could be created through the fusion of known HP1F and HP1S subtypes in the way that the earlier model23 proposed (Fig. 3c); for the earlier model to be true, HP2 would need to have arisen from a fusion of HP1S with a hypothetical diverged HP1 allele (containing Form L of the highly diverged region) that no longer segregates at an appreciable frequency in human populations. Alternatively, we propose that HP2 could be much older than previously thought, allowing these (non-allelic) sequences the time to diverge strongly from each other as paralogous sequence variants on an HP2 allele. HP2 does have all the sequences required to form HP1 alleles by simple non-allelic homologous recombination (NAHR) between the two tandem copies of the two-exon segment on HP2FS (Fig. 3d).
Flanking SNP haplotypes also suggested that HP2 did not arise from recombination between HP1F and HP1S. All HP1F alleles segregate with SNP haplotype B and almost all HP1S alleles segregate with SNP haplotype A (Supplementary Fig. 5). If HP2 had been created from non-allelic recombination between these two alleles, SNP haplotype B would be proximal to HP2 and SNP haplotype A would be distal (Fig. 3c); however, characteristic HP2 SNP haplotypes persist across the CNV region and do not appear to involve such recombinant haplotypes (Fig. 3d). (See Supplementary Fig. 6 for our complete model of HP structural evolution).
An alternative model would be that HP2 is in fact the ancestral allele in humans, and that HP1 alleles arose (and may continue to arise) by simple exonic deletions (due to NAHR) on an HP2 background. HP2-to-HP1 deletions have been observed at low frequency in the somatic and sperm cells of homozygous HP2 individuals29, demonstrating that the HP gene is prone to this type of structural mutation.
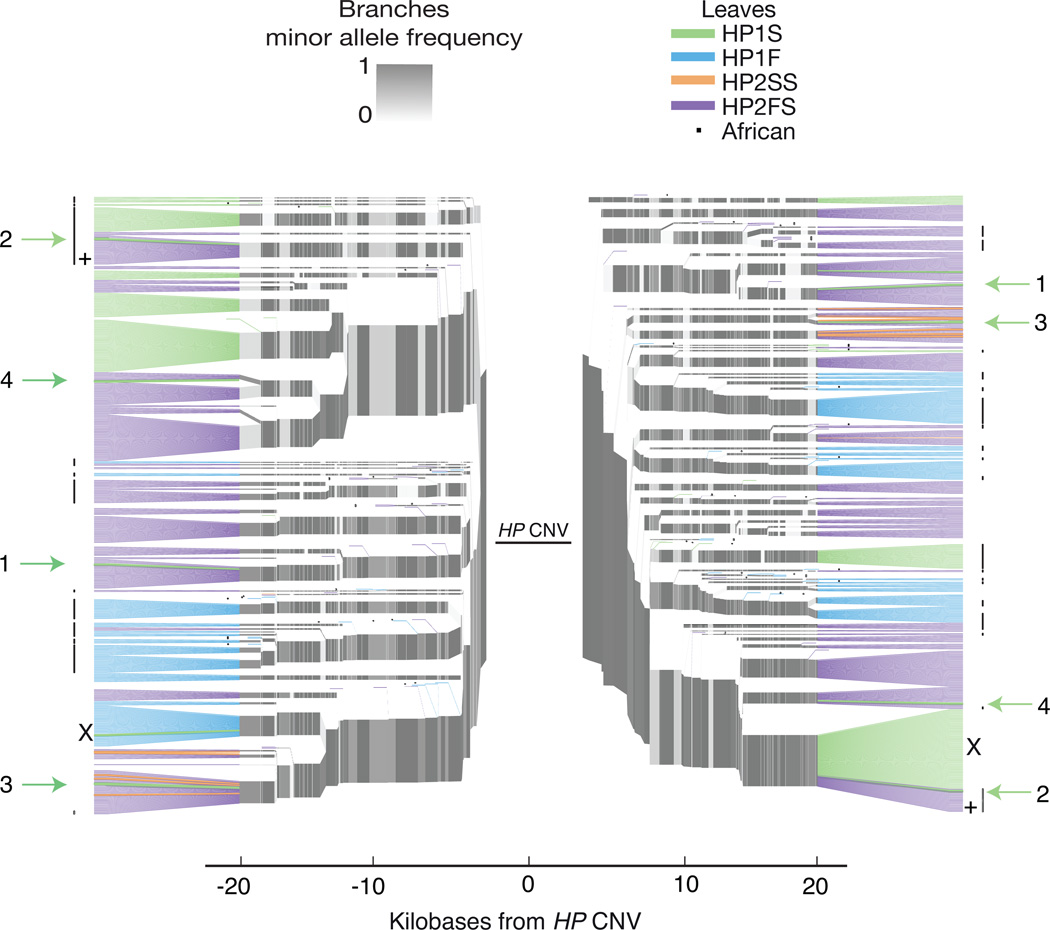
We sought to use information from long SNP haplotypes to further evaluate the alternative hypothesis that HP2-to-HP1 deletions gave rise to the structural variation at HP. If HP2-to-HP1 deletions occur intra-chromosomally (or between sister chromatids) and are transmitted to offspring, then rare HP1 subtypes might segregate on SNP haplotypes that are usually associated with common HP2 subtypes. While the short (10 kb) haplotypes immediately around HP cluster into a small number of groups (Fig. 2), we found that longer SNP haplotypes have much more information and cluster into a larger number of smaller groups (Fig. 4). A dendrogram analysis of these longer haplotypes revealed that several common HP2FS-flanking SNP haplotypes also contain rare (singleton) HP1S alleles (Fig. 4, Methods). Four of these rare HP1S structures segregate with SNP haplotypes that are identical to common HP2FS SNP haplotypes for at least 20 kb on either side of the CNV. The HP2FS and HP1S alleles from the same SNP haplotype branch also share derived mutations within the CNV region, consistent with shared ancestry (Supplementary Fig. 7). These observations indicate that these four HP1S alleles likely result from recent exonic deletions that occurred on an earlier HP2 allele. We also identified a SNP haplotype that carries the HP2FS allele in 15/16 sampled Africans but had the HP1S allele in 16/16 sampled Europeans, consistent with a deletion event in an African ancestor whose descendants migrated to Europe (Fig. 4). We conclude that HP structural variation reflects a combination of ancient and recent deletions that continue to create HP1 alleles from HP2 alleles.
Figure 4. Lone HP1S structural alleles segregate on common HP2FS SNP haplotypes.
SNP haplotype data is shown for three European populations (CEU, IBS, TSI) and one African population (YRI) totaling to 528 haplotypes. SNPs on the left half of the plot exist to the left of the HP duplication (proximal to the centromere), whereas SNPs on the right half of the plot physically reside to the right of the duplication (distal to the centromere). Branch points represent markers at which the depicted haplotypes diverge due to mutation and/or recombination with other haplotypes. The structures are represented on the leaves in order to clarify their relationships to SNP haplotypes, but the CNV and the paralogous gene conversion physically reside within the gap at center of the plot. The African individuals are identified with a dot after the leaf. Arrows with numbers indicate HP1 alleles segregating with the standard HP2 SNP haplotypes for at least 20 kb on both sides of the CNV. The + identifies the SNP haplotype branch which carries HP2FS in almost all sampled Africans, but HP1S in all sampled Europeans. This SNP haplotype is identical downstream of the CNV and differs by a single nucleotide upstream of the CNV. The X indicates the single haplotype observed in this study with apparent recombination in the CNV region (B/A in Figure 2). This recombination event appears to be recent because it is identical to standard haplotypes for at least 20 kb on either side of the CNV.
In order for common HP1 alleles to be derived deletions, HP2 would have to be ancient. The HP2 allele has not been observed in non-human primates, prompting the earlier model23 that it was a derived, recent30 allele. However, high-coverage genome sequences from ancient hominins are now available. We found that the Homo neandertalensis31 and Homo denisova32 genomes both have many sequence reads containing the breakpoint sequence that is present on HP2 but not on HP1, and that they also contain all other sequences that define the HP2FS subtype (Supplementary Table 1). The presence of HP2FS in neandertals, denisovans, and both modern and ancient33 African humans (Fig. 3b, Supplementary Table 1) indicates that HP2 arose prior to the divergence of these hominins 400 to 600 KYA34. (An earlier study, which assumed that HP2 was the derived allele, estimated the age of HP2 at less than 100 KY30, but this is contradicted by the ancient hominin genome sequences31,32.) SNP haplotypes further support the idea that HP2 is an ancient structural form: unlike HP1, HP2 segregates on all four common human SNP haplotypes identified at this locus (A, B, C, D in Fig. 2).
HP structural alleles can be imputed from SNP haplotypes
It is important to understand how complex, recurring variation contributes to human phenotypes. The gene-conversion history and limited LD between the HP CNV and surrounding SNPs have made it challenging to study this structural variation. We sought to develop a way to integrate HP structural variation into large-scale genetic studies whose large sample sizes enable robust analysis of relationships to phenotypes. We hypothesized that although HP structural mutations have occurred many times among human ancestors, the subset of these mutations that are old and common today might segregate on characteristic SNP haplotypes in many different individuals. Indeed, the above analysis of highly specific SNP haplotypes showed that such haplotypes usually segregate with a characteristic HP structural allele (Fig. 4).
To test this hypothesis, we phased HP structural alleles with SNP haplotypes to create reference chromosomes for imputation35–37 (Supplementary Dataset 1). To measure the efficacy of imputation (using Beagle35) we implemented a series of leave-one-out trials, in each of which we removed an individual’s HP gene structure from the reference panel and attempted to infer what structure was present based on the surrounding SNP haplotype and the rest of the reference panel (Methods, Supplementary Note). Although no individual SNP had “tagged” HP CNV status (HP2 vs. HP1) with high accuracy (maximum r2 = 0.44), we were able to impute HP CNV status from multi-SNP haplotypes in both African and European population samples with high accuracy (r2 = 0.94 in a European (CEU, IBS, TSI) population sample, r2 = 0.92 in a Yoruba (YRI) sample), using only SNPs present on common SNP genotyping arrays (Table 1, Supplementary Tables 2–5, Supplementary Fig. 8). We believe this result reflects that, despite recurring mutation at HP, most HP1 alleles trace back to a few ancient mutations in common ancestors (more-recent deletion events likely reduce the efficacy of imputation but are more rare) (Table 1). Our imputation approach allows HP structural variation to be incorporated into large genetic studies using existing SNP data.
Table 1. Imputation of HP structural features from surrounding SNPs.
This table shows the correlation (r2) between HP structural alleles (as identified by direct molecular analysis) and predictions from imputation from the SNP haplotypes, using SNPs on the Illumina Omni2.5 array. The correlation between each structural feature and the most strongly correlated individual SNP is also displayed. The CEU, IBS, and TSI populations were merged into a single European population for this analysis.
| Europeans (CEU, TSI, IBS) | |||
|---|---|---|---|
| Subtype | Imputation (r2) | Tag SNP (r2) | |
| HP1S | 0.94 | 0.86 | rs217181 |
| HP1F | 0.98 | 0.83 | rs9302635 |
| HP2FS | 0.94 | 0.40 | rs217181 |
| HP2SS | 0.75 | 0.58 | rs34914030 |
| HP1 vs. HP2 | 0.94 | 0.44 | rs217181 |
Haptoglobin and blood cholesterol levels
Both total cholesterol levels and LDL cholesterol levels associate strongly (p = 3×10−24 and 2×10−22, respectively, in a cohort of >100,000 individuals18) with the SNP rs2000999, which is within 15 kb of HP. Given that the HP protein binds to multiple types of cholesterol molecules19–22 and that the HP1/HP2 difference has at least a modest correlation with this SNP (r2 = 0.14), we hypothesized that the recurring structural variation that causes the HP1/HP2 difference could be responsible for the association of cholesterol levels to variation in this region.
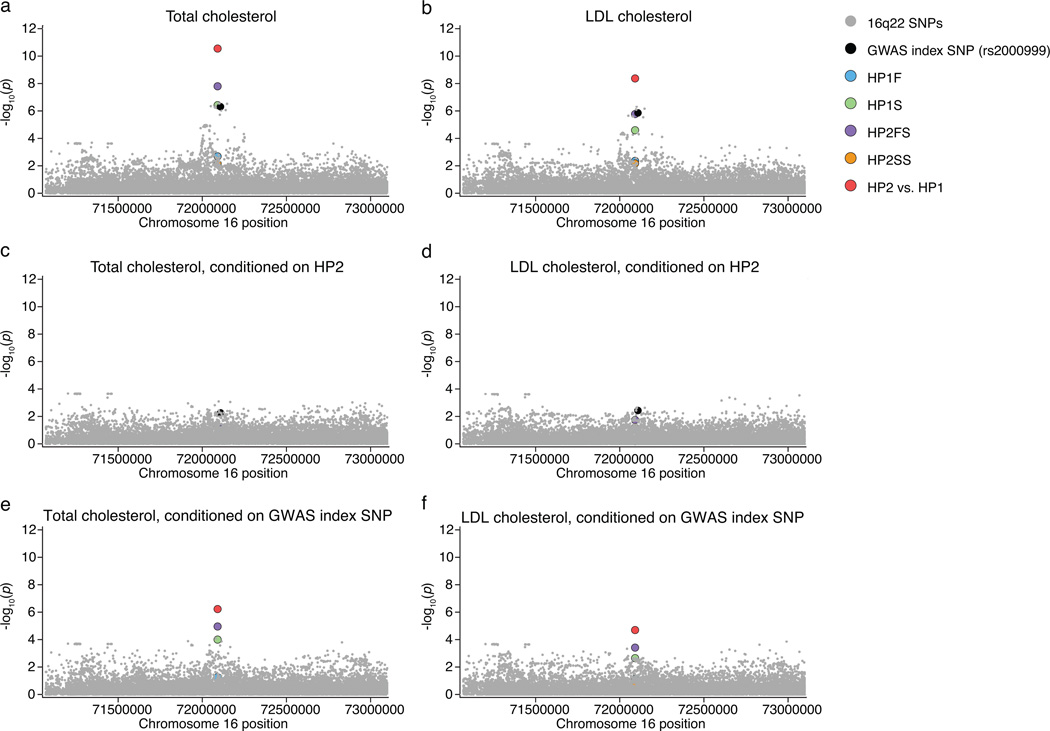
We were able to obtain genome-wide SNP data from 22,288 individuals of European descent with cholesterol measurements (Methods, Supplementary Note, Supplementary Table 6). In this sample we found that the GWAS index SNP (rs2000999) associated as expected with total cholesterol levels (p = 5.15×10−8) and LDL cholesterol levels (p = 1.43×10−7) (Fig. 5a,b). We used our approach to impute the most likely HP subtypes in each individual’s genome. The imputed HP2 state associated with cholesterol phenotypes much more strongly (p = 2.8×10−11 for total cholesterol levels and p = 4.3×10−9 for LDL cholesterol levels) than any SNP in the HP region did (Fig. 5a,b). Furthermore, in analyses controlling for the HP1/HP2 difference, the association at the index SNP was reduced to p = 0.006 for total cholesterol levels and to p = 0.004 for LDL cholesterol levels (Fig. 5c,d) (notably, this was still a nominally positive association, which we further explore below), while the HP1/HP2 variant continued to associate more strongly with cholesterol levels (p = 5.95×10−7 to total cholesterol levels and p = 2.02×10−5 to LDL cholesterol levels) in analyses controlling for the GWAS index SNP (Fig. 5e–f).
Figure 5. The HP2 allele associates with increased total cholesterol levels and increased LDL cholesterol levels.
The imputed structural variants and all regional SNPs imputed from 1,000 Genomes are shown for this analysis of 22,288 individuals. (a,b) The HP2 variant is the highest regional association to both total cholesterol levels (p = 2.79×10−11) and LDL cholesterol levels (p = 4.3×10−9). (c,d) Conditioning on the HP2 variant causes most of the signal to disappear. (e,f) Conditioning on the GWAS index SNP (rs2000999) only has a moderate effect on the association.
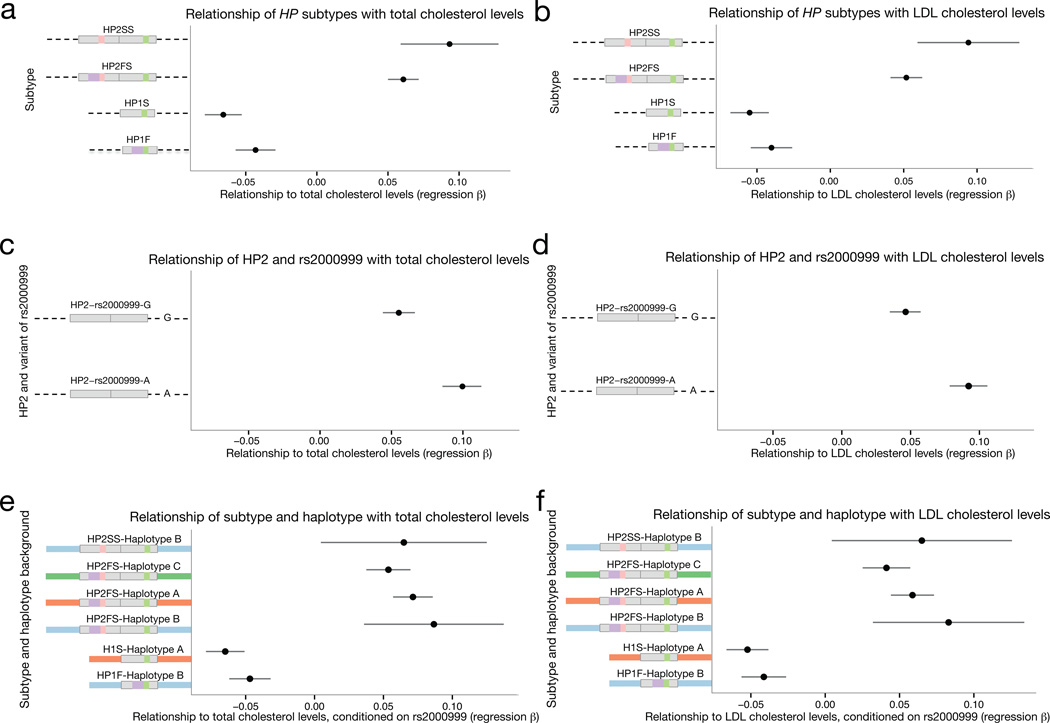
Both HP2 subtypes (HP2FS and HP2SS) were associated with increased cholesterol levels (Fig. 6a,b). While the HP1F and HP1S subtypes segregate on very different SNP haplotype backgrounds (Supplementary Fig. 5), they associated with similar levels of protection from elevated cholesterol levels (Fig. 6a,b), further supporting the idea that HP structural variation (rather than nearby sequence variation) is the primary driver of the association to these structural alleles.
Figure 6. The rs2000999-A allele on the HP2 background associated with a greater increase in total cholesterol levels and LDL cholesterol levels.
(a,b) The regression beta of HP1 and HP2 alleles with total and LDL cholesterol levels is shown with the standard error is shown for this analysis of 22,288 individuals. (c,d) The regression beta of each allele of rs2000999 with total and LDL cholesterol levels is shown with the standard error. (e,f) The regression beta of each HP subtype and total and LDL cholesterol levels separated by SNP haplotype background is plotted with the standard error. The beta for each HP1 allele was calculated by a comparison with HP2 alleles only, and the beta of HP2 alleles was calculated through a comparison with HP1 alleles only (See Methods and Supplementary Note).
The GWAS index SNP, rs2000999, is located in a strong enhancer sequence for hepatocytes38,39 (the primary source of HP), and the derived allele of this variant associates with reduced HP expression40,41. Though the above analysis more strongly implicated the HP structural variation than this SNP in cholesterol levels, we hypothesized that both the CNV and the rs2000999 variant might affect cholesterol levels through their respective effects on haptoglobin structure and abundance. The derived rs2000999-A allele is present almost exclusively on HP2 haplotypes (D’ = 0.96), so we examined the effect of each rs2000999 allele on the HP2 background. (The LD between rs2000999 and HP subtypes is shown in Supplementary Table 7). We found that while all HP2 alleles associated with an increase of total and LDL cholesterol levels when compared to HP1 (Fig. 6a,b), the effect was modestly enhanced for HP2 alleles with the derived rs2000999-A allele (Fig. 6c,d). When we corrected for the effect of rs2000999, HP2 alleles on all European SNP haplotype backgrounds (A–C as shown in Fig. 2) associated with similarly elevated cholesterol levels (Fig. 6e,f). We believe that the impact of rs2000999 on HP expression explains the residual nominal association that is present at this SNP in analyses conditioning on HP1/HP2. The imputation efficacy of the HP1/HP2 difference is similar for each SNP haplotype background (haplotype A: r2 = 0.93, haplotype B: r2 = 0.95, haplotype C: r2 = 0.95 using SNPs on the Illumina Omni2.5 array), indicating that imperfect imputation is unlikely to have strongly biased the association toward rs2000999 or any other SNP.
This analysis indicates that the association of cholesterol phenotypes to SNPs near HP reflects a complex allelic architecture arising from multiple variants and historical mutations (structural and single-nucleotide) at the locus. The status of rs2000999 as the lead (index) SNP at this locus likely reflects a combination of (i) a true genetic effect of this SNP arising from an effect on HP expression levels and explaining a ~1.49 mg/dl increase in total cholesterol; and (ii) partial LD (r2 = 0.14) to a larger effect (2.11 mg/dl increase in total cholesterol) arising from HP structural variation that changes the encoded protein (Supplementary Table 8).
Discussion
We presented multiple lines of evidence that recurrent deletions in HP2 have created new HP1 alleles, a phenomenon which likely explains the low LD between individual SNPs and HP1/HP2. We also found that HP is polymorphic for paralogous gene conversion from HPR, which has obscured the CNV from analysis by earlier sequencing and array-based CNV studies. While recurring deletions and paralogous gene conversion have made studying this structural variation historically challenging, we demonstrated that HP subtypes can be imputed from SNP haplotypes with high accuracy, an approach that should make it possible to resolve longstanding uncertainty about how genetic variation at HP relates to many human phenotypes. We used this imputation strategy to study HP variation in 22,288 individuals and showed that a complex allelic architecture, shaped most strongly by the HP CNV and also by a cis-acting expression effect, is likely responsible for the strong association of cholesterol levels with 16q22.2 in GWAS18.
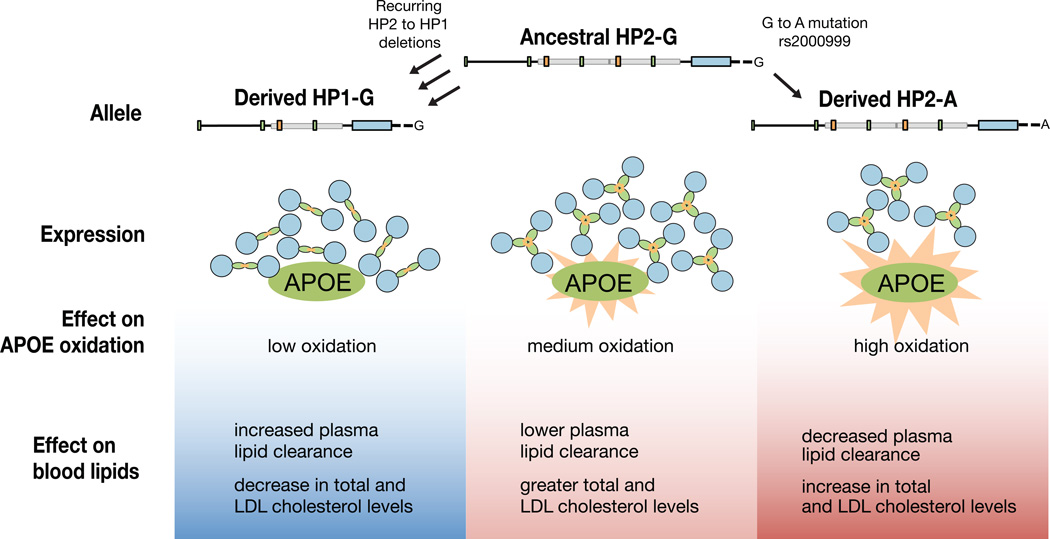
Haptoglobin interacts with the APOE protein, which is critical to maintaining low total cholesterol and LDL cholesterol levels42. Oxidation of APOE impairs its ability to clear plasma lipids43. The HP protein directly binds APOE20,22 and serves as an APOE antioxidant20. The HP2 form of the protein is a less efficient antioxidant than HP16, a potential mechanism for the association we observe. Decreased HP levels due to rs2000999-A may have a similar phenotypic effect, but by reducing the level (rather than changing the protein structure) of HP. HP2 and rs2000999-A could contribute to increased total and LDL cholesterol levels by providing insufficient antioxidant activity for APOE (Figure 7).
Figure 7. A model for the influence of HP genetic polymorphisms on total and LDL cholesterol levels.
Because HP serves as an antioxidant for bound APOE20,22 and HP1 has greater antioxidant activity than HP26, we propose that HP1 alleles (arising from HP2-to-HP1 deletions) lessen the oxidative burden on APOE, allowing it to more effectively clear plasma lipids. Conversely, the rs2000999-A allele decreases HP expression40,41 and thus antioxidant protection for APOE, contributing to elevated cholesterol levels.
We found that imputation could be used to extend the analysis of a complex CNV locus to very large samples (n=22,288) for which SNP data were available. The large sample was critical for resolving multiple effects that are in partial LD, and made it possible to appreciate (at high levels of significance) effects that were not apparent in an earlier, smaller study44. There are currently controversies about the HP CNV’s role in heart disease, cancer, malaria, Crohn’s disease, and numerous other human phenotypes. Our approach to imputing complex structural alleles, and the imputation resource we make available here (Supplementary Dataset 1), should make it possible to resolve these questions in a definitive way using large existing SNP data sets. A similar approach might be useful at the hundreds of other loci affected by complex and multi-allelic CNVs.
GWAS has identified thousands of genetic variants that associate with genetically complex traits. At almost all of these loci the responsible, functional variants have yet to be found, and the underlying allelic architectures are unknown. A particularly intriguing question involves the extent to which the underlying allelic architectures will turn out to be simple (e.g. a single, responsible functional variant) or complex. Haptoglobin appears to offer an early example of a locus at which an association signal arises from the combined effects of many different common functional alleles, with different kinds of effects – a set of many alleles that affect protein structure, and an additional allele that affects expression level. It will be interesting and important to understand how widespread such allelic complexity is in human biology.
Online Methods
Genotyping HP structural variants
To determine the copy number of the HP CNV and the other structural polymorphisms, we used a droplet-based digital PCR method26 to measure copy number at 4 locations (boundaries A,B,D,E in Supplementary Fig. 4b). We designed a pair of PCR primers and a dual-labeled fluorescence-FRET oligonucleotide probe to the sequence of each HP boundary and to a two-copy control locus. Intermediate copy number calls were repeated with triplicate measurements. We used a PCR assay for boundary C to verify the consistency of this boundary in HP2SS haplotypes. Only individuals predicted to carry the HP2SS haplotype based on ddPCR measurements produced an amplicon. A sufficient number of assays were designed such that no single incorrect copy number measurement would mistakenly identify a diploid subtype pair as another subtype pair (see Supplementary Table 9). Allelic copy numbers were determined based on a bi-allelic copy number model for each sequence boundary (Supplementary Table 9). Hardy-Weinberg Equilibrium of HP subtypes (Supplementary Table 10) and faithful transmission of HP subtypes in trios (Supplementary Table 11) were verified. One likely 3-copy allele was found as well as two rare mutations that interfere with assay amplification (Supplementary Table 12). Phasing confirmation of recent deletion alleles was performed with Drop-Phase45, which is further discussed below (Supplementary Table 13). Primer sequences are provided in Supplementary Table 14).
SNP haplotype analysis for HP CNV and subtypes
HP structural variants were phased with SNP haplotypes from the 1,000 Genomes Project Phase I data, and was used to show short haplotypes in four common clusters (Fig. 2) and longer closely related haplotypes in a dendrogram (Fig. 4). Haplotypes in Figure 2 were clustered with the k-means method using upstream SNP haplotypes only. All recent deletion alleles (HP1S subtype) as shown in Figure 4 were from individuals who have two standard HP2FS SNP haplotypes. Each deletion allele was phased onto the correct SNP haplotype using the Drop-Phase technique45, a new method for phasing based on the idea that physically linked sequences are more frequently partitioned into the same droplets (Supplementary Table 13). HP structural variants were also phased with SNPs from common SNP genotyping arrays to evaluate the potential for these variants to be imputed from existing GWAS data. Phasing and encoding for the structural alleles are further discussed in the Supplementary Note.
Population sequencing in the CNV region
We Sanger sequenced the CNV region of 27 human haplotypes segregating on diverse SNP haplotype backgrounds, and the analogous region of four great apes: chimpanzee, bonobo, gorilla, and orangutan. Individual human haplotypes were sequenced by targeting a single structural allele from HP1/HP2 heterozygotes. Primers for the human HP2 allele target the HP2 breakpoint. HP1 haplotypes were obtained with size selection through gel extraction. HP1 sequencing primers were designed to be compatible with the chimpanzee, gorilla, and orangutan reference genomes and were also used to sequence the corresponding region in each great ape. All primer pairs are specific to HP and do not amplify haptoglobin related protein (HPR) or primate haptoglobin (HPP). The hg19 reference genome supplied the human HPR sequence. The chimpanzee and gorilla HP genes were sequenced in samples NS03489 and PR00107 respectively (DNA provided by Coriell Cell Repositories). The sequence for the Chimpanzee HPR and HPP genes was supplied by previously sequenced clones (GenBank: M84462.1, M84463.1) See Supplementary Figures 2–3 for additional great apes and results by sample and see Supplementary Table 14 for primers.
Creating and testing imputation reference panels
We evaluated the efficacy of imputation for the HP CNV as well as HP subtypes (HP1F, HP1S, HP2FS, HP2SS) using reference panels composed of experimentally determined HP structural alleles and SNPs ascertained the 1000 Genomes Project13 and HapMap28. Separate reference panels were created and tested from each of the following SNP datasets: Illumina Omni2.5, HapMap3 (Illumina 1M and Affymetrix 6.0), and Illumina 1M and Affymetrix 6.0 individually (Table 1, Supplementary Tables 2–5). Separate reference panels were created and tested for European and African populations due to differences in SNP haplotype backgrounds for HP subtypes (Figure 4). We performed a series of “leave one out” trials to evaluate the efficacy of imputation for HP structural variants. See Supplementary Note for more information.
Imputation of HP structural variation into cohorts for cholesterol association study
A reference panel composed of encoded HP structural alleles and SNPs surrounding the CNV region from the Illumina OMNI, Illumina 1M, and Affymetrix 6.0 arrays was developed and used to impute HP structural variation into cohorts with cholesterol information using Beagle (v2.3.1) imputation software. See Supplementary Note for further detail.
Association analysis
The association between imputed HP structural variants and the four lipids traits (total cholesterol (TC), low-density lipoprotein (LDL), high density lipoprotein (HDL), and triglycerides (TRIGS) was performed in 6 studies of 22,288 individuals of European ancestry. Each lipid trait was regressed on age and gender, and inverse-normal transformed prior to analysis. Linear regression was performed to test the association between imputed structural variants or SNPs in the locus and lipid trait, assuming an additive genetic model using PLINK46 (v1.07). The imputed HP structural variants and genotypes were analyzed as dosages to account for imputation uncertainty, poorly imputed variants discarded (INFO<0.4). All analyses were adjusted for 10 study specific principal components. Study specific results were combined via inverse-variance fixed effects meta-analysis method implemented in METAL47. Sensitivity and specificity phenotype analyses were performed to assess the influence of type 2 diabetes (condition or removal of samples) and cholesterol lowering/statin medication use (recalculating values, condition or removal of samples). All analyses were performed using baseline lipid measurements for cohorts with longitudinal follow-up. See Supplementary Figure 8 for HDL and triglyceride association results and the Supplementary Note for more information.
Code availability
The following packages were used to analyze data and are publicly available online: Beagle (v2.3.1), PLINK (v1.07), SHAPEIT2 (version 2.644), IMPUTE2 (version 2.3), METAL, SMARTPCA. The following custom scripts are available upon request: R scripts used to format data, to perform linear regression analyses, and to cluster haplotypes in Fig. 2, a PYTHON script used to cluster haplotypes in Fig. 4, and PERL scripts used to format data.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Human Genome Research Institute (R01 HG 006855, to SM). The Yerkes Center (Grant No. P51OD011132) provided primate DNA samples. R.M.S. was supported by an NIH-NHLBI K99 award (#1K99HL122515-01A1) and advanced postdoctoral fellowship award from the Juvenile Diabetes Research Foundation (JDRF # 3-APF-2014-111-A-N). We thank Christina Usher for comments on the manuscript and work on the figures.
Footnotes
Accession codes. Sequence data are available on GenBank under accessions KT923758-KT923784.
Author Contributions
L.M.B., S.A.M., and R.E.H. designed the experiments for understanding HP structural evolution. R.M.S., L.M.B., and G.P. performed imputation and association analyses of cholesterol cohorts. L.M.B. performed computational analyses of HapMap and 1000 Genomes Project data, constructed the imputation reference panels, and performed all laboratory experiments. L.M.B. and S.A.M. wrote the manuscript. All authors contributed to interpretations of data and to revisions of the manuscript.
The authors declare no competing financial interests.
References
- 1.Allison AC, Rees WA. The binding of haemoglobin by plasma proteins (haptoglobins) British medical journal. 1957;2:1137–1143. doi: 10.1136/bmj.2.5054.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langlois MR, Delanghe JR. Biological and clinical significance of haptoglobin polymorphism in humans. Clinical Chemistry. 1996;42:1589–1600. [PubMed] [Google Scholar]
- 3.Smithies O, Walker NF. Genetic control of some serum proteins in normal humans. Nature. 1955;176:1265–1266. doi: 10.1038/1761265a0. [DOI] [PubMed] [Google Scholar]
- 4.Wejman JC, Hovsepian D, Wall JS, Hainfeld JF, Greer J. Structure and assembly of haptoglobin polymers by electron microscopy. J Mol Biol. 1984;174:343–368. doi: 10.1016/0022-2836(84)90342-5. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen MJ, Moestrup SK. Receptor targeting of hemoglobin mediated by the haptoglobins: roles beyond heme scavenging. Blood. 2009;114:764–771. doi: 10.1182/blood-2009-01-198309. [DOI] [PubMed] [Google Scholar]
- 6.Melamed-Frank M. Structure-function analysis of the antioxidant properties of haptoglobin. Blood. 2001;98:3693–3698. doi: 10.1182/blood.v98.13.3693. [DOI] [PubMed] [Google Scholar]
- 7.Tripathi A, et al. Identification of human zonulin, a physiological modulator of tight junctions, as prehaptoglobin-2. Proceedings of the National Academy of Sciences. 2009;106:16799–16804. doi: 10.1073/pnas.0906773106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smithies O, Connell GE, Dixon GH. Inheritance of haptoglobin subtypes. Am. J. Hum. Genet. 1962;14:14–21. [PMC free article] [PubMed] [Google Scholar]
- 9.Shindo S. Haptoglobin subtyping with anti-haptoglobin alpha chain antibodies. Electrophoresis. 1990;11:483–488. doi: 10.1002/elps.1150110609. [DOI] [PubMed] [Google Scholar]
- 10.Martosella J, Zolotarjova N. Multi-component immunoaffinity subtraction and reversed-phase chromatography of human serum. Methods Mol. Biol. 2008;425:27–39. doi: 10.1007/978-1-60327-210-0_3. [DOI] [PubMed] [Google Scholar]
- 11.Cahill LE, et al. Currently available versions of genome-wide association studies cannot be used to query the common haptoglobin copy number variant. J. Am. Coll. Cardiol. 2013;62:860–861. doi: 10.1016/j.jacc.2013.04.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy AP, et al. Haptoglobin phenotype and prevalent coronary heart disease in the Framingham offspring cohort. Atherosclerosis. 2004;172:361–365. doi: 10.1016/j.atherosclerosis.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 15.Koch W, et al. Genotyping of the common haptoglobin Hp 1/2 polymorphism based on PCR. Clinical Chemistry. 2002;48:1377–1382. [PubMed] [Google Scholar]
- 16.Soejima M, Koda Y. TaqMan-Based Real-Time PCR for Genotyping Common Polymorphisms of Haptoglobin (HP1 and HP2) Clinical Chemistry. 2008;54:1908–1913. doi: 10.1373/clinchem.2008.113126. [DOI] [PubMed] [Google Scholar]
- 17.Zethelius B, et al. Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. N. Engl. J. Med. 2008;358:2107–2116. doi: 10.1056/NEJMoa0707064. [DOI] [PubMed] [Google Scholar]
- 18.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salvatore A, et al. Haptoglobin Binding to Apolipoprotein A-I Prevents Damage from Hydroxyl Radicals on Its Stimulatory Activity of the Enzyme Lecithin-Cholesterol Acyl-Transferase †. Biochemistry. 2007;46:11158–11168. doi: 10.1021/bi7006349. [DOI] [PubMed] [Google Scholar]
- 20.Salvatore A, Cigliano L, Carlucci A, Bucci EM, Abrescia P. Haptoglobin binds apolipoprotein E and influences cholesterol esterification in the cerebrospinal fluid. J. Neurochem. 2009;110:255–263. doi: 10.1111/j.1471-4159.2009.06121.x. [DOI] [PubMed] [Google Scholar]
- 21.Spagnuolo MS, et al. Analysis of the haptoglobin binding region on the apolipoprotein A-I-derived P2a peptide. J. Pept. Sci. 2013;19:220–226. doi: 10.1002/psc.2487. [DOI] [PubMed] [Google Scholar]
- 22.Cigliano L, Pugliese CR, Spagnuolo MS, Palumbo R, Abrescia P. Haptoglobin binds the antiatherogenic protein apolipoprotein E - impairment of apolipoprotein E stimulation of both lecithin:cholesterol acyltransferase activity and cholesterol uptake by hepatocytes. FEBS J. 2009;276:6158–6171. doi: 10.1111/j.1742-4658.2009.07319.x. [DOI] [PubMed] [Google Scholar]
- 23.Maeda N, Yang F, Barnett DR, Bowman BH, Smithies O. Duplication within the haptoglobin Hp2 gene. Nature. 1984;309:131–135. doi: 10.1038/309131a0. [DOI] [PubMed] [Google Scholar]
- 24.McEvoy SM, Maeda N. Complex events in the evolution of the haptoglobin gene cluster in primates. J. Biol. Chem. 1988;263:15740–15747. [PubMed] [Google Scholar]
- 25.Hardwick RJ, et al. Haptoglobin (HP) and Haptoglobin-related protein (HPR) copy number variation, natural selection, and trypanosomiasis. Human genetics. 2013;133:69–83. doi: 10.1007/s00439-013-1352-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindson BJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asakawa J, Kodaira M, Nakamura N, Satoh C, Fujita M. Chimerism in humans after intragenic recombination at the haptoglobin locus during early embryogenesis. Proc Natl Acad Sci USA. 1999;96:10314–10319. doi: 10.1073/pnas.96.18.10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez S, et al. Molecular and Population Analysis of Natural Selection on the Human Haptoglobin Duplication. Annals of Human Genetics. 2012;76:352–362. doi: 10.1111/j.1469-1809.2012.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer M, Kircher M, Gansauge MT, Li H, Racimo F. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llorente MG, et al. Ancient Ethiopian genome reveals extensive Eurasian admixture throughout the African continent. Science. 2015;350:820–822. doi: 10.1126/science.aad2879. [DOI] [PubMed] [Google Scholar]
- 34.Scally A, Durbin R. Revising the human mutation rate: implications for understanding human evolution. Nat Rev Genet. 2012;13:745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 35.Browning SR. Missing data imputation and haplotype phase inference for genome-wide association studies. Human genetics. 2008;124:439–450. doi: 10.1007/s00439-008-0568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu. Rev. of Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nature Biotechnology. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Froguel P, et al. A genome-wide association study identifies rs2000999 as a strong genetic determinant of circulating haptoglobin levels. PLoS ONE. 2012;7:e32327. doi: 10.1371/journal.pone.0032327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soejima M, et al. Genetic factors associated with serum haptoglobin level in a Japanese population. Clin. Chim. Acta. 2014;433:54–57. doi: 10.1016/j.cca.2014.02.029. [DOI] [PubMed] [Google Scholar]
- 42.Ishibashi S, Herz J, Maeda N, Goldstein JL, Brown MS. The two-receptor model of lipoprotein clearance: tests of the hypothesis in ‘knockout’ mice lacking the low density lipoprotein receptor, apolipoprotein E, or both proteins. Proc Natl Acad Sci USA. 1994;91:4431–4435. doi: 10.1073/pnas.91.10.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Cao Z, Tian L, Garvey WT, Cheng G. VPO1 mediates ApoE oxidation and impairs the clearance of plasma lipids. PLoS ONE. 2013;8:e57571. doi: 10.1371/journal.pone.0057571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guthrie PAI, et al. Complexity of a complex trait locus: HP, HPR, haemoglobin and cholesterol. Gene. 2012;499:8–13. doi: 10.1016/j.gene.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 45.Regan JF, et al. A rapid molecular approach for chromosomal phasing. PLoS ONE. 2015;10:e0118270. doi: 10.1371/journal.pone.0118270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.