Abstract
The increasing epidemic of obesity and its comorbidities has spurred research interest in adipose biology and its regulatory functions. Recent studies have revealed that the mechanistic target of rapamycin (mTOR) signaling pathway plays a critical role in the regulation of adipose tissue function, including adipogenesis, lipid metabolism, thermogenesis, and adipokine synthesis/secretion. Given the extreme importance of mTOR signaling in controlling energy homeostasis, it is not unexpected that deregulated mTOR signaling is associated with obesity and related metabolic disorders. In this review, we highlight the current advances in the roles of the mTOR signaling pathway in adipose tissue. We also provide a more nuanced view of how the mTOR signaling pathway regulates adipose tissue biology and function. Finally, we describe approaches to modulate the activity and tissue specific function of mTOR that may pave the way towards counteracting obesity and related metabolic diseases.
Keywords: the mechanistic target of rapamycin (mTOR), adipogenesis, lipid metabolism, thermogenesis, adipokine
Adipose tissue: Regulation and function at a glance
With an escalating global epidemic of an overweight/obese population and increasing awareness of the role of adipose tissue in regulating energy homeostasis, interests in better understanding adipose function and regulation are rapidly rising [1]. Adipose tissue was originally viewed mainly as an energy storage depot, but it is now increasingly appreciated as a multi-functional tissue. Except for the two major types of fat (the white and brown), it has been shown the presence of another subtype of adipose tissue, the so-called beige/brite (brown in white) fat, which is provoked by prolonged cold exposure, adrenergic signaling or genetic manipulation [2, 3]. While located in the anatomical sites characteristic of white adipose tissue (WAT), beige adipose tissue shares some characteristics of classic brown adipose tissue (BAT) such as the presence of multilocular lipid droplets, high mitochondria content, and the expression of thermogenic genes such as the uncoupling protein 1 (UCP1). However, beige adipocytes come from progenitors different from that of brown adipocytes and express some unique surface markers such as Cd137 and Tmem26 [4]. In response to overnutrition, adipose tissue expands its mass to store extra energy through hypertrophy and/or hyperplasia in an effort to prevent ectopic lipid deposition and lipotoxicity in other tissues. Adipose tissue may reduce its lipid store via increased lipolysis, leading to the release of fatty acids into circulation upon energy deficit [5]. In addition to lipid mobilization, adipose tissue is responsible for synthesizing and secreting numerous metabolites and adipokines, which allow adipose tissue to communicate with other tissues/organs to regulate a myriad of physiological functions [6-8]. Another special feature of adipose tissue, mainly found in BAT and beige fat, is the ability to dissipate chemical energy into heat (thermogenesis) through uncoupling respiration [9]. Recent identification of BAT in human adults offers an intriguing new approach to improve metabolic homeostasis [10-12]. The ability of fat to regulate its size and metabolism in response to environmental signals could be exploited to combat obesity and related metabolic disorders. Thus, dissecting the signaling pathways that contribute toward these functional differences will be extremely important.
The mechanistic target of rapamycin (mTOR) is a well-known signaling node that integrates environmental nutrients, growth factors, cellular energy status and other cellular cues into a variety of anabolic processes, cytoskeleton dynamics and autophagy [13]. Dysregulation in mTOR signaling is implicated in various diseases such as obesity, type 2 diabetes, cancer and aging [14]. A considerable body of evidence is emerging to suggest that mTOR signaling is a key regulator of adipose tissue biology and function. Here we review current knowledge on mTOR signaling in modulating various adipose tissue functions including adipogenesis, lipogenesis, lipolysis, thermogenesis, and endocrine function, with an emphasis on animal model studies. We also discuss the potential link of dysregulated mTOR signaling pathway to metabolic diseases, and describe promising strategies of inhibiting mTOR in the prevention and treatment of obesity and its comorbidities.
mTOR and its signaling network regulation
mTOR is a phosphoinositide 3-kinase (PI3K)-like serine/threonine protein kinase that controls protein and lipid synthesis, cell size, proliferation, differentiation, autophagy and metabolism according to intracellular and extracellular cues [15, 16]. mTOR is the catalytic core of two distinct multiprotein complexes, mTOR complex 1 (mTORC1) and 2 (mTORC2), which differ in their components, regulation, function and sensitivity to rapamycin. In addition to mTOR, mTORC1 consists of regulatory associated protein of mTOR (Raptor), Akt/PKB substrate 40 kDa (PRAS40), mammalian lethal with SEC13 protein 8 (mLST8), Tti1/Tel2 complex and DEP domain containing mTOR-interacting protein (Deptor). Besides having the same components found in mTORC1 (mTOR, mLST8, Deptor and the Tti1/Tel2 complex), mTORC2 contains three unique elements, namely Raptor-independent companion of mTOR (Rictor), mammalian stress-activated protein kinase-interacting protein (mSin1) and protein observed with Rictor-1 and -2 (PROTOR1/2).
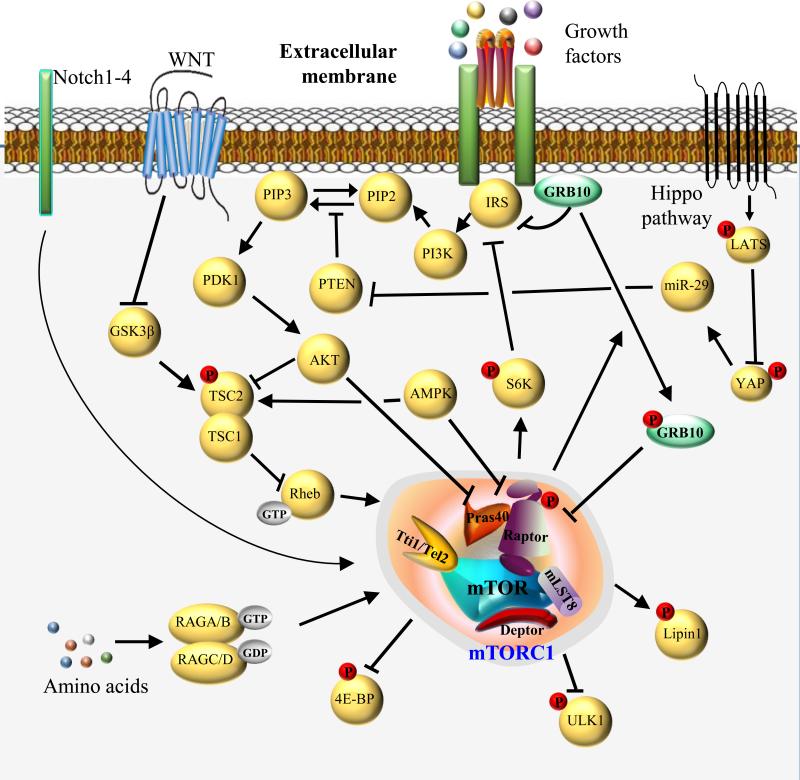
mTORC1 is the central integrator of multiple inputs such as growth factors, amino acids, cellular energy status, stress and oxygen. Growth factors stimulate mTORC1 through the activation of the canonical PI3K-Akt signaling pathway. For example, upon insulin stimulation, the PI3K-dependent activation of Akt leads to the phosphorylation and inhibition of the tuberous sclerosis complex (TSC1/2), resulting in the activation of the small GTPase Rheb (RAS homologue enriched in brain), an upstream activator of mTOR [17]. Amino acids activate mTORC1 by targeting mTOR to the lysosomal surface, where mTORC1 can encounter its activator Rheb, via a Rag-Ragulator complex-dependent mechanism [18]. Upon activation, mTORC1 regulates ribosomal biogenesis, cap-dependent translation, lysosomal biogenesis, lipid synthesis, autophagy, and thermogenesis by direct phosphorylation of a number of substrates including ribosomal S6 kinase 1/2 (S6K), eIF4E-binding protein 1/2 (4E-BP-1), transcription factor EB (TFEB1), Lipin1, UNC-51-like kinase 1 (Ulk1) and growth factor receptor-bound protein-10 (Grb10) [19-22]. Moreover, mTORC1 signaling can promote nucleotide biosynthesis by promoting the expression of genes involved in the pentose phosphate pathway [23] and pyrimidine biosynthesis via S6K1-mediated activation of CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase) [24, 25]. The mTOR substrates are in turn to fine-tune proper growth factors and mTOR signaling network through negative feedback mechanisms. For example, S6K directly phosphorylates insulin receptor substrate-1 (IRS-1) and hinders its association with the insulin receptor (IR) [26, 27]. In addition, Grb10 was recently identified as a negative regulator of the mTORC1 signaling pathway through a phosphorylation-dependent feedback mechanism [28, 29] (Figure 1).
Figure 1. Overview of mTOR complex 1 and its signaling network.
mTORC1 consists of mTOR, regulatory associated protein of mTOR (Raptor), Akt/PKB substrate 40 kDa (PRAS40), mammalian lethal with SEC13 protein 8 (mLST8), Tti1/Tel2 complex and DEP domain containing mTOR-interacting protein (Deptor). In response to growth factors, activation of the classical PI3K-Akt pathway leads to the phosphorylation and inhibition of TSC2. Subsequent activation of Rheb-GTP increases mTORC1 activity toward its various substrates, including 4E-BP, ULK1, Lipin1, S6K, and Grb10. Akt activation also promotes mTORC1 activity by inhibiting PRAS40, a negative regulator of mTORC1. Phosphorylation of Grb10 by mTORC1 switches its binding affinity from the insulin receptor to Raptor, thereby destabilizing mTORC1 through a novel negative feedback mechanism. Similarly, S6K can negatively feedback on the insulin signaling pathway by inducing the degradation of IRS. Amino acids activate mTORC1 signaling through GTP-loaded RAS-related GTP-binding Protein (RAG) A or B and GDP-loaded RAG C or D complexes. In addition, AMPK suppresses mTORC1 signaling by activating TSC and inhibiting Raptor. WNT signaling stimulates mTORC1 by inhibiting GSK3β-mediated TSC2 activation. Promotion of Hippo signaling triggers a kinase cascade that phosphorylates and inhibits Yes-associated protein (YAP) by large tumor suppressor (LATS) kinases. Meanwhile, YAP is able to regulate expression of microRNA miR-29 which can activate mTOR signaling through PTEN suppression. Notch signaling also regulates mTOR activity in liver.
In addition to the classical inputs mentioned above, mTORC1 is also regulated by WNT, Hippo and Notch signaling pathways (see Glossary). WNT signaling has been shown to promote mTORC1 signaling by inhibiting glycogen synthase kinase (GSK3β)-mediated TSC2 activation [30, 31]. Both mTORC1 and mTORC2 are regulated by Hippo signaling through phosphatase and tensin homolog (PTEN), which is repressed by Yes-associated protein (YAP) and the microRNA miR-29 [32]. In addition, hyperactivation of Notch signaling increases Raptor protein expression and its interaction with mTOR, leading to increased lipogenesis in the liver [33]. Intriguingly, the mice with adipose tissue-specific inactivation of the Notch signaling pathway seem to phenocopy the adipose tissue-specific raptor knockout mice, displaying elevated energy expenditure, browning of WAT, and resistance to high-fat diet induced obesity [34]. However, whether or not mTOR responds to Notch signaling in adipose tissue awaits further investigation.
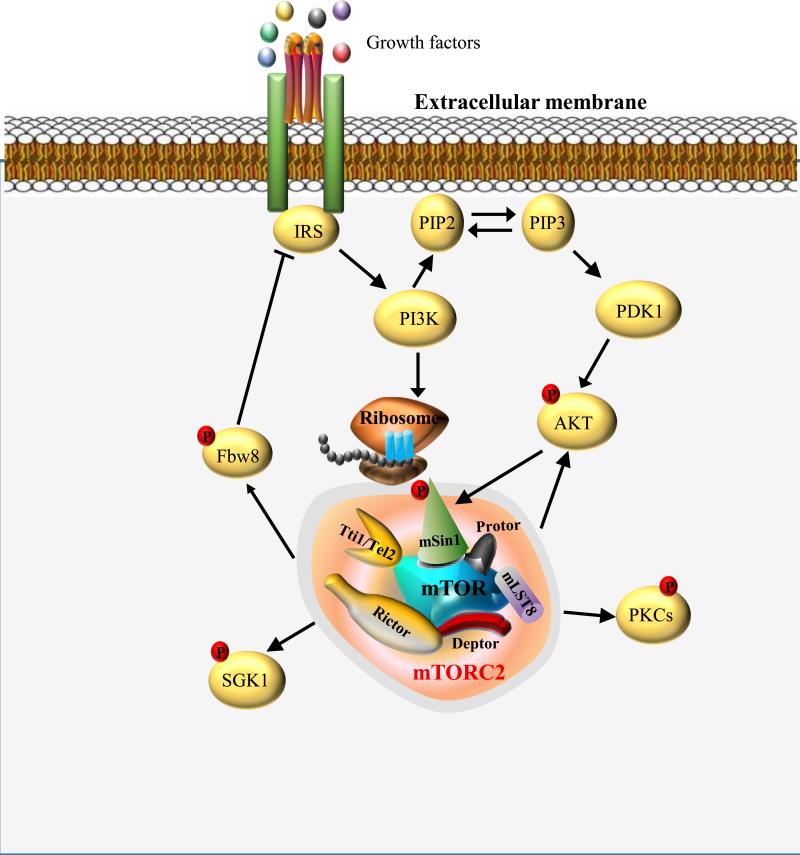
Unlike mTORC1, mTORC2 signaling is insensitive to nutrients, but responsive to growth factors mediated by PI3K. Insulin-stimulated PI3K signaling promotes mTORC2-ribosome binding, and the association of mTORC2 with ribosomes is essential for mTORC2 activation [35]. In addition, growth factor-dependent activation of Akt can directly phosphorylate SIN1 and enhance the activity of mTORC2, which in turn, results in increased feedback phosphorylation on Akt [36, 37]. mTORC2 regulates cell survival, proliferation, metabolism and cytoskeleton organization by promoting the phosphorylation of several AGC kinases such as serum- and glucocorticoid-induced protein kinase 1 (SGK1), protein kinase Cs and Akt [38]. Furthermore, mTORC2 negatively feeds back to IRS-1 via control of the Cullin 7 ubiquitin ligase substrate-targeting subunit Fbw8 to orchestrate proper mTORC2 signaling [39] (Figure 2).
Figure 2. Overview of mTOR complex 2 and its signaling network.
mTORC2 contains mTOR, mLST8, Deptor, Tti1/Tel2 complex, Raptor-independent companion of mTOR (Rictor), mammalian stress-activated protein kinase-interacting protein (mSin1) and protein observed with Rictor-1 and -2 (PROTOR1/2). Growth factor-stimulated PI3K signaling promotes mTORC2-ribosome binding and mTORC2 activation. Meanwhile, growth factor-dependent activation of Akt can directly phosphorylate mSin1 and enhance the activity of mTORC2. Upon activation, mTORC2 phosphorylates its downstream substrates including serum- and glucocorticoid-induced protein kinase 1 (SGK1), protein kinase Cs and Akt. Furthermore, mTORC2 negatively feeds back to IRS through Fbw8.
Role of mTOR signaling in adipogenesis
Adipogenesis is one of the most intensively studied models of cell differentiation. The Ser/Thr kinase Akt is well known for its essential role in adipocyte differentiation through multiple downstream signaling pathways [40]. By inhibiting FOXO1, Akt induces PPARγ expression and subsequent adipocyte differentiation [41]. Akt has also been shown to regulate adipogenesis via interplaying with the mTOR signaling pathway [42]. Inhibition of mTORC1 signaling genetically or with rapamycin impairs adipogenesis while increasing mTORC1 signaling promotes adipogenesis [43, 44], suggesting that mTORC1 is a positive regulator of adipogenesis. mTORC1 signaling has been implicated in promoting the three main steps of adipogenesis, namely lineage commitment, clonal expansion and terminal differentiation of preadipocytes to mature adipocytes, through distinct effectors. S6K1 has been shown to be involved in the commitment of embryonic stem cell to early adipocyte progenitors, but is dispensable for terminal adipocyte differentiation [45]. Consistently, mesenchymal stem cells lacking raptor exhibited a reduced capacity to form lipid-laden adipocytes but enhanced osteogenic differentiation capacity [46]. mTORC1 has also been implicated in hormonal induction of clonal expansion through the action of CCAAT/enhancer-binding protein-β and –δ (C/EBP-β and -δ). Furthermore, mTORC1 promotes the terminal differentiation of preadipocytes to mature adipocytes by inhibiting its direct substrate eukaryotic translation initiation factor 4E-binding proteins (4E-BPs) and activating PPAR-γ, a master regulator of adipocyte differentiation, lipogenesis and adipocyte function [47, 48]. mTORC1 also controls adipogenesis through another direct substrate, Lipin 1, which has a cell autonomous function in both white and brown adipocyte development and maintenance [49]. However, up-regulation of Deptor, a suppressor of mTOR signaling, promoted adipogenesis through activation of the proadipogenic Akt/PKB-PPAR-γ axis, indicating the mechanism underlying regulation of adipogenesis by the mTOR signaling pathway is far more complex than anticipated [50]. Intriguingly, while mTORC1 is essential for white adipocyte differentiation, this signaling pathway is only required for the first stage of brown adipogenesis. Consistent with this, subsequent inhibition of the mTOR-p70S6K1 signaling pathway by AMPK through Raptor inhibition and TSC2 activation has been shown to be indispensable for brown adipocyte differentiation [51, 52].
In contrast to mTORC1, the function of mTORC2 in adipogenesis is much less known. Adipose tissue-specific knockout of the rictor gene showed no effects on adipocyte cell size or overall adipose tissue mass, suggesting mTORC2 is dispensable for adipogenesis [53, 54]. However, in these studies the rictor gene was knocked out only in mature adipocytes or during the terminal phase of adipocyte differentiation, which may not be able to reveal the exact role of mTORC2 in early adipocyte differentiation. In fact, several recent studies demonstrated that mTORC2-mediated phosphorylation of Akt1 promoted adipogenesis by suppressing the expression of FoxC2, and Rictor-null mouse embryonic fibroblasts (MEFs) are incapable of differentiating into adipocytes [55-57]. In addition, Rictor-deficient brown adipocyte precursor cells are unable to differentiate and synthesize lipid droplets in vitro [58]. However, while these findings suggest that the mTORC2 signaling pathway is essential for adipocyte differentiation in vitro, loss of Rictor/mTORC2 in the Myf5 lineage (muscle, brown and some white adipocyte precursors) led to development of PPARγ and UCP1 positive BAT, though with smaller size [58]. Thus, it is possible that a compensatory signaling pathway may function in vivo to rescue impaired adipogenesis caused by the disruption of a signaling pathway normally involved in the regulation of such an important biological event.
Implication of mTOR signaling in lipogenesis
In cultured 3T3-L1 adipocytes, mTORC1 promotes lipogenesis and lipid homeostasis by activating sterol regulatory element-binding protein (SREBP) [59, 60], a key transcription factor that activates more than 30 genes dedicated to the synthesis and uptake of fatty acids, sterols, triglycerides and phospholipids [61]. However, the mechanism by which mTORC1 activates SREBP-1 is still under debate. S6K has been shown to mediate the regulatory effect of mTORC1 on SREBP-1 in TSC1/2-null mouse embryonic fibroblasts (MEFs), but it is not required for inducing SREBP processing in other cellular context [62, 63]. On the other hand, mTORC1 has been shown to promote the phosphorylation of lipin1 and blocks its nuclear translocation, leading to the activation of SREBP-1 in the nucleus [64]. Additionally, lipin1 can convert phosphatidic acid to diacylglycerol to positively regulate triacylglycerol synthesis [65]. Furthermore, the expression and activation of PPARγ, the critical stimulator of fatty acid uptake, synthesis, esterification and storage in the newly formed adipocytes, is also controlled by mTORC1 signaling [44, 66]. Hence mTORC1 may control lipogenesis through multiple effectors including SREBP-1, Lipin 1 and PPARγ. However, while these findings reveal a role of the mTORC1 signaling pathway in regulating lipogenesis in cultured adipocytes, the effects of mTORC1 on lipogenic gene expression are still incompletely understood in vivo [67]. Fat-specific knockout of raptor, a positive regulator of the mTORC1 signaling pathway, had little effect on the expression of PPARγ, C/EBPα and SREBP, despite a lean phenotype of these mice [68]. Consistent with this, activation of the mTORC1 signaling pathway in adipose tissue by fat-specific knockout of Grb10 had little effect on the expression of lipogenic enzymes such as acetyl-CoA carboxylase and fatty acid synthase [28]. Altogether, these results reveal either that adipose tissue mTORC1 signaling pathway does not regulate lipogenesis in vivo, or that the regulation of lipogenesis by mTORC1 signaling in vivo may be masked due to compensatory effects from other signaling mechanisms.
While a great deal of efforts has been made to elucidate the function of the mTORC1 signaling pathway in the regulation of lipid metabolism in adipocytes, much less is known about the role of the mTORC2 signaling pathway in modulating lipid synthesis. Conditionally deleting rictor in the brown adipocyte precursors reduced lipogenesis through Akt2 signaling and shifted BAT metabolism to a more oxidative state [58], suggesting that the mTORC2 signaling pathway may play a role in regulating lipogenesis. Consistent with this finding, liver-specific knockout of rictor led to reduced SREBP1 levels and impaired lipogenesis [69]. However, deletion of the rictor gene in the mature adipocytes, while increased lipolysis, had little effect on lipogenesis in mice [53]. One possible explanation for these distinct results could be that the regulation of lipogenesis by the mTORC2 signaling pathway may be time-dependent and/or tissue-specific. Further studies will be needed to test these possibilities.
Impact of mTOR signaling on lipolysis
Available evidence has indicated that the mTORC1 signaling pathway may regulate lipid metabolism via inhibition of lipolysis. Inhibition of the mTORC1 signaling pathway by rapamycin elevated phosphorylation of hormone sensitive lipase (HSL) and increased β-adrenergic agonist-induced lipolysis in adipocytes [70]. By contrast, overexpression of the mTOR activator Rheb in 3T3-L1 adipocytes reduced the expression levels of triacylglycerol lipase (ATGL) and HSL, leading to reduced lipolysis and increased de novo lipogenesis [67]. Mechanistically, mTORC1 suppresses lipolysis in adipocytes via the immediate-early response transcription factor, Egr1, which directly inhibits ATGL gene expression [71]. Very recently, the translation but not transcription of Egr1 was found to be regulated via the mTORC1-4E-BP-mediated axis, uncovering yet another mechanistic connection between mTORC1 and regulation of lipid homeostasis [72]. Furthermore, recent data also show that lipoprotein lipase (LPL) and triglyceride breakdown are inhibited by mTORC1 signaling, although the mechanism is yet to be fully defined [73, 74]. In mice, knockout of the mTORC1 downstream effectors, 4E-BP1 and 4E-BP2, decreased lipolysis while deleting S6K greatly increased lipolysis [75]. Moreover, up-regulation of mTORC1 signaling by fat-specific knockout of Grb10 significantly suppressed lipolysis and potentiated diet-induced obesity in mice [28]. Overall, these results demonstrate an inhibitory role of the mTORC1 signaling pathway in the regulation of lipolysis in adipocytes.
Unlike mTORC1, the role of mTORC2 in regulating lipolysis is less clear. One study showed that adipose-specific knockout of rictor in mice led to elevated serum levels of glycerol and free fatty acids, the products of lipolysis, under fasting condition. Additionally, rictor-null fat cells exhibited increased HSL phosphorylation concomitant with upregulated protein kinase A (PKA) activity and decreased suppression of lipolysis by insulin, suggesting that mTORC2 signaling inhibits lipolysis by regulating PKA and HSL activities. However, how mTORC2 modifies PKA activity is unknown [53]. In contrast, another study showed that fat-specific knockout of rictor in mice had no effect on free fatty acids level [54]. The reason for this discrepancy is currently unclear, but could be due to different substrains of the mouse models (129S6 vs. 129S1/Svlmj) and/or distinct methodology of assessing free fatty acid levels (under fasting vs. feeding condition). Consistent with the inhibitory role of mTORC2 signaling on lipolysis, liver-specific knockout of rictor led to increased lipolysis and mitochondrial oxidation in adipose tissue in mice, maybe through a non-cell autonomous mechanism [69]. Nevertheless, while both mTORC1 and mTORC2 have been shown to regulate lipolysis in adipose tissue, a recent study demonstrates that the products of lipolysis inhibit both mTORC1 and mTORC2 via complex dissociation [76], highlighting an interesting negative feedback mechanism through which proper lipolysis may be fine-tuned by mTOR signaling.
Effect of mTOR signaling on thermogenesis
Brown fat is specialized in dissipating chemical energy into heat via uncoupled respiration (non-shivering thermogenesis). This process, which can be activated by a variety of stimuli such as cold exposure and adrenergic agonists, is mediated by cAMP/PKA-dependent up-regulation of UCP-1 [3]. However, clusters of UCP-1-expressing adipocytes with thermogenic capacity also develop within white adipose depots upon stimulation (the beige fat) [4]. An increasing body of evidence reveals that the mTOR signaling pathway plays an important role in regulating thermogenic function in BAT and beige fat. Activation of mTORC1 signaling by removal of Tsc1 gene inhibits the expression of thermogenic genes such as Ucp-1 and Pgc-1α in BAT [77]. Interestingly, mTORC1 signaling pathway negatively controls thermogenic function not only in brown fat but also in beige fat. For example, augmentation of adipose mTORC1 signaling by fat-specific disrupting Grb10 expression decreases core body temperature and cold tolerance in mice, and attenuates cold-induced thermogenic genes expression in BAT and inguinal WAT [28]. By contrast, WAT but not BAT of fat-specific raptor knockout mice displayed elevated expression of Ucp-1, type 2deiodinase (dio2) and cidea, indicating a negative regulation of beige fat development by the mTORC1 signaling pathway [68]. Along similar lines, white adipocytes from S6K1−/− mice gained some characteristics of brown adipocytes including increased UCP-1 expression, multilocular lipids, mitochondrial size and number [78]. Nevertheless, how the mTORC1 signaling pathway regulates thermogenic function in BAT and beige fat remains enigmatic. As UCP-1 functions as a H+/fatty acid symporter, fatty acids produced from lipolysis were revealed to allosterically activate UCP-1-mdiated uncoupling [79, 80]. Thus, the inhibition of lipolysis by mTOR signaling may partially account for reduced UCP-1-mediated uncoupling and impaired thermogenic function observed in mTORC1-hyperactive animal models, and vice versa. Despite a common ability to undergo thermogenesis, brown and beige adipocytes have distinct gene signatures, come from distinguishing progenitors, and express different Ucp-1 levels under unstimulated conditions [81]. In addition, a number of factors are associated with induction of beige but not brown adipocytes. For instance, activation of type 2 innate lymphoid cells promotes beige fat biogenesis in an IL-4/13-dependent manner [82]. However, administration of IL-4 or knockout of the Il4/13 has little effect on UCP1 protein in BAT of thermoneutral mice, revealing distinct mechanisms involved in the regulation of the thermogenic gene expression in BAT and beige fat [83]. Therefore, more efforts are in need to understand whether mTORC1 signaling regulates thermogenic functions in BAT and beige fat through similar or distinct mechanisms.
In contrast to mTORC1, little is known thus far about the effect of mTORC2 signaling on thermogenic function in brown or beige fat. Myf5 lineage-specific deficiency of rictor increased diet-induced thermogenesis, upregulated genes involved in thermogenesis and mitochondrial biogenesis such as Ucp-1, Pgc-1α, Dio2, Tfam and C/ebpβ in BAT [58], indicating a negative regulation of thermogenesis by mTORC2 signaling. Upon cold exposure or adrenergic stimulation the glucose uptake in BAT is also greatly enhanced [84, 85]. Interestingly, mTORC2 but not mTORC1 has a novel role in β 3-adrenoceptor-stimulated glucose uptake in BAT, in which mTORC2 stimulates translocation of GLUT1 to the plasma membrane and increases glucose uptake, independent of classic insulin-PI3K-Akt pathway [86]. Fatty acids stimulate UCP-1 activity and supply fuel for BAT thermogenesis. However, BAT also utilizes glucose as an important fuel source and maintains whole body glucose homeostasis [87]. The essential role of mTORC2 in mediatingβ3-adrenoceptor-stimulated glucose uptake in BAT makes it an appealing target for treatment of diabetes and other metabolic disorders. A recent study revealed mTORC2 localized to mitochondria-associated endoplasmic reticulum (ER) membrane (MAM) upon growth factor stimulation and inhibited mitochondrial inner membrane potential [88]. However, whether the special localization of mTORC2 to MAM and its regulation of mitochondrial physiology are associated with the effect of mTORC2 signaling on mitochondrial uncoupling and thermogenesis is unknown. Thus, how mTORC2 signaling regulates thermogenic function in adipose tissue warrants further investigation. Given the unique capacity of dissipating energy as heat, mTOR signaling-based brown and beige fat activation could be an important avenue towards enabling therapeutic prevention of obesity and related metabolic diseases.
Regulation of the endocrine function of adipose tissue by mTOR
Adipose tissue is now well recognized as a highly active metabolic and endocrine organ. A growing body of evidence suggests that mTOR may play a key role in regulating the endocrine function of adipose tissue. mTORC1 was found to regulate insulin-, leucine-, and dexamethasone-stimulated leptin production in isolated rat adipocytes [89, 90]. Leptin expression was also increased in the mTORC1 consecutive activating cells [42]. Interestingly, fat-specific raptor knockout mice displayed decreased serum leptin levels [68]. However, the reduced serum leptin levels could be due to the substantially less adipose mass observed in raptor null mice. Further investigations are therefore in need to elucidate potential function of mTORC1 signaling in leptin production in vivo. Besides the alterations in adipose tissue per se, the physiology of non-adipose organs and the whole body metabolism have also been modified in fat-specific raptor knockout mice. For example, the muscle insulin sensitivity was greatly enhanced, glucose tolerance and energy expenditure were elevated, but the physical activity was decreased in fat-specific raptor knockout mice (Table 1). Although decreased circulating leptin levels could partially account for the reduced physical activity observed in raptor fat-null mice [91, 92], it is likely that the levels of other unidentified adipokines may also be changed due to inactivation of mTORC1 signaling in fat, through which adipose tissue communicates with other organs and coordinates whole body energy homeostasis. In contrary, serum leptin levels in adipose tissue-specific rictor knockout mice were unchanged, indicating that mTORC2 signaling may not participate in leptin synthesis and/or secretion [53, 54]. mTOR signaling has also been shown to regulate the synthesis and secretion of adiponectin, but the effect remains controversial. Activation of mTORC1 in Tsc2-deficient mouse embryonic fibroblasts (MEFs) correlates with higher adiponectin levels [42], while treating 3T3-L1 with rapamycin had no effect on insulin- and amino acid-stimulated adiponectin production and secretion [93]. However, mice with adipose-specific depletion of raptor have lower plasma adiponectin levels [68], indicating that mTORC1 signaling may be responsible for adiponectin biosynthesis and/or secretion in vivo. Nonetheless, it is currently unclear how mTORC1 signaling regulates adiponectin production. There is some data suggesting a role of the mTORC2 signaling pathway in the regulation of adiponectin biosynthesis and/or secretion, although the results remain controversial. In one study, it has been found that fat-specific knockout of rictor reduced serum adiponectin levels in mice [54]. In another study, however, no effect in serum adiponectin levels was observed in fat-specific rictor-null mice [53]. Intriguingly, these fat-specific rictor knockout mice exhibited alterations in non-fat tissues and whole body physiology, including enlarged non-adipose tissues, hyperinsulinemia, insulin resistance in muscle and liver, as well as hepatosteatosis (Table 1). However, considering the endocrine function of adipose tissue, it is not unexpected that fat-specific inactivation of mTORC2 signaling will change the adipose secretome and in turn affects non-fat tissue physiology and whole body metabolism. For example, reduced adiponectin levels in fat-specific rictor null mice may contribute to insulin resistance in muscle and liver. Although the potential adipokines are yet to be identified, the rictor-deleted adipose tissue could control pancreatic insulin production and hepatic insulin-like growth factor-1 secretion through these secreting factors and eventually lead to whole body metabolic alterations [54]. Fibroblast growth factor 21 (FGF21) is another important adipokine produced in adipose tissue, in addition to liver and skeletal muscle [94]. mTORC1 signaling has been found to regulate FGF21 expression in the liver [95], although its role in regulating FGF21 production in adipose tissue remains to be determined. To date more than 600 potentially secretory proteins have been identified in adipose tissue that play key roles in regulating metabolism and energy homeostasis [96]. Nonetheless, whether and how mTOR signaling controls the expression and secretion of these adipokines remains unknown. Better understanding the roles of mTOR signaling in orchestrating the endocrine function of adipose tissue should promote the development of novel adipokine-based pharmacological treatment strategies and diagnostic tools.
Table 1.
The effects of altered mTOR signaling on adipose functions and whole body metabolism
| Mouse models | mTOR signaling alterations | Phenotypes | References |
|---|---|---|---|
| Fabp4-raptor−/− mice (raptor gene is deleted by FABP4-Cre) | Inactivation of mTORC1 signaling mainly in adipose tissue | White adipocytes size and number ↓ Mitochondrial uncoupling in WAT ↑ Diet-induced obesity ↓ Plasma leptin level ↓ Glucose tolerance in HFD ↑ Muscle insulin sensitivity ↑ Energy expenditure ↑ Physical activity ↓ |
[61] |
| Adiponectin-Grb10−/− mice (Grb10 gene is deleted by Adipoq-Cre) | Hyper-activation of mTORC1 signaling specifically in adipose tissue | Lipolysis and fatty acid oxidation ↓ Diet-induced obesity ↑ Glucose and insulin tolerance in HFD ↓ Hepatosteatosis ↑ Thermogenesis ↓ Energy expenditure ↓ |
[24] |
| Fabp4-Tsc1−/− mice (Tsc1 gene is deleted by FABP4-Cre) | Hyper-activation of mTORC1 signaling mainly in adipose tissue | Abnormal mitochondrial structure in BAT ↑ mtDNA content in BAT ↓ Thermogenic genes expression in BAT ↓ White adipocyte morphological properties in BAT ↑ |
[70] |
| Fabp4-rictor−/− mice (rictor gene is deleted by FABP4-Cre) | Inactivation of mTORC2 signaling mainly in adipose tissue | Body size and lean mass ↑ Serum adiponectin level ↓ Circulating insulin level ↑ Insulin sensitivity ↓ Serum insulin-like growth factor 1 ↑ Hepatosteatosis ↑ |
[47] |
| Fabp4-rictor−/− mice (rictor gene is deleted by FABP4-Cre) | Inactivation of mTORC2 signaling mainly in adipose tissue | Non-adipose organ weight ↑ Circulating insulin level ↑ Systemic insulin sensitivity ↓ Glucose metabolism ↓ Lipolysis ↑ Lipid accumulation in skeletal muscle ↑ Hepatosteatosis ↑ |
[46] |
| Myf5-rictor−/− mice (rictor gene is deleted by Myf5-Cre) | Inactivation of mTORC2 signaling mainly in muscle, brown and some white adipocyte precursors | BAT, retroperitoneal and anterior subcutaneous WAT mass ↓ Lipogenesis in BAT ↓ Mitochondrial activity in BAT ↑ Diet induced obesity ↓ Thermogenesis ↑ Hepatosteatosis in HFD ↓ Glucose tolerance in HFD ↑ |
[51] |
| S6K1−/− mice | Inactivation of mTORC1 signaling in the whole body | Diet induced obesity ↓ Early adipocyte differentiation ↓ Mitochondrial content and metabolic rate ↑ Insulin sensitivity ↑ |
[38, 71] |
| 4E-BP1 and 2−/− mice | Hyper-activation of mTORC1 signaling in the whole body | Adipocyte differentiation ↑ Diet-induced obesity ↑ Lipolysis ↓ Insulin resistance ↑ Hepatosteatosis ↑ |
[41] |
Tissue-specific mTOR signaling in obesity/diabetes and therapeutic prospects
Dysregulation of the mTOR pathway has been linked to a number of pathological conditions such as obesity, diabetes, cancer, autoimmune disorders, neurodegenerative diseases, and aging. mTORC1 is highly active in tissues of obese and high-fat-diet fed rodents [97]. In humans, the mTORC1 signaling effector S6K is upregulated in visceral fat of human subjects with obesity and insulin resistance [98]. In addition, single nucleotide polymorphisms (SNPs) analysis revealed that common genetic variation in Raptor is associated with overweight/obesity in American men of Japanese ancestry [99]. Although the mechanism of hyperactivation of mTOR signaling in the setting of obesity and diabetes is still elusive, elevated branched-chain amino acids in obese and type 2 diabetic patients may lead to elevated mTORC1 signaling, which may further exacerbate hyperlipidemia and hyperinsulinemia [100].
Rapamycin was originally used as an antifungal and immunosuppressive agent, and subsequently discovered to suppress cell proliferation by inhibiting the functions of TOR signaling [101, 102]. While hyperactivation of the mTORC1 signaling pathway in obese mice and humans appears to play a role in developing insulin resistance and diabetes, inhibition of mTOR pathway by rapamycin has been demonstrated to have both beneficial and detrimental effects on insulin sensitivity and whole body metabolism [103, 104]. Interestingly, a recent study showed that the duration of rapamycin treatment may have differential effects on metabolism, providing a likely explanation for previously conflicting reports on the roles of rapamycin [105]. Short-term (2 weeks) rapamycin treatment causes hyperlipidemia, insulin resistance and promotes hepatic gluconeogenesis, while prolonged rapamycin (20 weeks) treatment leads to beneficial metabolic alterations including reduced adiposity, increased insulin sensitivity, improved lipid profile and higher energy expenditure [105]. However, an even longer rapamycin treatment (52 weeks) causes diabetes in male mice, which could be protected by estradiol [106]. Despite inhibition of mTORC1 by acute rapamycin treatment, chronic rapamycin administration impairs the integrity of mTORC2 and disrupts its role in Akt phosphorylation and hepatic gluconeogenesis inhibition, leading to impaired whole body insulin sensitivity and diabetic phenotype [107]. Consistently, two recent studies showed that suppressing mTORC2 signaling decreases the lifespan of male mice and that the rapamycin-mediated metabolic improvement and lifespan extension are dose and sex dependent [108, 109].
Although rapamycin has been clinically used to suppress immune rejection following transplant surgery and treatment of renal cell carcinoma, it has numerous side effects including suppression of the immune system, dermatological adverse events and reduction of male fertility [110]. The newly developed anti-cancer mTOR inhibitors, such as mTOR kinase inhibitors and dual PI3K/mTOR inhibitors, impair cell growth and proliferation to a much better degree than rapamycin and have been tested in clinical trials [111]. However, as these compounds have strong inhibitions of both mTORC1 and mTORC2, it is likely they will have undesirable side effects if they are used chronically for suppressing mTOR function in obesity and diabetes. Therefore, strategies of normalizing mTORC1 activity to the physiological range rather than completely blocking its activity by using sub-optimal doses of rapamycin and mild mTOR inhibitors, could reduce adiposity, improve energy metabolism and limit the inhibition of mTORC2. In addition, inhibition of mTORC1 downstream effectors could represent another interesting approach to combat obesity without too many side effects. Although S6K1 inhibitors are now being developed, these compounds will require long-term testing before their application in treating obesity and metabolic diseases [112, 113]. Moreover, the emerging evidence of genetic interventions of mTOR signaling in distinct tissues elicits diverse effects [114, 115]. For example, muscle-specific deficiency of Raptor or mTOR led to muscular dystrophy or myopathy associated with impaired oxidative metabolism and reduced mitochondrial function, whereas adipose tissue-specific knockout of raptor or Myf5+ cell-specific knockout of rictor contributed to increased energy expenditure, better metabolic profiles and protection from diet-induced obesity in mice [116]. Therefore, fat-specific manipulation of mTOR activity may pave a new road towards therapeutic prevention of obesity and its comorbidities.
Concluding Remarks
Fat is a highly adaptive tissue with multiple functions and involved in maintaining systemic energy homeostasis. mTOR complexes are the crucial signaling nodes involved in various anabolic and catabolic processes such as promoting lipid, protein and nucleotide synthesis as well as suppressing lipolysis and β-oxidation in response to diverse cellular, nutrient and environmental cues. In nutrient abundant circumstances, mTOR signaling stimulates adipose tissue expansion and thus prevents ectopic lipid accumulation and lipotoxicity in non-adipose tissue through promoting adipogenesis and lipogenesis as well as inhibiting lipolysis (Figure 3). By contrast, mTOR will exert opposite action in adipose tissue under nutrient deficient conditions. In addition, mTOR signaling can regulate adipokine and cytokine synthesis and/or secretion, through which adipose tissue cross talks to other organs and thus orchestrates a large number of physiological processes. Although great strides have been made towards understanding how mTOR signaling regulates adipose tissue function, we have only reached the tip of iceberg with regards to the precise mechanisms through which mTOR signaling pathway exerts its actions on each physiological process in adipose tissue. Correcting dysregulated mTOR signaling represents a promising therapeutic strategy in fighting against obesity and associated diseases. Given that the mTORC1 and mTORC2 signaling pathways have different effects in distinct tissues and organs, selective and tissue-specific targeting of these signaling pathways may lead to effective and highly specific therapeutic interventions. Despite the great progress that has been made, a number of outstanding questions remain to be addressed (see Outstanding Questions). Answers to these questions will provide novel insights into the mechanisms linking mTOR signaling to lipid metabolism and adipose tissue function.
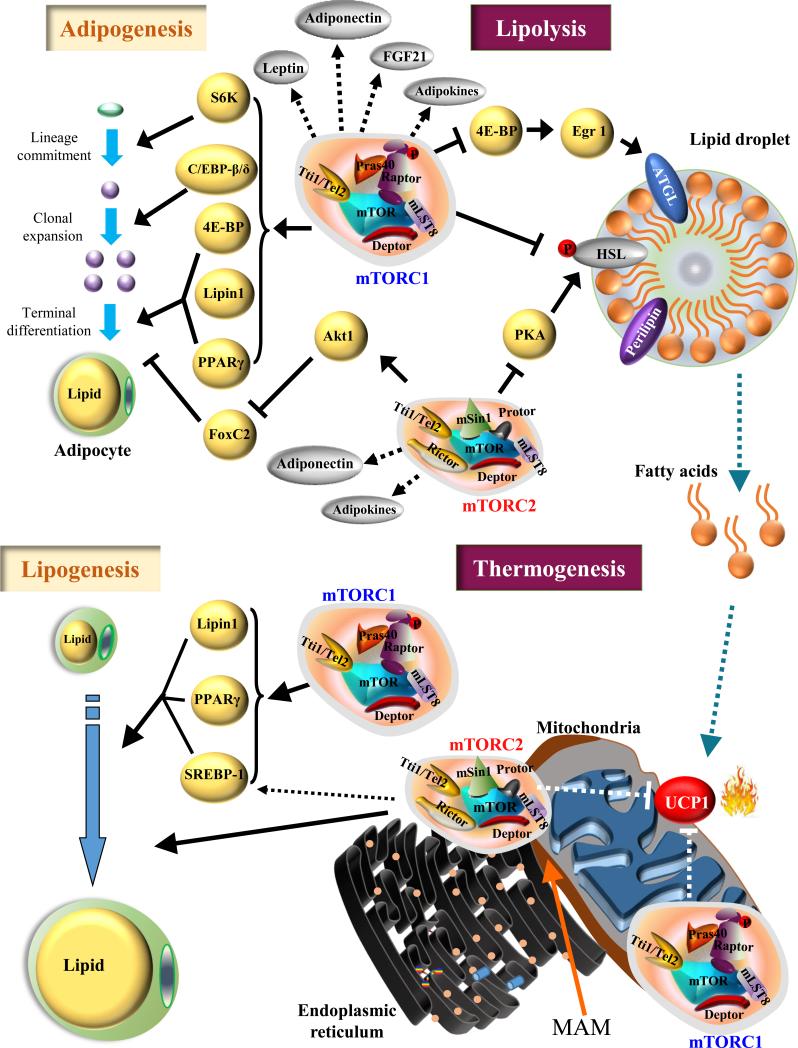
Figure 3. The roles of mTORC1 and mTORC2 in the adipose functions.
mTORC1 signaling promotes the main steps of adipogenesis, including lineage commitment, clonal expansion and terminal differentiation of preadipocytes to mature adipocytes, through S6K, C/EBP-β and -δ, 4E-BP, Lipin1, and PPAR-γ, respectively. mTORC2 stimulates adipogenesis by activating Akt1 and inhibiting FoxC2. mTORC1 may regulate endocrinal function of adipose tissue via promoting production of Leptin, Adiponectin and other adipokines. Similarly, mTORC2 may also be involved in adipokine synthesis and/or secretion. mTORC1 controls lipogenesis through multiple effectors including SREBP-1, Lipin 1, and PPARγ. mTORC2 stimulates lipogenesis in adipose tissue and may regulate SREBP-1 levels. mTORC1 suppresses lipolysis by inhibiting phosphorylation of HSL as well as reducing ATGL expression through suppression of 4E-BP regulated Egr1 translation. Moreover, mTORC2 suppresses lipolysis by regulating PKA activity. Inhibition of lipolysis and free fatty acid production by mTOR signaling may lead to reduced UCP1, and thermogenesis. mTORC2 is localized to MAM and may regulate mitochondrial respiration and thermogenesis. Dashed arrows represent unclear or hypothetical signaling pathway. Solid arrows represent well-established signaling pathway.
Highlights.
● Adipose tissue is a multi-functional organ displaying enormous plasticity by altering tissue size as well as phenotypic and metabolic functions in response to environmental signals.
● The mechanistic target of rapamycin (mTOR) signaling pathway regulates adipose biology and function, including adipogenesis, lipid metabolism, thermogenesis, and adipokine synthesis/secretion.
● Deregulated mTOR signaling is associated with obesity and related metabolic disorders, and strategies of appropriate modulation of the activity and tissue specific function of mTOR signaling pave the way towards counteracting obesity and its comorbidities.
Outstanding questions.
Does mTOR signaling use a similar or distinct mechanism to regulate mitochondrial uncoupling and thermogenesis in brown and beige adipose tissue?
How is the mTOR signaling pathway specifically regulated in distinct cells or tissues?
The roles of mTORC2 signaling in adipose tissue function are still incompletely understood and it is likely that our knowledge will be improved with the identification and characterization of more mTORC2 substrates and mTORC2-specific inhibitors.
Dysregulation of the mTOR pathway has been implicated in a spectrum of pathological conditions. What is the connection between mTOR dysregulation and metabolic dysfunction, cancer, autoimmune diseases, neurodegenerative disorders, and aging?
The action of mTOR signaling in regulating adipokine synthesis and secretion has significant importance in adipose biology, but the underlying mechanisms remain largely unknown. Further investigations are in need to better understand how mTOR signaling coordinates endocrine function of adipose tissue.
Obesity elicits an immune response characterized by myeloid cell recruitment to adipose tissue. Whether and how do mTORC1 and mTORC2 signaling regulate immune response in adipose tissue?
Acknowledgements
This work is supported by grants from the NIH (DK100697 to FL and R01 DK080344 to LQD), the Basic Research Program of China (#2014CB910501 to FL) and National Nature Science Foundation of China (81500662 to HC). We thank Mr. Christopher Cervantes for proofreading the manuscript, and Dr. Guangdi Li for assistance in the graphic presentation.
Abbreviation
- Akt
protein kinase B
- AMPK
5’AMP-activated protein kinase
- ATGL
adipose triacylglycerol lipase
- C/EBP-β and -δ
CCAAT/enhancer-binding protein –β and –δ
- 4E-BP
elF4E-binding protein
- Egr 1
early growth response transcription factor 1
- Grb10
growth factor receptor-bound protein 10
- HSL
hormone sensitive lipase
- IRS
insulin receptor substrate
- MAM
mitochondria-associated endoplasmic reticulum membrane
- PPARγ
peroxisome proliferator-activated receptor γ
- Rheb
Ras-homolog enriched in brain
- S6K
ribosomal S6 kinase
- TSC
tuberous sclerosis complex
- ULK1
UNC-51-like kinase 1
Glossary
- AMPK
5’ adenosine monophosphate-activated protein kinase is a serine/threonine heterotrimeric kinase composed of a catalytic α subunit and two regulatory subunits, β and γ. AMPK is sensitive to the AMP to ATP ratio and is activated by an increasing AMP concentration and by the upstream kinases including liver kinase B1 (LKB1) and calcium/calmodulin (CaM) kinase (CaMKK). AMPK mediates the metabolic response to environmental or dietary changes and has a crucial role in both cellular and whole-body energy status.
- AGC kinase
AGC kinase is a subgroup of Serine/Threonine protein kinases that are most related to protein kinase A, protein kinase G and protein kinase C based on sequence alignments of their catalytic kinase domain. The AGC family contains 60 protein kinases mediating diverse and important cellular functions.
- C/EBP
CCAAT/enhancer-binding proteins belong to a family of transcription factors which interact with the CCAAT box motif in several gene promoters. In the process of adipogenesis, C/EBP-β and-δ are transiently induced during the early stages of adipocyte differentiation, while C/EBP-α is upregulated during the terminal stages of adipogenesis. Each of them plays an important role in adipogenesis.
- db/db mice
db/db mice are homozygous for a point mutation in the leptin receptor gene. Because of the deficiency of leptin receptor activity, the mice display obesity, diabetes and dyslipidemia.
- FoxC2
a member of the fork head box (FOX) family of transcription factors. FoxC2 is recognized as a regulator of vascular formation and remodeling. It is also involved in cancer metastases. FoxC2 was also found to inhibit white adipocyte differentiation, counteract obesity, hypertriglyceridemia, and diet-induced insulin resistance.
- Hippo signaling pathway
Hippo signaling which was first discovered in the fruit fly, is a highly conserved signaling network that control cell proliferation, differentiation, and cell death. The Hippo signaling pathway is composed of a core kinase cascade initiating from Hippo (Mst1 and Mst2 in mammals) to the phosphorylation of a Yki (YAP and TAZ in mammals), which leads to change of the subcellular localization of Yki from the nucleus, where it acts as a transcriptional activator, to the cytoplasm. Hippo pathway plays key roles in organ size control, regeneration and cancer development.
- Myf5
myogenic factor 5 is a protein with a key role in regulating muscle differentiation or myogenesis. By comparing the gene expression profiles of preadipocytes, brown preadipocytes were found to have a myogenic-like transcriptional signature including expression of Myf5. However, a subset of white adipocytes also comes from Myf5 expressing precursors.
- Notch signaling pathway
The Notch signaling pathway, which is an evolutionarily conserved pathway important for cell–cell communication and cell-fate determination during development, plays an important role in tumorigenesis, central nervous system function, cardiovascular function and energy metabolism. Notch ligands such as Delta-like (Dll) and Jagged (Jag) bind to Notch receptors and induce proteolytic cleavage and release of the Notch receptor intracellular domain, which enters the cell nucleus to modify gene expression.
- PGC-1α
peroxisome proliferator-activated receptor gamma coactivator 1-alpha, is a transcriptional coactivator that regulates the genes involved in energy metabolism. PGC-1α is a regulator of mitochondrial biogenesis and function.
- PPAR-γ
peroxisome proliferator-activated receptor γ is a nuclear receptor protein that functions as the transcriptional factor. PPAR-γ plays a key role in adipogenesis and lipid uptake in adipocytes. In addition, PPAR-γ activation by synthetic full agonists drives browning of white adipose tissue.
- PTEN
phosphatase and tensin homolog is a tumor suppressing gene and mutations of this gene contribute to the development of many cancers. The protein encoded by this gene is a phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase. It contains a tensin-like domain as well as a catalytic domain. It negatively regulates intracellular levels of phosphatidylinositol-3,4,5-trisphosphate and Akt/PKB signaling pathway.
- WNT signaling pathway
Wnt signaling begins with Wnt proteins bind to the N-terminal extra-cellular cycteine-rich domain of a Frizzled family receptor. The canonical Wnt pathway involves β-catenin while noncanonical pathway is independent of it. Wnt signaling pathway plays important roles in embryonic development, cell proliferation, cell migration, cancer and diabetes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sethi JK, Vidal-Puig AJ. Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J Lipid Res. 2007;48:1253–1262. doi: 10.1194/jlr.R700005-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rutkowski JM, et al. The cell biology of fat expansion. J Cell Biol. 2015;208:501–512. doi: 10.1083/jcb.201409063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fasshauer M, Bluher M. Adipokines in health and disease. Trends Pharmacol Sci. 2015;36:461–470. doi: 10.1016/j.tips.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Bluher M, Mantzoros CS. From leptin to other adipokines in health and disease: facts and expectations at the beginning of the 21st century. Metabolism. 2015;64:131–145. doi: 10.1016/j.metabol.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 8.Wang GX, et al. The brown fat secretome: metabolic functions beyond thermogenesis. Trends Endocrinol Metab. 2015;26:231–237. doi: 10.1016/j.tem.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMillan AC, White MD. Induction of thermogenesis in brown and beige adipose tissues: molecular markers, mild cold exposure and novel therapies. Curr Opin Endocrinol Diabetes Obes. 2015;22:347–352. doi: 10.1097/MED.0000000000000191. [DOI] [PubMed] [Google Scholar]
- 10.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cypess AM, et al. Activation of human brown adipose tissue by a beta3-adrenergic receptor agonist. Cell Metab. 2015;21:33–38. doi: 10.1016/j.cmet.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanssen MJ, et al. Short-term cold acclimation improves insulin sensitivity in patients with type 2 diabetes mellitus. Nat Med. 2015;21:863–865. doi: 10.1038/nm.3891. [DOI] [PubMed] [Google Scholar]
- 13.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornu M, et al. mTOR in aging, metabolism, and cancer. Curr Opin Genet Dev. 2013;23:53–62. doi: 10.1016/j.gde.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Albert V, Hall MN. mTOR signaling in cellular and organismal energetics. Curr Opin Cell Biol. 2015;33:55–66. doi: 10.1016/j.ceb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Lamming DW, Sabatini DM. A Central role for mTOR in lipid homeostasis. Cell metabolism. 2013;18:465–469. doi: 10.1016/j.cmet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garami A, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 18.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu PP, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnuson B, et al. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 22.Huang K, Fingar DC. Growing knowledge of the mTOR signaling network. Semin Cell Dev Biol. 2014;36:79–90. doi: 10.1016/j.semcdb.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robitaille AM, et al. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 25.Ben-Sahra I, et al. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah OJ, et al. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 27.Harrington LS, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu M, et al. Grb10 promotes lipolysis and thermogenesis by phosphorylation-dependent feedback inhibition of mTORC1. Cell metabolism. 2014;19:967–980. doi: 10.1016/j.cmet.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B, Liu F. Feedback regulation of mTORC1 by Grb10 in metabolism and beyond. Cell Cycle. 2014;13:2643–2644. doi: 10.4161/15384101.2014.954221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoki K, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 31.Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15:155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- 32.Tumaneng K, et al. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol. 2012;14:1322–1329. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pajvani UB, et al. Inhibition of Notch uncouples Akt activation from hepatic lipid accumulation by decreasing mTorc1 stability. Nat Med. 2013;19:1054–1060. doi: 10.1038/nm.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bi P, et al. Inhibition of Notch signaling promotes browning of white adipose tissue and ameliorates obesity. Nature medicine. 2014;20:911–918. doi: 10.1038/nm.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zinzalla V, et al. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Humphrey SJ, et al. Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 2013;17:1009–1020. doi: 10.1016/j.cmet.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang G, et al. A Positive Feedback Loop between Akt and mTORC2 via SIN1 Phosphorylation. Cell Rep. 2015;12:937–943. doi: 10.1016/j.celrep.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 38.Su B, Jacinto E. Mammalian TOR signaling to the AGC kinases. Crit Rev Biochem Mol Biol. 2011;46:527–547. doi: 10.3109/10409238.2011.618113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Destefano MA, Jacinto E. Regulation of insulin receptor substrate-1 by mTORC2 (mammalian target of rapamycin complex 2). Biochem Soc Trans. 2013;41:896–901. doi: 10.1042/BST20130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baudry A, et al. PKBalpha is required for adipose differentiation of mouse embryonic fibroblasts. J Cell Sci. 2006;119:889–897. doi: 10.1242/jcs.02792. [DOI] [PubMed] [Google Scholar]
- 41.Nakae J, et al. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell. 2003;4:119–129. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang HH, et al. Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PloS one. 2009;4:e6189. doi: 10.1371/journal.pone.0006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JE, Chen J. regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes. 2004;53:2748–2756. doi: 10.2337/diabetes.53.11.2748. [DOI] [PubMed] [Google Scholar]
- 44.Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr Biol. 2009;19:R1046–1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carnevalli LS, et al. S6K1 plays a critical role in early adipocyte differentiation. Dev Cell. 2010;18:763–774. doi: 10.1016/j.devcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin SK, et al. Brief report: the differential roles of mTORC1 and mTORC2 in mesenchymal stem cell differentiation. Stem Cells. 2015;33:1359–1365. doi: 10.1002/stem.1931. [DOI] [PubMed] [Google Scholar]
- 47.Lefterova MI, et al. PPARgamma and the global map of adipogenesis and beyond. Trends Endocrinol Metab. 2014;25:293–302. doi: 10.1016/j.tem.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Bacquer O, et al. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest. 2007;117:387–396. doi: 10.1172/JCI29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nadra K, et al. Cell autonomous lipin 1 function is essential for development and maintenance of white and brown adipose tissue. Mol Cell Biol. 2012;32:4794–4810. doi: 10.1128/MCB.00512-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laplante M, et al. DEPTOR cell-autonomously promotes adipogenesis, and its expression is associated with obesity. Cell Metab. 2012;16:202–212. doi: 10.1016/j.cmet.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernandez-Veledo S, et al. Role of energy- and nutrient-sensing kinases AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) in adipocyte differentiation. IUBMB Life. 2013;65:572–583. doi: 10.1002/iub.1170. [DOI] [PubMed] [Google Scholar]
- 52.Vila-Bedmar R, et al. Adenosine 5′-monophosphate-activated protein kinase-mammalian target of rapamycin cross talk regulates brown adipocyte differentiation. Endocrinology. 2010;151:980–992. doi: 10.1210/en.2009-0810. [DOI] [PubMed] [Google Scholar]
- 53.Kumar A, et al. Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes. 2010;59:1397–1406. doi: 10.2337/db09-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cybulski N, et al. mTOR complex 2 in adipose tissue negatively controls whole-body growth. Proc Natl Acad Sci U S A. 2009;106:9902–9907. doi: 10.1073/pnas.0811321106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis KE, et al. The forkhead transcription factor FoxC2 inhibits white adipocyte differentiation. J Biol Chem. 2004;279:42453–42461. doi: 10.1074/jbc.M402197200. [DOI] [PubMed] [Google Scholar]
- 56.Gerin I, et al. On the role of FOX transcription factors in adipocyte differentiation and insulin-stimulated glucose uptake. J Biol Chem. 2009;284:10755–10763. doi: 10.1074/jbc.M809115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao Y, et al. BSTA promotes mTORC2-mediated phosphorylation of Akt1 to suppress expression of FoxC2 and stimulate adipocyte differentiation. Sci Signal. 2013;6:ra2. doi: 10.1126/scisignal.2003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hung CM, et al. Rictor/mTORC2 loss in the Myf5 lineage reprograms brown fat metabolism and protects mice against obesity and metabolic disease. Cell Rep. 2014;8:256–271. doi: 10.1016/j.celrep.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Danai LV, et al. Map4k4 suppresses Srebp-1 and adipocyte lipogenesis independent of JNK signaling. J Lipid Res. 2013;54:2697–2707. doi: 10.1194/jlr.M038802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Porstmann T, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell metabolism. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shao W, Espenshade PJ. Expanding roles for SREBP in metabolism. Cell Metab. 2012;16:414–419. doi: 10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li S, et al. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci U S A. 2010;107:3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peterson TR, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitra MS, et al. Mice with an adipocyte-specific lipin 1 separation-of-function allele reveal unexpected roles for phosphatidic acid in metabolic regulation. Proc Natl Acad Sci U S A. 2013;110:642–647. doi: 10.1073/pnas.1213493110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guntur KV, et al. Map4k4 negatively regulates peroxisome proliferator-activated receptor (PPAR) gamma protein translation by suppressing the mammalian target of rapamycin (mTOR) signaling pathway in cultured adipocytes. J Biol Chem. 2010;285:6595–6603. doi: 10.1074/jbc.M109.068502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chakrabarti P, et al. Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes. 2010;59:775–781. doi: 10.2337/db09-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Polak P, et al. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008;8:399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 69.Hagiwara A, et al. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 70.Soliman GA, et al. mTORC1 inhibition via rapamycin promotes triacylglycerol lipolysis and release of free fatty acids in 3T3-L1 adipocytes. Lipids. 2010;45:1089–1100. doi: 10.1007/s11745-010-3488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chakrabarti P, et al. Insulin inhibits lipolysis in adipocytes via the evolutionarily conserved mTORC1-Egr1-ATGL-mediated pathway. Mol Cell Biol. 2013;33:3659–3666. doi: 10.1128/MCB.01584-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh M, et al. 4E-BPs control fat storage by regulating the expression of Egr1 and ATGL. J Biol Chem. 2015 doi: 10.1074/jbc.M114.631895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uno K, et al. A hepatic amino acid/mTOR/S6K-dependent signalling pathway modulates systemic lipid metabolism via neuronal signals. Nat Commun. 2015;6:7940. doi: 10.1038/ncomms8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ricoult SJ, Manning BD. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep. 2013;14:242–251. doi: 10.1038/embor.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Um SH, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 76.Mullins GR, et al. Catecholamine-induced lipolysis causes mTOR complex dissociation and inhibits glucose uptake in adipocytes. Proc Natl Acad Sci U S A. 2014;111:17450–17455. doi: 10.1073/pnas.1410530111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiang X, et al. Tuberous sclerosis complex 1-mechanistic target of rapamycin complex 1 signaling determines brown-to-white adipocyte phenotypic switch. Diabetes. 2015;64:519–528. doi: 10.2337/db14-0427. [DOI] [PubMed] [Google Scholar]
- 78.Um SH, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 79.Fedorenko A, et al. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell. 2012;151:400–413. doi: 10.1016/j.cell.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chakrabarti P, Kandror KV. The role of mTOR in lipid homeostasis and diabetes progression. Curr Opin Endocrinol Diabetes Obes. 2015;22:340–346. doi: 10.1097/MED.0000000000000187. [DOI] [PubMed] [Google Scholar]
- 81.Dempersmier J, Sul HS. Shades of brown: a model for thermogenic fat. Front Endocrinol (Lausanne) 2015;6:71. doi: 10.3389/fendo.2015.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee MW, et al. Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell. 2015;160:74–87. doi: 10.1016/j.cell.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Qiu Y, et al. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell. 2014;157:1292–1308. doi: 10.1016/j.cell.2014.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shimizu Y, et al. Sympathetic activation of glucose utilization in brown adipose tissue in rats. J Biochem. 1991;110:688–692. doi: 10.1093/oxfordjournals.jbchem.a123642. [DOI] [PubMed] [Google Scholar]
- 85.Orava J, et al. Different Metabolic Responses of Human Brown Adipose Tissue to Activation by Cold and Insulin. Cell Metabolism. 2011;14:272–279. doi: 10.1016/j.cmet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 86.Olsen JM, et al. Glucose uptake in brown fat cells is dependent on mTOR complex 2-promoted GLUT1 translocation. Journal of Cell Biology. 2014;207:365–374. doi: 10.1083/jcb.201403080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Townsend KL, Tseng YH. Brown fat fuel utilization and thermogenesis. Trends Endocrinol Metab. 2014;25:168–177. doi: 10.1016/j.tem.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Betz C, et al. Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc Natl Acad Sci U S A. 2013;110:12526–12534. doi: 10.1073/pnas.1302455110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bradley RL, Cheatham B. Regulation of ob gene expression and leptin secretion by insulin and dexamethasone in rat adipocytes. Diabetes. 1999;48:272–278. doi: 10.2337/diabetes.48.2.272. [DOI] [PubMed] [Google Scholar]
- 90.Roh C, et al. Nutrient-sensing mTOR-mediated pathway regulates leptin production in isolated rat adipocytes. Am J Physiol Endocrinol Metab. 2003;284:E322–330. doi: 10.1152/ajpendo.00230.2002. [DOI] [PubMed] [Google Scholar]
- 91.Tang GB, et al. Intracerebroventricular administration of leptin increase physical activity but has no effect on thermogenesis in cold-acclimated rats. Sci Rep. 2015;5:11189. doi: 10.1038/srep11189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huo L, et al. Leptin-dependent control of glucose balance and locomotor activity by POMC neurons. Cell Metab. 2009;9:537–547. doi: 10.1016/j.cmet.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blumer RM, et al. Regulation of adiponectin secretion by insulin and amino acids in 3T3-L1 adipocytes. Metabolism. 2008;57:1655–1662. doi: 10.1016/j.metabol.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 94.Itoh N. FGF21 as a Hepatokine, Adipokine, and Myokine in Metabolism and Diseases. Front Endocrinol (Lausanne) 2014;5:107. doi: 10.3389/fendo.2014.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cornu M, et al. Hepatic mTORC1 controls locomotor activity, body temperature, and lipid metabolism through FGF21. Proc Natl Acad Sci U S A. 2014;111:11592–11599. doi: 10.1073/pnas.1412047111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lehr S, et al. Adipokines: a treasure trove for the discovery of biomarkers for metabolic disorders. Proteomics Clin Appl. 2012;6:91–101. doi: 10.1002/prca.201100052. [DOI] [PubMed] [Google Scholar]
- 97.Khamzina L, et al. Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats: possible involvement in obesity-linked insulin resistance. Endocrinology. 2005;146:1473–1481. doi: 10.1210/en.2004-0921. [DOI] [PubMed] [Google Scholar]
- 98.Catalan V, et al. Expression of S6K1 in human visceral adipose tissue is upregulated in obesity and related to insulin resistance and inflammation. Acta Diabetol. 2015;52:257–266. doi: 10.1007/s00592-014-0632-9. [DOI] [PubMed] [Google Scholar]
- 99.Morris BJ, et al. Genetic variation in the raptor gene is associated with overweight but not hypertension in American men of Japanese ancestry. Am J Hypertens. 2015;28:508–517. doi: 10.1093/ajh/hpu188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dann SG, et al. mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med. 2007;13:252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 101.Sehgal SN. Sirolimus: its discovery, biological properties, and mechanism of action. Transplant Proc. 2003;35:7S–14S. doi: 10.1016/s0041-1345(03)00211-2. [DOI] [PubMed] [Google Scholar]
- 102.Li J, et al. Rapamycin: one drug, many effects. Cell Metab. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang SB, et al. Rapamycin ameliorates age-dependent obesity associated with increased mTOR signaling in hypothalamic POMC neurons. Neuron. 2012;75:425–436. doi: 10.1016/j.neuron.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Houde VP, et al. Chronic rapamycin treatment causes glucose intolerance and hyperlipidemia by upregulating hepatic gluconeogenesis and impairing lipid deposition in adipose tissue. Diabetes. 2010;59:1338–1348. doi: 10.2337/db09-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fang Y, et al. Duration of rapamycin treatment has differential effects on metabolism in mice. Cell Metab. 2013;17:456–462. doi: 10.1016/j.cmet.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schindler CE, et al. Chronic rapamycin treatment causes diabetes in male mice. Am J Physiol Regul Integr Comp Physiol. 2014;307:R434–443. doi: 10.1152/ajpregu.00123.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lamming DW, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lamming DW, et al. Depletion of Rictor, an essential protein component of mTORC2, decreases male lifespan. Aging Cell. 2014;13:911–917. doi: 10.1111/acel.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miller RA, et al. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell. 2014;13:468–477. doi: 10.1111/acel.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hartford CM, Ratain MJ. Rapamycin: something old, something new, sometimes borrowed and now renewed. Clin Pharmacol Ther. 2007;82:381–388. doi: 10.1038/sj.clpt.6100317. [DOI] [PubMed] [Google Scholar]
- 111.Chiarini F, et al. Current treatment strategies for inhibiting mTOR in cancer. Trends Pharmacol Sci. 2015;36:124–135. doi: 10.1016/j.tips.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 112.Qin J, et al. Development of organometallic S6K1 inhibitors. J Med Chem. 2015;58:305–314. doi: 10.1021/jm5011868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pearce LR, et al. Characterization of PF-4708671, a novel and highly specific inhibitor of p70 ribosomal S6 kinase (S6K1). Biochem J. 2010;431:245–255. doi: 10.1042/BJ20101024. [DOI] [PubMed] [Google Scholar]
- 114.Hu F, Liu F. Targeting tissue-specific metabolic signaling pathways in aging: the promise and limitations. Protein Cell. 2014;5:21–35. doi: 10.1007/s13238-013-0002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang J, Liu F. Tissue-specific insulin signaling in the regulation of metabolism and aging. IUBMB Life. 2014;66:485–495. doi: 10.1002/iub.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bentzinger CF, et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]