Abstract
Aims
To demonstrate standardized methods for spiking pathogens into human matrices for evaluation and comparison among diagnostic platforms.
Methods and Results
This study presents detailed methods for spiking bacteria or protozoan parasites into whole blood and virus into plasma. Proper methods must start with a documented, reproducible pathogen source followed by steps that include standardized culture, preparation of cryopreserved aliquots, quantification of the aliquots by molecular methods, production of sufficient numbers of individual specimens and testing of the platform with multiple mock specimens. Results are presented following the described procedures that showed acceptable reproducibility comparing in-house real-time PCR assays to a commercially available multiplex molecular assay.
Conclusions
A step by step procedure has been described that can be followed by assay developers who are targeting low prevalence pathogens.
Significance and Impact of Study
The development of diagnostic platforms for detection of low prevalence pathogens such as biothreat or emerging agents is challenged by the lack of clinical specimens for performance evaluation. This deficit can be overcome using mock clinical specimens made by spiking cultured pathogens into human matrices. To facilitate evaluation and comparison among platforms, standardized methods must be followed in the preparation and application of spiked specimens.
Keywords: Mock specimens, molecular quantification, standardized methods, pathogen spiking, Standardization, Methods, Spiking, Quantification, PCR, Detection
Introduction
Diagnostics are a crucial component of the public health strategy to reduce the burden of infectious diseases. A series of diagnostic devices/platforms are being developed to detect National Institute of Allergy and Infectious Diseases (NIAID) Category A-C priority biodefense pathogens as well as other emerging agents, however, development of diagnostics for these Medical Counter Measure-related pathogens is complicated by the low prevalence of many of these pathogens. As such, there may not be sufficient clinical samples available to conduct clinical sensitivity studies required for U.S. Food and Drug Administration (FDA) clearance. To address this regulatory scientific challenge, the FDA could consider use of mock (spiked) clinical samples to support clinical sensitivity studies for the evaluation of diagnostic devices that detect low prevalence pathogens. It is recommended that investigators consult with the FDA regarding this approach prior to undertaking this type of study. Please refer to the “Highly Multiplexed Microbiological/Medical Countermeasure In Vitro Nucleic Acid Based Diagnostic Devices - Guidance for Industry and Food and Drug Administration Staff” document (http://www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm327293.htm) for additional information regarding the potential use of spiked clinical samples. Development of general methods for generating spiked blood and plasma samples will assist diagnostics developers with performance evaluation studies when clinical specimens are not available. To facilitate evaluation and comparison among platforms, standardized methods must be followed in the preparation and application of spiked specimens. These methods could also be used to generate mock (spiked) samples using other clinical matrices such as nasal wash or cerebrospinal fluid. This study was performed to present optimized methods and test them sufficiently to validate the methods for evaluation of molecular diagnostic devices.
Materials and Methods
In developing these methods, model blood-borne organisms representing two Gram-positive bacteria including a potential bioterror bacterium, a Gram-negative bacterium that is a surrogate for another potential bioterror bacterium, a protozoan parasite, and an emerging virus, were used. This breadth of organism types allowed identification of common as well as unique procedures for each pathogen.
The common work flow began by performing repeated growth curves for each well-characterized source organism (bacterium, parasite and virus) under optimal conditions. Characterization of the source organism can include traditional tests routinely performed to identify a culture isolate and/or by obtaining the genomic sequence of the organism. Performing repeated growth curves was intended to identify the initial inoculum, time in culture and cell density at which the culture was in mid-exponential growth. Each of these parameters was specific to the species and strain in culture.
Bacteria and parasites were harvested at the mid-exponential growth point because the culture would be most uniform under these conditions. The harvested cells were aliquoted into vials and frozen under viable cryopreservation conditions that were appropriate for that species.
For viruses, the viral stock was a culture-supernatant collected from infected susceptible cells. The initial inoculum (multiplicity of infection) was allowed to grow for a pre-determined number of days and supernatant was harvested at post-infection collection time and subjected to low-speed centrifugation to clear cell debris from the supernatant. This entire process must be optimized for each viral agent. The supernatant was aliquoted into a large number of single-use vials and frozen under cryopreservation conditions.
After the frozen stock aliquots were prepared, the next step was to determine the mean molecular concentration of pathogen copy number in the frozen aliquots by performing replicates of quantitative PCR on at least four aliquots. Note: if aliquots are stored for >6 months, an aliquot from this set should be tested to ensure the sample concentration has remained stable and unaffected by storage conditions.
To prepare spiked specimens, the quantity of the aliquot suspension that was necessary to achieve the final target concentration was added into the human matrix such as blood or plasma. Throughout this study, healthy human blood was obtained anonymously under the FDA IRB approved protocol 03–120B. Whole blood was collected at the NIH Department of Transfusion Medicine (http://cc.nih.gov/dtm/our_services.html) aseptically by venipuncture, from de-identified, screened, healthy donors into 50mL polypropylene screw cap tubes and is distributed within several hours to the FDA lab, heparinized and stored at 4°C for no more than 30 days before spiking and extraction. Other mock specimens could be similarly prepared with nasal wash, cerebro-spinal fluid or other relevant human matrix. The spiked specimens were mixed and a diagnostic sample was processed as a patient sample would be. Spiking was done with healthy donor matrix creating enough unique specimens for statistical analysis. A second set of spiked specimens was made using human matrix acquired from patients with fever or other signs and symptoms of disease as recommended by FDA/Center for Devices and Radiological Health (“www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm327294.pdf,” accessed 1/7/2016). The patient’s condition need not be identical to the symptoms of an infection with the pathogen under investigation; rather the general alteration of the blood or plasma seen in a symptomatic patient is the important quality. The primary criterion was body temperature greater than 98.6°F. These specimens were obtained anonymously from BioReclaimationIVT under approval by Schulman Associates IRB, which was further reviewed and approved by the FDA IRB under protocol 14–072B. The patient matrix can pose challenges to the diagnostic device not seen with the healthy donor matrix.
More specific methods will be described below for a collection of model organisms: Bacillus anthracis, Staphylococcus aureus, Yersinia pseudotuberculosis, Leishmania donovani and Dengue virus type 2.
Bacillus anthracis (Anthrax Spore Vaccine strain)
Determination of the Growth Curve
All steps were performed using standard benchtop microbiology aseptic technique (Collins et al. 2004). Bacteria were streaked from a well-characterized stock onto a Luria-Bertani agar plate and incubated at 37°C overnight. To initiate the growth in culture, a single colony from the agar plate was inoculated into 10ml of Superbroth (Quality Biological Inc., Gaithersburg, MD) in a sterile 50ml conical tube and incubated at 37°C overnight with shaking at 220 rpm. The following morning, when the bacteria culture has reached saturated, stationary culture, it is necessary to dilute the culture at least 1/20 (v/v) to restart exponential growth. Approximately 0.5ml of the overnight culture was inoculated into 10ml of Superbroth to achieve an OD600 of 0.1 in a 50ml conical tube. This was incubated at 37°C with shaking and the OD600 measured every hour until the optical density did not increase at the maximal rate (growth approaching stationary phase). Each of the samples was read immediately and was not placed on ice prior to measuring the OD600. Measurement of the growth curve was repeated twice more. We found the region of the growth curve where the slope of a line drawn through the plot of OD vs time in incubation was constant, indicating a uniform growth rate. We determined the time in culture and the optical density that was in the center of the linear portion of the growth curve (exponential phase). Our results for B. anthracis (Anthrax Spore Vaccine was obtained from Colorado Serum Co., Denver, CO) were 2.5 hours to an OD600 of 1.0.
Preparation of Cryopreserved Aliquots
Liquid culture harvested at the chosen mid-exponential growth stage was mixed with sterile 100% glycerol to make the suspension 20% (v/v) glycerol. With frequent mixing to maintain a uniform suspension, 0.5ml per tube was aliquoted into sterile labelled tubes that were tightly capped, making at least 50 aliquots. Tubes were placed immediately in crushed dry ice and stored at −80°C.
Determination of Molecular Pathogen Concentration of Frozen Aliquots
The chosen method of quantification based on real-time PCR achieved high reproducibility and less variability among laboratories compared to culture based methods. The pathogen concentration was calculated by PCR detection of a limiting dilution of the nucleic acid extracted from the frozen aliquot and the concentration was expressed as PCR Detectible Units (PDU) per ml.
Steps included thawing a frozen aliquot, extracting total DNA from 3 separate 100µL samples of the aliquot suspension by a chosen method (DNeasy kit, catalogue #69504, Qiagen, Valencia, CA, following the manufacturer’s protocol was used for this step with all bacteria and eukaryotes in this study). The measurement was repeated with at least 3 other frozen aliquots of B. anthracis from the same batch. A series of 10-fold serial dilutions was made for each extracted DNA sample. Quantitative real-time PCR was performed on all the extracts of all the aliquots in the range of dilutions that span from 100% detectible down to below detectability. Our results for B. anthracis were between 1/104 and 1/107 dilutions of the DNA solution extracted from the frozen aliquots. Our assay was a Taqman style reaction targeting the Protective Antigen Gene on the pXO1 plasmid, which was specific for B. anthracis (Table 1).
Table 1.
Real-time PCR assays and cycling conditions for detection of model blood borne pathogens. All reactions were performed with a Bio-Rad CFX96 Real-Time PCR System. The four DNA genome pathogen reactions had primer concentrations of 200nM in the final reaction volume and the reagent mix was Premix Ex Taq, catalogue #RR039A, Takara Bio, Kusatsu, Shiga, Japan.
| Organism | Forward Primer | Probe | Reverse Primer |
|---|---|---|---|
| B. anthracis | CAGAATCAAGTTCCCAGGGG | TCTCCTGAAAAATGGAGCACGGC | TCGGATAAGCTGCCACAAGG |
| Cycling profile | (1) 95°, 1min; (2)
95°, 15 sec; (3) 60°, 1 min; step 2>step 3, 40
cycles Probe concentration: 488nM* |
||
| S. aureus | TAGGTGGCAAGCGTTATCCG | TTGAGCCGTGGGCTTTCACATCAGAC | TCCAGTTTCCAATGACCCTCC |
| Cycling profile | (1) 95°, 1min; (2)
95°, 15 sec; (3) 60°, 1 min; step 2>step 3, 40
cycles Probe concentration: 400nM* |
||
| Y. pseudotuberculosis | GTAGTTTACTACTTTGCCGG | CCGCATGACCTCGCAAGAGC | GATTGAGCGTATTAAACTCA |
| Cycling profile | (1) 95°, 1min; (2) 95°, 15
sec; (3) 60°, 1 min; step 2>step 3, 40
cycles Probe concentration: 269nM* |
||
| L. donovani | CCTATTTTACACCAACCCCCAGT | RAAARKKVRTRCAGAAAYCCCGT | GGGTAGGGGCGTTCTGCGAAA |
| Cycling profile | (1) 95°, 2.5min; (2)
95°, 10 sec; (3) 56°, 30 sec; step 2>step 3, 40
cycles Probe concentration: 400nM* |
||
| Dengue virus type 2 | CAGGYTATGGCACTGTCACGAT | CTCTCCGAGAACRGGCCTCGACTTCAA | CCATYTGCAGCAACACCATCTC |
| Cycling profile | (1) 60°, 30min; (2)
95°, 10min; (3) 95°, 15 sec; (4) 60°, 1 min;
step 3>step 4, 45 cycles Primer concentration: 500nM, Probe concentration: 250nM*, reaction mix: ABI RNA- to-Ct 1-Step Kit, catalogue #4393463 (Life Technologies Corporation, Grand Island, NY) |
||
The measurement of the concentration of a Taqman probe is complicated by the chemically attached fluorophore and quencher. Therefore the optimal probe concentration in the final mix is chosen by titration.
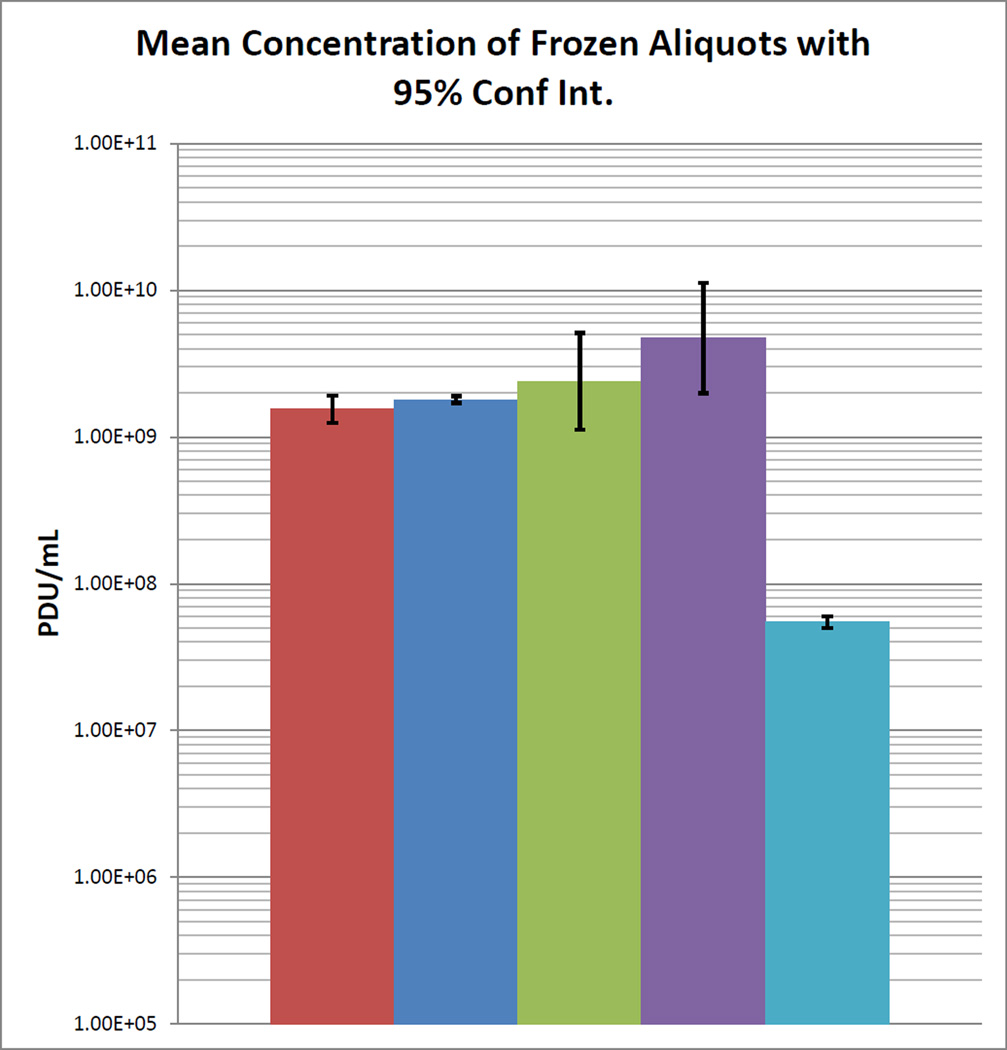
The lowest dilution detectible for each DNA extraction was used to express the PDU ml−1 in the frozen aliquot. For example, if 1/106 was the last dilution detectible, then there are 106 PDU in the undiluted volume of the nucleic acid sample corresponding to the amount of template added to the PCR reaction. If the volume of template was 1µL, then the final concentration calculated for the extracted nucleic acid sample would be 1×106 PDU µl−1, better expressed as 1×109 PDU ml−1. The concentration in the frozen aliquot was calculated by the ratio of eluted volume to extracted volume times the PDU ml−1 of the nucleic acid sample (eluted sample). For example in our procedure, the nucleic acid from 100 µL of aliquot suspension was eluted in 200µL indicating a 2/1 ratio. Thus there are 1×109 PDU ml−1 × 2 = 2.0 × 109 PDU ml−1 in the frozen aliquot. The values for each extraction of a frozen aliquot were used to evaluate the variability among the extractions for each aliquot. The mean PDU ml−1 was calculated for each aliquot. Considering that the means arise from 10-fold dilutions, the mean and standard deviation among the aliquots are best calculated on the log transformed aliquot means. This standard deviation was used to calculate the 95% confidence interval (Sokal and Rohlf 1981) of the log transformed data (Mean +/− 1.96 × SD/N0.5). The antilog of the mean of all the aliquot PDU ml−1 log values and the antilog of the 95% confidence interval were the parameters of the frozen aliquots. The 95% confidence interval of the mean values for the aliquots should be less than twice the mean to insure sufficient reproducibility among aliquots. Once this condition was met, the mean of the PDU ml−1 for the measured aliquots was used as the value for all remaining cryopreserved aliquots that were prepared as the same batch. In our study, the B. anthracis aliquot average (Figure 1) was 1.54 × 109 PDU ml−1 (1.25×109−1.91 ×109, 95% Confidence Interval).
Fig 1.
Bar graph showing the mean concentration of the set of frozen aliquots
for each species was determined in PCR Detectible Units per milliliter. The
95% confidence intervals of the mean are indicated by error
bars. B. anthracis,
B. anthracis,  S. aureus,
S. aureus,  Y. pseudotuberculosis,
Y. pseudotuberculosis,  L. donovani,
L. donovani,  Dengue 2
Dengue 2
Preparation of Spiked Specimens
An initial study was performed to determine the detectible range in the spiked specimens for a given extraction method and assay. An initial estimate was made from the range of concentrations detectible when the pure frozen aliquot was the starting material. Therefore an appropriate quantity of a thawed aliquot was diluted in fresh human blood to a range of concentrations in PDU ml−1 that, based on previous analytical studies, was detectible by the diagnostic test and included some dilutions in the series that should be below the limit-of-detection for the diagnostic test. The process of dilution to the desired concentration involved a number of dilution steps from the highly concentrated frozen stock. Therefore a few steps were made in appropriate buffer such as Phosphate Buffered Saline (PBS). However, a dilution used as a specimen should have pathogen added such that the specimen volume was 99.9% blood. This was accomplished by making 2 ten-fold dilutions into blood before the final ten-fold dilution into the specimen. Three samples of each specimen in this initial dilution panel were then extracted using the Qiagen DNA Blood Mini Kit (catalog #51104, Qiagen, Valencia, CA) or its equivalent and an undiluted sample assayed by PCR. These results indicated the detectible range near the limit of detection of the assay and platform. Further evaluation of the platform and assay was done with multiple specimens in the detectible range prepared from the additional frozen aliquots in the same manner.
To assess whether the diagnostic test had similar performance in ill patients, analysis of a series was performed by spiking blood samples obtained from individuals with symptoms similar to the very early stage of an infection (fever, malaise or flu-like symptoms). The study was performed as described above using the same frozen aliquots of pathogen spiked in the same concentrations tested for healthy donor specimens, the only difference being the source of donor matrix.
Staphylococcus aureus (ATCC # 29213)
Determination of the growth curve
A loop dipped in a vial of bacteria obtained from the American Type Culture collection (ATCC, Manassas, VA, USA) was streaked on an LB plate then the procedure followed was identical to that described above for B. anthracis. In our culture of S. aureus, the mid-exponential growth point was OD600 of 1.0, reached 2 hours after the initial inoculation.
Preparation of cryopreserved aliquots
After harvesting the culture when it reached OD600 of 1.0 in about 2 hours, the procedure described for B. anthracis was followed.
Determination of molecular pathogen concentration of frozen aliquots
We tested the frozen aliquots and calculated the PDU ml−1 proceeding as described above for B. anthracis using an appropriate PCR primer and probe set to detect S. aureus. Our assay was a Taqman style reaction targeting the 16S rRNA gene, which detected all Gram-positive bacteria (Table 1).
In our study, the S. aureus aliquot average (Figure 1) was 1.8 × 109 PDU ml−1 for all 4 tested aliquots, thus no confidence interval was calculated.
Preparation of spiked specimens
Procedures for preparing spiked specimens were the same as described above for B. anthracis.
Yersinia pseudotuberculosis ATCC #29833, (Pfeiffer) Smith and Thal strain, Surrogate for Y. pestis
Determination of the Growth Curve
A loop dipped in a vial of bacteria obtained from the ATCC was streaked on an LB plate then the procedure followed was identical to that described above (37°C incubation) for B. anthracis. In our culture, Y. pseudotuberculosis reached an OD600 of 1.0 in 4 hours.
Preparation of Cyopreserved Aliquots
After harvesting the culture when it reached OD600 of 1.0 in about 5 hours, the procedure described for B. anthracis was followed.
Determination of Molecular Pathogen Concentration of Frozen Aliquots
We tested the frozen aliquots and calculated the PDU ml−1 proceeding as described above for B. anthracis using an appropriate PCR primer and probe set to detect Y. pseudotuberculosis. Our assay was a Taqman style reaction targeting the 16S rRNA gene (Table 1), which detected Y. pseudotuberculosis and Y. pestis (Drygin Iu et al. 1995) but does not react with all other Gram-negative bacteria according to Drygin Iu et al.
In our study, the Y. pseudotuberculosis aliquot average (Figure 1) was 2.39 × 109 PDU ml−1 (1.12 × 109−5.11 × 109, 95% Confidence Interval).
Preparation of Spiked Specimens
Procedures for preparing spiked specimens were the same as described above for B. anthracis.
Leishmania donovani (LD1S2D strain, CBER laboratory culture) (Debrabant et al. 2004)
This strain of the protozoan parasite has been characterized by sequencing multiple gene segments and aligning to database genomic sequence (Duncan et al. 2001; Selvapandiyan et al. 2001; Debrabant et al. 2002; Goyal et al. 2006; Duncan et al. 2009; Gannavaram et al. 2011; Gannavaram et al. 2012; Lakhal-Naouar et al. 2012; Gannavaram et al. 2014). Frozen aliquots of live organisms of this strain are available from the authors or any of the laboratories that are referenced above.
Determination of the Growth Curve
All steps were performed using sterile technique in a biosafety cabinet. A cryopreserved aliquot was thawed and placed in a plastic 25cm2 culture flask with 5ml promastigote medium (Debrabant et al. 2004). Incubation occurred at 26°C for 24 hours. The proportion of viable cells in the culture, indicated by their motility, was assessed by periodically observing the cells in the flask using a microscope. Culture growth continued for several passages until greater than 90% of the parasites appeared motile. A dye exclusion test was performed to assess cell viability. One dye exclusion method (Chan et al. 2012) that worked well with the Leishmania cells required mixing acridine orange (AO, Sigma-Aldrich #A6014, St. Louis, MO, USA) and propidium iodide (PI, Sigma-Aldrich #287075, St. Louis, MO, USA) with an aliquot of cells from the culture and observing them with a fluorescent microscope. All cells will be fluorescent green by taking up the AO. Only the dead cells will take up the PI, appearing fluorescent red. When parasites were greater than 90% viable, we proceeded with documenting the growth curve. Parasites were inoculated into 35ml of culture medium in a 225cm2 flask at the concentration of 1×106 cells/ml. During incubation at 26°C, samples were taken every 6–12 hours to measure the cell density. Measurement was made by automated microscopic image analysis (Vision Cellometer, Nexcelom Bioscience, Lawrence, MA). A hemocytometer and microscope or a Beckman/Coulter particle counter could also be used. We determined the time in culture and the cell concentration that were in the center of the linear portion of the growth curve (exponential phase). Our results for L. donovani were 55 hours and 4×107 cells ml−1.
Preparation of Cryopreserved Aliquots
Liquid culture harvested at the chosen mid-exponential stage (30ml) was centrifuged for 10 minutes at 2500 rpm, the liquid removed and the cells resuspended in 30 ml cryopreservation medium (standard culture medium + Fetal Bovine Serum + Glycerol, mixed in a 6:3:1 ratio and sterile filtered). The suspension was aliquoted into sterile cryotubes, 0.5 ml each, mixing the suspension continually to maintain a uniform concentration. It was stored at room temperature for 30–60 minutes, then stored at −80°C overnight and transferred to a liquid nitrogen freezer for long-term storage.
Determination of Molecular Pathogen Concentration of Frozen Aliquots
The L. donovani concentration was calculated by PCR detection of a limiting dilution of the nucleic acid extracted from the frozen aliquot and the concentration was expressed as PCR Detectible Units (PDU) per ml. Steps included thawing a frozen aliquot and extracting total DNA from 3 separate 100µL samples of the aliquot liquid by a chosen method (the Qiagen DNeasy kit, catalogue #69504, Qiagen, Valencia, CA, following the manufacturer’s protocol). The process was repeated with at least 3 other frozen aliquots of L. donovani from the same batch. Each extracted DNA sample was diluted in 10-fold serial steps. All the extracts of all the aliquots in the range of dilutions that span from 100% detectible down to below detectability were tested by quantitative real-time PCR. Our results for L. donovani were between 1/105 and 1/107 dilutions of the DNA solution extracted from the frozen aliquots. Our assay was a Taqman style reaction targeting the minicircle DNA structure in the kinetoplast (Table 1). The primers bound in conserved regions and the probe was synthesized with degenerate bases to permit amplification of most species of Leishmania (Selvapandiyan et al. 2009).
In our study, the L. donovani aliquot average (Figure 1) was 4.73 × 109 PDU ml−1 (2.74×109−6.51 ×109, 95% Confidence Interval).
Preparation of Spiked Specimens
Procedures for preparing spiked specimens were similar to those described above for B. anthracis.
Dengue virus type 2 (Strain New Guinea C, ATCC # VR-1584)
Growth Conditions
All steps were performed using sterile technique. Dengue virus type 2, strain New Guinea C (NGC), an RNA (+) sense genome virus was used as seed to infect susceptible Aedes albopictus C6/36 cells (# CRL-1660, ATCC). Cells were grown to about 80% confluence in T-150 culture flasks and incubated at 32°C in an atmosphere of 5% CO2, in minimum essential media (MEM, Life Technologies Corp., Grand Island, NY, USA) plus 2% fetal bovine serum (FBS, Technologies Corp.) supplemented with L-glutamine (584 mg/L), Fungizone (2.5 mg/L) and Gentamicin (66 mg/L). The number of C6/36 cells in a flask to be inoculated was estimated by counting the cells in a separate T-150 flask seeded and grown to 80% confluence in parallel. Cells were trypsinized and the cell suspension counted in a Countess Automated Cell Counter (Invitrogen, Grand Island, NY). DENV inoculum was prepared by diluting the viral seed to give a multiplicity of infection (MOI) = 0.01 in MEM, added to the C6/36 cell cultures and incubated for 1 hour with gentle rocking every 15 min. After incubation, 20 ml of MEM + 2% FBS were added to each flask, the cells were returned to the incubator and observed daily for the appearance of cytopathic effect. Supernatants were harvested at peak production of DENV NGC virus. The peak production was defined to be at day 5 post-infection based on multi-step viral growth curves carried out in our laboratory. The cell culture supernatants were clarified to remove cell debris by centrifugation at 1,000 g for 10 minutes at 4°C.
Preparation of Cryopreserved Aliquots
After collecting the supernatant from centrifugation, the virus suspension in culture medium was aliquoted 100µL per tube into sterile labelled tubes that were tightly capped, making at least 50 aliquots. Tubes were placed immediately in crushed dry ice and transferred to a –80°C freezer for long-term storage.
Determination of Molecular Pathogen Concentration of Frozen Aliquots
After a minimum of 24 hours post-freezing, aliquots of the viral suspension were thawed and DENV RNA loads were determined using an appropriate quantitative molecular assay. In our study, the frozen aliquots were extracted with a QIAamp MinElute Virus Spin Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The RNA sample was quantified with our TaqMan assay (Table 1, ABI Taqman RT-PCR Mix, Life Technologies, Carlsbad, CA) by limiting dilution.
The concentration in the aliquot was expressed as PCR Detectible Units (PDU) per ml following the calculations as described for B. anthracis. Our result for the Dengue virus type2 aliquots was 5.5 × 107 PDU ml−1 for each of the four tested aliquots (Figure 1); thus no confidence interval was calculated.
Preparation of Spiked Specimens
Most diagnostic and blood donor screening devices for detection of viruses are currently optimized for using human plasma as the sample type. Therefore these spiked specimens were prepared using plasma samples. Frozen aliquots of the virus stock were diluted into fresh human plasma to at least three different concentrations in PDU ml−1 that included one near the limit of detection for the diagnostic test that resulted in 100% positive detection and two more 10-fold dilutions below that. Three samples of each specimen in this dilution panel were then extracted using the QIAamp MinElute Virus Spin Kit (Qiagen) or equivalent and tested for detectability with the platform and assay being evaluated. Further evaluation of the platform and assay were done with multiple copies of the specimens in this range prepared from the additional frozen aliquots in the same manner.
To assess whether the diagnostic test had similar performance in ill patients, analysis of a dilution series was performed using plasma samples obtained from individuals with early symptoms of infection (fever, malaise or flu-like symptoms).
Results
B. anthracis
In our study analyzing blood samples from healthy human donors spiked with B. anthracis, using the real-time PCR pXO1 assay (Table 1) bacteria were 100% detectible (10/10) at 105 PDU ml−1 (Table 2), 55.6% detectible (5/9, one of the 10 specimens resulted in an invalid amplification curve) at 104 PDU ml−1 and 10% (1/10) at 103. Negative control samples extracted from unspiked blood were below cut off in 80% of the replicates with the same PCR assay (8/10).
Table 2.
Healthy donor blood or plasma spiked specimen testing by the FDA laboratory and by Diatherix Laboratories. The numbers of replicates that met each condition based on Real-Time PCR (FDA) or TEM-PCR (Diatherix) are shown.
| Spiked Pathogen, concentration | Pathogen identified | Diatherix Result | FDA Result | ||
|---|---|---|---|---|---|
| Pos | Neg | Pos | Neg | ||
| B. anthracis, 103 ml−1 | B. anthracis | 1 | 9 | 1 | 9 |
| B. anthracis, 104 ml−1 | B. anthracis | 8 | 2 | 5 | 4 |
| B. anthracis, 105 ml−1 | B. anthracis | 10 | 0 | 10 | 0 |
| S. aureus 104 ml−1 | S. aureus | 3 | 7 | 1 | 9 |
| S. aureus 105 ml−1 | S. aureus | 10 | 0 | 10 | 0 |
| S. aureus 106 ml−1 | S. aureus | 10 | 0 | 10 | 0 |
| Y. pseudotuberculosis, 102 ml−1 | Y. pseudotuberculosis | 5 | 5 | 2 | 8 |
| Y. pseudotuberculosis, 103 ml−1 | Y. pseudotuberculosis | 8 | 2 | 9 | 1 |
| Y. pseudotuberculosis, 104 ml−1 | Y. pseudotuberculosis | 10 | 0 | 10 | 0 |
| Leishmania, 103 ml−1 | Leishmania | 4 | 6 | 6 | 4 |
| Leishmania, 104 ml−1 | Leishmania | 10 | 0 | 10 | 0 |
| Leishmania, 105 ml−1 | Leishmania | 10 | 0 | 10 | 0 |
| Unspiked blood | Negative | 2 | 18 | 4 | 36 |
| Dengue Virus type 2, 10 ml−1 | Dengue Virus | 9 | 1 | 10 | 0 |
| Dengue Virus type 2, 102 ml−1 | Dengue Virus | 10 | 0 | 10 | 0 |
| Dengue Virus type 2, 103 ml−1 | Dengue Virus | 10 | 0 | 10 | 0 |
| Unspiked plasma | Negative | 0 | 10 | 0 | 10 |
The same specimens were sent out with coded labels to a commercial diagnostic biotechnology laboratory, Diatherix Laboratories, Huntsville, AL, for independent testing using Target enriched multiplex PCR (TEM-PCR), a proprietary method (US 7, 851,148 B2) developed by Diatherix Laboratories. This amplification technology in combination with detection on bar-coded magnetic beads (BMB, Applied Biocodes, Santa Fe Springs, CA) could simultaneously detect B. anthracis, Staphylococcus aureus, Yersinia pseudotuberculosis, Leishmania donovani and Dengue virus type 2 tested in this study. The TEM-PCR/BMB assay identified 10/10 (100%) spiked specimens correctly at 105 PDU ml−1 (Table 2), at 104 PDU ml−1, 8/10 (80%) and at 103 1/10 (10%). One of the 20 Negative control samples extracted from unspiked blood had above cutoff reactivity for B. anthracis (95% specificity).
Blood samples were obtained commercially (BioreclamationIVT, Hicksville, NY, USA) from patients with fever and signs and symptoms of flu-like illness. Spiked specimens were made following the same procedure used for healthy blood donors reproducing the same range of concentrations used with healthy donor blood. With B. anthracis spiked at 105 PDU ml−1, 6/7 (85.7%) were detected (Table 3), 2/7 (28.6%) at 104 PDU ml−1 and 0/7 (0%) at 103 PDU ml−1. Negative control specimens made with the patient donor blood resulted in none reactive out of 10 controls (100% specificity).
Table 3.
Symptomatic donor spiked specimen testing by the FDA laboratory and by Diatherix Laboratories. The numbers of replicates that met each condition based on Real-Time PCR (FDA) or TEM-PCR (Diatherix) are shown. FP=false positive
| Spiked Pathogen,
concentration PDU ml−1 |
Pathogen identified | Diatherix Result | FDA Result | |||
|---|---|---|---|---|---|---|
| Pos | Neg | FP | Pos | Neg | ||
| B. anthracis, 103 ml−1 | B. anthracis | 1 | 6 | 0 | 7 | |
| B. anthracis, 104 ml−1 | B. anthracis | 6 | 1 | 2 | 5 | |
| B. anthracis, 105 ml−1 | B. anthracis | 7 | 0 | 1 | 6 | 1 |
| S. aureus 104 ml−1 | S. aureus | 3 | 4 | 4 | 3 | |
| S. aureus 105 ml−1 | S. aureus | 5 | 2 | 7 | 0 | |
| S. aureus 106 ml−1 | S. aureus | 7 | 0 | 1 | 7 | 0 |
| Y. pseudotuberculosis, 102 ml−1 | Y. pseudotuberculosis | 1 | 6 | 0 | 7 | |
| Y. pseudotuberculosis, 103 ml−1 | Y. pseudotuberculosis | 5 | 2 | 6 | 1 | |
| Y. pseudotuberculosis, 104 ml−1 | Y. pseudotuberculosis | 7 | 0 | 7 | 0 | |
| Leishmania, 103 ml−1 | Leishmania | 1 | 6 | 6 | 1 | |
| Leishmania, 104 ml−1 | Leishmania | 4 | 3 | 6 | 1 | |
| Leishmania, 105 ml−1 | Leishmania | 7 | 0 | 7 | 0 | |
| Unspiked blood | Negative | 0 | 20 | 3 | 37 | |
| Dengue Virus type 2, 10 ml−1 | Dengue Virus | 5 | 2 | 0 | 7 | |
| Dengue Virus type 2, 102 ml−1 | Dengue Virus | 7 | 0 | 6 | 1 | |
| Dengue Virus type 2, 103 ml−1 | Dengue Virus | 7 | 0 | 7 | 0 | |
| Unspiked plasma | Negative | 0 | 10 | 0 | 10 | |
The same specimens were sent out with coded labels to Diatherix Laboratories for independent testing. Using their multiplex nucleic acid test that is capable of detecting B. anthracis, spiked specimens at 105 PDU ml−1 were all identified correctly 7/7 (100%) though one specimen spiked with B. anthracis showed above cutoff reactivity with both B. anthracis and Y. pseudotuberculosis (Table 3). This type of result that only occurs with multiplex assays is both true positive for B. anthracis and false positive for Y. pseudotuberculosis. At 104 PDU ml−1, 6/7 (85.7%) were correctly identified and at 103 PDU ml−1, 1/7 (14.3%). Negative control samples extracted from unspiked blood were all correctly identified 20/20 (100%).
S. aureus
In our study analyzing blood samples from healthy human donors spiked with S. aureus, using our real-time PCR assay targeting the 16S rRNA gene (Table 1) the pathogen was 100% detectible (10/10) at 106 PDU ml−1 (Table 2), 100% detectible (10/10) at 105 PDU ml−1 and 10% detectible (1/10) at 104 PDU ml−1. Negative control samples extracted from unspiked blood were 10/10 (100%) below cut off with the same PCR assay.
The same specimens were sent out with coded labels to Diatherix Laboratories, for independent testing. Using their multiplex nucleic acid test that is capable of detecting S. aureus, at 106 PDU ml−1 10/10 (100%) were identified correctly (Table 2), at 105 PDU ml−1, 10/10 (100%) were identified correctly and at 104 PDU ml−1, 3/10 (30%), showing good correspondence with the in house results. Negative control samples extracted from unspiked blood were correctly identified 20/20 (100%).
Blood samples were obtained commercially through BioreclamationIVT from patients with fever and signs and symptoms of flu-like illness. Spiked specimens were made following the same procedure used for healthy blood donors reproducing the same range of concentrations. With S. aureus spiked at 106 PDU ml−1, 7/7 (100%) were detected (Table 3), 7/7 (100%) at 105 PDU ml−1 and 4/7 (57.1%) at 104 PDU ml−1. Negative control specimens made with the patient donor blood resulted in 2 reactive out of 10 controls (80% specificity).
The same specimens were sent out with coded labels to Diatherix Laboratories for independent testing. Using their multiplex nucleic acid test that is capable of detecting S. aureus, spiked specimens at 106 PDU ml−1 were all identified correctly 7/7 (100%) though one specimen spiked with S. aureus showed above cutoff reactivity with both S. aureus and Y. pseudotuberculosis (Table 3). This type of result that only occurs with multiplex assays is both true positive for S. aureus and false positive for Y. pseudotuberculosis. At 105 PDU ml−1, 5/7 (71.4%) were correctly identified and at 104 PDU ml−1, 3/7 (42.9%). Negative control samples extracted from unspiked blood were correctly identified 20/20 (100%).
Y. pseudotuberculosis
In our study analyzing blood samples from healthy human donors spiked with Y. pseudotuberculosis, using our real-time PCR assay targeting a 16S rRNA sequence unique to Yersinia species (Table 1) the pathogen was 100% detectible (10/10) at 104 PDU ml−1, 90% detectible (9/10) at 103 PDU ml−1 and 20% detectible (2/10) at 102 PDU ml−1 (Table 2). Among negative control samples extracted from unspiked blood, 2 were above cut off with the same PCR assay (2/10, 20%).
The same specimens were sent out with coded labels to Diatherix Laboratories for independent testing. Using their multiplex nucleic acid test that is capable of detecting Y. pseudotuberculosis, at 104 PDU ml−1, 10/10 (100%) were identified correctly and at 103 PDU ml−1, 8/10 (80%) were identified correctly and at 102 PDU ml−1, 5/10 (50%), showing good correspondence with the in-house results (Table 2). Negative control samples extracted from unspiked blood were correctly identified 19/20 (95%).
Blood samples from patients with fever and signs and symptoms of flu-like illness were spiked following the same procedure used for healthy blood donors reproducing the same range of concentrations. With Y. pseudotuberculosis spiked at 104 PDU ml−1, 7/7 (100%) were detected (Table 3), 6/7 (85.7%) at 103 PDU ml−1 and 0/7 (0%) at 102 PDU ml−1. Negative control specimens made with the patient donor blood resulted in none reactive out of 10 controls (100% specificity).
The same specimens made with blood from patients with fever and signs and symptoms of flu-like illness were sent out with coded labels to Diatherix Laboratories for independent testing. Using their multiplex nucleic acid test that is capable of detecting Y. pseudotuberculosis, spiked specimens at 104 PDU ml−1 (Table 3) were all identified correctly 7/7 (100%). At 103 PDU ml−1, 5/7 (71.4%) were correctly identified and at 102 PDU ml−1, 1/7 (14.3%). Negative control samples extracted from unspiked blood were correctly identified 20/20 (100%).
L. donovani
In our study analyzing blood samples from healthy human donors spiked with L. donovani, using our real-time PCR assay targeting the minicircle (Table 1) the pathogen was 100% detectible (10/10) at 105 PDU ml−1 (Table 2), 100% detectible (10/10) at 104 PDU ml−1 and 60% detectible (6/10) at 103 PDU ml−1. Negative control samples extracted from unspiked blood were all below cut off with the same PCR assay (10/10).
The same specimens were sent out with coded labels to Diatherix Laboratories for independent testing. Using their multiplex nucleic acid test that is capable of detecting L. donovani, 10/10 (100%) were identified correctly at 105 PDU ml−1, at 104 PDU ml−1, 10/10 (100%) were identified correctly and at 103 PDU ml−1, 4/10 (40%), showing good correspondence with our results (Table 2). Negative control samples extracted from unspiked blood were identified as negative for L. donovani, 20/20 (100%).
Blood samples from patients with fever and signs and symptoms of flu-like illness were spiked following the same procedure used for healthy blood donors reproducing the same range of concentrations. With L. donovani spiked at 105 PDU ml−1, 7/7 (100%) were detected (Table 3), 6/7 (85.7%) at 104 PDU ml−1 and 6/7 (85.7%) at 103 PDU ml−1. Negative control specimens made with the patient donor blood resulted in one reactive out of 10 controls (90% specificity). The same specimens were sent out with coded labels to Diatherix Laboratories for independent testing. Using their multiplex nucleic acid test that is capable of detecting L. donovani, spiked specimens at 105 PDU ml−1 (Table 3) were all identified correctly 7/7 (100%). At 104 PDU ml−1, 4/7 (57.1%) were correctly identified and at 103 PDU ml−1, 1/7 (14.3%). Negative control samples extracted from unspiked blood were correctly identified 20/20 (100%).
Dengue Virus
In our study, the Dengue virus type 2 frozen stock diluted into fresh human plasma to 103 PDU ml−1 was 100% detected (10/10) with our real-time PCR assay targeting the envelope region (Table 1) at an average cycle quantification (Cq) value of 28.6 (Table 2), 102 PDU ml−1 was 100% detected (10/10) at an average Cq value of 32.1 and 10 PDU ml−1 was 100% detected (10/10) at an average Cq value of 36.4. Though we failed to prepare specimens with virus concentrations below the limit of detection in this assay, the log/linear increase in the Cq value shows the device’s response to decreasing pathogen and the detection at another 10-fold dilution, 1 PDU ml−1, would not likely be 100%. Negative control samples extracted from unspiked plasma were all below cut off (average Ct value 44.0) with the same PCR assay (19/19).
The same specimens were sent with coded labels to Diatherix Laboratories for independent testing. Using their multiplex nucleic acid test that is capable of detecting Dengue virus type 2, 100% (Table 2) were detected (10/10) at 103 PDU ml−1 at an average mean fluorescence index (MFI) of 24,136. At 102 PDU ml−1, 10/10 (100%) were identified correctly with an average MFI of 22,864. At 10 PDU ml−1, 9/10 (80%) were detected with an average MFI of 14,147. As with the in-house RT-PCR, this range of virus concentration did not reach far enough below the limit of detection of this assay; however a declining response showed that one more dilution probably would. Negative control samples extracted from unspiked blood were correctly identified 10/10 (100%).
Plasma samples from patients with fever and signs and symptoms of flu-like illness were spiked following the same procedure used for healthy donors reproducing the same range of concentrations. With Dengue virus type 2 spiked at 103 PDU ml−1, 7/7 (100%) were detected (Table 3), 6/7 (85.7%) at 102 PDU ml−1 and 0/7 (0%) at 10 PDU ml−1. Negative control specimens made with the patient donor plasma resulted in none reactive out of 10 controls (100% specificity).
The same specimens were sent out with coded labels to Diatherix Laboratories for independent testing. Using their multiplex nucleic acid test that is capable of detecting Dengue virus type 2, spiked specimens at 103 PDU ml−1 (Table 3) were all identified correctly 7/7 (100%). At 102 PDU ml−1, 7/7 (100%) were correctly identified and at 10 PDU ml−1, 5/7 (71.4%). Negative control samples extracted from unspiked blood were correctly identified 10/10 (100%).
Discussion
A step by step procedure has been described that can be followed by assay developers who are targeting low prevalence pathogens. These types of assays and platforms are essential for detection of biothreat and emerging agents; yet clinical specimens to test their performance are difficult or impossible to obtain. Mock clinical specimens made by spiking cultured pathogens into the human matrix are a solution to that difficulty (FDA/CDRH 2014). If each developer is free to choose the most convenient procedure for making spiked specimens that may differ substantially from other developers, evaluation of the relative performance will be difficult. Therefore, the intention of this study was to describe standardized methods and demonstrate how the spiked specimens could be used in evaluating an in-house real-time PCR assay and a commercial, multiplex assay.
The correspondence of our results with spiked specimens prepared by these methods and the results obtained using a commercially available multiplex assay to test the same specimens at another location demonstrates the reproducibility, practicality and utility of this approach to generate mock clinical specimens. Thus following these methods, specimens can be prepared that could be used to assess the sensitivity, specificity and reproducibility of a diagnostic device to detect low prevalence viral, bacterial or protozoan pathogens in plasma, whole blood, or other human matrices. The mock specimens were made in blood or plasma from healthy donors. Mock specimens were also made from these matrices obtained from patients with fevers and other flu-like symptoms. The importance of evaluating the performance with sick donor as well as healthy donor specimens is illustrated by the lower sensitivity and specificity of these two assay platforms that occurred with the sick donor specimens. Documenting the performance with the sick donor specimens comes closer to reproducing the true clinical specimens that the assay may be required to test.
Though the use of standardized methods to make and use mock specimens will substantially improve the evaluation and comparison of new devices intended to detect biothreat and emerging low prevalence pathogens, the performance measurements will not be absolute. Absolute comparison would require the establishment of internationally recognized reference standard material as the World Health Organization has done for common pathogens such as HIV (Morris et al. 2011) or Trypanosoma cruzi (Otani et al. 2011). As the cited documents will testify, establishment, storage and distribution of a reference standard is a time-consuming and costly venture only achieved when there is a broad consensus supporting the undertaking. At this time, the establishment of standardized spiking methods is an important first step.
In conclusion, device developers are encouraged to systematically follow the methods described here to evaluate the performance of new assays and platforms. Tabulated results from testing multiple, independently spiked specimens are intended for description of the characteristics of the new device.
Acknowledgments
We wish to acknowledge that this work was supported by NIAID through an Interagency Agreement AAI13005-001 awarded to CBER/FDA. Members of the NIAID/ Office of Biodefense, Research Resources, and Translational Research, Division of Microbiology and Infectious Diseases aided in the editing of the text. Yukiko Kozaki LEP/DETTD/CBER graciously assisted with statistical analysis. Healthy human blood and plasma was obtained thanks to the NIH Clinical Center, Department of Transfusion Medicine.
Footnotes
Conflict of interest
No conflict of interest is declared for any of the authors
References
- Chan LL, Wilkinson AR, Paradis BD, Lai N. Rapid Image-based Cytometry for Comparison of Fluorescent Viability Staining Methods. J Fluoresc. 2012;22:1301–1311. doi: 10.1007/s10895-012-1072-y. [DOI] [PubMed] [Google Scholar]
- Collins CH, Lyne PM, Grange JM, Falkinham JO., III . Microbiological Methods. London: Arnold, A member of the Hodder Headline Group; 2004. [Google Scholar]
- Debrabant A, Joshi MB, Pimenta PFP, Dwyer DM. Generation of Leishmania donovani axenic amastigotes: their growth and biological characteristics. Intl J Parasitology. 2004;34:205–217. doi: 10.1016/j.ijpara.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Debrabant A, Lee N, Pogue GP, Dwyer DM, Nakhasi HL. Expression of calreticulin P-domain results in impairment of secretory pathway in Leishmania donovani and reduced parasite survival in macrophages. International journal for parasitology. 2002;32:1423–1434. doi: 10.1016/s0020-7519(02)00134-0. [DOI] [PubMed] [Google Scholar]
- Drygin Iu F, Sakhuriia IB, Vorob’ev II, Dikhanov GG, Podladchikova ON. [Isolation of Yersinia pestis and Yersinia pseudotuberculosis by amplifying 16S rRNA and its genes] Mol Biol (Mosk) 1995;29:1326–1335. [PubMed] [Google Scholar]
- Duncan R, Alvarez R, Jaffe C, Wiese M, Klutch M, Shakarian A, Dwyer D, Nakhasi H. Early response gene expression during differentiation of cultured Leishmania donovani . Parasit Res. 2001;87:897–906. doi: 10.1007/s004360100464. [DOI] [PubMed] [Google Scholar]
- Duncan R, Dey R, Tomioka K, Hairston H, Selvapandiyan A, Nakhasi HL. Biomarkers of Attenuation in the Leishmania donovani Centrin Gene Deleted Cell Line-Requirements for Safety in a Live Vaccine Candidate. Open Parasitology. 2009;3:14–23. [Google Scholar]
- FDA/CDRH. Highly Multiplexed Microbiological/Medical Countermeasure In Vitro Nucleic Acid Based Diagnostic Devices. 2014 www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm327294.pdf.
- Gannavaram S, Connelly PS, Daniels MP, Duncan R, Salotra P, Nakhasi HL. Deletion of mitochondrial associated ubiquitin fold modifier protein Ufm1 in Leishmania donovani results in loss of beta-oxidation of fatty acids and blocks cell division in the amastigote stage. Mol Microbiol. 2012;86:187–198. doi: 10.1111/j.1365-2958.2012.08183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannavaram S, Davey S, Lakhal-Naouar I, Duncan R, Nakhasi HL. Deletion of Ubiquitin Fold Modifier Protein Ufm1 Processing Peptidase Ufsp in L. donovani Abolishes Ufm1 Processing and Alters Pathogenesis. PLoS Negl Trop Dis. 2014;8:e2707. doi: 10.1371/journal.pntd.0002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannavaram S, Sharma P, Duncan RC, Salotra P, Nakhasi HL. Mitochondrial Associated Ubiquitin Fold Modifier-1 Mediated Protein Conjugation in Leishmania donovani. PLoS One. 2011;6:e16156. doi: 10.1371/journal.pone.0016156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal N, Duncan R, Selvapandiyan A, Debrabant A, Baig MS, Nakhasi HL. Cloning and characterization of angiotensin converting enzyme related dipeptidylcarboxypeptidase from Leishmania donovani. Mol Biochem Parasitol. 2006;145:147–157. doi: 10.1016/j.molbiopara.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Lakhal-Naouar I, Jardim A, Strasser R, Luo S, Kozakai Y, Nakhasi HL, Duncan RC. Leishmania donovani argininosuccinate synthase is an active enzyme associated with parasite pathogenesis. PLoS Negl Trop Dis. 2012;6:e1849. doi: 10.1371/journal.pntd.0001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani M, Hockley J, Bracho CG, Rijpkema S, Luquetti AO, Duncan R, Rigsby P, Albajar-Viñas P, Padilla A. Evaluation of two International Reference Standards for antibodies to Trypanosoma cruzi in a WHO collaborative study. World Health Organization. 2011 www.who.int.
- Selvapandiyan A, Dey R, Nylen S, Duncan R, Sacks D, Nakhasi HL. Intracellular replication-deficient Leishmania donovani induces long lasting protective immunity against visceral leishmaniasis. J Immunol. 2009;183:1813–1820. doi: 10.4049/jimmunol.0900276. [DOI] [PubMed] [Google Scholar]
- Selvapandiyan A, Duncan R, Debrabant A, Bertholet S, Sreenivas G, Negi NS, Salotra P, Nakhasi HL. Expression of a mutant form of Leishmania donovani centrin reduces the growth of the parasite. J Biol Chem. 2001;276:43253–43261. doi: 10.1074/jbc.M106806200. [DOI] [PubMed] [Google Scholar]
- Sokal R, Rohlf F. Biometry: The Principles and Practice of Statistics in Biological Research. New York: W. H. Freeman and Company; 1981. [Google Scholar]
- Morris CL, Heath AB WHO. International collaborative study to establish the 3rd WHO international standard for HIV-1 NAT Assays. 2011 [Google Scholar]