Abstract
Domestic animals are unique models for biomedical research due to their long history (thousands of years) of strong phenotypic selection. This process has enriched for novel mutations that have contributed to phenotype evolution in domestic animals. The characterization of such mutations provides insights in gene function and biological mechanisms. This review summarizes genetic dissection of about 50 genetic variants affecting pigmentation, behaviour, metabolic regulation, and the pattern of locomotion. The variants are controlled by mutations in about 30 different genes, and for 10 of these our group was the first to report an association between the gene and a phenotype. Almost half of the reported mutations occur in non-coding sequences, suggesting that this is the most common type of polymorphism underlying phenotypic variation since this is a biased list where the proportion of coding mutations are inflated as they are easier to find. The review documents that structural changes (duplications, deletions, and inversions) have contributed significantly to the evolution of phenotypic diversity in domestic animals. Finally, we describe five examples of evolution of alleles, which means that alleles have evolved by the accumulation of several consecutive mutations affecting the function of the same gene.
Keywords: Comparative genomics, domestic animals, mutation detection

Winner of the Rudbeck Award 2013, at the Medical Faculty of Uppsala University for his pioneering studies of the pathogenesis of many non-communicable diseases by means of molecular and animal genetics.
Introduction
During the last 30 years I have used domestic animals as models for biomedical research. Why domestic animals, when there are well-established animal models such as mouse, zebra fish, and Drosophila? The unique feature of domestic animals is their long history of selective breeding. Ever since the first animals were domesticated (1) humans have changed their gene pools by favouring animals that could survive and reproduce in captivity, and that provided useful commodities to humans such as food, skin and fur, transportation, and company. Domestication and breeding is an evolutionary process in which gene variants with favourable phenotypic effects are enriched. A major aim in animal genomics as well as in most genome projects is to reveal genotype–phenotype relationships and to study their underlying molecular mechanisms.
In my PhD thesis defended in December 1983 I expressed the vision that the emerging methods of molecular genetics will unleash the full potential of domestic animals as models for genetic studies of phenotypic traits. I started this research programme the following year as a guest post-doctoral fellow at the Department of Cell Research in Uppsala led by Per A. Peterson and Lars Rask—two of the pioneers applying molecular genetics for biomedical research in Sweden. Furthermore, they led a strong research programme on major histocompatibility complex (MHC) genes and their role in the human immune system. This area was and still is of considerable interest in domestic animals because immune response and disease resistance are of major importance in animal breeding. Thus, during my first years as an independent researcher I characterized the MHC class II region in cattle and demonstrated the presence of extensive genetic diversity at this locus (2), as well as providing evidence that genetic polymorphism at the cattle DRB3 locus is maintained by balancing selection (3). However, by the end of the 1980s, after PCR was invented, it became possible to do genome research, and since then my research team has been working in the field of genetics and genomics. Table I provides a comprehensive list of phenotypic traits in various domestic animals where we have identified the underlying gene and in most cases the specific mutation(s) causing a phenotypic effect.
Table I.
Genes and mutations associated with phenotypic traits in domestic animals.
| Phenotype | Gene | Mutation identified | Type of mutationa | Ref. |
|---|---|---|---|---|
| Horse | ||||
| Chestnut coat colour | MC1R | Yes | Missense | (48) |
| Roan coat colour | KIT | No | Non-coding | (49) |
| Silver coat colour | PMEL | Yes | Missense | (50) |
| Greying with age | STX17 | Yes | 4.6 kb duplication | (51) |
| Gait | DMRT3 | Yes | Nonsense | (44) |
| Pig | ||||
| Dominant white colour | KIT | Yes | Duplication + splice mutation | (13,52–55) |
| Patch coat colour | KIT | Yes | Duplication | (13,52,54,55) |
| Belt coat colour | KIT | Yes | Duplication(s) | (55,56) |
| Dominant black colour | MC1R | Yes | Missense | (57) |
| Recessive red colour | MC1R | Yes | Missense | (57) |
| Black spotting | MC1R | Yes | 2 bp insertion + missense | (15) |
| Black-and-tan coat colour | ASIP | No | Non-coding | (58) |
| Hypercholesterolemia | LDLR | Yes | Missense | (5) |
| Muscle glycogen content | PRKAG3 | Yes | Missense | (27) |
| Muscle growth | IGF2 | Yes | SNP | (37) |
| Chicken | ||||
| Dominant black colour | MC1R | Yes | Missense | (59) |
| Buttercup colour | MC1R | Yes | Missense | (59) |
| Dominant white colour and feather pecking | PMEL | Yes | 9 bp insertion | (16,60) |
| Dun colour | PMEL | Yes | 15 bp deletion | (16) |
| Smoky colour | PMEL | Yes | 9 bp insertion + 12 bp deletion | (16) |
| Silver | SLC45A2 | Yes | Missense | (61) |
| Sex-linked imperfect albinism | SLC45A2 | Yes | 1 bp insertion | (61) |
| Sex-linked barring | CDKN2A/B | Yes | Non-coding + missense | (62) |
| Dark brown colour | SOX10 | Yes | 8.3 kb deletion | (12) |
| Yellow skin | BCDO2 | No | Non-coding | (63) |
| Fibromelanosis | EDN3 | Yes | Duplications | (64) |
| Silky feather | PDSS2 | Yes | SNP | (65) |
| Pea-comb | SOX5 | Yes | CNV | (66) |
| Rose-comb | MNR2 | Yes | 7.4 Mb inversion | (4) |
| Sperm motility | CCDC108 | Yes | 7.4 Mb inversion | (4) |
| Duplex comb | EOMES | Yes | 20 kb duplication | (67) |
| Crested | HOXC8 | No | Non-coding | (68) |
| Growth | SH3RF2 | Yes | Deletion | (14) |
| Dog | ||||
| Leukocyte adhesion deficiency | ITGB2 | Yes | Missense | (6) |
| White spotting (S)b | MITF | Yes | Non-coding | (7,69) |
| Dorsal hair ridge | Multiple | Yes | 133 kb duplication | (70) |
| Sensory ataxic neuropathy | mt_tRNATyr | Yes | 1 bp deletion | (71) |
| Shar-pei fever | HAS2 | Yes | Duplication | (72) |
| Cattle | ||||
| Fishy off-flavour | FMO3 | Yes | Nonsense | (8) |
| Dominant red colour | COPA | Yes | Missense | (73) |
| Water buffalo | ||||
| White spotting | MITF | Yes | Nonsense | (74) |
| White spotting | MITF | Yes | Splice site | (74) |
| Japanese quail | ||||
| Cinnamon colour | SLC45A2 | Yes | Missense | (61) |
| Sex-linked imperfect albinism | SLC45A2 | Yes | Splice site | (61) |
aMutations in coding sequences are in bold.
bThere are four alleles at this locus: Solid (wild-type), Irish spotting, Piebald, and Extreme white. Our data suggest that these are caused by different combinations of non-coding mutations.
CNV = copy number variation.
My vision to use domestic animals as models for biomedical research by taking advantage of the advances in molecular genetics and genomics has been shown to be a fruitful approach that has resulted both in new basic knowledge and in practical applications. However, during the first 20 years of my career it was not easy to convince funding bodies that this was an approach worth supporting. My research applications were sometimes rejected by funding bodies supporting agricultural research because the approach was considered of limited practical interest and was rather to be classified as basic research, and the applications were consistently rejected by the Swedish Research Council (supporting basic research) because they were considered too applied.
Research on the genetic basis for diseases and disorders in domestic animals is of outmost importance for veterinary medicine and animal breeding in order to keep disease incidence at a minimum level. However, domestic animals are less important as models for human disease for several reasons. Firstly, genetic variants causing disease usually occur at a low frequency because there is strong selection to eliminate unfit animals, in particular in the animals we use for food production. Secondly, the clinical characterization of disease is rarely as advanced in animals as in human medicine. Thirdly, thanks to the development of powerful methods for genetic studies in humans the need for using animals to identify candidate genes for human disease has become less important. However, when one finds mutations in domestic animals in genes associated with human disease, the domestic animal can be used as a model to develop disease prevention and new treatments.
The major merit of domestic animals as models for biomedical research relates to the rapid phenotypic evolution that has occurred during the course of animal domestication. Here domestic animals give a unique possibility to gain insight into molecular mechanisms underlying phenotypic change. The aim with this review is to give examples of important discoveries that we have made by studying some of the traits under selection in domestic animals.
General lessons
Table I lists 50 phenotypes in various domestic animals for which my group has been involved in a study that has resulted in the detection of the underlying gene and in most cases the underlying mutation(s). This list involves about 30 different genes. The number of genes is lower than the number of traits because some genes are associated with multiple phenotypes. For instance, we have identified mutations in the melanocortin 1 receptor (MC1R) gene underlying six different variant alleles affecting pigmentation in horse, pig, and chicken. For about 10 of the genes listed in Table I our study was the first in any organism that has documented an association between that gene and a phenotype. These studies have therefore contributed to the functional annotation of the vertebrate genome. For instance, the paper on our finding that a 7.4 Mb inversion disrupts the coiled-coil domain containing 108 gene (CCDC108) which leads to poor sperm motility in homozygous Rose-comb roosters (4) is still the only publication on the CCDC108 gene in any organism. It is likely that loss-of-function mutations affecting this sperm protein gene are also causing reduced sperm motility and reduced fertility in some human males.
Only a handful of the traits listed in Table I are obvious disorders. One example is a missense mutation in the low-density lipoprotein receptor gene (LDLR) that we identified in a strain of pigs used as models for hypercholesterolemia in humans (5). Other examples include leukocyte adhesion deficiency in dogs caused by a missense mutation in the integrin beta 2 (ITGB2) gene (6), sensory ataxic neuropathy in dogs caused by a one base pair deletion in the mitochondrial genome (7), and fishy off-flavour in cattle, a condition where homozygous cows produce milk that smells of rotten fish, caused by a missense mutation in the flavin containing monooxygenase 3 (FMO3) gene (8). Mutations in the corresponding genes cause very similar diseases or disorders in humans. In the three last-mentioned cases a diagnostic DNA test has been developed and used to reduce or eliminate disease. Furthermore, a colony of Irish setters carrying the mutation causing leukocyte adhesion deficiency was subsequently used as a model for human gene therapy (9). Sick dogs were cured by introducing a functional copy of the ITGB2 gene ex vivo into hematopoietic stem cells which were then introduced to the affected animal, an example of how a big animal model could be very valuable for the development of new therapeutic strategies in human medicine.
A long-standing question in biology is the relative importance of coding and non-coding mutations for explaining phenotypic variation and disease (10). Almost 50% of the mutations listed in Table I are non-coding, and this strongly suggests that non-coding mutations by far dominate among the mutations underlying the phenotypic diversity present in domestic animals. This conclusion is based on the fact that this is a biased estimate inflating the proportion of coding changes because they are much easier to find. Firstly, changes in coding sequences often lead to alleles with more striking phenotypic effects. Secondly, it is much more straightforward to interpret the functional consequence of changes in coding sequence than in non-coding sequence. These conclusions are consistent with data from human genetics showing that whereas a majority of mutations causing severe inherited disorders affects coding sequences, a majority of sequence variants associated with increased risk for a multifactorial disorder occurs in non-coding sequences (11).
Our characterization of the genetic basis for a phenotypic trait has revealed the importance of structural changes (deletions, duplications, and inversion). Table I lists about 10 examples of structural changes being the causal mutation for a phenotypic trait. A common theme here is that the structural change leads to altered regulation of one or several of the genes directly affected by the structural change or located in the close vicinity of the structural change. This may occur because a regulatory element is deleted (e.g. Dark brown colour in chicken) (12), or duplicated (e.g. Greying with age in horses) (13), or because a gene has been translocated to another position and is there influenced by another constellation of regulatory elements (e.g. Rose-comb in chicken) (4).
Another interesting finding is that we have documented evolution of alleles, which means the accumulation of multiple consecutive causal mutations in the same gene. The genetic literature on the identification of causal mutations is largely based on studies of monogenic disorders in humans or mutations causing monogenic phenotypes in experimental organisms. These are almost always due to a single hit. However, our data from domestic animals gives a different picture because animal domestication has a sufficiently long history (about 10,000 years) to allow evolution of alleles. We have so far documented five examples where multiple causal mutations contribute to a phenotype: Dominant white colour in pigs (14), Black spotting in pigs (15), Smoky colour in chicken (16), the Rose-comb2 allele in chicken (4), and White-spotting in dogs (7). It appears plausible that a considerable portion of the phenotypic diversity that occurs in natural populations, including the one underlying the risk to develop multifactorial disorders in humans, is due to allelic variants that differ by multiple causal differences rather than single mutations with large effects.
Pigment cell biology
Many of the phenotypes listed in Table I are related to pigmentation. Pigmentation has been used throughout the history of genetics as a model to study how genes act and interact because they often show a simple monogenic inheritance that also facilitates the identification of casual genes and mutations. Furthermore, the phenotypic readout is precise, allowing the scoring of subtle phenotypic differences.
A striking difference between domestic animals and their wild ancestors is the amazing coat colour diversity, as species in the wild in most cases show very modest variation. In fact, coat colour was one of the first traits that changed after domestication, and colour variants in domestic animals are mentioned in some of our earliest written records from the Ur III dynasty in Mesopotamia dated to about 5,000 years before present (17). So why did the coat colour change in our domestic animals? Most importantly, humans have actively selected for colour variation among domestic animals because 1) this allowed us to distinguish our prized domesticated animals from their wild ancestors at a time when gene flow often occurred; 2) selection against camouflage facilitated animal husbandry; and 3) we and apparently our ancestors appreciate diversity of colour and therefore have kept animals carrying novel phenotypes as long as the variant is not associated with deleterious effects that reduce their utility. Relaxed purifying selection has most likely also contributed to the rich coat colour diversity in domestic animals.
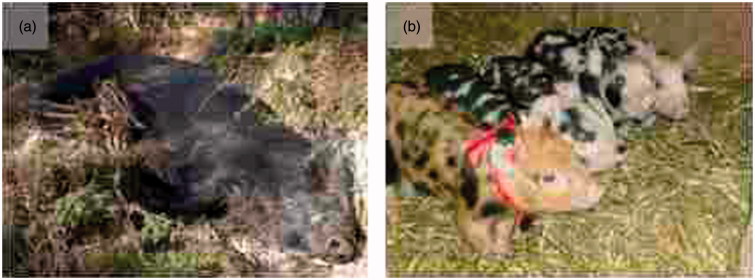
Our study comparing genetic variation in the melanocortin 1 receptor (MC1R) gene among wild and domestic pigs illustrates the striking difference in selection pressure in the wild and at the farm (18). MC1R is expressed at the cell surface of melanocytes and has a critical role in determining pigmentation in vertebrates because it controls pigment switching, with the absence or presence of MC1R signalling being associated with the synthesis of red and black pigment, respectively (19). Mutations in MC1R are clearly the most common reason for different colour morphs both in domestic animals and in wild species, most likely because the function of MC1R is largely restricted to the pigment cell, which means that mutations in this gene are not associated with strong negative effects on other traits. MC1R mutations are causing the dominant black, recessive red, and black spotting coat colour variants in pigs (Table I). A striking difference between wild boars and domestic pigs is that the wild boar piglets are striped whereas piglets carrying MC1R mutations are not (Figure 1). The striping pattern is a camouflage colour requiring MC1R pigment switching, and this mechanism is disrupted by these mutations (18). In our study we sequenced the entire MC1R coding sequence from European and Chinese domestic pigs as well as European and Asian wild boars. We identified seven different sequence variants among the wild boars, and all were synonymous, and thus all tested wild boars expressed an identical MC1R protein sequence despite the fact that European and Asian wild boars are classified as different subspecies that diverged about 1 million years ago (20). This indicates strong purifying selection to maintain camouflage colour in the wild. In contrast, 9 out of 10 sequence variants detected among domestic pigs changed the protein sequence consistent with strong selection to change colour. In this study we analysed 51 different pig breeds from Europe and China, and almost all breeds carried MC1R mutations. The only exception was the Mangalica pig from Hungary that carried the MC1R wild-type allele, and their piglets are in fact striped like the wild boar!
Winner of the Rudbeck Award 2013, at the Medical Faculty of Uppsala University for his pioneering studies of the pathogenesis of many non-communicable diseases by means of molecular and animal genetics.
The black spotting phenotype in pigs (Figure 1B) is a particularly interesting variant because this allele is the result of two consecutive mutations (15). Firstly, it carries the D121N missense mutation leading to a constitutively active receptor causing the dominant black colour variant. In addition, the black spotting allele is associated with a two base pair insertion at codon 23 (nt67insCC) causing a frameshift and thus a complete loss-of-function. As explained above, lack of MC1R signalling is expected to lead to only red/yellow pigmentation, so how can these pigs show black spots? By sequencing MC1R mRNA isolated from black spots we were able to demonstrate that this is caused by somatic mutations that restore the reading frame. The most likely explanation why this happens at a high frequency is that the insertion of two cytosine nucleotides occurs in a stretch of six cytosines and this results in a mononucleotide repeat (CCCCCCCC) that is somatically unstable. This illustrates why studies on pigmentation have been so rewarding because it would have been extremely challenging to reveal such a somatically unstable mutation if the gene for instance affected insulin secretion unless one characterized the release from individual cells. The MC1R black spotting allele is one of our examples of ‘evolution of alleles’ by the accumulation of several consecutive mutations affecting the same gene.
Behaviour
Behaviour is another trait that has changed dramatically after animal domestication. Changes in behaviour were required for the animals to survive and reproduce in captivity. There is a huge potential to study the genetic basis for variation in behaviour in dogs due to the complex interaction between humans and dogs that has evolved since domestication (21). There is also a very fascinating diversity in behaviour among breeds where dogs have been bred for various tasks such as herding, hunting, retrieving various objects, guarding, or just for pleasure as a companion to humans. However, so far, little progress has been made in identifying specific genes underlying variation in dog behaviour. A possible reason is that behaviour has a very complex genetic background with many genes involved.
We have used the rabbit as a model to study the genetic basis for domestication (22). There are three main reasons why the rabbit is a good model for studies of domestication. Firstly, domestication is relatively recent, only about 1,400 years ago. Secondly, we know where rabbit domestication took place (Southern France), and at that time wild rabbits (Oryctolagus cuniculus) were restricted to Southern France and the Iberian Peninsula. Thirdly, the area where domestication happened is still densely populated with wild rabbits that can be sampled for genetic studies. Thus, we can make very precise comparisons of allele frequency differences between domestic rabbits and relevant populations of wild rabbits. This is more difficult for other domestic animals. For instance, domestication of the wolf happened a long time ago (15,000 years before present or earlier) (1), and at that time wolves were spread across the entire Northern hemisphere. It is also possible that the population(s) of wolves that contributed mostly to dog domestication has become extinct due to human expansion. Furthermore, since domestication there has probably been a considerable amount of gene flow between wolves and dogs. This complex demographic history blurs the picture because it is difficult to deduce whether an observed difference in allele frequency between dogs and contemporary wolves is caused by selection, genetic drift, or because there is a genetic difference between contemporary wolves and the wolf population(s) that contributed to dog domestication.
We carried out whole-genome sequencing of 14 population samples of wild rabbits and 6 breeds of domestic rabbits (22). We sequenced pools of animals from each population, and each pool comprised 10 to 20 animals. The rabbit is one of the most polymorphic mammals sequenced so far, and the nucleotide diversity measured as the average number of nucleotide substitutions per 1,000 base pairs between two random chromosomes is nine times higher in rabbits (about 9) than in humans (about 1). We identified a total of 50 million SNPs in rabbits and compared the allele frequency of these in wild and domestic rabbits. This analysis revealed strong signatures of selection and the major conclusions were: 1) rabbit domestication has a highly polygenic basis involving many hundreds of genes; 2) non-coding changes dominate largely over changes in coding sequence; 3) we observed very few complete fixations but rather shifts in allele frequencies consistent with a polygenic basis where the majority of loci have small phenotypic effects; and 4) sequence variants in the vicinity of genes with an established role in brain and neuronal development were highly enriched among those showing the strongest differentiation between wild and domestic rabbits. This implies that changes in genes affecting behaviour have played a prominent role during rabbit domestication. This result makes perfect sense because the wild rabbit has a very strong flight response making it extremely difficult to keep them in captivity, whereas domestic rabbits are well adapted to a life in captivity. In fact, Charles Darwin wrote in On the Origin of Species that ‘… hardly any animal is more difficult to tame than the young of the wild rabbit; scarcely any animal is tamer than the young of the tame rabbit …’ (23). We postulated that tame behaviour in rabbits and other domestic animals has a truly complex genetic background and evolved by shifts in allele frequencies at many loci rather than by critical changes at a few domestication loci.
Metabolic traits
The metabolism of our domestic animals has often been drastically altered by our need to use them for food production. A good example is layer chicken that have been selected to produce more than 300 eggs during a year without being mated to a rooster, whereas its wild ancestor, the red junglefowl female, normally produces one clutch of eggs after mating. We have in particular studied the altered metabolism and body composition in pigs. Since the 1940s there has been a drastic increase in pig muscle growth and a corresponding decrease in fat deposition due to the consumer’s demand for lean meat. This was achieved after procedures to measure body composition (the relative proportion of protein and fat in the carcass) were introduced and powerful statistical methods for calculating breeding values were developed. We have identified two major loci that have responded to the strong selection for lean meat: the RN locus affecting glycogen content in skeletal muscle, and the IGF2 locus affecting muscle growth.
The RN story started in France where researchers noted that there was a meat quality problem in Hampshire pigs since a large proportion of the individuals produced meat with an unusually low pH (measured 24 h after slaughter), reduced water-holding capacity, and reduced yield of cured cooked ham. This had a large effect on pig production worldwide since it is a common practice to use Hampshire pigs as a sire line mated to a dam line, which means that a large proportion of the pigs used for meat production would have a Hampshire male as the father. Further research revealed that these effects on meat quality were due to a 70% increase in glycogen content in skeletal muscle and that this phenotype showed a simple monogenic inheritance with two alleles, RN− (high glycogen) and rn+ (normal glycogen) (24). The fact that liver glycogen was normal in mutant pigs suggested that the mutation underlying this phenotype affected the function of a muscle-specific isoform not expressed in the liver.
In the mid-1990s we and others decided to attempt to identify the gene underlying the RN phenotype by positional cloning, but this was very challenging at the time because there was no genome assembly available from any vertebrate. The first step was to map the locus to a region on pig chromosome 15 by classical linkage analysis using pedigree data (25). We then investigated whether the corresponding region in the more well-studied human genome harboured any candidate genes with a known role in glycogen metabolism, but that was not the case. We then carried out a very laborious procedure where we isolated the entire region harbouring the RN locus in overlapping bacterial artificial chromosomes (BACs) (26). The BACs were used to isolate new genetic markers and for further fine mapping that eventually resulted in the assignment of the locus to a region of about 100 kb present in a single BAC. We sequenced this entire BAC and identified four genes. One of these was particularly interesting because it encoded a homolog to the regulatory γ-chain of the SNF4 kinase in yeast that has a key role in carbohydrate metabolism including glycogen metabolism. This kinase named AMP-activated protein kinase (AMPK) in vertebrates is a heterotrimeric protein composed of a catalytic α-chain and non-catalytic β- and γ-chains. At the time two α-chain genes (PRKAA1 and PRKAA2), two β-chain genes (PRKAB1 and PRKAB2), and two γ-chain genes (PRKAG1 and PRKAG2) had been identified in humans. We therefore named our newly discovered isoform PRKAG3 (27). The fact that AMPK is an important energy-sensing enzyme, activated by high AMP–low ATP, made it an excellent positional candidate gene for the RN phenotype. Northern blot analysis provided very strong support for this notion because it revealed that PRKAG3 is a muscle-specific isoform not expressed in the liver, in perfect agreement with the RN phenotype. Furthermore, sequence analysis revealed an R225Q missense mutation at a site that is extremely well conserved among AMPK γ-chains in eukaryotes. Genetic analyses in many thousands of pigs have provided conclusive evidence that R225Q is the causal mutation for the RN phenotype.
The year after our discovery, another group reported the identification of a quantitative trait locus (QTL) for glycogen content that mapped to the PRKAG3 region, and they could exclude the presence of the R225Q mutation (28). The data strongly suggested that the causal mutation for this phenotype (lower glycogen than the wild-type) was a missense mutation V224I at the neighbouring residue. Transfection experiments in COS cells demonstrated that the wild-type allele V224R225 showed low kinase activity at low AMP and was activated by high AMP, whereas the V224Q225 (RN-) allele was constitutively active, and the third allele I224R225 showed low activity and could not be induced by high AMP (29). Thus, the result in this assay is a perfect match in relation to the glycogen content in skeletal muscle V224Q225 > V224R225 > I224R225. The explanation for the strong effects of these missense mutations became apparent a few years later when it was demonstrated that these two residues are located in the binding pocket for AMP and ATP that regulates AMPK activity (30).
To explore further the functional significance of the PRKAG3 isoform we created transgenic mice overexpressing the wild-type (R225) or mutant (Q225) forms in skeletal muscle as well as a PRKAG3 knock-out mouse (29). The transgenic mutant but not the transgenic wild-type mouse showed excess glycogen content in skeletal muscle and thus replicated the pig phenotype. Surprisingly, the knock-out mice showed normal glycogen levels and normal glycogen utilization during exercise. However, re-synthesis of glycogen after exercise was impaired, and in vitro tests on skeletal muscle showed defective AMPK-mediated glucose uptake (29). Taken together the combined pig and mouse data show that the AMPK isoform containing the muscle-specific γ3-chain has a key role in monitoring glycogen content in white skeletal muscle and promotes glycogen re-synthesis after exercise by activating glucose uptake and fatty oxidation (29). In humans, a rare naturally occurring R225W mutation affecting the same residue as the pig R225Q mutation has been reported (31,32). Individuals carrying this mutation showed about 90% higher glycogen content in skeletal muscle, about 30% lower intramuscular triacylglycerol content, and no obvious impairment in glucose metabolism, in agreement with the pig and transgenic mouse phenotypes associated with the R225Q mutation.
The PRKAG3 protein is a validated drug target for treatment of type II diabetes. It is plausible that a drug activating the AMPK isoform involving PRKAG3 would have a positive effect on blood glucose levels by activating insulin-independent glucose uptake and promoting fatty acid oxidation mimicking the effects of exercise. However, the project is challenging because it is more difficult to activate a kinase than to inactivate it; in the latter case one ‘only’ needs to find a small molecule that interferes with protein function.
The discovery of the mutation in IGF2 encoding insulin-like growth factor II was made using our intercross between Large White domestic pigs and the European wild boar (33). This intercross was initiated already 1989 with the ambition to develop a comprehensive linkage map for pigs and to map loci underlying phenotypic traits. The pedigree comprised 200 F2 progeny that were carefully phenotyped for a range of traits including body composition and weight of internal organs. A genetic analysis revealed a paternally expressed QTL with major effects on muscle growth, subcutaneous fat depth, and the size of the heart that mapped to the IGF2 region (34). F2 progeny that had inherited the domestic pig allele showed 3%–4% more muscle, lower subcutaneous fat depth, and had a bigger heart. The fact that both this QTL and IGF2 showed paternal expression and that IGF2 is an important growth factor strongly suggested that the causal mutation(s) for this QTL affected IGF2 function.
Sequence analysis revealed that the IGF2 coding sequence was identical in wild boar and domestic pigs, suggesting a regulatory mutation. A problem with genetic analysis of quantitative traits or multifactorial disorders is that there is no simple one-to-one relationship between genotype and phenotype because each QTL controls only a fraction of the variance. However, in domestic animals it is possible to transform a quantitative trait to a Mendelian trait by progeny testing. In collaboration with Michel Georges (University of Liège, Belgium), who had independently identified the IGF2 QTL in an intercross between Large White and Piétrain pigs (35), we used pedigree analysis to identify individual sires that segregated for the QTL and then sorted their chromosomes as Q (for high muscle growth) and q (wild-type). Sequence analysis of the entire IGF2 region as well as the flanking regions including the insulin gene showed that the mutation(s) underlying the QTL must be located within the IGF2 region since all Q chromosomes were identical-by-descent (IBD) for a 29 kb region within IGF2. The problem was that the sequence divergence between the Q and q chromosomes was as high as 1%, meaning that there were about 300 sequence differences between the two types of chromosomes. A 1% sequence difference is exceptionally high between two alleles from the same species (cf. orthologous sequences from human and chimpanzee show on average a 1.2% sequence divergence), and we therefore hypothesized that the Q allele may originate from Asian pigs since we had shown a few years before that European and Asian pigs were domesticated from different subspecies of wild boars and that there has been a considerable import of Asian pigs into Europe during the eighteenth and nineteenth centuries (36). We therefore started a collaboration with Alan Archibald and Chris Haley (The Roslin Institute, Edinburgh, UK) who had developed an intercross between Chinese Meishan pigs and European Large White pigs. Statistical and sequence analysis showed that the Meishan pigs carried chromosomes classified as q, and it turned out that these differed from the Q allele by a single base change (C to G) in intron 3 of IGF2, providing conclusive genetic evidence that this must be the causal mutation (37). This was the first study in any organism that revealed the causal mutation for a multifactorial trait or disorder as a single base change in a non-coding region. This major achievement, accomplished long before there was a pig genome assembly, is a beautiful illustration of how powerful genetic studies in domestic animals can be by combining extensive pedigree analysis and the rapid evolution that makes it sometimes possible to identify the wild-type ancestral chromosome for a mutant chromosome. It is still an open question whether the IGF2 mutation arose in Asian wild boars before domestication, in Asian domestic pigs prior to introgression to European pigs, or in Europe after introgression of a wild-type Asian chromosome. This mutation has gone through a selective sweep in modern meat-producing pigs, and a very high proportion of all pigs used for meat production worldwide carry this mutation.
The IGF2 mutation occurs at an evolutionary conserved CpG island in intron 3; 16 base pairs involving the mutated site show 100% sequence identity among 18 out of 18 mammalian species (38). Gel shift experiments revealed that the mutation disrupts the interaction with a nuclear factor present in mouse C2C12 myoblast cells, and a Luciferase reporter assay in the same cell type showed that the wild-type sequence but not the mutant sequence supports repression of transcription from the endogenous IGF2 promoter (37). This result was in perfect agreement with expression analysis showing upregulated IGF2 expression in postnatal skeletal and cardiac muscle from mutant chromosomes but not in prenatal muscle or in liver. Thus, the mutation knocks out the interaction with a repressor, and the effect of the mutation is both tissue- and stage-specific.
The remaining big question after the identification of the IGF2 mutation was which transcription factor binds the wild-type sequence. The mutation did not disrupt the recognition sequence for a known factor, suggesting that it is an unusual site for a known factor or that an uncharacterized factor binds to the site. The latter turned out to be the correct answer when we 6 years later in collaboration with researchers at the Broad Institute (Cambridge, USA) were able to fish out the factor from preparations of SILAC-labelled C2C12 nuclear proteins using biotin-labelled wild-type and mutant oligonucleotides (39). The protein that we named ZBED6 was not just an uncharacterized transcription factor but a previously unknown protein. The reason why it was not annotated as a protein in the human and mouse genomes was that it is encoded by a domesticated DNA transposon located in an intron of another gene, ZC3H11A. Bioinformatic analysis indicated that this DNA transposon must have integrated in the genome more than 200 million years ago, before the split between monotremes and other mammals, but the open reading frame is only maintained in placental mammals. The data imply that ZBED6 is an innovation in placental mammals that evolved after the divergence of the ancestors of marsupials and placental mammals. The extremely high sequence conservation of the two DNA-binding BED domains among placental mammals (39) suggests that sequence changes in these regions are not tolerated and that ZBED6 has evolved an essential function. ChIP-seq analysis revealed about 2,500 putative ZBED6 binding sites in mouse C2C12 cells, and the consensus binding motif GCTCG was in perfect agreement with the wild-type sequence in pig IGF2 intron 3 that has changed to GCTCA in mutant pigs (39). ZBED6 sites are found in GC-rich sequences close to promoters with a peak downstream of the start of transcription. An analysis of histone marks in mouse C2C12 cells showed that ZBED6 primarily binds active promoters (40). Further characterization of ZBED6 in C2C12 mouse myoblasts, mouse and human islet cells, as well as in human colorectal cancer cell lines has demonstrated the following about ZBED6: 1) it regulates IGF2 expression in a variety of cell types; 2) it has profound effects on transcriptional regulation; and 3) it acts as a transcriptional modulator that fine-tunes the expression of many genes including IGF2 (40–42). The working hypothesis is that the essential function of ZBED6 is to provide metabolic flexibility as illustrated by the pig IGF2 mutation. Wild boars have a better ability to store fat when muscle growth is not needed, whereas domestic pigs carrying the IGF2 mutation tend to shunt available energy to muscle growth.
Patterns of locomotion
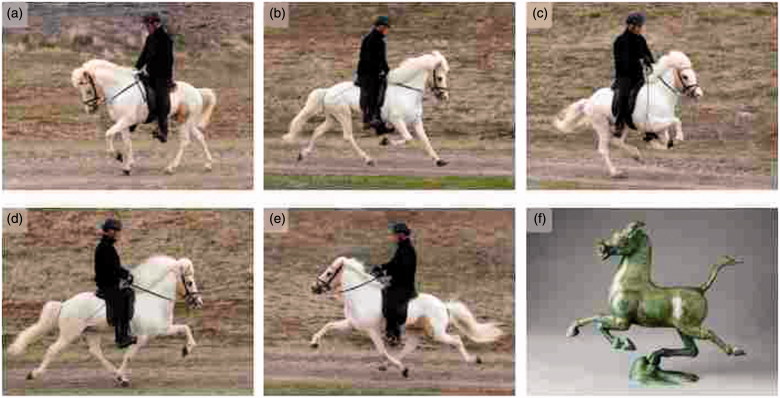
The horse is particularly well suited for genetic studies of patterns of locomotion because the gait of horses is critical for the different ways we use horses, e.g. as draught horses for transporting heavy loads, as riding horses for fast transport, or as trotting horses in front of light carriages. All horses can perform the three gaits walk, trot, and gallop (Figure 2A–C). However, some horses can perform alternative gaits. Icelandic horses are a good example since they can perform two alternative gaits, tölt and pace (Figure 2D and E). Tölt is a four-beat ambling gait that is almost as fast as trot but gives a much smoother ride as the horse always has one foot on the ground. The ability to perform the tölt is an important reason for the popularity of this breed. Pace is a two-beat lateral gait whereas trot is a two-beat diagonal gait (Figure 2B and E). Pace is faster than tölt, as fast as trot, but slower than the gallop.
Figure 2.
Gaits in horses. (a–e): Different gaits performed by an Icelandic horse. (a): walk; (b): trot; (c): gallop; (d): tölt; and (e): pace. Photos: Freyja Imsland. (f): ‘Bronze pacing horse poised on a swallow with wings outstretched’, bronze sculpture from Eastern Han Dynasty, about 200 AD. Photo: Erik Cornelius, Museum of Far Eastern Antiquities, Stockholm.
We decided to explore the genetic basis for gait variations in Icelandic horses because half of the population is classified as four-gaited (walk, trot, gallop, and tölt) and the other half is five-gaited (walk, trot, gallop, tölt, and pace). It was also known that this variation was not solely a consequence of training because the ability to pace has a high heritability (43). However, a high heritability does not necessarily imply a simple, monogenic inheritance. For instance, human height has a similar high heritability but is affected by hundreds of genes each with a tiny effect. We carried out a genome-wide association analysis using 40 five-gaited and 30 four-gaited Icelandic horses and a SNP-chip comprising about 50,000 SNPs evenly spread across the genome. Remarkably, the statistical analysis revealed a single marker on chromosome 23 that showed a highly significant association (44). Further analysis quickly revealed a single base change causing a premature stop codon in DMRT3 encoding doublesex and mab-3 related transcription factor 3, which turned out to be the causative mutation. All five-gaited Icelandic horses were homozygous mutant (AA) whereas a majority of four-gaited horses were heterozygous CA, about 40% were homozygous mutant (AA), and a few were homozygous wild-type (CC) (44). It is still an open question whether lack of training or genetic modifiers explain why some homozygous mutants do not perform pace. Further analysis showed that horses used for harness racing (trot or pace) have a very high frequency of the Gait keeper mutation, suggesting that, in addition to promoting pace, it also inhibits the transition from trot/pace to gallop and thereby allows horses to trot and pace at very high speed without galloping, which is the natural gait at high speed but not allowed in harness racing. The Gait keeper mutation has a very strong positive effect on racing performance (44), and a diagnostic test for this mutation is now used for horse breeding.
We performed a worldwide screen of 4,396 horses representing 141 breeds for the presence of the DMRT3 mutation (45). It is present in breeds spread across Eurasia and is also widely spread in breeds originating from North and South America. It is absent from breeds used as draught horses and in breeds where the gallop is the most important gait, such as Thoroughbred horses. The worldwide distribution indicates that horses carrying the Gait keeper mutation have been highly appreciated most likely because of their ability to offer a smooth ride when horses were the only means of long-distance transport and for trotting at high speed in front of small carriages. The worldwide distribution in combination with the presence of ancient sculptures showing horses with alternate gaits (Figure 2F) suggests that the Gait keeper mutation arose more than 2,000 years before present.
The DMRT3 mutation is another example where our genetic analysis allowed us to identify the causal mutation without any prior knowledge about gene function. When we discovered this locus the function of the DMRT3 protein was unknown. It belongs to a small family of transcription factors, and the most well-studied member (DMRT1) has a critical role in sexual development in vertebrates, so the assumption was that the closely related DMRT3 protein had a similar role. We explored the function of DMRT3 in collaboration with Klas Kullander and his group at Uppsala University that has a strong expertise in spinal cord neurobiology. DMRT3 is expressed in a specific subset of neurons in the spinal cord (now named DMRT3 neurons) in mice and horses (44). These neurons were classified as interneurons as their axons crossed the mid-line of the spinal cord and they are inhibitory neurons that make direct contacts with motor neurons. Thus, these characteristics are in perfect agreement with the genetic data suggesting that these neurons play a critical role in co-ordinating muscle contractions during locomotion. Interestingly, another group had previously developed a Dmrt3 knock-out mouse but did not notice any striking phenotype (44). In the light of the horse data and the characterization of the DMRT3 neurons we decided to characterize the pattern of locomotion in these mice that appeared to move normally in the cage. A detailed characterization of the pattern of locomotion on a TreadScan, limb co-ordination of newborn mice, and fictive locomotion of isolated spinal cords revealed severe defects in limb co-ordination in the Dmrt3 knock-out mice (44). Despite severe disturbances in the co-ordination of limb development at birth, the Dmrt3 null mice are able to move normally under unstressed conditions in the cage implying that the locomotor network in the spinal cord to a large extent can compensate for the loss of DMRT3. The horse and mouse data together with the functional characterization demonstrate that DMRT3 neurons have a critical role for co-ordinating limb movements in vertebrates. It is likely that the horse mutation has a more severe effect than the null mutation present in the knock-out mouse. The horse mutation causes a premature stop codon, and the mutant protein contains only 300 out of the 474 residues in the full-length protein. The mutant protein may constitute a dominant negative form containing the DNA-binding domain but with defect protein–protein interaction. The horse mutation is partially dominant with a clear phenotypic effect in heterozygotes, albeit milder than the one in homozygotes, whereas no significant phenotypic effect was found in the mice heterozygous for the null allele.
No DMRT3 mutation in humans has as yet been described, but since homozygotes for the horse mutation and the mouse knock-out are fully viable it is very likely that humans lacking functional DMRT3 expression exist. The prediction is that they have a mild defect in the co-ordination of limb movements. In fact, there are humans that tend to pace rather than performing the diagonal movement of legs and arms, but no investigation has yet been carried out if this has a genetic basis. However, there is a previously described 225 kb deletion upstream of DMRT3 that causes a dominant form of a congenital neurodegenerative disease resembling cerebral palsy (46). It is possible that this is due to a defect in DMRT3 function. This large deletion disrupts the KANK1 gene, and one of the deletion breakpoints occurs just upstream of the DMRT1 gene, which in turn is located just upstream of DMRT3. Thus, it is possible that the deleted region contains negative regulatory elements and the elimination of these leads to overexpression or ectopic expression of DMRT3 and/or DMRT1; altered expression of DMRT1 may disturb DMRT3 function since they are expected to interact with identical or very similar DNA sequences (47). It is likely that a disturbed development of DMRT3 neurons, which are classified as inhibitory neurons controlling the firing of motor neurons, may have severe deleterious effects on limb co-ordination. This hypothesis can be tested using animal models carrying targeted mutations mimicking the human mutation.
Concluding remarks
Genetic and genomic studies of domestic animals are well justified due to their agricultural importance. This review illustrates how research on domestic animals can also contribute with new basic knowledge concerning gene function and biological mechanisms. Genetic studies on domestic animals give a complimentary view on genotype–phenotype relationships compared with what we have learned from studies on humans and experimental organisms. The reason is that domestication and breeding are evolutionary processes with strong phenotypic selection over thousands of years. The fact that this is the most extensive genetic screen that has been accomplished ensures that studies of domestic animals will continue to enrich the field of biomedicine.
Acknowledgements
I thank all my co-workers and co-investigators for their contribution to the work described here. I thank Freyja Imsland for help in preparing the figures.
Declaration of interest
The author reports no conflicts of interest. Work in the author’s laboratory was supported by grants from Formas, the Swedish Research Council, Knut and Alice Wallenberg foundation, and the European Research Council.
References
- 1.Larson G, Piperno DR, Allaby RG, Purugganan MD, Andersson L, Arroyo-Kalin M, et al. Current perspectives and the future of domestication studies. Proc Natl Acad Sci USA. 2014;111:6139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson L, Böhme J, Rask L, Peterson PA.. Genomic hybridization of bovine class II major histocompatibility genes: 1. Extensive polymorphism of DQ alpha and DQ beta genes. Anim Genet. 1986;17:95–112. [DOI] [PubMed] [Google Scholar]
- 3.Andersson L, Sigurdardóttir S, Borsch C, Gustafsson K.. Evolution of MHC polymorphism: extensive sharing of polymorphic sequence motifs between human and bovine DRB alleles. Immunogenetics. 1991;33:188–93. [DOI] [PubMed] [Google Scholar]
- 4.Imsland F, Feng CG, Boije H, Bed’hom B, Fillon V, Dorshorst B, et al. The Rose-comb mutation in chickens constitutes a structural rearrangement causing both altered comb morphology and defective sperm motility. PLoS Genet. 2012;8:e1002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasler-Rapacz J, Ellegren H, Fridolfsson AK, Kirkpatrick B, Kirk S, Andersson L, et al. Identification of a mutation in the low density lipoprotein receptor gene associated with recessive familial hypercholesterolemia in swine. Am J Med Genet. 1998;76:379–86. [PubMed] [Google Scholar]
- 6.Kijas J, Bauer JTR, Gäfvert S, Marklund S, Trowald-Wigh G, Johannisson A, et al. A missense mutation in the b-2 integrin gene (ITGB2) causes canine leukocyte adhesion deficiency. Genomics. 1999;61:101–7. [DOI] [PubMed] [Google Scholar]
- 7.Baranowska Körberg I, Sundström E, Meadows JRS, Rosengren Pielberg G, Gustafson U, Hedhammar Å, et al. A simple repeat polymorphism in the MITF-M promoter is a key regulator of white spotting in dogs. PLoS One. 2014;9:e104363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lundén A, Marklund S, Gustafsson V, Andersson L.. A nonsense mutation in the FMO3 gene underlies fishy off-flavor in cow’s milk. Genome Res. 2003;12:1885–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer TRJ, Hai M, Tuschong L, Burkholder T, Gu Y, Sokolic R, et al. Correction of the disease phenotype in canine leukocyte adhesion deficiency using ex vivo hematopoietic stem cell gene therapy. Blood. 2006;108:3313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King MC, Wilson AC.. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–16. [DOI] [PubMed] [Google Scholar]
- 11.Gusev A, Lee SH, Trynka G, Finucane H, Vilhjálmsson Bjarni J, Xu H, et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95:535–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunnarsson U, Kerje S, Bed’hom B, Sahlqvist AS, Ekwall O, Tixier-Boichard M, et al. The Dark brown plumage color in chickens is caused by an 8.3 kb deletion upstream of SOX10. Pigment Cell Melanoma Res. 2011;24:268–74. [DOI] [PubMed] [Google Scholar]
- 13.Pielberg G, Olsson C, Syvänen A-C, Andersson L.. Unexpectedly high allelic diversity at the KIT locus causing dominant white color in the domestic pig. Genetics. 2002;160:305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubin C-J, Zody MC, Eriksson J, Meadows JRS, Sherwood E, Webster MT, et al. Whole genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–91. [DOI] [PubMed] [Google Scholar]
- 15.Kijas JMH, Moller M, Plastow G, Andersson L.. A frameshift mutation in MC1R and a high frequency of somatic reversions cause black spotting in pigs. Genetics. 2001;158:779–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerje S, Sharma P, Gunnarsson U, Kim H, Bagchi S, Fredriksson R, et al. The Dominant white, Dun and Smoky color variants in chicken are associated with insertion/deletion polymorphisms in the PMEL17 gene. Genetics. 2004;168:1507–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeder MA. Of kings and shepherds: specialized animal economy in Ur III Mesopotamia In: Stein G, Rothman M, editors. Chiefdoms and early states in the Near East: The organizational dynamics of complexity Madison, WI: Prehistory Press; 1994. p. 175–91. [Google Scholar]
- 18.Fang M, Larson G, Soares Ribeiro H, Li N, Andersson L.. Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 2009;5:e1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaelin CB, Barsh GS.. Genetics of pigmentation in dogs and cats. Ann Rev Anim Biosci. 2013;1:125–56. [DOI] [PubMed] [Google Scholar]
- 20.Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, et al. Pig genomes provide insight into porcine demography and evolution. Nature. 2012;491:393–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacLean EL, Hare B.. Dogs hijack the human bonding pathway. Science. 2015;348:280–1. [DOI] [PubMed] [Google Scholar]
- 22.Carneiro M, Rubin C-J, Di Palma F, Albert FW, Alföldi J, Barrio AM, et al. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science. 2014;345:1074–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darwin C. On the origins of species by means of natural selection or the preservation of favoured races in the struggle for life. London: John Murray; 1859. [Google Scholar]
- 24.Le Roy P, Naveau J, Elsen JM, Sellier P.. Evidence for a new major gene influencing meat quality in pigs. Genet Res. 1990;55:33–44. [DOI] [PubMed] [Google Scholar]
- 25.Mariani P, Lundström K, Gustafsson U, Enfält AC, Juneja RK, Andersson L.. A major locus (RN) affecting muscle glycogen content is located on pig chromosome 15. Mamm Genome. 1996;7:52–4. [DOI] [PubMed] [Google Scholar]
- 26.Jeon J-T, Amarger V, Robic A, Rogel-Gaillard C, Bongcam-Rudloff E, Paul S, et al. Comparative analysis of a BAC contig of the porcine RN region and the human transcript map: implications for the cloning of trait loci. Genomics. 2001;72:297–303. [DOI] [PubMed] [Google Scholar]
- 27.Milan D, Jeon JT, Looft C, Amarger V, Thelander M, Robic A, et al. A mutation in PRKAG3 associated with excess glycogen content in pig skeletal muscle. Science. 2000;288:1248–51. [DOI] [PubMed] [Google Scholar]
- 28.Ciobanu D, Bastiaansen J, Malek M, Helm J, Woollard J, Plastow G, et al. Evidence for new alleles in the protein kinase adenosine monophosphate-activated g3-subunit gene associated with low glycogen content in pig skeletal muscle and improved meat quality. Genetics. 2001;159:1151–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnes BR, Marklund S, Steiler TL, Walter M, Hjälm G, Amarger V, et al. The AMPK-gamma3 isoform has a key role for carbohydrate and lipid metabolism in glycolytic skeletal muscle. J Biol Chem. 2004;279:38441–7. [DOI] [PubMed] [Google Scholar]
- 30.Xiao B, Heath R, Saiu P, Leiper FC, Leone P, Jing C, et al. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature. 2007;449:496–500. [DOI] [PubMed] [Google Scholar]
- 31.Costford SR, Kavaslar N, Ahituv N, Chaudhry SN, Schackwitz WS, Dent R, et al. Gain-of-function R225W mutation in human AMPKγ3 causing increased glycogen and decreased triglyceride in skeletal muscle. PLoS One. 2007;2:e903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crawford SA, Costford SR, Aguer C, Thomas SC, deKemp RA, DaSilva JN, et al. Naturally occurring R225W mutation of the gene encoding AMP-activated protein kinase (AMPK)γ3 results in increased oxidative capacity and glucose uptake in human primary myotubes. Diabetologia. 2010;53:1986–97. [DOI] [PubMed] [Google Scholar]
- 33.Andersson L, Haley CS, Ellegren H, Knott SA, Johansson M, Andersson K, et al. Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science. 1994;263:1771–4. [DOI] [PubMed] [Google Scholar]
- 34.Jeon J-T, Carlborg Ö, Törnsten A, Giuffra E, Amarger V, Chardon P, et al. A paternally expressed QTL affecting skeletal and cardiac muscle mass in pigs maps to the IGF2 locus. Nat Genet. 1999;21:157–8. [DOI] [PubMed] [Google Scholar]
- 35.Nezer C, Moreau L, Brouwers B, Coppieters W, Detilleux J, Hanset R, et al. An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nat Genet. 1999;21:155–6. [DOI] [PubMed] [Google Scholar]
- 36.Giuffra E, Kijas JMH, Amarger V, Carlborg Ö, Jeon J-T, Andersson L.. The origin of the domestic pig: independent domestication and subsequent introgression. Genetics. 2000;154:1785–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Laere AS, Nguyen M, Braunschweig M, Nezer C, Collette C, Moreau L, et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003;425:832–6. [DOI] [PubMed] [Google Scholar]
- 38.Andersson L, Andersson G, Hjälm G, Jiang L, Lindblad-Toh K, Lindroth AM, et al. ZBED6 – the birth of a new transcription factor in the common ancestor of placental mammals. Transcription. 2010;1:144–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markljung E, Jiang L, Jaffe JD, Mikkelsen TS, Wallerman O, Larhammar M, et al. ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol. 2009;7:e1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang L, Wallerman O, Younis S, Rubin C-J, Gilbert ER, Sundström E, et al. ZBED6 modulates the transcription of myogenic genes in mouse myoblast cells. PLoS One. 2014;9:e94187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Jiang L, Wallerman O, Engström U, Ameur A, Gupta RK, et al. Transcription factor ZBED6 affects gene expression, proliferation, and cell death in pancreatic beta cells. Proc Natl Acad Sci USA. 2013;110:15997–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akhtar Ali M, Younis S, Wallerman O, Gupta R, Andersson L, Sjöblom T.. Transcriptional modulator ZBED6 affects cell cycle and growth of human colorectal cancer cells. Proc Natl Acad Sci USA. 2015;112:7743–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Albertsdóttir E, Eriksson S, Sigurdsson A, Arnason T.. Genetic analysis of ‘breeding field test status’ in Icelandic horses. J Anim Breed Genet. 2011;128:124–32. [DOI] [PubMed] [Google Scholar]
- 44.Andersson LS, Larhammar M, Memic F, Wootz H, Schwochow D, Rubin CJ, et al. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature. 2012;488:642–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Promerová M, Andersson LS, Juras R, Penedo MCT, Reissmann M, Tozaki T, et al. Worldwide frequency distribution of the ‘Gait keeper’ mutation in the DMRT3 gene. Anim Genet. 2014;45:274–82. [DOI] [PubMed] [Google Scholar]
- 46.Lerer I, Sagi M, Meiner V, Cohen T, Zlotogora J, Abeliovich D.. Deletion of the ANKRD15 gene at 9p24.3 causes parent-of-origin-dependent inheritance of familial cerebral palsy. Hum Mol Genet. 2005;14:3911–20. [DOI] [PubMed] [Google Scholar]
- 47.Murphy MW, Zarkower D, Bardwell VJ.. Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol Biol. 2007;8:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marklund L, Moller MJ, Sandberg K, Andersson L.. A missense mutation in the gene for melanocyte-stimulating hormone receptor (MC1R) is associated with the chestnut coat color in horses. Mamm Genome. 1996;7:895–9. [DOI] [PubMed] [Google Scholar]
- 49.Marklund S, Moller M, Sandberg K, Andersson L.. Close association between sequence polymorphism in the KIT gene and the roan coat color in horses. Mamm Genome. 1999;10:283–8. [DOI] [PubMed] [Google Scholar]
- 50.Brunberg E, Andersson L, Cothran G, Sandberg K, Mikko S, Lindgren G.. A missense mutation in PMEL17 is associated with the Silver coat color in the horse. BMC Genet. 2006;7:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosengren Pielberg G, Golovko A, Sundström E, Curik I, Lennartsson J, Seltenhammer MH, et al. A cis-acting regulatory mutation causes premature hair greying and susceptibility to melanoma in the horse. Nat Genet. 2008;40:1004–9. [DOI] [PubMed] [Google Scholar]
- 52.Johansson Moller M, Chaudhary R, Hellmen E, Hoyheim B, Chowdhary B, Andersson L.. Pigs with the dominant white coat color phenotype carry a duplication of the KIT gene encoding the mast/stem cell growth factor receptor. Mamm Genome. 1996;7:822–30. [DOI] [PubMed] [Google Scholar]
- 53.Marklund S, Kijas J, Rodriguez-Martinez H, Rönnstrand L, Funa K, Moller M, et al. Molecular basis for the dominant white phenotype in the domestic pig. Genome Res. 1998;8:826–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giuffra E, Törnsten A, Marklund S, Bongcam-Rudloff E, Chardon P, Kijas JMH, et al. A large duplication associated with Dominant White color in pigs originated by homologous recombination between LINE elements flanking KIT. Mamm Genome. 2002;13:569–77. [DOI] [PubMed] [Google Scholar]
- 55.Rubin C-J, Megens H, Martinez Barrio A, Maqbool K, Sayyab S, Schwochow D, et al. Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci U S A. 2012;109:19529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giuffra E, Evans G, Törnsten A, Wales R, Day A, Looft H, et al. The Belt mutation in pigs is an allele at the Dominant white (I/KIT) locus. Mamm Genome. 1999;10:1132–6. [DOI] [PubMed] [Google Scholar]
- 57.Kijas JMH, Wales R, Törnsten A, Chardon P, Moller M, Andersson L.. Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics. 1998;150:1177–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drögemüller C, Giese A, Martins-Wess F, Wiedemann S, Andersson L, Brenig B, et al. The mutation causing the black-and-tan pigmentation phenotype of Mangalitza pigs maps to the porcine ASIP locus but does not affect its coding sequence. Mamm Genome. 2006;17:58–66. [DOI] [PubMed] [Google Scholar]
- 59.Kerje S, Lind J, Schütz K, Jensen P, Andersson L.. Melanocortin 1-receptor (MC1R) mutations are associated with plumage colour in chicken. Anim Genet. 2003;34:241–8. [DOI] [PubMed] [Google Scholar]
- 60.Keeling L, Andersson L, Schütz KE, Kerje S, Fredriksson R, Carlborg Ö, et al. Feather-pecking and victim pigmentation. Nature. 2004;431:645–6. [DOI] [PubMed] [Google Scholar]
- 61.Gunnarsson U, Hellström AR, Tixier-Boichard M, Minvielle F, Bed’hom B, Ito S, et al. Mutations in SLC45A2 cause plumage color variation in chicken and Japanese quail. Genetics. 2007;175:867–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hellström AR, Sundström E, Gunnarsson U, Bed’Hom B, Tixier-Boichard M, Honaker CF, et al. Sex-linked barring in chickens is controlled by the CDKN2A/B tumour suppressor locus. Pigment Cell Melanoma Res. 2010;23:521–30. [DOI] [PubMed] [Google Scholar]
- 63.Eriksson J, Larson G, Gunnarsson U, Bed’hom B, Tixier-Boichard M, Strömstedt L, et al. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 2008;4:e1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dorshorst B, Molin A-M, Johansson A, Strömstedt L, Pham M-H, Chen C, et al. A complex genomic rearrangement involving the Endothelin 3 locus causes dermal hyperpigmentation in the chicken. PLoS Genet. 2011;7:e1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng C, Gao Y, Dorshorst B, Song C, Gu X, Li Q, et al. A cis-regulatory mutation of PDSS2 causes silky-feather in chickens. PLoS Genet. 2014;10:e1004576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wright D, Boije H, Meadows JRS, Bed’hom B, Gourichon D, Vieaud A, et al. Copy number variation in intron 1 of SOX5 causes the Pea-comb phenotype in chickens. PLoS Genet. 2009;5:e1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dorshorst B, Harun-Or-Rashid M, Bagherpoor AJ, Rubin C-J, Ashwell C, Gourichon D, et al. A genomic duplication is associated with ectopic eomesodermin expression in the embryonic chicken comb and two duplex-comb phenotypes. PLoS Genet. 2015;11:e1004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y, Gao Y, Imsland F, Gu X, Feng C, Liu R, et al. The crest phenotype in chicken is associated with ectopic expression of HOXC8 in cranial skin. PLoS One. 2012;7:e34012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NHC, Zody MC, Andersson N, et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–8. [DOI] [PubMed] [Google Scholar]
- 70.Salmon Hillbertz NHC, Isaksson M, Karlsson EK, Hellmén E, Rosengren Pielberg G, Savolainen P, et al. A duplication of FGF3, FGF4, FGF19 and ORAOV1 causes hair ridge and predisposition to dermoid sinus in Ridgeback dogs. Nat Genet. 2007;38:1318–20. [DOI] [PubMed] [Google Scholar]
- 71.Baranowska I, Hultin Jäderlund K, Nennesmo I, Holmqvist E, Heidrich N, Larsson N-G, et al. Sensory ataxic neuropathy in golden retriever dogs is caused by a deletion in the mitochondrial tRNATyr gene. PLoS Genet 2009;5:e1000499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olsson M, Meadows JRS, Truvé K, Rosengren Pielberg G, Puppo F, Mauceli E, et al. A novel unstable duplication upstream of HAS2 predisposes to a breed-defining skin phenotype and a periodic fever syndrome in Chinese Shar-Pei dogs. PLoS Genet. 2011;7:e1001332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dorshorst B, Henegar C, Liao X, Sällman Almén M, Rubin C-J, Ito S, et al. Dominant red coat color in Holstein cattle is associated with a missense mutation in the coatomer protein complex, subunit alpha (COPA) gene. PLoS One. 2015;10:e0128969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yusnizar Y, Wilbe M, Herlino AO, Sumantri C, Rachman Noor R, Boediono A, et al. MITF mutations are associated with white spotted coat color in swamp buffalo. Anim Genet. 2015;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]