Abstract
Biopersistence of carbon nanotubes, graphene oxide (GO) and several other types of carbonaceous nanomaterials is an essential determinant of their health effects. Successful biodegradation is one of the major factors defining the life span and biological responses to nanoparticles. Here, we review the role and contribution of different oxidative enzymes of inflammatory cells - myeloperoxidase, eosinophil peroxidase, lactoperoxidase, hemoglobin, and xanthine oxidase - to the reactions of nanoparticle biodegradation. We further focus on interactions of nanomaterials with hemoproteins dependent on the specific features of their physico-chemical and structural characteristics. Mechanistically, we highlight the significance of immobilized peroxidase reactive intermediates vs diffusible small molecule oxidants (hypochlorous and hypobromous acids) for the overall oxidative biodegradation process in neutrophils and eosinophils. We also accentuate the importance of peroxynitrite-driven pathways realized in macrophages via the engagement of NADPH oxidase- and NO synthase-triggered oxidative mechanisms. We consider possible involvement of oxidative machinery of other professional phagocytes such as microglial cells, myeloid-derived suppressor cells, in the context of biodegradation relevant to targeted drug delivery. We evaluate the importance of genetic factors and their manipulations for the enzymatic biodegradation in vivo. Finally, we emphasize a novel type of biodegradation realized via the activation of the “dormant” peroxidase activity of hemoproteins by the nano-surface. This is exemplified by the binding of GO to cyt c causing the unfolding and “unmasking” of the peroxidase activity of the latter. We conclude with the strategies leading to safe by design carbonaceous nanoparticles with optimized characteristics for mechanism-based targeted delivery and regulatable life-span of drugs in circulation.
Keywords: carbon nanotubes, graphene oxide, peroxidase activity, oxidants, free radicals, phagocytes
Introduction
Advances in the development, production and applications of nanomaterials inevitably lead to numerous chemical and biochemical interactions at the interfaces of biological systems with nanoparticles that result in a variety of responses by the former as well as modifications of the latter. One can imagine that during the repetitive cycles of these interactions both the materials and the organisms are affected resulting in alterations that may change the consequences and the meaning of the interfacing partners. One of the first important modifications of the nanoparticles in biological systems is the formation of protein-lipid “corona” (Monopoli et al, 2012; Kapralov et al., 2012; Tenzer et al., 2013). The composition and properties of “corona” are dependent on the local microenvironments in biological fluids, tissues and cells, thus determining the specificity of nanoparticles-evoked reactions (Walkey et al., 2012; Sacchetti et al., 2013; Cai et al., 2013; Treuel et al., 2015). The physicochemical characteristics of the corona can also undergo marked changes due to metabolic conversions in the body and also via chemical reactions catalyzed by active ingredients of nanoparticles (Gao et al., 2014; Ma et al., 2015; Docter et al., 2015). This, in turn, triggers strongly modified biological responses (Dutta et al., 2007; Wang et al., 2013). Among the constituents of the protein corona, there may be enzymes contributing to the biodegradation process such as peroxidases, isoforms of CYP450, lysosomal hydrolases, etc.
Overall, nanoparticles tend to either be readily degradable or resistant to degradation in terms of their sensitivity to biodegradation. The first group includes nano-liposomes and polymeric nano-arrangements (such as dendrimers, micelles) (Kamaly et al., 2012; Xie et al., 2014; Chuan et al., 2015). Effective degradation of these nanomaterials is particularly important in the context of drug delivery aimed at the achievement of the prolonged circulation of the nano-vehicle with the payload (Owens et al., 2006; Loos et al., 2014; Pérez-Herrero et al., 2015). The second group comprises carbon-based nanoparticles with sp2 hybridization of carbon atoms (i.e. carbon nanotubes, nanohorns and graphene family materials) (Kotchey et al., 2013a; Zhang et al., 2014a). These nanomaterials are more persistent and can either display prolonged life-time at the sites of their entry or migrate to distant locations (Liu et al., 2008; Yang et al., 2008; Shvedova et al., 2012a, 2014). The mechanisms underlying the resistance of the latter type of nanoparticles to biodegradation are important not only for their biomedical applications but also in regards to unintended exposures in occupational and environmental settings.
While studies of nanoparticles degradation have been conducted essentially from the time of their discovery and initial applications (mostly in the field of anticancer therapy (McDevitt et al., 2007; Liu et al., 2009; Burke et al., 2009; Liang and Chen, 2010)), the discovery of in vivo biodegradation of carbonaceous nanomaterials by enzymatic machinery of inflammatory cells (Kagan et al., 2010; Shvedova et al., 2012a) and enhancement of the enzymatic degradation of carbon nanotubes by surface modification caused a new wave of interest to this issue (Ali-Boucetta and Kostarelos, 2013; Orecchioni et al., 2014; Sureshbabu et al., 2015). This was mostly driven by exploration of new approaches to regulate the life-time of nanoparticles in desirable ways: increasing the circulation time of drug nano-carriers and enhancing the biodegradation process of nanomaterials causing inflammatory responses and toxicity after inadvertent exposures (Liu et al., 2010; Sacchetti et al., 2013; Shvedova et al., 2012b). Notably, a variety of microbial biodegradation enzymatic mechanisms have been described with the emphasis on their potential role in biodegradation of environmental nanoparticles (Zhang et al., 2013).
Enzymatic oxidative degradation of carbonaceous nanoparticles
The chemical oxidative degradation of pristine carbonaceous materials using strong acids and oxidants (such as mixtures of sulfuric acid and hydrogen peroxide, different chemical generators of hydroxyl radicals) has been known for quite some time (Liu et al., 1998; Zhang et al., 2003; Allen et al., 2009). However, the biological relevance of these oxidative processes remained elusive in spite of the fact that the catabolic pathways for oxidative degradation of different organic molecules in the body (e.g., by different P450 isoforms) have been well characterized (Hrycay and Bandiera, 2015; Olsen et al., 2015). One of the first indications that biologically relevant peroxidase reactions may be responsible for degradation of nanomaterials came from experiments with single-walled carbon nanotubes (SWCNTs) by a plant enzyme, horseradish peroxidase (HRP) (Allen et al., 2008). Subsequent detailed studies of the mechanisms and the reaction products (Allen et al., 2009; Zhao et al., 2011) demonstrated that other bio-peroxidases, particularly those present in inflammatory cells, can also effectively oxidatively “metabolize” carbonaceous nanomaterials (Kagan et al., 2010; Kotchey et al., 2012). Indeed, a number of different oxidative enzymes have been tested and found effective as a mechanism of nanoparticle biodegradation (Kotchey et al., 2013a). The list of enzymes includes myeloperoxidase (MPO), eosinophil peroxidase (EPO), lactoperoxidase, hemoglobin and xanthine oxidase (Table 1). Contrary to HRP, another plant metallo-enzyme Mn peroxidase, was shown to degrade pristine but not carboxylated SWCNTs (Zhang et al., 2014b). These studies also established that two types of reactive intermediates – those formed within the protein (particularly oxo-ferryl iron (Fe4+=O) of heme-peroxidases (Compound I)) as well as freely diffusable low molecular weight oxidants such as hypochlorous and hypobromous acids (HOCl and HOBr) – can be responsible for the oxidative modification of carbonaceous nanomaterials, (Table 2) (Sutherland et al., 1993; Kagan et al., 2010; Vlasova et al., 2011). The relative contribution of these two types of oxidants to the overall degradation process may vary dependently on the type of enzyme, conditions (particularly pH), pro-/anti-inflammatory status, etc (Kotchey et al., 2013b; Vlasova et al., 2012). In all cases, however, the presence of catalytic metals is necessary for triggering the degradation process.
Table 1.
Enzymatic degradation of carbonaceous nanomaterials with sp2 hybridization by in vivo relevant oxidative systems.
| Biological system | Oxidative equivalents | Carbonaceous nanomaterials |
References |
|---|---|---|---|
| Enzymes + H2O2 | |||
| Myeloperoxidase (MPO) |
Compound I, Compound II, |
c-SWCNTs PEG-SWCNTs |
Kagan et al., 2010; Vlasova et al., 2012; |
| HOCl, HOBr | PEG-SWCNTs | Bhattacharya et al., 2014 |
|
| Nitrogen-doped CNT cups |
Zhao et al., 2014 | ||
| Graphene oxide | Kurapati et al., 2015 | ||
| Carbon nanohorns | Zhang et al., 2015 | ||
| Eosinophil peroxidase (EPO) |
Compound I, Compound II, |
c-SWCNTs | Andón et al., 2013 |
| HOBr; (low HOCl) | |||
| Lactoperoxidase (LPO) |
Compound I, | c-SWCNTs, PEG-SWCNTs ox-SWCNTs and |
Vlasova et al., 2011, 2012 |
| HOBr, HOSCN | ox-SWCNTs+lung surfactant |
Bhattacharya et al., 2015 | |
| Xanthine oxidase + NO | |||
| (without H2O2) | ONOO | c-SWCNTs | Kagan et al., 2014 |
| Xanthine oxidase | ox-MWCNTs, f-MWCNTs | ||
| Hemoglobin | O2•− + H2O2 → HO• | Sureshbabu et al., 2015 | |
| Tyr-O•, HOO•, HO• | c-SWCNTs, PEG-SWCNTs | ||
| Cytochrome c | Tyr-O•, HO• | Graphene oxide | Vlasova et al., 2011, 2012 |
| Phagolysosomal simulant fluid + H2O2 |
n.d. | ox-SWCNTs, ox- MWCNTs |
the present paper Russier et al., 2010 |
| Murine bronchoalveolar lavage fluid + H2O2 + NaSCN |
LPO, MPO, HOSCN | ox-SWCNTs | Bhattacharya et. al., 2015 |
|
Neutrophil extracellular traps + H2O2 + NaBr |
MPO, HOCl, HOBr | ox-SWCNTs | Farrera et al., 2014 |
|
Human neutrophils + fMLP and cytochalasin B + fMLP and cytochalasin B + serum opsonized zymosan |
MPO, HOCl | c-SWCNTs+IgG PEG-SWCNTs HSA-SWCNTs |
Kagan et al., 2010; |
| Bhattacharya et al., 2014 | |||
| Macrophages: | ROS, ONOO- | Lu et al., 2014 | |
| Human monocyte-derived | c-SWCNTs+IgG | ||
| THP-1 | c-SWCNTs | ||
| THP-1 | MWCNTs-NH2 | Kagan et al., 2010 | |
| RAW 264.7, THP-1 | Carbon nanohorns | Kagan et al., 2014 | |
| Elgrabli et al., 2015 | |||
|
Murine eosnophils + cytochalasin B and PAF |
EPO, HOBr | ox-SWCNTs | Zhang et al., 2015 |
|
Myeloid-derived suppressor cells |
MPO, HOCl | Nitrogen-doped CNT cups |
Andón et al., 2013 |
| Primary microglia | n.d. (ROS, RNS) | ox-MWCNTs, MWCNT- | Zhao et al., 2014 |
| Experimental animals | NH2, | ||
| -Pharyngeal aspiration | ox-MWCNT-NH2 | Bussy et al., 2016 | |
| w/t and MPO k/o mice -Pharyngeal aspiration |
MPO, HOCl | c-SWCNTs | |
| w/t and NADPH-oxidase- deficient mice |
ONOO- | c-SWCNTs | Shvedova et al, 2012 |
| -Stereotactic administration |
Kagan et al, 2014 | ||
| C57BL/6 mice | (ROS) | MWCNT-NH3+ | |
| Nunes et al., 2012 | |||
ROS – reactive oxygen species; RNS – reactive nitrogen species; n.d. – not determined.
Table 2.
Standard reduction potentials for redox couples of oxidants which are capable to oxidize and degrade carbonaceous nanomaterials in vivo (E° > 0.5 V). pH 6.5-7.0, 25°C.
| Redox couple | Reduction potentials (E°, V) |
Reference |
|---|---|---|
| Myeloperoxidase: | ||
| Compound I/compound II | 1.35 | Furtmuller et al., 2003 |
| Compound II/ferric enzyme | 0.97 | Furtmuller et al., 2003 |
| Eosinophyl peroxidase | n.d. | |
| Lactoperoxidase: | ||
| Compound I/compound II | 1.14 | Furtmuller et al., 2005 |
| Compound II/ferric enzyme | 1.04 | Furtmuller et al., 2005 |
| Cytochrome P450 | ||
| Compound I/Compound II | 1.35 | Koppenol, 2007 |
| HO•, H+/H2O | 2.31 | Buettner, 1993 |
| RO•, H+/ROH | 1.60 | Buettner, 1993 |
| HOO•, H+/H2O2 | 1.06 | Buettner, 1993 |
| ROO•, H+/H2O2 | 1.00 | Buettner, 1993 |
| Trp•, H+/Trp | 1.05 | DeFillipis et al., 1989 |
| Tyr-O•, H+/Tyr-OH | 0.94 | DeFillipis et al., 1989 |
| ONOOH, H+ /•NO2, H2O | 1.40 | Koppenol et al., 1992 |
| ONOOH, H+ /NO2−, H2O | 1.20 | Koppenol et al., 1992 |
| HOCl, H+/Cl−, H2O | 1.48 | Vanýsek, 2011 |
| HOBr, H+/Br−, H2O | 1.33 | Vanýsek, 2011 |
| HOSCN, H+/SCN−, H2O | 0.56 | Arnhold et al., 2006 |
E° = 0.5V – reduction potential of the valence band for SWCNTs (Gratzel, 2001; Choi et al., 2002).
Numerous studies emphasized the effects of diversified organic compounds, particularly hydrophobic molecules, with the expression of different isoforms of CYP450 and their activity (reviewed in Zanger and Schwab, 2013; Hrycay and Bandiera, 2015). Moreover, particular carbonaceous materials can also cause robust changes in the functions of CYP450 system (Ji et al., 2009; Fröhlich et al., 2010; Che Abdullah et al, 2014; etc.). This catalytic responsiveness of CYP450 has lead to the development and applications of carbonaceous nanoparticles complexes with these hemoproteins as CNT-conjugated P450-biosensors (Pauwels et al., 2010; Carrara et al., 2011; Baj-Rossi et al., 2012).
Surprisingly, to the best of our knowledge there have been no studies demonstrating the propensity of CYP450 to biodegrade nanoparticles. This may represent an interesting direction of research because highly reactive Compound I is generated during P450 catalyzed metabolism of xenobiotics or direct interaction of these hemoproteins with H2O2 or organic hydroperoxides (peroxidase shunt) (Table 1) (Krest et al., 2013; Shoji and Watanabe, 2014).
In contrast to highly likely and straightforward capacities of CYP450 to be involved in the degradation of carbonaceous nanoparticles, the potential role of catalase is less obvious. In contrast to widely opened and solvent exposed active sites, characteristic of most hemoprotein-based peroxidases, the heme of catalases is deeply buried into the protein structure and connected with the surface via a very long and narrow access channel (Vidossich et al., 2012). Catalase compound I is accessible only for very small molecules to effectively fulfill the function of oxoferryl iron to either oxidize H2O2 or one of the aromatic residues of the protein thus leading to the formation of tyrosyl of tryptophanyl radicals (Kirkman and Gaetani, 2007; Alfonso-Prieto et al., 2012). The peroxidase activity of native catalase is relatively low. It is possible, however, that catalase monomers formed upon dissociation of multi-meric protein and/or their structural re-arrangements on the surface of nano-materials that will lead to the heme exposure and appearance of peroxidase-like activity with a typical catalytic competence to oxidize phenolic compounds (Horozova et al., 1998). Interestingly, this type of peroxidase activity of catalase has been explored in reactions of controllable degradation of C3N4 to obtain biocompartible fluorescent N-C dots (Liu et al., 2014).
Pseudo-peroxidase degradation by adventitious transition metals present in nanomaterials
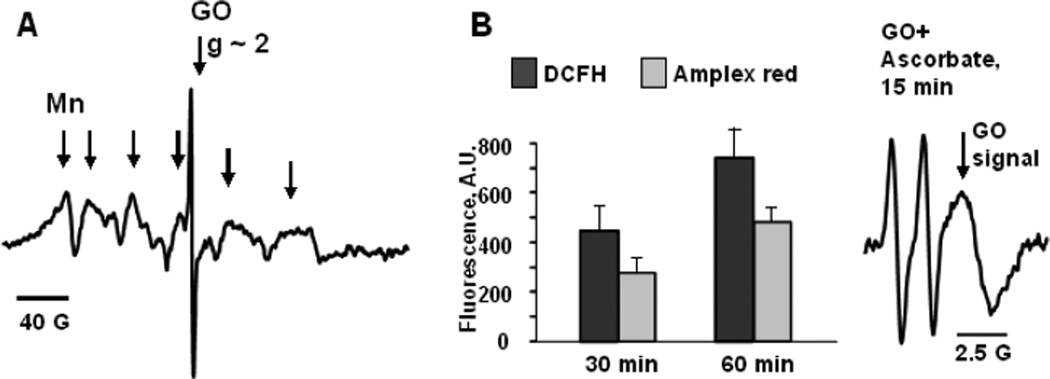
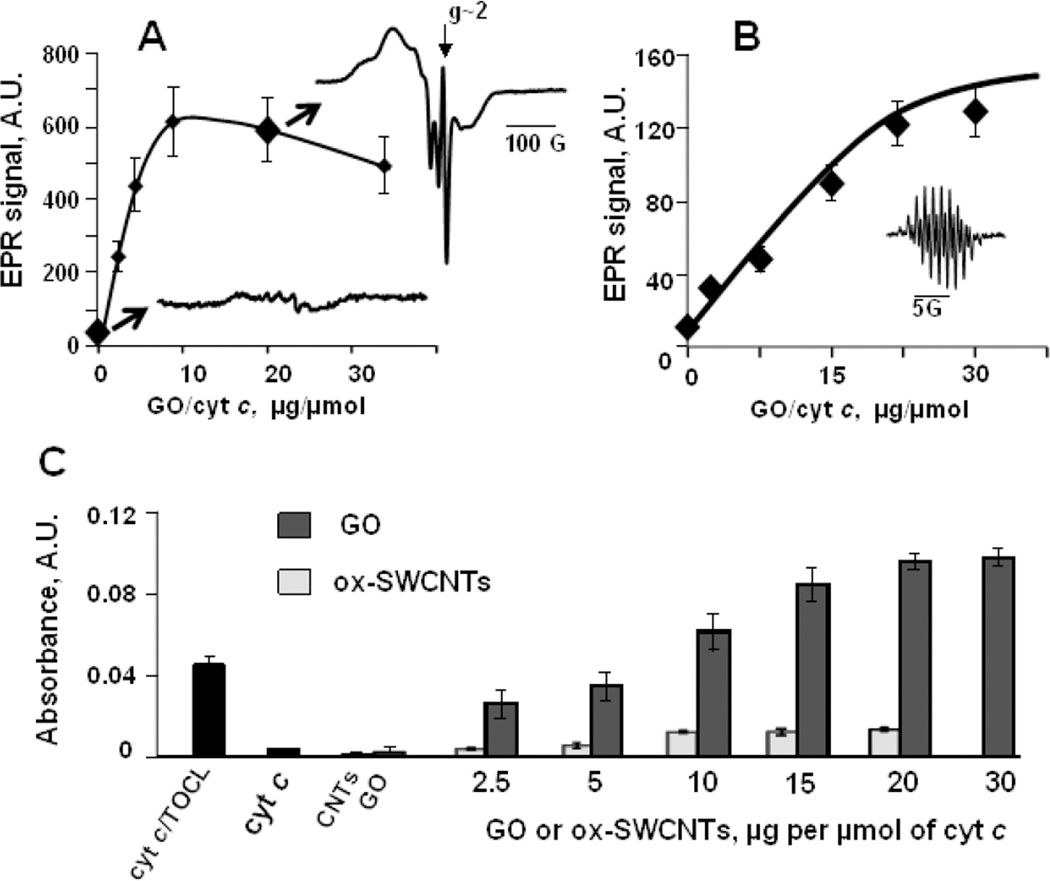
The production of carbonaceous nanomaterials is often associated with the employment of significant amounts of transition metal catalysts, including iron, copper, manganese, etc (Laurent et al., 1998; Panish et al., 2013). The presence of these metals should be inevitably associated with the pseudo-peroxidase function inherent to nano-materials. Moreover, electron donor-acceptor specificity of nano-environments may be conducive to the unusual peroxidase-like activities of metals not traditionally associated with redox catalysis such as gold, silver, etc (Garg et al., 2015; Tao et al., 2013). Indeed, many studies have documented the peroxidase-like activities of a variety of metal-containing nanoparticles inherent to their structure or present as adventitious metals (Gao et al., 2014; Kagan et al., 2006a). Intrinsic catalytic activity of graphene oxide (GO) may be associated with its paramagnetic properties (Song et al., 2010; Su et al., 2012; Garg et al., 2015). As an illustration, we present EPR spectra of GO samples demonstrating the presence of Mn(II) inclusions with paramagnetic propensities and narrow paramagnetic signal of GO structure (Fig. 1A). Notably, the Mn-containing GO samples displayed peroxidase-like activity as revealed by their ability to oxidize typical peroxidase substrates Amplex Red and dichlorofluorescein (DCFH) (Fig. 1B). This type of peroxidase activities associated with integrated or adventitious metals in nanoparticles can act as an important biodegradation factor. It may act as a self-propelled biodegradation mechanism, including one that may be intentionally built-in as a self-biodegradation factor.
Fig. 1. Paramagnetic properties of GO are accountable for its peroxidase-like activity.
A) – EPR spectrum of GO measured at 77K is a composition of a narrow EPR spectrum at g~2.0 which is due to intrinsic paramagnetic properties of GO and hexa-component EPR spectrum of manganese embedded into carbon structure.
B) – GO can oxidize peroxidase substrates characterized by low reduction potentials: Amplex red, dichlorofluorescein (DCFH), and ascorbate (n=3). H2O2 (100 µM) was added to the solutions of substrates (50 µM) and GO (0.1 mg/ml) and formation of fluorescent products was detected at 30 or 60 min. In the absence of GO fluorescence responses from Amplex red or DCFH were lower than 10 A.U. EPR signal of ascorbate (1 mM) could be measured in the absence of H2O2 but not in the absence of GO (0.5 mg/ml).
Awakening of dormant peroxidase activity during interactions of hemoproteins with nano-surfaces
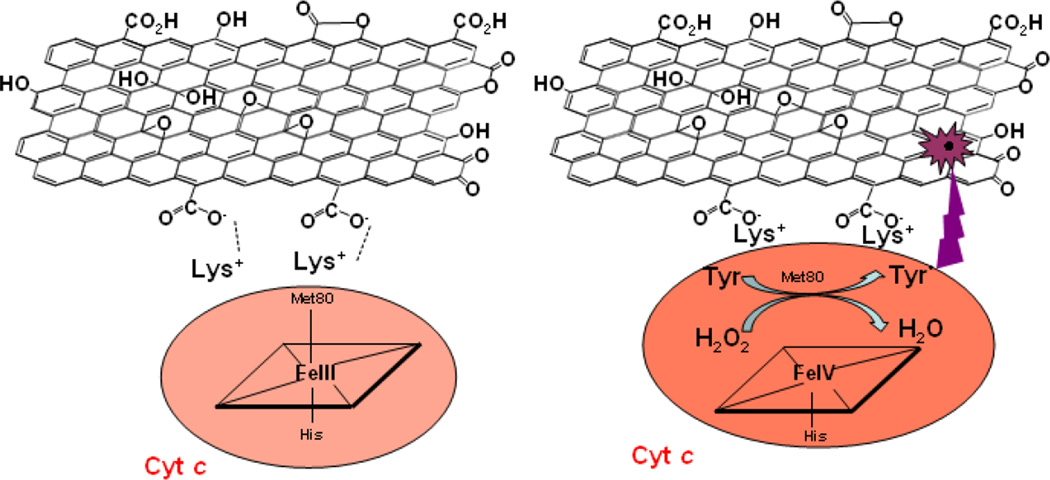
Specific interactions of nanomaterials with hemoproteins that can trigger the conversion of hexa- to penta-coordinated states of the heme iron in “dormant” peroxidases are of particular interest in the context of biodegradation (Kagan et al., 2004; Kapralov 2007). As a typical example, one can consider interactions of cytochrome c (cyt c), the hexa-coordinated hemoprotein in the intermembrane space of mitochondria, with GO. The negative charges on GO’s surface favor binding to basic proteins such as cyt c that has eight positive charges on its surface (Fig. 2) (Koppenol et al., 1982; Yang et al., 2013). Extensive previous work established a very peculiar behavior of cyt c upon its interactions with negatively charged phospholipids, particularly, cardiolipin, a unique doubly-charged phospholipid of mitochondria (Kagan et al. 2005; Kapralov et al., 2011). During this interaction, the protein undergoes structural rearrangements leading to its conversion from the hexa-coordinated to the penta-coordinated electron configuration resulting in “unmasking” of its peroxidase activity (Vlasova et al., 2006). This phenomenon has been extensively studied in mitochondria, cells, and tissues and its role in the execution of apoptotic cell death program and consequences for tissue damage have been well established (Kagan et al., 2006b; Hüttemann et al., 2011). Notably, these specific interactions of hemoproteins with nano-surfaces are meaningful in the context of degradation of nanomaterials. The schema of these interactions and several sets of experimental data illustrate the pathways and significance of GO binding of cyt c resulting in unfolding of the protein, weakening of the heme iron/Met80 sulfur bond and low to high spin transition leading to peroxidase activation (Fig.3). The respective transitions of cyt c can be characterized by spectral changes (Figs. 4A, 4B) (Jiang et al., 2014). Accordingly, cyt c displays significant peroxidase activity upon its binding to the GO surface (Fig. 5) (Yang et al., 2013; Hua et al., 2012). Most importantly, the peroxidase reactive intermediates of cyt c can directly oxidize GO causing its degradation detectable by visible-NIR spectroscopy, TEM and XPS (Fig. 6 and Table 3). Thus, by unfolding and activating cyt c into a peroxidase, GO inflicts self-degradation. It is likely that cyt c/GO interactions represent a prototypical example of a very interesting new type of biodegradation reactions triggered by interactions of different hemoproteins with charged nano-surfaces. This new type of biodegradation reaction may lead to the design and development of new generations of nano-platforms for drug delivery as well as for modulating the physicochemical characteristics of nanomaterials. Indeed, oxidative modifications of GO have been associated with the changes of its conducting-semiconducting characteristics (Kotchey et al., 2011). In this regard, the entire family of globins, particularly a recently discovered cytoglobin (Kawada et al., 2001; Beckerson et al., 2015), can represent a promising instrument for the controlled and targeted modification/degrade action of carbonaceous nanomaterials (Ascenzi et al., 2013).
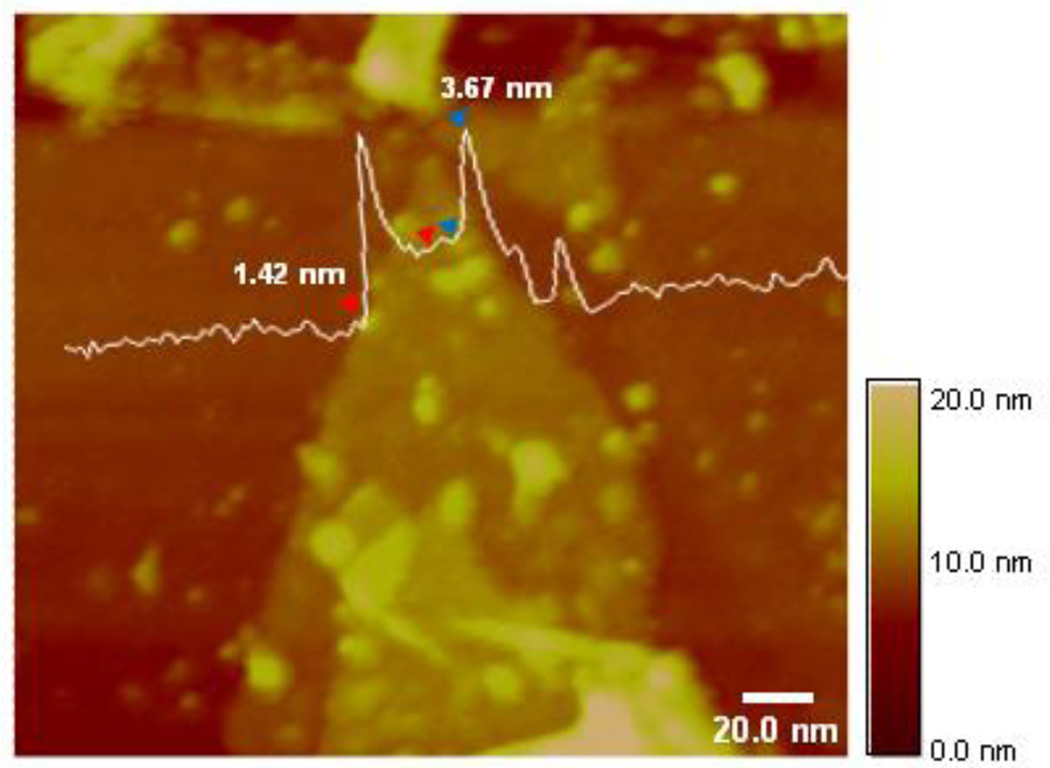
Fig. 2.
AFM evidence for cyt c binding with GO. 5 µM cyt c were added to a suspension of GO (0.2 mg/ml) in 20 mM Na-phosphate buffer, pH 7.4 and tapping mode AFM height analysis was performed. Average GO height is 1.5 nm, and height of cyt c ranges from 3.3 to 3.7 nm.
Fig. 3.
Cyt c binding and peroxidase activation upon interaction with GO. Eight positive charges on the surface of cyt c (Lys+) can electrostatically interact with the negatively charged oxygen-containing functional groups of GO. This interaction of cyt c with GO leads to protein unfolding associated with the “awakening” of dormant peroxidase activity. This peroxidase activation of cyt c can trigger – in the presence of oxidizing equivalents (H2O2, organic hydroperoxides) -oxidative modification/degradation of GO.
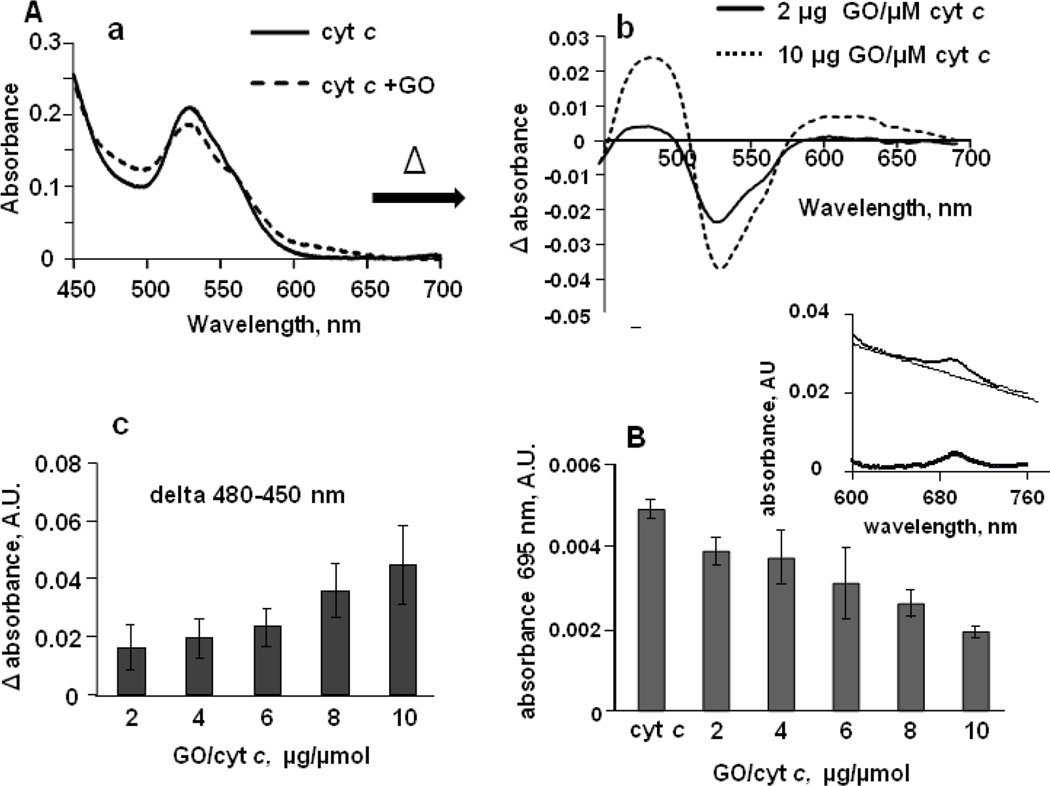
Fig. 4. Absorption spectra of cyt c illustrating GO-induced formation of high-spin form of heme-iron and weakening of the heme iron/Met80 sulfur bond.
A) - Effect of GO on the formation of high-spin iron state of the hemoprotein. a) - Optical absorption spectra of cyt c in the absence (solid line) and in the presence (dashed line) of GO; b) - differential absorption spectrum (obtained by subtracting spectrum of cyt c from spectrum of cyt c incubated with GO) shows positive features at 480–495 and 610–625 nm accompanied by a clear trough at 700 nm indicative of high-spin ferric heme; c) Effect of GO on the height of a peak at 480 nm (75 µM cyt c, n=3).
B) - Dependence of the absorbance of the Fe-S(Met80) bond (λ 695 nm) on GO/cyt c ratio. Characteristic absorbance band at 695 nm is associated with an axial coordination of the heme iron by the sulfur atom of Met80 in cyt c active site. In the presence of GO, loss of absorbance band at 695 nm was observed indicating the disruption or weakening of the Fe-S(Met80) coordination. Insert - optical absorption spectrum of cyt c at region around 695 nm before and after subtracting of baseline. The absorbance was measured in 20 mM HEPES buffer containing 100 µM DTPA (pH 7.4) using 50 µM of cyt c, n=7-9.
Fig. 5. Binding of GO to cyt c causes unfolding of the protein and increases the accessibility of cyt c active site.
A) - GO facilitates cyt c nitrosylation as evidenced by low temperature EPR spectroscopy. Magnitude of the signal of nitrosylated cyt c depends on GO/cyt c ratio revealing the increase of the accessibility of cyt c active site towards nitric oxide upon enzyme binding to GO (Vlasova et al., 2006). Inserts are EPR spectra of cyt c nitrosylated in the absence (blue) and in the presence (red) of GO. EPR spectrum of nitrosylated cyt c/GO complexes includes pronounced narrow GO spectrum at g~2. Reduced cyt c (100 µM, 500 µM ascorbate) was incubated with GO at 37°C for 5 min in 50 mM sodium phosphate buffer, pH 7.4, containing 100 µM DTPA, then 750 µM PAPANONOate was added and incubation was continued for additional 15 min. The reaction was stopped by freezing the samples in liquid nitrogen. EPR spectra of nitrosylated cyt c were measured at 77K.
Binding to GO confers peroxidase activity on cyt c: B) - Magnitude of EPR spectrum of etoposide-phenoxyl radicals (insert) generated in the peroxidase reaction of cyt c/GO complexes in 2 min after addition of 100 µM H2O2 (7.5 µM cyt c) (Kagan et al., 1999); C) - Oxidation of peroxidase substrate ABTS by cyt c/GO. Oxidation of ABTS to yield the product with absorbance maximum at 410 nm was performed at reagent concentrations: 5 µM cyt c, 1 mM ABTS and 100 µM H2O2. The effect of GO on cyt c peroxidase activity is compared to the effects of TOCL (blue bar) and ox-SWCNTs.
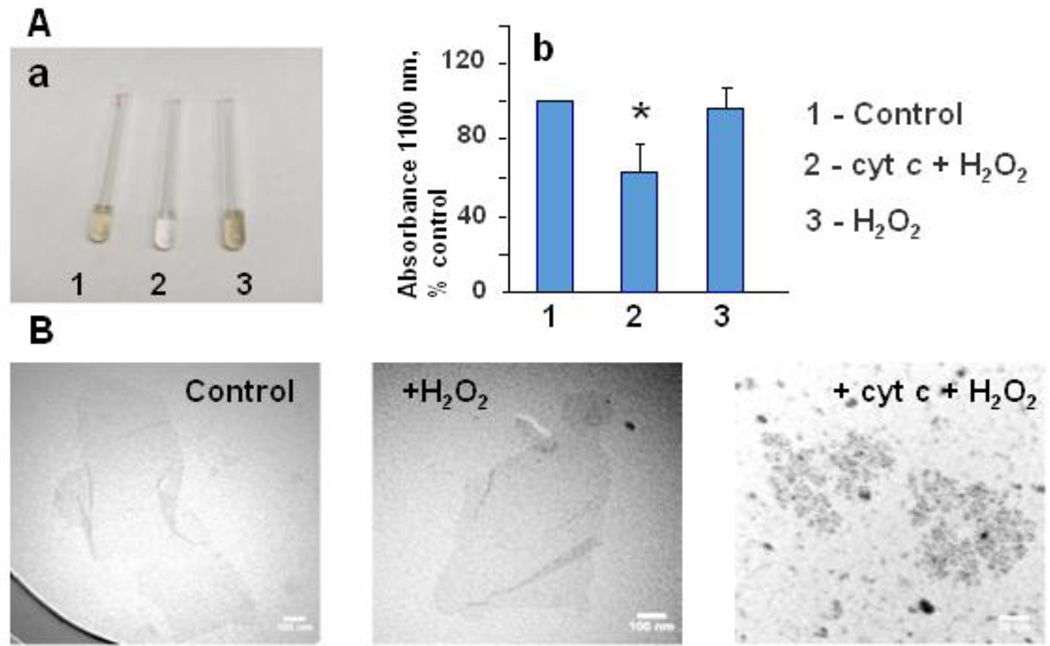
Fig. 6.
Activation of cyt c into peroxidase caused biodegradation of GO. GO (100 µg/ml) was suspended in 50 mM Na-phosphate buffer, containing 100 µM DTPA, pH 7.4. Cyt c (2.5 µM) was added once a day and micro aliquotes of H2O2 (50 µM after each addition) were added 5 times a day during 5 days. a) - Visual evidence of GO degradation as a result of treatment with cyt c and H2O2 (sample 2). Pictures of GO samples were taken in 6 days of incubation; b) -optical absorbance of the sonicated samples at 1100 nm. For each sample, data are averaged for three independent suspensions, * p<0.05 versus control. c) - TEM images of untreated GO (control), GO incubated with H2O2, and GO treated with cyt c and H2O2.
Table 3.
X-Ray Photoelectron Spectroscopy (XPS) characterization of GO samples. Carbon/oxygen content (calculated from survey spectra) and relative concentrations of functional groups (calculated from hi-res C1s spectra) in graphene oxide samples
| Control | + H2O2 | + cyt c + H2O2 |
|
|---|---|---|---|
| C:O | 1.72 | 2.34 | 1.52 |
| C−O (%) | 18.19 | 17.20 | 28.05 |
| C=O (%) | 0.23 | 0 | 0.01 |
| O−C=O (%) | 1.40 | 1.72 | 0.15 |
| C−C (%) | 80.17 | 81.07 | 71.79 |
Alternatives (non-peroxidase) to enzymatic oxidative degradation of nanomaterials
In addition to peroxidase-based mechanisms, other oxidative metabolic reactions may contribute to the biodegradation process. Among the physiologically relevant mechanisms, peroxynitrite generating reactions have been identified as potent mechanisms of nanoparticle biodegradation (Kagan et al., 2014). Oxidative biodegradation of SWCNTs via superoxide/NO• → peroxynitrite-driven pathways of activated macrophages facilitate clearance of nanoparticles from the lung. This particular pathway includes two enzymatic components producing NO• and superoxide radicals, respectively – iNOS and NADPH oxidase. Interestingly, another generator of superoxide radicals, xanthine oxidase, can also contribute to degradation of oxidized multi-walled carbon nanotubes (ox-MWCNTs) (Table 1) (Sureshbabu et al., 2015).
Oxidative degradation of nanoparticles by inflammatory cells
Among many encounters with the gateway cells of the body, interactions of nanoparticles with immune/inflammatory cells are of particular interest for at least two reasons. Inflammatory cells are “armed” with different types of generators of reactive oxygen and nitrogen species (ROS and RNS, respectively) as well as hypochlorous acid. These chemical agents have strong oxidizing redox potentials enabling their reactivity towards the carbonaceous surface of nanoparticles (Sutherland et al., 1993; Davies et al., 2008) (Table 2). Notably, oxygenation (along with nitration, chlorination) of the lipid-protein “corona” of nanoparticles generates clusters of hydrophilic and/or negatively charged functionalities recognizable by inflammatory cell receptors, thus triggering a vicious cycle of interactions leading to a severe inflammatory response (Konduru et al., 2009; Shvedova et al., 2005, 2012b). In professional phagocytes, the major events include engulfment, uptake, possible intracellular metabolism, and digestion (Nauseef, 2007; Dale et al., 2008; Elgrabli et al., 2008). Two major factors – enzymatic machinery generating reactive intermediates and a source of oxidizing equivalents (e.g., H2O2, lipid hydroperoxides) – are required for the effective degradation of carbonaceous nanomaterials. While oxidative enzymes are always constitutively expressed in certain types of immune cells, the levels of their expression may be increased many-fold by pro-inflammatory conditions (Brüne et al, 2013; Mangge et al., 2014), including those triggered by the nanoparticles (Lee et al., 2012; Bussy et al., 2012).
Neutrophils are the first line of responders to pro-inflammatory stimulation and their `response reaction includes the activation of MPO-driven pathways. In the context of biodegradation, two mechanisms – immobilized reactive intermediates of the protein itself and highly diffusible small molecule oxidants such as hypochlorous acid HOCl – have been identified as components of the oxidative process (Nauseef et al., 2007, Arnhold and Flemmig, 2010). The role and contribution of these two factors into the overall degradation may depend on the specific conditions (e.g., the presence of sufficient amounts of Cl− ions) in the microenvironment. In vitro, activation of neutrophils (by fMLP and cytochalasin B or by serum opsonized zymosan) is required to achieve significant levels of oxidative biodegradation (Kagan et al., 2010; Lu et al., 2014). Normally occurring opsonization of particles in the body –stimulating their uptake by phagocytes - can be mimicked by their functionalization with immunoglobulins to facilitate the particles’ uptake by neutrophils. Under these conditions, neutrophils respond by oxidative burst detectable by the generation of superoxide anion radicals which dismutate to yield H2O2. The latter is required to feed the peroxidase reaction of MPO, thus causing oxidative biodegradation of nanoparticles. The efficiency of this pathway has been documented for SWCNTs (including their PEGylated forms) and GO (Kagan et al., 2010; Bhattacharya et al., 2014; Kurapati et al., 2015). Not only active MPO but also sufficiently high activity of NADPH oxidase – generating superoxide radicals - is necessary to maintain the degradation process. The essentiality of this function of NADPH oxidase has been demonstrated both pharmacologically (using its inhibitors) as well as genetically (using NADPH oxidase deficient animals (Shvedova et al., 2008; Kagan et al., 2014)).
Eosinophils another class of immune cells - combat multicellular parasitic organisms engaging a specialized peroxidase, EPO, capable of generating hypobromous acid (HOBr) and low levels of hypochlorous acid at acidic pH (Davies 2008). In murine eosinophils activated by cytochalasin B plus platelet-activating factor (PAF), the oxidizing enzyme, EPO, is released to cause extracellular degradation of SWCNTs (Andón et al., 2013).
Macrophages employ the complex of reactions leading to the production of peroxynitrite, another potent oxidant capable of oxidative degradation of carbonaceous nanomaterials. Two enzymatic systems - NADPH oxidase and NO synthase - produce superoxide radicals and NO•, respectively. Both of these molecules are not reactive enough to oxidatively biodegrade nanoparticles. However, O2−• and NO• can effectively react to yield peroxynitrite, whose oxidizing potency is sufficient to cause biodegradation (Ischiropoulos et al., 1992). In vitro, effective biodegradation capacity has been demonstrated for several types of macrophages such as RAW 264.7, THP-1, and human monocyte-derived macrophages (Kagan et al., 2014; Elgrabli et al., 2008, 2015; Zhang et al., 2015) (Table 1). Because peroxynitrite-dependent oxidation reactions are independent of the direct binding of the reactive protein intermediates with nanoparticles, the degradation process driven by macrophages is independent on the specific positioning of the oxidative machinery on the surface of nanoparticles. As a result, different types of nanomaterials – oxidatively pre-modified as well as pristine - may undergo degradation by macrophages. It is also possible that macrophages “prime” the nano-objects to generate “oxidized” sites where released enzymes of neutrophils and eosinophils can selectively “land” to propagate the process initiated by macrophages.
It is possible that other types of immune cells – specific to particular organs and/or disease conditions – may be involved in biodegradation of carbonaceous nanomaterials. For example, microglial cells in the brain can act similar to macrophages and catalyze the reactions leading to peroxynitrite and ROS formation, hence effectively biodegrading nanoparticles (Goode et al., 2015; Bussy et al., 2016). The evidence for the occurrence and effectiveness of this pathway in the brain is accumulating (Nunes et al., 2012). Another example is myeloid-derived suppressor cells (MDSC) – a heterogenous population of immature cells from the myeloid lineage. As a result of an altered hematopoiesis, amounts of MDSCs can be highly increased in severe disease conditions, particularly chronic infections and cancer (Gabrilovich et al., 2012). These pathological conditions lead to over-expression of NADPH oxidase, iNOS and MPO in MDSC creating a highly pro-oxidant intracellular environment (Youn et al., 2012). These specific features of MDSC may be exploited for the targeted degradation of nanomaterials for optimized delivery of drugs (Zhao et al., 2015). MDSC-derived oxidants can open carbon nano-cups (NCNCs) loaded with antitumor drug paclitaxel and corked with gold nanoparticles (GNPs) and release paclitaxel (Figs. 7, 8).
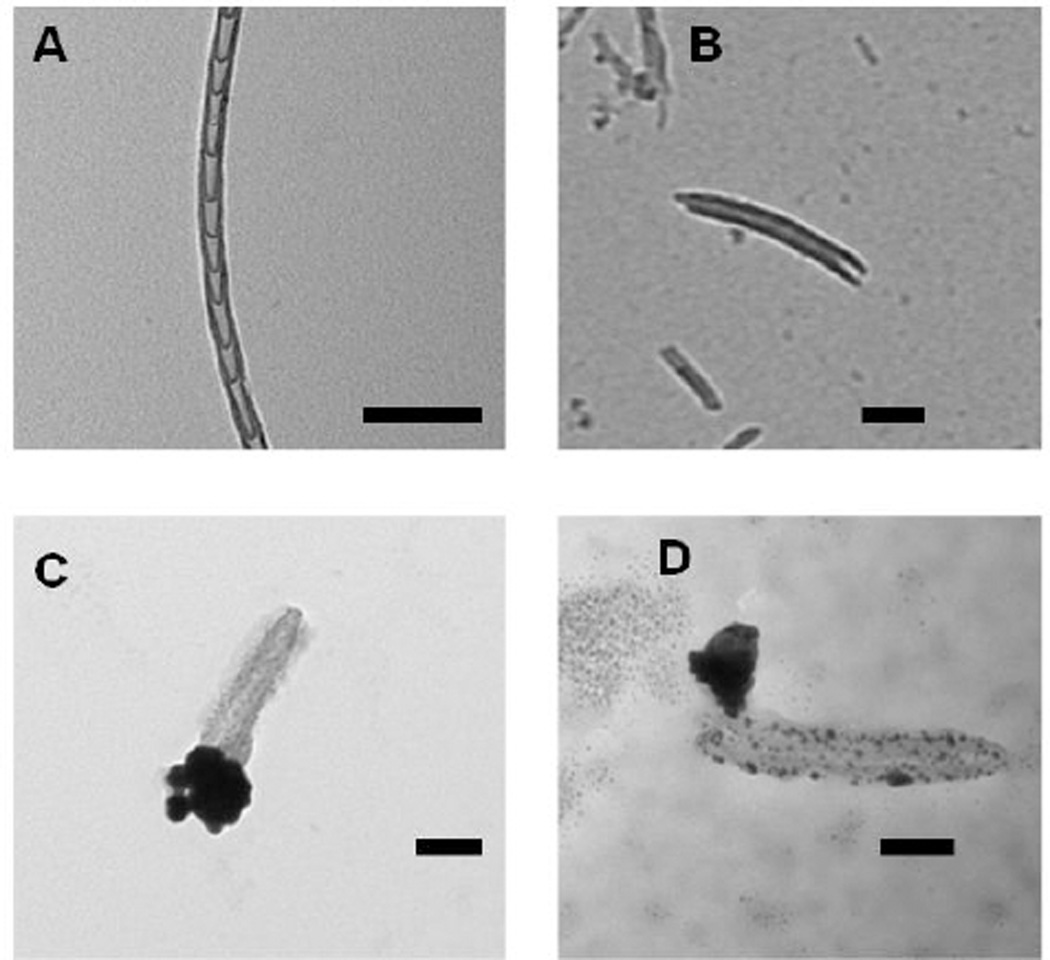
Fig. 7.
Transmission electron microscopy (TEM) images of nitrogen-doped carbon nanotube cups (NCNCs). A) NCNCs synthesized from chemical vapor deposition; B) NCNCs after separation through probe-tip sonication; C) separated NCNCs loaded with paclitaxel and corked with gold nanoparticles (GNPs) through citrate reduction of chloroauric acid; D) GNP corked NCNCs after incubation with myeloid-derived suppressor cells (MDSC), removal of the GNP cork and degradation of nanotube sidewalls is demonstrated in addition to release of loaded cargo. All scale bars are 100 nm.
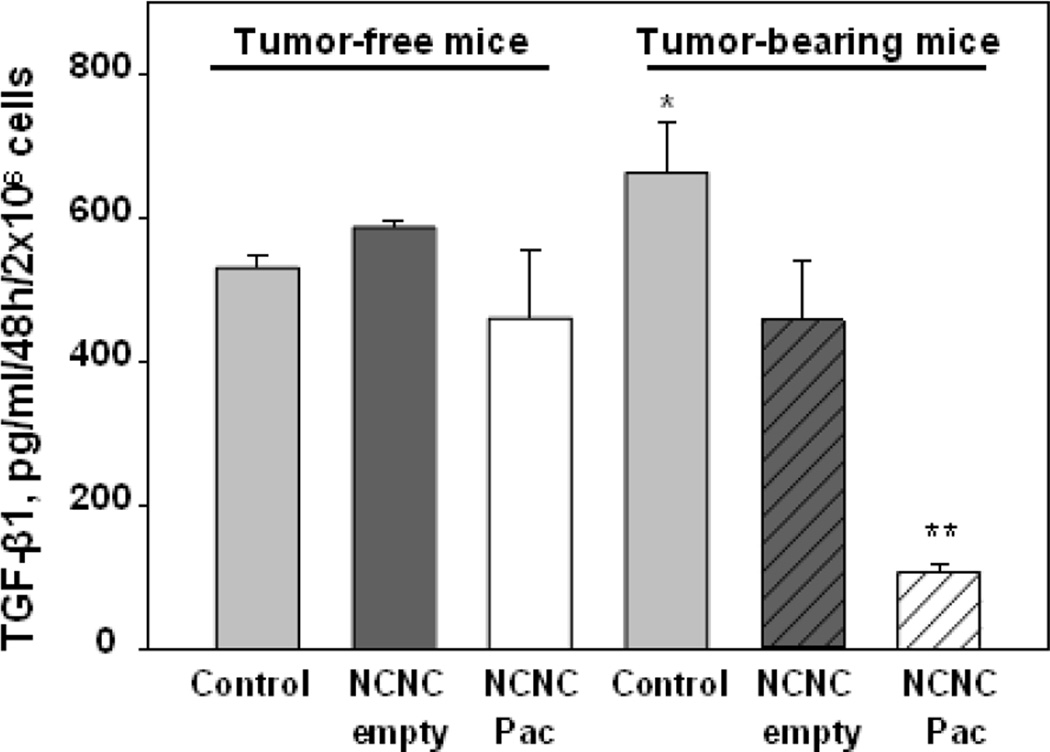
Fig. 8.
Nitrogen-doped carbon nanotube cups (NCNC) loaded with paclitaxel (Pac) block TGF-β production by tumor-associated MDSC. Bone marrow MDSC were sorted from tumor-free and B16-bearing mice (3 weeks), incubated with medium (Control), empty NCNC and NCNC/Pac. TGF-β was measured by ELISA. *, p<0.05 (vs controls in tumor-free mice; **, p<0.05 (vs NCNC Pac).
Biodegradation of nanomaterials by inflammatory cells in vivo : role in pulmonary inflammation and fibrosis
There is a common opinion about biopersistence of nanoparticles in the body. While extended circulation of drug nano-carriers with payloads may be desirable, the ineffective elimination of nanoparticles from the organs after unintentional exposure or as a result of therapeutic attempts seems to represent a serious problem. Poorly degradable nanomaterials can accumulate in organs and inside cells where they can cause detrimental effects (Fig. 9). Even with regards to carbonaceous nanomaterials that are readily susceptible to biodegradation, carbon nanotubes may remain inside macrophages in the spleen and liver for prolonged periods of time following parenteral administration (Yang et al., 2008, Clichici et al., 2014; Albini et al., 2015). Moreover, SWCNTs have been observed in the lungs of exposed mice up to one year after pharyngeal administration (Shvedova et al., 2014). Overall, however, high-aspect, bulky carbonaceous nanomaterials tend to have longer retention times within the tissues and are less effectively cleared than short functionalized particles that are readily taken up and degraded by phagocytes (Mercer et al., 2008; Murphy et al., 2011, Bussy et al., 2012). Notoriously, the appearance of nanoparticles in tissues triggers robust inflammatory responses. Given that immune cells can spend some of their pro-oxidant potential on biodegradation of the nanoparticles, and thus display a weakened immune response, studies of nanoparticle biodegradation in vivo and possible regulation of biodegradation in the context of inflammation in vivo became necessary (Shvedova et al., 2007; Kotchey et al., 2013b; Dumortier et al., 2013). These issues have also stimulated the concepts of creating safe-by-design nanoparticles as well as employment of inflammatory cells for targeted drug delivery (Sureshbabu et al., 2015; Zhao et al., 2015).
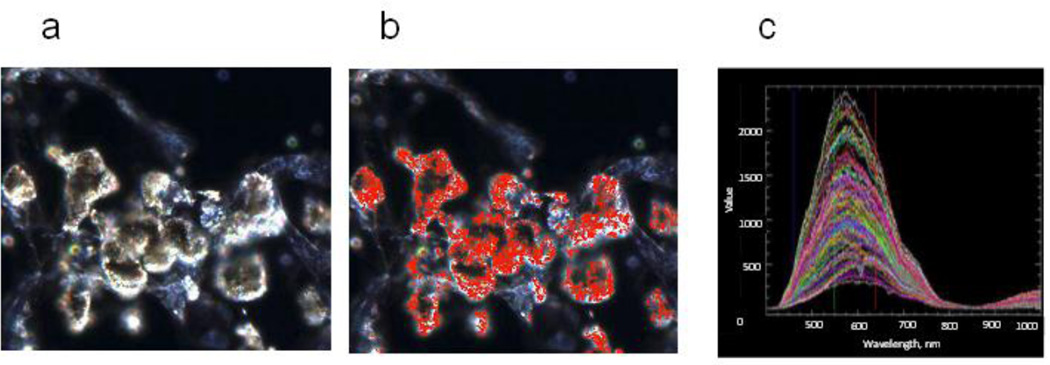
Fig. 9.
Hyperspectral images (CytoViva) of the lung tissue of mice at day 7 after pharyngeal aspiration of SWCNTs. The scanned images a) were collected at 40 × employing an Olympus BX-51 microscope and a 100 W quartz-halogen light source. The pixels that match the SWCNTs spectral profiles were mapped in red b). Spectral data c) was captured with the CytoViva spectrophotometer utilizing an integrated CCD camera.
In line with the ability to take up and biodegrade carbonaceous nanomaterials, professional phagocytes are believed to be mostly accountable for the clearance of the engulfed nanoparticles in vivo. Several studies established the association between the clearance of carbonaceous nanoparticles in the lungs and the amounts of neutrophils and macrophages in the respective tissues (Shvedova et al., 2005, 2012a). These correlational relationships imply that, indeed, biodegradation reactions taking place in inflammatory cells are substantial contributors to the overall elimination of nanoparticles from the tissues. In support of this conclusion, the data on the time course of inflammatory responses and SWCNTs elimination in mice with k/o MPO clearly demonstrated the dependence of these biomarkers on the genetic manipulations with the major biodegrading oxidative enzyme of neutrophils, MPO. Quantitative imaging clearly demonstrated the link between MPO-catalyzed degradation of nanoparticles and one of the hallmarks of the inflammation – pulmonary fibrosis – in wild type (WT) versus MPO k/o animals (Shvedova et al., 2012a). The role of NADPH oxidase as a supplier of superoxide for the subsequent reactions of dismutation (to generate H2O2 as a fuel for MPO) or with NO• (to produce peroxynitrite in macrophages) has been revealed in experiments with genetically manipulated animals. Clearance of SWCNTs was 10-fold less effective in NADPH oxidase-deficient mice (gp91phox(−/−) mice) vs WT animals. Photoacoustic imaging also documented significantly reduced rate of SWCNTs clearance in the lung of NADPH-deficient mice compared to WT control animals (Kagan et al., 2014). There are clear experimental indications that microglial cells - with their highly developed oxidative enzymatic machinery similar to that in macrophages – are primarily responsible for the biodegradative elimination of MWCNTs from the brain (Nunes et al., 2012; Bardi et al., 2013; Bussy et al., 2016).
Employment of enzymatic biodegradation of nano-containers for targeting inflammatory cells in cancer. Peroxidase degradation of payloads vs nano-containers – significance for drug delivery
Design and development of nano-platform based carriers for drug delivery represents one of the active fields for biomedical applications (Battigelli et al., 2013). In this context, effective timely degradation of drug carriers becomes particularly important but must be optimized with regards to nano-carrier vs drug payload degradation (Farokhzad et al., 2006). It has been well documented that oxidative enzymes of inflammatory cells, particularly MPO, can catalytically destroy different types of small organic molecules, including drugs, in circulation (Davies 2008). This wasteful drug metabolism may be exceptionally strong in pro-inflammatory conditions associated with increased amounts and activation of inflammatory cells. This raises the question of possible “protective” role of nano-carriers in preventing unnecessary degradation of payloads and preservation of their therapeutic potential. Notably, experimental assessments of nano-carrier vs drug degradation have not been adequately addressed. A recent study compared degradation of an antitumor drug, doxorubicin (DOX), in free form vs its conjugate with SWCNTs in the presence of MPO or ONOO-generating systems (Seo et al., 2015). The evaluations in simple biochemical enzymatic systems clearly demonstrated that the SWCNTs-associated drug molecules (DOX-SWCNTs) degraded more slowly than free DOX. Notably, cytostatic and cytotoxic effects of free DOX, but not nanotube-carried drug, on melanoma and lung carcinoma cell lines were abolished in the presence of tumor-activated MDSC known to express high levels of MPO, NADPH oxidase and iNOS thus providing enhanced myeloperoxidase- and peroxynitrite-induced conditions for biodegradation of organic molecules. Optimizing the balance between the degradation and resistance of the drug carrier and the payload towards the oxidants generated by inflammatory cells is critical to meet the needs for safety and prolonged circulation while orchestrating the stability and therapeutic effect of the drug. This strategy opens opportunities for exploring new parameters in biodegradation and developing controllable degradation properties by chemical modification of the surface of nanotubes. Further studies are necessary to elucidate important details relevant to the degradation characteristics of different drug nano-delivery systems in the context of achieving their optimized therapeutic potential.
Concluding remarks
The currently accelerating progress in nanotechnologies has already accepted the “safe-by-design” principle as a necessary requisite in the development of new nanomaterials. This principle has to include the “safe-by-biodegradation” component, providing for the optimized life-time and clearance of nanoparticles from the body. In this context, the broad applications of nitrogen-doped carbon nanomaterials offer a very important advantage in creating defects in the structure that can serve as “biodegradation” initiation centers. The introduction of principally new nanomaterials with unexpected features of their interactions with the intracellular mechanisms of genetic, epigenetic, and metabolomics regulation calls for new approaches to assessments of their toxic mechanisms. For example, a recently introduced spherical nucleic acids (Service, 2015) – nano-DNA arrays that can readily enter the cells, “seeks and binds” very low quantities of complementary DNA and RNA from pathogenic bacteria and viruses in the blood. As this affects the expression of selected genes, the resourcefulness in evaluating potential side effects, toxicity, and clearance become particularly important. New generations of nano-platforms are designed and used not only as effective delivery vehicles but also for the engagement of their unique nano-characteristics in manipulations of cell metabolism, cell-cell communications, and pathogen-host interactions. Oxidative modifications of these innovative materials by immune cells may result in the emergence of unforeseen propensities and, consequently, unexpected effects on cell functions. Exploration and utilization of targeted, enzymatically-catalyzed, oxidative modification/biodegradation of these new nanomaterials represent an exciting future area of research.
Highlights.
Nanoparticles can be degraded by oxidative enzymatic machinery of inflammatory cells.
Peroxidase-generated oxidants are the reactive species executing the biodegradation.
Unmasked by GO binding peroxidase activity of cyt biodegrades GO.
Professional phagocytes are accountable for the clearance of nanoparticles in vivo.
Carbonaceous nano-carriers of drugs protect against degradation of payloads.
Acknowledgments
This work was supported by grants from the NIH (PO1 HL114453, HL086884, U19AIO68021, ES020693; NS076511, NS061817, CA165065; NIOSH OH008282), the Human Frontier Science Program (HFSP-RGP0013/2014), and a Core grant to the UPCI Cancer Biomarkers Facility/Luminex Core Laboratory (P30CA047904); NSF CAREER Award No. 0954345, NIH R01ES019304.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that they have no conflict of interest.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
References
- Albini A, Pagani A, Pulze L, Bruno A, Principi E, Congiu T, Gini E, Grimaldi A, Bassani B, De Flora S, de Eguileor M, Noonan DM. Environmental impact of multi-wall carbon nanotubes in a novel model of exposure: systemic distribution, macrophage accumulation, and amyloid deposition. Int. J. Nanomedicine. 2015;10:6133–6145. doi: 10.2147/IJN.S85275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali-Boucetta H, Kostarelos K. Carbon nanotubes in medicine & biology - therapy and diagnostics. Adv. Drug Deliv. Rev. 2013;65:1897–1898. doi: 10.1016/j.addr.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Alfonso-Prieto M, Vidossich P, Rovira C. The reaction mechanisms of heme catalases: an atomistic view by ab initio molecular dynamics. Arch. Biochem. Biophys. 2012;525:121–130. doi: 10.1016/j.abb.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Allen BL, Kichambare PD, Gou P, Vlasova II, Kapralov AA, Konduru NV, Kagan VE, Star A. Biodegradation of single-walled carbon nanotubes through enzymatic catalysis. Nano Lett. 2008;8:3899–3903. doi: 10.1021/nl802315h. [DOI] [PubMed] [Google Scholar]
- Allen BL, Kotchey GP, Chen Y, Yanamala NV, Klein-Seetharaman J, Kagan VE, Star A. Mechanistic investigations of horseradish peroxidase-catalyzed degradation of single-walled carbon nanotubes. J. Am. Chem. Soc. 2009;131:17194–17205. doi: 10.1021/ja9083623. [DOI] [PubMed] [Google Scholar]
- Andón FT, Kapralov AA, Yanamala N, Feng W, Baygan A, Chambers BJ, Hultenby K, Ye F, Toprak MS, Brandner BD, Fornara A, Klein-Seetharaman J, Kotchey GP, Star A, Shvedova AA, Fadeel B, Kagan VE. Biodegradation of single-walled carbon nanotubes by eosinophil peroxidase. Small. 2013;9:2721–2729. doi: 10.1002/smll.201202508. 2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnhold J, Monzani E, Furtmuller PG, Zederbauer M, Casella L, Obinger C. Kinetics and thermodynamics of halide and nitrite oxidation by mammalian heme peroxidases. Eur. J. Inorg. Chem. 2006;2006:3801–3811. [Google Scholar]
- Arnhold J, Flemmig J. Human myeloperoxidase in innate and acquired immunity. Arch. Biochem. Biophys. 2010;500:92–106. doi: 10.1016/j.abb.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Ascenzi P, Marino M, Polticelli F, Coletta M, Gioia M, Marini S, Pesce A, Nardini M, Bolognesi M, Reeder BJ, Wilson MT. Non-covalent and covalent modifications modulate the reactivity of monomeric mammalian globins. Biochim. Biophys. Acta. 2013;1834:1750–1756. doi: 10.1016/j.bbapap.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Baj-Rossi C, De Micheli G, Carrara S. Electrochemical detection of anti-breast-cancer agents in human serum by cytochrome P450-coated carbon nanotubes. Sensors (Basel) 2012;12:6520–6537. doi: 10.3390/s120506520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardi G, Nunes A, Gherardini L, Bates K, Al-Jamal KT, Gaillard C, Prato M, Bianco A, Pizzorusso T, Kostarelos K. Functionalized carbon nanotubes in the brain: cellular internalization and neuroinflammatory responses. PLoS One. 2013;8:e80964. doi: 10.1371/journal.pone.0080964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battigelli A, Ménard-Moyon C, Da Ros T, Prato M, Bianco A. Endowing carbon nanotubes with biological and biomedical properties by chemical modifications. Adv. Drug Deliv. Rev. 2013;65:1899–1920. doi: 10.1016/j.addr.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Beckerson P, Wilson MT, Svistunenko DA, Reeder BJ. Cytoglobin ligand binding regulated by changing haem-co-ordination in response to intramolecular disulfide bond formation and lipid interaction. Biochem. J. 2015;465:127–137. doi: 10.1042/BJ20140827. [DOI] [PubMed] [Google Scholar]
- Bhattacharya K, Sacchetti C, El-Sayed R, Fornara A, Kotchey GP, Gaugler JA, Star A, Bottini M, Fadeel B. Enzymatic ‘stripping’ and degradation of PEGylated carbon nanotubes. Nanoscale. 2014;6:14686–14690. doi: 10.1039/c4nr03604b. [DOI] [PubMed] [Google Scholar]
- Bhattacharya K, El-Sayed R, Andón FT, Mukherjee SP, Gregory J, Li H, Zhao Y, Seo W, Fornara A, Brandner B, Toprak MS, Klaus Leifer K, Star A, Fadeel B. Lactoperoxidase-mediated degradation of single- walled carbon nanotubes in the presence of ulmonary surfactant. Carbon. 2015;91:506–517. [Google Scholar]
- Brüne B, Dehne N, Grossmann N, Jung M, Namgaladze D, Schmid T, von Knethen A, Weigert A. Redox control of inflammation in macrophages. Antioxid. Redox Signal. 2013;19:595–637. doi: 10.1089/ars.2012.4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner GR. The pecking order of free radicals and antioxidants: lipid peroxidation, alpha-tocopherol, and ascorbate. Arch. Biochem. Biophys. 1993;300:535–543. doi: 10.1006/abbi.1993.1074. [DOI] [PubMed] [Google Scholar]
- Burke A, Ding X, Singh R, Kraft RA, Levi-Polyachenko N, Rylander MN, Szot C, Buchanan C, Whitney J, Fisher J, Hatcher HC, D’Agostino R, Jr, Kock ND, Ajayan PM, Carroll DL, Akman S, Torti FM, Torti SV. Long-term survival following a single treatment of kidney tumors with multiwalled carbon nanotubes and near-infrared radiation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12897–12902. doi: 10.1073/pnas.0905195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussy C, Paineau E, Cambedouzou J, Brun N, Mory C, Fayard B, Salomé M, Pinault M, Huard M, Belade E, Armand L, Boczkowski J, Launois P, Lanone S. Critical role of surface chemical modifications induced by length shortening on multi-walled carbon nanotubes-induced toxicity. Part. Fibre Toxicol. 2012;9:46. doi: 10.1186/1743-8977-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussy C, Hadad C, Prato M, Bianco A, Kostarelos K. Intracellular degradation of chemically functionalized carbon nanotubes using a long-term primary microglial culture model. Nanoscale. 2016;8:590–601. doi: 10.1039/c5nr06625e. [DOI] [PubMed] [Google Scholar]
- Cai X, Ramalingam R, Wong HS, Cheng J, Ajuh P, Cheng SH, Lam YW. Characterization of carbon nanotube protein corona by using quantitative proteomics. Nanomedicine: NBM. 2013;9:583–593. doi: 10.1016/j.nano.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Carrara S, Cavallini A, Erokhin V, De Micheli G. Multi-panel drugs detection in human serum for personalized therapy. Biosens. Bioelectron. 2011;26:3914–3919. doi: 10.1016/j.bios.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Che Abdullah CA, Azad CL, Ovalle-Robles R, Fang S, Lima MD, Lepró X, Collins S, Baughman RH, Dalton AB, Plant NJ, Sear RP. Primary liver cells cultured on carbon nanotube substrates for liver tissue engineering and drug discovery applications. ACS Appl. Mater. Interfaces. 2014;6:10373–10380. doi: 10.1021/am5018489. [DOI] [PubMed] [Google Scholar]
- Choi HC, Shim M, Bangsaruntip S, Dai H. Spontaneous reduction of metal ions on the sidewalls of carbon nanotubes. J. Am. Chem. Soc. 2002;124:9058–9059. doi: 10.1021/ja026824t. [DOI] [PubMed] [Google Scholar]
- Chuan L, Jia Z, Yu-Jiao Z, Shu-Fang N, Jun C, Qian W, Shao-Ping N, Ze-Yuan D, Ming-Yong X, Shu W. Biocompatible and biodegradable nanoparticles for enhancement of anti-cancer activities of phytochemicals. Chin. J. Nat. Med. 2015;13:641–652. doi: 10.1016/S1875-5364(15)30061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clichici S, Biris AR, Catoi C, Filip A, Tabaran F. Short-term splenic impact of single-strand DNA functionalized multi-walled carbon nanotubes intraperitoneally injected in rats. J. Appl. Toxicol. 2014;34:332–344. doi: 10.1002/jat.2883. [DOI] [PubMed] [Google Scholar]
- Dale DC, Boxer L, Liles WC. The phagocytes: neutrophils and monocytes. Blood. 2008;112:935–945. doi: 10.1182/blood-2007-12-077917. [DOI] [PubMed] [Google Scholar]
- Davies MJ, Hawkins CL, Pattison DI, Rees MD. Mammalian heme peroxidases: from molecular mechanisms to health implications. Antioxid. Redox Signal. 2008;10:1199–1234. doi: 10.1089/ars.2007.1927. [DOI] [PubMed] [Google Scholar]
- DeFillipis MR, Murthy CP, Faraggi M, Klapper MH. Pulse radiolytic measurement of redox potentials: the tyrosine and tryptophan radicals. Biochemistry. 1989;28:4847–4853. doi: 10.1021/bi00437a049. [DOI] [PubMed] [Google Scholar]
- Docter D, Westmeier D, Markiewicz M, Stolte S, Knauer SK, Stauber RH. The nanoparticle biomolecule corona: lessons learned - challenge accepted? Chem. Soc. Rev. 2015;44:6094–6121. doi: 10.1039/c5cs00217f. [DOI] [PubMed] [Google Scholar]
- Dumortier H. When carbon nanotubes encounter the immune system: desirable and undesirable effects. Adv. Drug Deliv. Rev. 2013;65:2120–2126. doi: 10.1016/j.addr.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Dutta D, Sundaram SK, Teeguarden JG, Riley BJ, Fifield LS, Jacobs JM, Addleman SR, Kaysen GA, Moudgil BM, Weber TJ. Adsorbed proteins influence the biological activity and molecular targeting of nanomaterials. Toxicol. Sci. 2007;100:303–315. doi: 10.1093/toxsci/kfm217. [DOI] [PubMed] [Google Scholar]
- Elgrabli D, Floriani M, Abella-Gallart S, Meunier L, Gamez C, Delalain P, Rogerieux F, Boczkowski J, Lacroix G. Biodistribution and clearance of instilled carbon nanotubes in rat lung. Part. Fibre Toxicol. 2008;5:20. doi: 10.1186/1743-8977-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgrabli D, Dachraoui W, Ménard-Moyon C, Liu XJ, Bégin D, Bégin-Colin S, Bianco A, Gazeau F, Alloyeau D. Carbon nanotube degradation in macrophages: live nanoscale monitoring and understanding of biological pathway. ACS Nano. 2015;9 doi: 10.1021/acsnano.5b03708. 10113-101124. [DOI] [PubMed] [Google Scholar]
- Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo . Proc. Natl. Acad. Sci. U. S. A. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrera C, Bhattacharya K, Lazzaretto B, Andón FT, Hultenby K, Kotchey GP, Star A, Fadeel B. Extracellular entrapment and degradation of single-walled carbon nanotubes. Nanoscale. 2014;6:6974–6983. doi: 10.1039/c3nr06047k. [DOI] [PubMed] [Google Scholar]
- Fröhlich E, Kueznik T, Samberger C, Roblegg E, Wrighton C, Pieber TR. Size-dependent effects of nanoparticles on the activity of cytochrome P450 isoenzymes. Toxicol. Appl. Pharmacol. 2010;242:326–332. doi: 10.1016/j.taap.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Furtmüller PG, Arnhold J, Jantschko W, Pichler H, Obinger C. Redox properties of the couples compound I/compound II and compound II/native enzyme of human myeloperoxidase. Biochem. Biophys. Res. Commun. 2003;301:551–557. doi: 10.1016/s0006-291x(02)03075-9. [DOI] [PubMed] [Google Scholar]
- Furtmüller PG, Arnhold J, Jantschko W, Zederbauer M, Jakopitsch C, Obinger C. Standard reduction potentials of all couples of the peroxidase cycle of lactoperoxidase. J. Inorg. Biochem. 2005;99:1220–1229. doi: 10.1016/j.jinorgbio.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Giglio KM, Nelson JL, Sondermann H, Travis AJ. Ferromagnetic nanoparticles with peroxidase-like activity enhance the cleavage of biological macromolecules for biofilm elimination. Nanoscale. 2014;6:2588–2593. doi: 10.1039/c3nr05422e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg B, Bisht T, Ling Y-C. Graphene-based nanomaterials as efficient peroxidase mimetic catalysts for biosensing applications: an overview. Molecules. 2015;20:14155–14190. doi: 10.3390/molecules200814155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode AE, Gonzalez Carter DA, Motskin M, Pienaar IS, Chen S, Hu S, Ruenraroengsak P, Ryan MP, Shaffer MS, Dexter DT, Porter AE. High resolution and dynamic imaging of biopersistence and bioreactivity of extra and intracellular MWNTs exposed to microglial cells. Biomaterials. 2015;70:57–70. doi: 10.1016/j.biomaterials.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grätzel M. Photoelectrochemical cells. Nature. 2001;414:338–344. doi: 10.1038/35104607. [DOI] [PubMed] [Google Scholar]
- Horozova E, Dimcheva N, Jordanova Z. Peroxidase-like activity of catalase immobilized on carbon materials. Z. Naturforsch. 1998;53c:863–866. [Google Scholar]
- Hrycay EJ, Bandiera CM. Monooxygenase, peroxidase and peroxygenase properties and reaction mechanisms of cytochrome P450 enzymes. Adv. Exp. Med. Biol. 2015;851:1–61. doi: 10.1007/978-3-319-16009-2_1. [DOI] [PubMed] [Google Scholar]
- Hua B-Y, Wang J, Wang K, Li X, Zhu X-J, Xia X-H. Greatly improved catalytic activity and direct electron transfer rate of cytochrome C due to the confinement effect in a layered self-assembly structure. Chem. Commun. 2012;48:2316–2318. doi: 10.1039/c2cc17516a. [DOI] [PubMed] [Google Scholar]
- Hüttemann M, Pecina P, Rainbolt M, Sanderson TH, Kagan VE, Samavati L, Doan JW, Lee I. The multiple functions of cytochrome c and their regulation in life and death decisions of the mammalian cell: From respiration to apoptosis. Mitochondrion. 2011;11:369–381. doi: 10.1016/j.mito.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischiropoulos H, Zhu L, Beckman JS. Peroxynitrite formation from macrophage-derived nitric oxide. Arch. Biochem. Biophys. 1992;298:446–451. doi: 10.1016/0003-9861(92)90433-w. [DOI] [PubMed] [Google Scholar]
- Ji Z, Zhang D, Li L, Shen X, Deng X, Dong L, Wu M, Liu Y. The hepatotoxicity of multi-walled carbon nanotubes in mice. Nanotechnology. 2009;20:445101. doi: 10.1088/0957-4484/20/44/445101. [DOI] [PubMed] [Google Scholar]
- Jiang J, Bakan A, Kapralov AA, Silva KI, Huang Z, Amoscato AA, Peterson J, Garapati VK, Saxena S, Bayir H, Atkinson J, Bahar I, Kagan VE. Designing inhibitors of cytochrome c/cardiolipin peroxidase complexes: mitochondria-targeted imidazole-substituted fatty acids. Free Radic. Biol. Med. 2014;71:221–230. doi: 10.1016/j.freeradbiomed.2014.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan VE, Yalowich JC, Borisenko GG, Tyurina YY, Tyurin VA, Thampatty P, Fabisiak JP. Mechanism-based chemopreventive strategies against etoposide-induced acute myeloid leukemia: free radical/antioxidant approach. Mol. Pharmacol. 1999;56:494–506. doi: 10.1124/mol.56.3.494. [DOI] [PubMed] [Google Scholar]
- Kagan VE, Borisenko GG, Tyurina YY, Tyurin VA, Jiang J, Potapovich AI, Kini V, Amoscato AA, Fujii Y. Oxidative lipidomics of apoptosis: redox catalytic interactions of cytochrome c with cardiolipin and phosphatidylserine. Free Radic. Biol. Med. 2004;37:1963–1985. doi: 10.1016/j.freeradbiomed.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- Kagan VE, Tyurina YY, Tyurin VA, Konduru NV, Potapovich AI, Osipov AN, Kisin ER, Schwegler-Berry D, Mercer R, Castranova V, Shvedova AA. Direct and indirect effects of single walled carbon nanotubes on RAW 264.7 macrophages: role of iron. Toxicol. Lett. 2006a;165:88–100. doi: 10.1016/j.toxlet.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Kagan VE, Tyurina YY, Bayir H, Chu CT, Kapralov AA, Vlasova II, Belikova NA, Tyurin VA, Amoscato A, Epperly M, Greenberger J, Dekosky S, Shvedova AA, Jiang J. The “pro-apoptotic genies” get out of mitochondria: oxidative lipidomics and redox activity of cytochrome c/cardiolipin complexes. Chem. Biol. Interact. 2006b;163:15–28. doi: 10.1016/j.cbi.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Kagan VE, Konduru NV, Feng W, Allen BL, Conroy J, Volkov Y, Vlasova II, Belikova NA, Yanamala N, Kapralov A, Tyurina YY, Shi J, Kisin ER, Murray AR, Franks J, Stolz D, Gou P, Klein-Seetharaman J, Fadeel B, Star A, Shvedova AA. Carbon nanotubes degraded by neutrophil myeloperoxidase induce less pulmonary inflammation. Nat. Nanotechnol. 2010;5:354–359. doi: 10.1038/nnano.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan VE, Kapralov AA, St Croix CM, Watkins SC, Kisin ER, Kotchey GP, Balasubramanian K, Vlasova II, Yu J, Kim K, Seo W, Mallampalli RK, Star A, Shvedova AA. Lung macrophages “digest” carbon nanotubes using a superoxide/peroxynitrite oxidative pathway. ACS Nano. 2014;8:5610–5621. doi: 10.1021/nn406484b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farok hzad OC. Targeted polymeric therapeutic nanoparticles: design, development and clinical translation. Chem. Soc. Rev. 2012;41:2971–3010. doi: 10.1039/c2cs15344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapralov AA, Kurnikov IV, Vlasova II, Belikova NA, Tyurin VA, Basova LV, Zhao Q, Tyurina YY, Jiang J, Bayir H, Vladimirov YA, Kagan VE. The hierarchy of structural transitions induced in cytochrome c by anionic phospholipids determines its peroxidase activation and selective peroxidation during apoptosis in cells. Biochemistry. 2007;46:14232–14244. doi: 10.1021/bi701237b. [DOI] [PubMed] [Google Scholar]
- Kapralov AA, Yanamala N, Tyurina YY, Castro L, Samhan-Arias A, Vladimirov YA, Maeda A, Weitz AA, Peterson J, Mylnikov D, Demicheli V, Tortora V, Klein-Seetharaman J, Radi R, Kagan VE. Topography of tyrosine residues and their involvement in peroxidation of polyunsaturated cardiolipin in cytochrome c/cardiolipin peroxidase complexes. Biochim. Biophys. Acta. 2011;1808:2147–2155. doi: 10.1016/j.bbamem.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapralov AA, Feng WH, Amoscato AA, Yanamala N, Balasubramanian K, Winnica DE, Kisin ER, Kotchey GP, Gou P, Sparvero LJ, Ray P, Mallampalli RK, Klein-Seetharaman J, Fadeel B, Star A, Shvedova AA, Kagan VE. Adsorption of surfactant lipids by single-walled carbon nanotubes in mouse lung upon pharyngeal aspiration. ACS Nano. 2012;6:4147–4156. doi: 10.1021/nn300626q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada N, Kristensen DB, Asahina K, Nakatani K, Minamiyama Y, Seki S, Yoshizato K. Characterization of a stellate cell activation-associated protein (STAP) with peroxidase activity found in rat hepatic stellate cells. J. Biol. Chem. 2001;276:25318–25323. doi: 10.1074/jbc.M102630200. [DOI] [PubMed] [Google Scholar]
- Konduru NV, Tyurina YY, Feng W, Basova LV, Belikova NA, Bayir H, Clark K, Rubin M, Stolz D, Vallhov H, Scheynius A, Witasp E, Fadeel B, Kichambare PD, Star A, Kisin ER, Murray AR, Shvedova AA, Kagan VE. Phosphatidylserine targets single-walled carbon nanotubes to professional phagocytes in vitro and in vivo . PLoS One. 2009;4:e4398. doi: 10.1371/journal.pone.0004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Margoliash E. The asymmetric distribution of charges on the surface of horse cytochrome c . J. Biol. Chem. 1982;257 443-4437. [PubMed] [Google Scholar]
- Koppenol WH, Moreno JJ, Pryor WA, Ischiropoulos H, Beckman JS. Peroxynitrite, a cloaked oxidant formed by nitric oxide and superoxide. Chem. Res. Toxicol. 1992;5:834–842. doi: 10.1021/tx00030a017. [DOI] [PubMed] [Google Scholar]
- Koppenol WH. Oxygen activation by cytochrome P450: a thermodynamic analysis. J. Am. Chem. Soc. 2007;129:9686–9690. doi: 10.1021/ja071546p. [DOI] [PubMed] [Google Scholar]
- Kotchey GP, Allen BL, Vedala H, Yanamala N, Kapralov AA, Tyurina YY, Klein-Seetharaman J, Kagan VE, Star A. The enzymatic oxidation of graphene oxide. ACS Nano. 2011;5:2098–2108. doi: 10.1021/nn103265h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotchey GP, Hasan SA, Kapralov AA, Ha SH, Kim K, Shvedova AA, Kagan VE, Star A. A natural vanishing act: the enzyme-catalyzed degradation of carbon nanomaterials. Acc. Chem. Res. 2012;45:1770–1781. doi: 10.1021/ar300106h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotchey GP, Zhao Y, Kagan VE, Star A. Peroxidase-mediated biodegradation of carbon nanotubes in vitro and in vivo . Adv. Drug Deliv. Rev. 2013a;65:1921–1932. doi: 10.1016/j.addr.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotchey GP, Gaugler JA, Kapralov AA, Kagan VE, Star A. Effect of antioxidants on enzyme-catalysed biodegradation of carbon nanotubes. J. Mater. Chem. B Mater. Biol. Med. 2013b;1:302–309. doi: 10.1039/C2TB00047D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krest CM, Onderko EL, Yosca TH, Calixto JC, Karp RF, Livada J, Rittle J, Green MT. Reactive intermediates in cytochrome p450 catalysis. J. Biol. Chem. 2013;288:17074–17081. doi: 10.1074/jbc.R113.473108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman HN, Gaetani GF. Mammalian catalase: a venerable enzyme with new mysteries. Trends Biochem. Sci. 2007;32:44–50. doi: 10.1016/j.tibs.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Kurapati R, Russier J, Squillaci MA, Treossi E, Ménard-Moyon C, Del Rio-Castillo AE, Vazquez E, Samorì P, Palermo V, Bianco A. Dispersibility-dependent biodegradation of graphene oxide by myeloperoxidase. Small. 2015;11:3985–3994. doi: 10.1002/smll.201500038. [DOI] [PubMed] [Google Scholar]
- Laurent C, Flahaut E, Peigney A, Rousset A. Metal nanoparticles for the catalytic synthesis of carbon nanotubes. New J. Chem. 1998;22:1229–1237. [Google Scholar]
- Liang F, Chen B. A review on biomedical applications of single-walled carbon nanotubes. Curr. Med. Chem. 2010;17:10–24. doi: 10.2174/092986710789957742. [DOI] [PubMed] [Google Scholar]
- Liu J, Rinzler AG, Dai HJ, Hafner JH, Bradley RK, Boul PJ, Lu A, Lverson T, Shelimov K, Huffman CB, Rodriguez-Macias F, Shon YS, Lee TR, Colbert DT, Smalley RE. Fullerene pipers. Science. 1998;280:1253–1255. doi: 10.1126/science.280.5367.1253. [DOI] [PubMed] [Google Scholar]
- Liu Z, Davis C, Cai W, He L, Chen X, Dai H. Circulation and long-term fate of functionalized, biocompatible single-walled carbon nanotubes in mice probed by Raman spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1410–1415. doi: 10.1073/pnas.0707654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Tabakman S, Welsher K, Dai H. Carbon nanotubes in biology and medicine: in vitro and in vivo detection, imaging and drug delivery. Nano Res. 2009;2:85–120. doi: 10.1007/s12274-009-9009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hurt RH, Kane AB. Biodurability of single-walled carbon nanotubes depends on surface functionalization. Carbon. 2010;48:1961–1969. doi: 10.1016/j.carbon.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Liu J, Kong W, Li H, Huang H, Liu Y, Kang Z. One-step catalase controllable degradation of C3N4 for N-doped carbon dot green fabrication and their bioimaging applications. J. Mater. Chem. B. 2014;2:5768–5774. doi: 10.1039/c4tb00772g. [DOI] [PubMed] [Google Scholar]
- Loos C, Syrovets T, Musyanovych A, Mailänder V, Landfester K, Nienhaus GU, Simmet T. Functionalized polystyrene nanoparticles as a platform for studying bio-nano interactions. Beilstein J. Nanotechnol. 2014;5:2403–2412. doi: 10.3762/bjnano.5.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N, Li J, Tian R, Peng YY. Binding of human serum albumin to single-walled carbon nanotubes activated neutrophils to increase production of hypochlorous acid, the oxidant capable of degrading nanotubes. Chem. Res. Toxicol. 2014;27:1070–1077. doi: 10.1021/tx5001317. [DOI] [PubMed] [Google Scholar]
- Ma Z, Bai J, Jiang X. Monitoring of the enzymatic degradation of protein corona and evaluating the accompanying cytotoxicity of nanoparticles. ACS Appl. Mater. Interfaces. 2015;7:17614–17622. doi: 10.1021/acsami.5b05744. [DOI] [PubMed] [Google Scholar]
- McDevitt MR, Chattopadhyay D, Kappel BJ, Jaggi JS, Schiffman SR, Antczak C, Njardarson JT, Brentjens R, Scheinberg DA. Tumor targeting with antibody-functionalized, radiolabeled carbon nanotubes. J. Nucl. Med. 2007. 2007;48:1180–1189. doi: 10.2967/jnumed.106.039131. [DOI] [PubMed] [Google Scholar]
- Mercer RR, Scabilloni J, Wang L, Kisin E, Murray AR, Schwegler-Berry D, Shvedova AA, Castranova V. Alteration of deposition pattern and pulmonary response as a result of improved dispersion of aspirated single-walled carbon nanotubes in a mouse model. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;294:L87–L97. doi: 10.1152/ajplung.00186.2007. [DOI] [PubMed] [Google Scholar]
- Monopoli MP, Aberg C, Salvati A, Dawson KA. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- Murphy FA, Poland CA, Duffin R, Al-Jamal KT, Ali-Boucetta H, Nunes A, Byrne F, Prina-Mello A, Volkov Y, Li S, Mather SJ, Bianco A, Prato M, Macnee W, Wallace WA, Kostarelos K, Donaldson K. Length-dependent retention of carbon nanotubes in the pleural space of mice initiates sustained inflammation and progressive fibrosis on the parietal pleura. Am. J. Pathol. 2011;178:2587–2600. doi: 10.1016/j.ajpath.2011.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauseef WM. How human neutrophils kill and degrade microbes: an integrated view. Immunol. Rev. 2007;219:88–102. doi: 10.1111/j.1600-065X.2007.00550.x. [DOI] [PubMed] [Google Scholar]
- Nunes A, Bussy C, Gherardini L, Meneghetti M, Herrero MA, Bianco A, Prato M, Pizzorusso T, Al-Jamal KT, Kostarelos K. In vivo degradation of functionalized carbon nanotubes after stereotactic administration in the brain cortex. Nanomedicine (Lond) 2012;7:1485–1494. doi: 10.2217/nnm.12.33. [DOI] [PubMed] [Google Scholar]
- Olsen L, Oostenbrink C, Jørgensen FS. Prediction of cytochrome P450 mediated metabolism. Adv. Drug Deliv. Rev. 2015;86:61–71. doi: 10.1016/j.addr.2015.04.020. [DOI] [PubMed] [Google Scholar]
- Orecchioni M, Bedognetti D, Sgarrella F, Marincola FM, Bianco A, Delogu LG. Impact of carbon nanotubes and graphene on immune cells. J. Transl. Med. 2014;12:138. doi: 10.1186/1479-5876-12-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DA, III, Nicholas A, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int. J. Pharmac. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Panich AM, Shames AI, Sergeev NA. Paramagnetic impurities in graphene oxide. Appl. Magn. Reson. 2013;44:107–116. [Google Scholar]
- Pauwels J, Hoogmartens J, Van Schepdael A. Application of carbon nanotubes for in-capillary incubations with cytochrome P450 enzymes. Electrophoresis. 2010;31:3867–3873. doi: 10.1002/elps.201000356. [DOI] [PubMed] [Google Scholar]
- Pérez-Herrero E, Fernandez-Medarde A. Advanced targeted therapies in cancer: Drug nanocarriers, the future of chemotherapy. Eur. J. Pharmac. Biopharmac. 2015;93:52–79. doi: 10.1016/j.ejpb.2015.03.018. [DOI] [PubMed] [Google Scholar]
- Russier J, Ménard-Moyon C, Venturelli E, Gravel E, Marcolongo G, Meneghetti M, Doris E, Bianco A. Oxidative biodegradation of single- and multi-walled carbon nanotubes. Nanoscale. 2011;3:893–896. doi: 10.1039/c0nr00779j. [DOI] [PubMed] [Google Scholar]
- Sacchetti C, Motamedchaboki K, Magrini A, Palmieri G, Mattei M, Bernardini S, Rosato N, Bottini N, Bottini M. Surface polyethylene glycol conformation influences the protein corona of polyethylene glycol-modified single-walled carbon nanotubes: potential implications on biological performance. ACS Nano. 2013;7:1974–1989. doi: 10.1021/nn400409h. [DOI] [PubMed] [Google Scholar]
- Seo W, Kapralov AA, Shurin GV, Shurin MR, Kagan VE, Star A. Payload drug vs. nanocarrier biodegradation by myeloperoxidase- and peroxynitrite-mediated oxidations: pharmacokinetic implications. Nanoscale. 2015;7:8689–8694. doi: 10.1039/c5nr00251f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service RF. MEDICAL NANOTECHNOLOGY. Spherical nucleic acids start rolling. Science. 2015;349:1150–1151. doi: 10.1126/science.349.6253.1150. [DOI] [PubMed] [Google Scholar]
- Shvedova AA, Kisin ER, Mercer R, Murray AR, Johnson VJ, Potapovich AI, Tyurina YY, Gorelik O, Arepalli S, Schwegler-Berry D, Hubbs AF, Antonini J, Evans DE, Ku B-K, Ramsey D, Maynard A, Kagan VE, Castranova V, Baron P. Unusual inflammatory and fibrogenic pulmonary responses to single-walled carbon nanotubes in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005;289:L698–L708. doi: 10.1152/ajplung.00084.2005. [DOI] [PubMed] [Google Scholar]
- Shvedova AA, Kisin ER, Murray AR, Gorelik O, Arepalli S, Castranova V, Young SH, Gao F, Tyurina YY, Oury TD, Kagan VE. Vitamin E deficiency enhances pulmonary inflammatory response and oxidative stress induced by single-walled carbon nanotubes in C57BL/6 mice. Toxicol. Appl. Pharmacol. 2007;221:339–348. doi: 10.1016/j.taap.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvedova AA, Kisin ER, Murray AR, Kommineni C, Castranova V, Fadeel B, Kagan VE. Increased accumulation of neutrophils and decreased fibrosis in the lung of NADPH oxidase-deficient C57BL/6 mice exposed to carbon nanotubes. Toxicol. Appl. Pharmacol. 2008;231:235–240. doi: 10.1016/j.taap.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Shvedova AA, Kapralov AA, Feng WH, Kisin ER, Murray AR, Mercer RR, St Croix CM, Lang MA, Watkins SC, Konduru NV, Allen BL, Conroy J, Kotchey GP, Mohamed BM, Meade AD, Volkov Y, Star A, Fadeel B, Kagan VE. Impaired clearance and enhanced pulmonary inflammatory/fibrotic response to carbon nanotubes in myeloperoxidase-deficient mice. PLoS ONE. 2012a;7:e30923. doi: 10.1371/journal.pone.0030923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvedova AA, Pietroiusti A, Fadeel B, Kagan VE. Mechanisms of carbon nanotube-induced toxicity: focus on oxidative stress. Toxicol. Appl. Pharmacol. 2012b;261:121–133. doi: 10.1016/j.taap.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvedova AA, Yanamala N, Kisin ER, Tkach AV, Murray AR, Hubbs A, Chirila MM, Keohavong P, Sycheva LP, Kagan VE, Castranova V. Long-term effects of carbon containing engineered nanomaterials and asbestos in the lung: one year postexposure comparisons. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014;306:L170–L182. doi: 10.1152/ajplung.00167.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji O, Watanabe Y. Peroxygenase reactions catalyzed by cytochromes P450. J. Biol. Inorg. Chem. 2014;19:529–539. doi: 10.1007/s00775-014-1106-9. [DOI] [PubMed] [Google Scholar]
- Song Y, Qu K, Zhao C, Ren J, Qu X. Graphene oxide: intrinsic peroxidase catalytic activity and its application to glucose detection. Adv. Mater. 2010;22:2206–2210. doi: 10.1002/adma.200903783. [DOI] [PubMed] [Google Scholar]
- Su C, Acik M, Takai K, Lu J, Hao SJ, Zheng Y, Wu P, Bao Q, Enoki T, Chabal YJ, Loh KP. Probing the catalytic activity of porous graphene oxide and the origin of this behaviour. Nat. Commun. 2012;3:1298. doi: 10.1038/ncomms2315. [DOI] [PubMed] [Google Scholar]
- Sureshbabu AR, Kurapati R, Russier J, Ménard-Moyon C, Bartolini I, Meneghetti M, Kostarelos K, Bianco A. Degradation-by-design: Surface modification with functional substrates that enhance the enzymatic degradation of carbon nanotubes. Biomaterials. 2015;72:20–28. doi: 10.1016/j.biomaterials.2015.08.046. [DOI] [PubMed] [Google Scholar]
- Sutherland K, Mahoneym JR, II, Coury AJ, Eatonil JW. Degradation of biomaterials by phagocyte-derived oxidants. J. Clin. Invest. 1993;92:2360–2367. doi: 10.1172/JCI116841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Lin Y, Huang Z, Ren J, Qu X. Incorporating graphene oxide and gold nanoclusters: a synergistic catalyst with surprisingly high peroxidase-like activity over a broad pH range and its application for cancer cell detection. Adv. Mater. 2013;25:2594–2599. doi: 10.1002/adma.201204419. [DOI] [PubMed] [Google Scholar]
- Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, Schlenk F, Fischer D, Kiouptsi K, Reinhardt C, Landfester K, Schild H, Maskos M, Knauer SK, Stauber RH. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013;8:772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- Treuel L, Docter D, Maskos M, Stauber RH. Protein corona - from molecular adsorption to physiological complexity. Beilstein J. Nanotechnol. 2015;6:857–873. doi: 10.3762/bjnano.6.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanýsek P. Electrochemical Series. In: Lide David R, editor. Handbook of Chemistry and Physics. 92nd. 2011. pp. 8–23. [Google Scholar]
- Vidossich P, Alfonso-Prieto M, Rovira C. Catalases versus peroxidases: DFT investigation of H2O2 oxidation in models systems and implications for heme protein engineering. J. Inorg. Biochem. 2012;117:292–297. doi: 10.1016/j.jinorgbio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Vlasova II, Tyurin VA, Kapralov AA, Kurnikov IV, Osipov AN, Potapovich MV, Stoyanovsky DA, Kagan VE. Nitric oxide inhibits peroxidase activity of cytochrome c.cardiolipin complex and blocks cardiolipin oxidation. J. Biol. Chem. 2006;281:14554–14562. doi: 10.1074/jbc.M509507200. [DOI] [PubMed] [Google Scholar]
- Vlasova II, Sokolov AV, Chekanov AV, Kostevich VA, Vasilyev VB. Myeloperoxidase-induced biodegradation of single-walled carbon nanotubes is mediated by hypochlorite. Rus. J. Bioorg. Chem. 2011;37:453–463. doi: 10.1134/s1068162011040157. [DOI] [PubMed] [Google Scholar]
- Vlasova II, Vakhrusheva TV, Sokolov AV, Kostevich VA, Gusev AA, Gusev SA, Melnikova VI, Lobach AS. PEGylated single-walled carbon nanotubes activate neutrophils to increase production of hypochlorous acid, the oxidant capable of degrading nanotubes. Toxicol. Appl. Pharmacol. 2012;264:131–142. doi: 10.1016/j.taap.2012.07.027. [DOI] [PubMed] [Google Scholar]
- Walkey CD, Chan WC. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012;41:2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- Wang F, Yu L, Monopoli MP, Sandin P, Mahon E, Salvati A, Dawson KA. The biomolecular corona is retained during nanoparticle uptake and protects the cells from the damage induced by cationic nanoparticles until degraded in the lysosomes. Nanomedicine: NBM. 2013;9:1159–1168. doi: 10.1016/j.nano.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Xie S, Tao Y, Pan Y, Qu W, Cheng G, Huang L, Chen D, Wang X, Liu Z, Yuan Z. Biodegradable nanoparticles for intracellular delivery of antimicrobial agents. J. Control. Release. 2014;187:101–117. doi: 10.1016/j.jconrel.2014.05.034. [DOI] [PubMed] [Google Scholar]
- Yang ST, Wang X, Jia G, Gu Y, Wang T, Nie H, Ge C, Wang H, Liu Yu. Long-term accumulation and low toxicity of single-walled carbon nanotubes in intravenously exposed mice. Toxicol. Lett. 2008;181:182–189. doi: 10.1016/j.toxlet.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhao C, Ju E, Ren J, Qu X. Contrasting modulation of enzyme activity exhibited by graphene oxide and reduced graphene. Chem. Commun. (Camb.) 2013;49:8611–8613. doi: 10.1039/c3cc44632h. [DOI] [PubMed] [Google Scholar]
- Youn J-I, Collazo M, Shalova IN, Biswas S-K, Gabrilovich DI. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Neu. J. Leukoc. Biol. 2012;91:167–181. doi: 10.1189/jlb.0311177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zou H, Qing Q, Yang Y, Li Q, Liu Z, Guo X, Du Z. Effect of chemical oxidation on the structure of single-walled carbon nanotubes. J. Phys. Chem. B. 2003;107:3712–3718. [Google Scholar]
- Zhang L, Petersen EJ, Habteselassie MY, Mao L, Huang Q. Degradation of multiwall carbon nanotubes by bacteria. Environ. Pollut. 2013;181:335–339. doi: 10.1016/j.envpol.2013.05.058. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Petibone D, Xu Y, Mahmood M, Karmakar A, Casciano D, Ali S, Biris AS. Toxicity and efficacy of carbon nanotubes and graphene: the utility of carbon-based nanoparticles in nanomedicine. Drug Metab. Rev. 2014a;46:232–246. doi: 10.3109/03602532.2014.883406. [DOI] [PubMed] [Google Scholar]
- Zhang C, Chen W, Alvarez PJ. Manganese peroxidase degrades pristine but not surface-oxidized (carboxylated) single-walled carbon nanotubes. Environ. Sci. Technol. 2014b;48:7918–7923. doi: 10.1021/es5011175. [DOI] [PubMed] [Google Scholar]
- Zhang M, Yang M, Bussy C, Iijima S, Kostarelos K, Yudasaka M. Biodegradation of carbon nanohorns in macrophage cells. Nanoscale. 2015;7:2834–2840. doi: 10.1039/c4nr06175f. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Allen BL, Star A. Enzymatic degradation of multiwalled carbon nanotubes. J. Phys. Chem. A. 2011;115:9536–9544. doi: 10.1021/jp112324d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Burkert SC, Tang Y, Sorescu DC, Kapralov AA, Shurin GV, Shurin MR, Kagan VE, Star A. Nano-gold corking and enzymatic uncorking of carbon nanotube cups. J. Am. Chem. Soc. 2015;137:675–684. doi: 10.1021/ja511843w. [DOI] [PMC free article] [PubMed] [Google Scholar]