Figure 5. Only a minor fraction of ERMS harbors MET amplification.
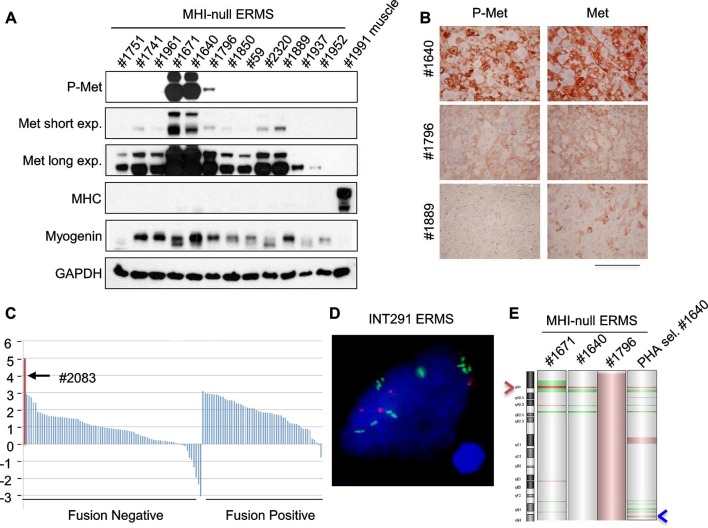
(A) Western blot analysis in a panel of primary murine MHI-null ERMS probed for the indicate proteins. (B) Immunohistochemical analysis of primary murine MHI-null ERMS shown in A, probed for Met and phosphorylated Met. (Scale bar = 100 µm). (C) Waterfall plot showing the MET Z-score expression from a panel of previously characterized human RMS tumors (Shern et al., 2014) (http://pob.abcc.ncifcrf.gov/cgi-bin/JK). The exceptionally high MET expression in Patient 2083 (highlighted) is associated with focal amplification of chromosome 7q31.2 (Shern et al., 2014). (D) FISH for MET in a human ERMS sample (INT291) characterized by small cluster signals (with an average of 9 signals per cell; MET is labeled in green and centromeres in red). (E) CGH analysis of murine MHI-null ERMS. Red indicates copy number gain, green indicates copy number loss. Arrowheads mark the Met (red) and Kras (blue) loci.
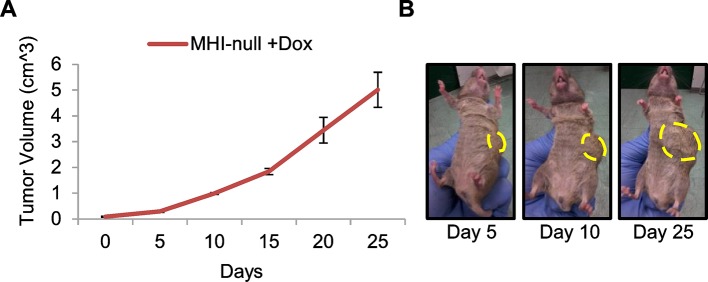
Figure 5—figure supplement 1. Transgenic Hgf downmodulation does not impair tumor growth.
Figure 5—figure supplement 2. Determination of the level of Met protein and phosphorylation in rhabdomyosarcomagenesis.
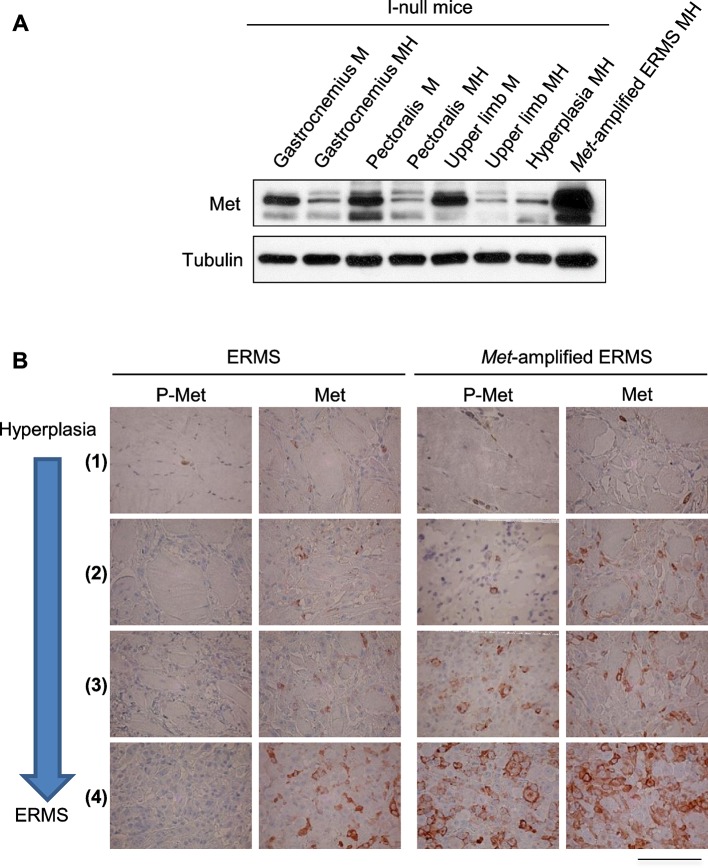
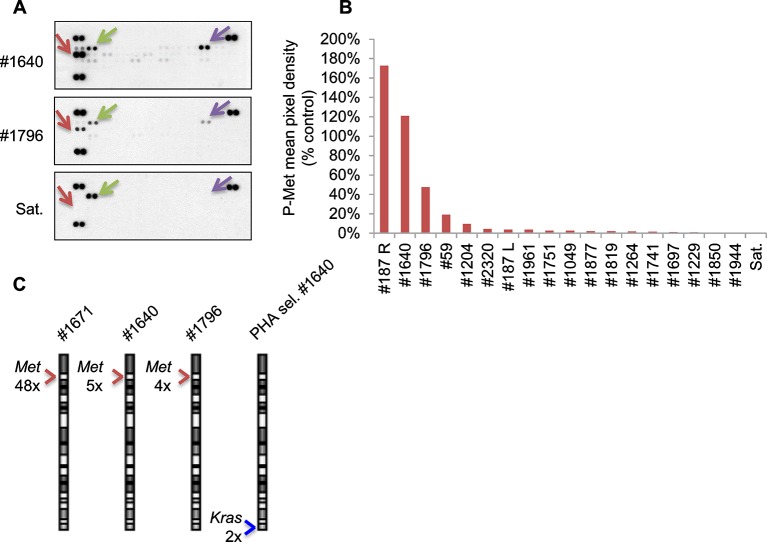
Figure 5—figure supplement 3. Only a minor fraction of murine ERMS displays Met activation and amplification.
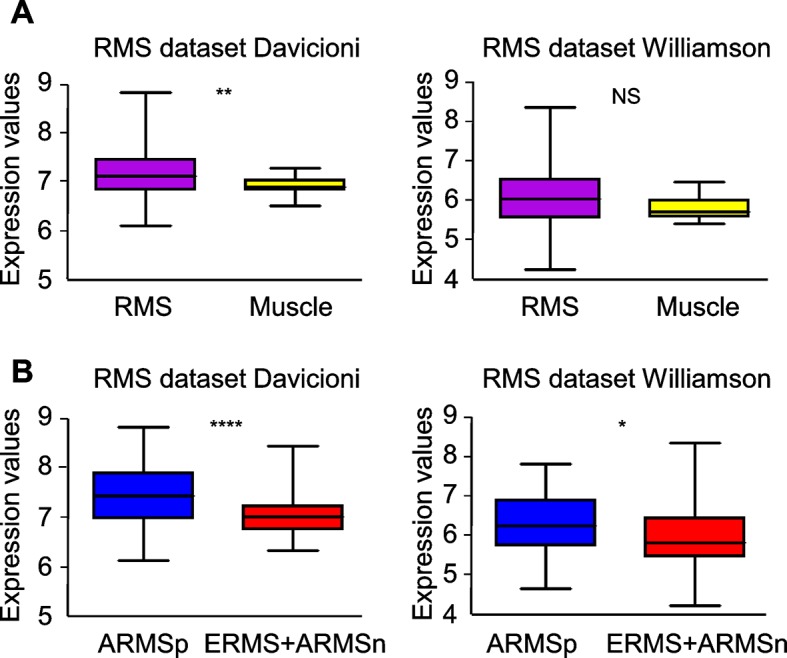
Figure 5—figure supplement 4. Evaluation of Met expression in human RMS datasets.