Abstract
The present study aimed to investigate elemene's effects on cell proliferation, apoptosis, and the cell cycle in the hepatocellular carcinoma (HCC) cell line, QYG7703, and to investigate GSTP1 gene methylation change in QGY7703 cells after being treated with elemene to explore whether elemene reversed the abnormal GSTP1 gene methylation. QGY7703 cells were treated with different elemene concentrations. Cell proliferation was measured with MTT assay, cell apoptosis and cell cycle were analyzed by flow cytometry, and GSTP1 gene methylation was analyzed by methlation-specific polymerase chain reaction. The cells' apoptotic rate increased dose-dependently with elemene concentration, and the difference was statistically significant (P<0.05). Elemene treatment arrested the cells in S phase, and thus the percentage of cells in G1 phase decreased while the cells in S phase increased dose-dependently, and the difference was statistically significant compared to the control group (P<0.05). All QGY7703 cells were identified to contain GSTP1 gene methylation before being treated with elemene and the methylation state decreased after treatment. In the present study, elemene induced cell apoptosis, inhibited the cell cycle, and reversed GSTP1 gene methylation in QGY7703 cells.
Keywords: elemene, QGY7703, cell apoptosis, methylation, GSTP1
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common type of cancer and the third cause of mortality induced by tumors in the world (1). China has the highest HCC prevalence in the world, and about 2/3 of all novel HCC cases occur there (2). Surgical resection is accepted as the preferred treatment method for early stage HCC, but a great proportion of patients are in the advanced stage when diagnosed and therefore miss the opportunity (3). Surgical treatment for these patients is associated with high recurrence and metastasis rates, so the 5-year survival rate is <50% (4). Currently, various anti-tumor agents including Sorafenib, Doxorubicin, cis-platinum and 5-fluorouracil have been used to treat HCC, which only increase patient survival by ~2-3 months and the severe toxic effects greatly affect patient's life quality (5,6) HCC's exact mechanisms remain unclear at present. Current knowledge suggests that HCC development is a multi-step process involving multiple oncogenes and tumor suppressor genes. Previous studies have demonstrated that abnormal tumor suppressor gene promoter methylation could inactivate the corresponding genes and thus promote tumor development and progression (7,8). The GSTP1 gene is a very important DNA damage repair gene that can inhibit the effects of cytotoxic agents and carcinogens and is regarded as a tumor suppressor gene (9). Previous studies have also demonstrated that hypermethylation of the CpG islands in the GSTP1 gene promoter is involved in the development and progression of multiple tumors, and thus could be used as a promising biomarker to help early screening, diagnosis, and patient prognosis (10–12).
DNA methylation of tumor suppressor genes is a reversible process. Reversing abnormal methylation to restore the normal corresponding gene expression has been accepted as a novel method to treat tumors (13,14). Methyltransferase inhibitors, including 5-aza-2-deoxycytidine (Aza), may reverse abnormal tumor suppressor gene methylation to restore gene expression and thus inhibit tumor growth. A number of clinical studies used 5-Aza to treat leukaemia and myelodysplastic syndrome (15). Although several previous studies demonstrated that nucleosides like 5-Aza could restore HCC-related gene expression and inhibit solid tumor cell growth, the severe side effects and lack of a safe and effective dose have restricted investigation in clinical studies (16). Identifying effective and less toxic demethylation agents is now a priority. Elemene is a traditional Chinese medicine (TCM) extracted from Curcuma zedoaria. As a non-cytotoxic anti-tumor agent, elemene has minor side effects but may inhibit tumor cell proliferation, induce apoptosis and differentiation, eliminate tumor cells, reverse multidrug resistance, and inhibit tumor metastasis, especially in HCC (17–19). Elemene's anti-tumor effects are very similar to tumor suppressor genes, but to the best of our knowledge, no previous study has investigated whether it could reverse tumor suppressor gene methylation and thus restore gene activity. In the present study, cultured HCC cell lines QGY7703 were treated with different elemene concentrations and the cell viability, apoptosis, and cell cycle were measured. GSTP1 gene methylation was also measured prior to and following treatment to investigate whether elemene inhibits or reverses the abnormal tumor suppressor gene methylation and the mechanisms involved in the anti-tumor effects.
Materials and methods
Materials
The HCC cell line QGY7703 was a gift from the cell bank of Suzhou University (Suzhou, China). Elemene injection was purchased from Hualijingang Pharmaceutical Co., LTD (Dalian, China), 5-Aza and MTT were purchased from Sigma-Aldrich (St. Louis, MO, USA), Annexin V-FITC apoptosis kits were purchased from Biyuntian Biotechnology Co., Ltd. (Shanghai, China), genomic DNA extraction kits were purchased from QIAGEN Biotechnology Co., Ltd. (Shanghai, China), EZ DNA Methylation-Direct Kit TM was purchased from ZYMO RESEARCH (USA), primers were purchased from Jierui Biotechnology Co., Ltd. (Shanghai, China), PCR Mixture 2xMix (BS-PCR002) was purchased from Bio-serve Company (Shanghai, China), and 100 bp DNA Marker was purchased from Takara Company (Japan).
Beckman Coulter Epics XL flow cytometer was from Beckman Coulter, Inc. (Brea, CA, USA), PCR Thermocycle Instrument (PTC200) was from MJ Research, Inc. (Waltham, MA, USA), Ultraviolet Spectrometry Photometer (NanoDrop2000) was from Thermo Fisher Scientific (Waltham, MA, USA), Gel Imaging System (JS-3000) was from Peiqing Technology Ltd. (Shanghai, China), and Multi-function Electrophoresis System (PowerBC-6002S1) was from Shennengbocai Biotechnology Co., Ltd. (Shanghai, China).
Cell proliferation inhibition
Cell proliferation inhibition induced by elemene was measured by MTT assay. In brief, QGY7703 cells in logarthmic growth phase were harvested and suspended, and the cell density was adjusted to 5×104/ml with plate count method (20). A total of 200 ml suspension was dispensed into each well of a 96-well plate and cultured for 24 h to allow adhesion. Then, the culture medium was discarded, elemene-containing culture medium was added, and the elemene concentration was adjusted to 20, 40, 80, and 160 µg/ml. For each concentration, 6 wells of cells were exposed. The final culture medium volume in each was not >200 µl. After the cells were cultured for 48 h, 20 µl MTT was added and incubated for 4 h, and then the supernatant was removed. A total of 150 µl DMSO was then added into each well, and the plate was oscillated on a shaker at low speed for 10 min to allow the crystals to dissolve. The absorbance (A) of each well was measured by ELISA Reader/Microplate Reader DR-200Bs (Huawen Machinery & Electronics Co., Wuxi, China). at 570 nm. Cell proliferation inhibition rate (%) was calculated as: (A in treatment group-A in control group)/(A in control group-A in blank control)x100.
QGY7703 cell apoptosis induced by elemene
QGY7703 cells were seeded into a 6-well plate at a density of 1×105 cells/ml, then complete DMEM was added until the final volume was 3 ml. Then, elemene-containing culture medium was added and the final elemene concentration was adjusted to 10, 20, or 30 µg/ml A blank control containing no elemene was also used. The cells were harvested, 50,000–100,000 suspended cells were centrifuged at 1,000 × g for 5 min, the supernatant was discarded, and 195 µl of Annexin V-FITC binding buffer was added to resuspend the cells (Annexin V-FITC Apoptosis Detection Kit C1062; Biyuntian Biotechnology Co., Ltd.). Then, 5 µl of Annexin V-FITC was added, and the cells were incubated at 20–25°C in the dark for 10 min. The cells were then centrifuged at 1000 × g for 5 min, the supernatant was discarded, and 190 µl of Annexin V-FITC binding buffer was added to resuspend the cells. Propidium iodide (PI) (10 µl; Annexin V-FITC Apoptosis Detection Kit C1062; Biyuntian Biotechnology Co., Ltd.) was added and mixed gently, and the cells were put on ice for 10 min in darkness. Cell apoptosis was measured using a Beckman Coulter Epics XL flow cytometer (Beckman Coulter, Inc., Brea, CA, USA) and FlowJo 7.6.3 software (FlowJo, LLC, Ashland, OR, USA).
Cell cycle measurement
QGY7703 cells were harvested and 50,000–100,000 suspended cells were centrifuged at 1000 × g for 5 min, the supernatant was discarded, and 70% ice-cold ethanol was used to fix the cells overnight (over 12 h) at 4°C. The cells were then centrifuged at 1000 × g for 5 min to remove the ethanol. After the cells were washed gently with precooled (4°C) PBS twice, 0.5 ml of precooled PBS was used to resuspend the cells. Then, 5 µl RNAse A (10 mg/ml, final concentration: 100 µg/ml; Qiagen19101 RNaseA; Qiagen GmbH, Hilden, Germany) was added to each well, and the cells were incubated at 37°C in darkness for 30 min, after which 50 µg/ml PI was added and incubated with the cells at 4°C in darkness for 30 min. The cell cycle was measured using a Beckman Coulter Epics XL flow cytometer. Each experiment was performed in triplicate.
GSTP1 gene methylation measurement
QGY7703 cells in the logarithmic growth phase were collected, and elemene was added to obtain a final concentration of 0, 40, 80, or 160 µg/m. DNA extracting kits (QIAGEN) were used according to instruction. Bisulfite conversion of the extracted DNA was then performed to convert the unmethylated C in CpG into U, while methylated C was not converted. Then, methylation specific polymerase chain reaction (MSP) was performed to measure DNA methylation. All these processes were performed in strict accordance with kit instructions.
MSP included the PCR of methylated primer sequences and unmethylated primer sequences. In the present study, primers were designed to amplify CpG rich regions using online software, MethPrimer (www.urogene.org/methprimer/). Two primer types, methylated (M) and unmethylated (U) primers, were used for the amplification. The primer sequences and PCR conditions are listed in Table I. The PCR conditions were as follows: pre-denaturation at 94°C for 3 min, denaturation at 94°C for 30 sec, annealing at 61°C for 30 sec, and extension at 72°C for 30 sec. After 40 cycles were completed, an additional extension at 72°C for 7 min was performed before the PCR was completed. A 30 µl reaction volume was used, which included PCR Mixture 2xMix 15 µl, U or M-Primer F 0.5 µl (10 µM), U or M-Primer R 0.5 µl (10 µM), Modified DNA 1–5 µl, and ddH2O. No DNA but ddH2O was added for blank control, while QGY7703 cells treated with 5-Aza were used as a positive control. Agarose gel electrophoresis (2%; catalog no., 111860; Biowest SAS, Nuaillé, France) was performed for all amplicons at 120 V for 30 min, and a gel image analysis system (JC-300; Shanghai Peiqing Science and Technology Co., Ltd, Shanghai, China) was used to analyze the results under ultraviolet excitation.
Table I.
Primers used for GSTP1 gene methylation-specific polymerase chain reaction.
| Gene | M/U | Sequences of the primers (5′-3′) | Annealing temperature | Product length |
|---|---|---|---|---|
| GSTP1 | M | (F)-TTCGGGGTGTAGCGGTCGTC | 61°C | 91 bp |
| (R)-GCCCCAATACTAAATCACGACG | ||||
| U | (F)-GATGTTTGGGGTGTAGTGGTTGTT | 55°C | 91 bp | |
| (R)-CCACCCCAATACTAAATCACAACA |
M, methylated; U, unmethylated.
Statistical analysis
SPSS software, version 15.0 (SPSS, Inc., Chicago, IL, USA) was used for the statistical analysis. Quantitative data with equal variances were described as means and standard divisions (SDs). Independent t-test was used to compare means between 2 groups, while one-way analysis of variances (one-way ANOVA) was used to compare means among 3 or more groups. P<0.05 was considered to indicate a statistically significant difference.
Results
QGY7703 proliferation inhibition by elemene
After being treated with elemene for 48 h, an MTT assay was used to evaluate adhesive QGY7703 cell viability. As shown in Table II and Fig. 1, elemene significantly inhibited the proliferation rate of QGY7703 cells, in a dose-dependent manner (P<0.01). The inhibitive effect increased with elemene concentration, and the IC50 of elemene was determined as 63±2.1 µg/ml by linear regression.
Table II.
Elemene's effects on QGY7703 cell proliferation (n=3, mean ± SD).
| Group (ug/ml) | A-value | Inhibition rate (%) |
|---|---|---|
| 0 | 0.47±0.038 | 0 |
| 20 | 0.46±0.035 | 3.2 |
| 40 | 0.33±0.030b | 30.3 |
| 80 | 0.10±0.111b | 78.4 |
| 160 | 0.01±0.007b | 96.9 |
All experiments were repeated 3 times, and similar results must be achieved. Each number in the table represents the mean value of these 3 experiments. Standard deviations were expressed as ±SD.
P<0.05
P<0.01, vs. control group.
Figure 1.
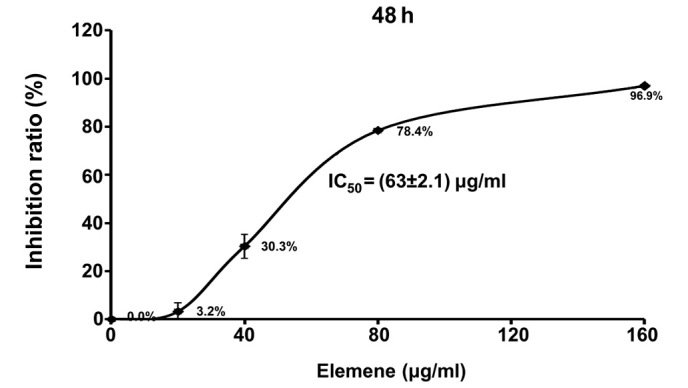
QGY7703 cell proliferation curve after elemene treatment.
Cell apoptosis
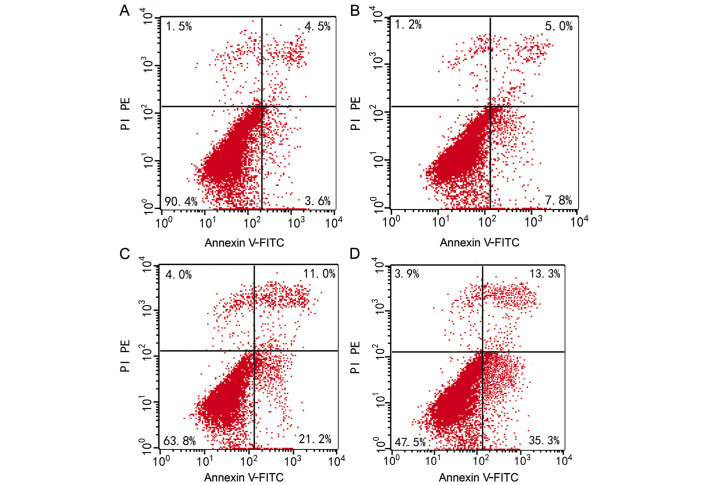
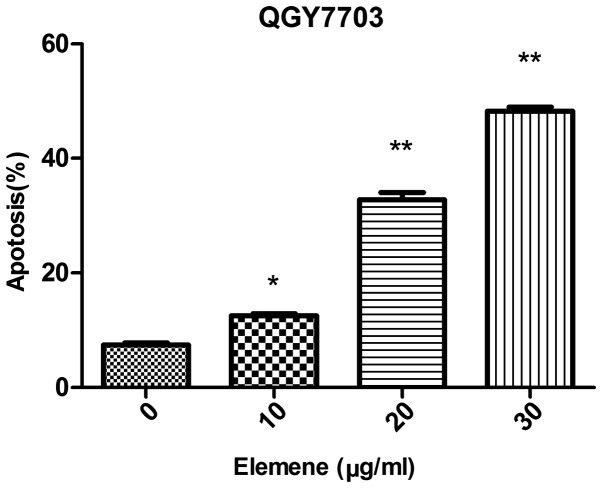
The apoptosis rate was 7.40±0.66% for QGY7703 cells prior to elemene treatment. Following treatment with different elemene concentrations, dose-dependent apoptosis rate increases were found. The apoptosis rate was 12.50±0.61%, 32.77±2.30%, and 48.27±1.23% for cells treated with 10, 20, and 30 µg/ml of elemene, respectively, and the difference was statistically significant when compared with the control group (10 µg/ml, P<0.05; 20 and 30 µg/ml, P<0.01) (Table III, Figs. 2 and 3).
Table III.
QGY7703 cell apoptosis rates (n=3, mean ± SD).
| Different concentrations of elemene (µg/ml) | ||||
|---|---|---|---|---|
| Apoptosis rate (%) | 0 | 10 | 20 | 30 |
| Early apoptosis rate | 3.37±0.17 | 7.37±0.37 | 21.60±1.10 | 36.20±0.79 |
| Necrosis and late apoptosis rate | 4.03±0.50 | 5.13±0.32 | 11.17±0.96 | 12.07±1.25 |
| Overall apoptosis rate | 7.40±0.66 | 12.50±0.61a | 32.77±2.30b | 48.27±1.23b |
All experiments were repeated 3 times, and similar results were achieved. Each number in the table represents the mean value of these 3 experiments. Standard deviations were expressed as ±SD. The overall apoptosis rate is significantly different between the control group and the elemene groups.
P<0.05 for 10 µg/ml vs. control group
P<0.01 for 20 and 30 µg/ml vs. control group.
Figure 2.
QGY7703 cell apoptosis rates after being treated with different elemene concentrations for 24 h. (A) Untreated QGY7703 cells; (B) cells treated with 10 µg/ml of elemene; (C) cells treated with 20 µg/ml of elemene; and (D) cells treated with 30 µg/ml of elemene. Normal cells are distributed in the lower left quadrant of the scatter diagram, while early apoptotic cells are distributed in the lower right quadrant, necrotic and late apoptotic cells are in the upper right quadrant, and detection error within the acceptable range is in the upper right quadrant.
Figure 3.
QGY7703 cell apoptosis rates after being treated with elemene for 24 h. *P<0.05 and **P<0.01 vs. control group.
QGY7703 cell cycles after elemene treatment
After being treated with 10, 20, and 30 µg/ml of elemene for 48 h (Table IV, Fig. 4), the cells in the G1 phase reduced gradually with elemene concentration, while cells in the S phase increased gradually, indicating elemene arrested the cells in S phase. The differences between the treated groups and control group were statistically significant (P<0.01).
Table IV.
QGY7703 cell cycle after being treated with elemene for 48 h (n=3, mean ± SD).
| Different concentrations of elemene (ug/ml) | ||||
|---|---|---|---|---|
| Cell cycle | 0 | 10 | 20 | 30 |
| G2/M | 8.39±0.08 | 9.45±0.08 | 8.77±0.73 | 8.57±0.49 |
| S | 27.04±0.45 | 35.05±0.32 | 36.57±0.57 | 38.43±0.60 |
| G1 | 65.56±0.41 | 55.53±0.13 | 54.66±0.33 | 53.00±0.15 |
Figure 4.
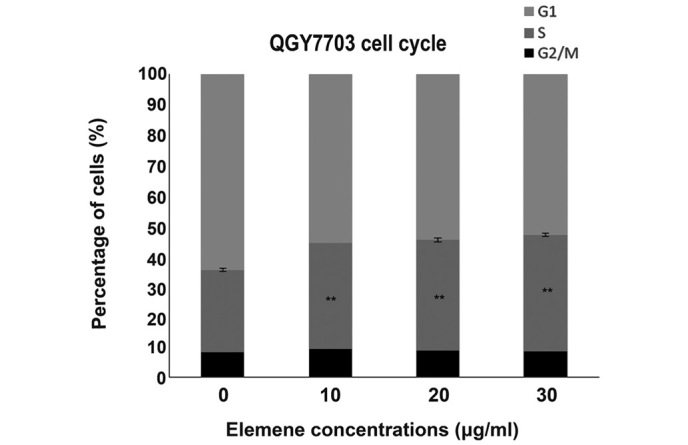
QGY7703 cell cycle after being treated with different elemene concentrations. **P<0.01 vs. control group.
GSTP1 gene methylation in QGY7703 cells
All GSTP1 genes in untreated QGY7703 cells were found to be methylated, but after being treated with 40 µg/ml or 160 µg/ml of elemene or 10 µmol/l of 5-Aza-dc, unmethylated GSTP1 genes were observed (Fig. 5, black arrow).
Figure 5.
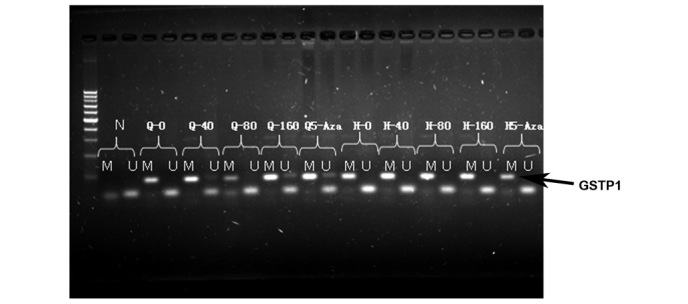
GSTP1 promoter methylation in the QGY7703 cells following treatment with various elemene concentrations. The black arrow shows the methylated bands. N, negative control; M, methylated; U, unmethylated; Q-0, untreated cells; Q-40, cells treated with 40 µg/ml of elemene; Q-80, cells treated with 80 µg/ml of elemene; Q-160, cells treated with 160 µg/ml of elemene; and Q5-Aza, cells treated with 10 µmol/l of 5-Aza, which was used as the positive control.
Discussion
Elemene injection with a major β-elemene component was approved as a second line of anticancer drug for clinical use in China in 1995 (21). The major characteristics that distinguish elemene, an effective TCM extraction, from other anticancer drugs is that its cytotoxic effects against non-cancerous normal cells are minimal (21). Previous studies have shown that elemene is effective for liver (22), ovarian (23), gastric (24), pulmonary (25), and breast (26) cancers and lymphosarcoma, particularly for patients with malignant pleural effusion and ascites (27). Elemene's major anti-tumor mechanisms include tumor cell proliferation inhibition, tumor cell elimination, cell apoptosis and differentiation induction, multidrug resistance reversion, tumor metastasis inhibition, and immunity up-regulation (17–19). Recently, several studies investigated elemene's anti-tumor mechanisms at the molecular and gene levels (28,29). However, to the best of our knowledge, no previous study has investigated whether elemene reverses tumor suppressor gene methylation and thus restores gene activity.
In the present study, an MTT assay was used to investigate the effects of different elemene concentrations on QGY7703 cell proliferation. After being treated with 20, 40, 80, and 160 µg/ml of elemene for 48 h, the results showed that elemene could significantly inhibit QGY7703 cell proliferation. Further analysis showed that the inhibition was dose-dependent, and the differences between each treatment group and the control group were statistically significant (P<0.05). The IC50 was 63 µg/ml. Flow cytometry was used to investigate elemene's effects on the cell cycle, and it was determined that elemene could effectively inhibit cells entering the G2/M phase, in accordance with the findings of Yang et al (30). In Yang's study, elemene could inhibit HL-60 and K562 cell lines entering the G2/M phase. The cell cycle arrest may be associated with intracellular free calcium ion concentration changes, immunoprophylatic effects, and P53 and Bcl-2 inhibition. In a study by Lee et al (23), the authors found that elemene could significantly inhibit the A2780 cell line growth and arrest cells in the G2 phase, which could be associated with the down-regulation of cyclin-dependent kinases (including CDC2, cyclin A, and cyclin B1). Another study also showed that elemene could inhibit malignant glioma cell line entering G1 phase from G0 phase (31). These findings showed that elemene could inhibit the proliferation of multiple tumor cells. However, the cells were found arrested in different phases, which could result from the different mechanisms involved in different tumor cells.
The present study also investigated elemene's effects on cell apoptosis in QGY7703 cells, and the results showed that it could significantly induce and may promote early apoptosis, and the effects were dose-dependent. Dai et al (32) treated HepG2 cells with elemene and found that it could significantly inhibit cell proliferation, promote cell apoptosis, and upregulate Fas/Fasl protein expression, and thus supposed that the apoptosis induced by elemene could be associated with Fas/Fasl. Other studies have also found that elemene could induce tumor cell apoptosis in pulmonary cancer, laryngeal cancer, leukemia, and glioma (33–36). The mechanisms involved in the apoptosis induction effects could be as follows: Influencing the expression of oncogenes and tumor suppressor genes, influencing MAPK/ERK and PI3K/Akt/mTOR signaling pathways, activating Caspase cascade, inducing mitochondrial damages, inducing oxidative damages, inhibiting telomerase activity, and altering intracellular Ca2+ concentration. These findings indicated that the pathway involved in elemene's apoptosis induction effects in different tumor cells could be different. Elemene may induce cell apoptosis by regulating various signaling pathways.
The present study further investigated GSTP1 gene methylation in QGY7703 cells treated with different elemene concentrations and compared the results with the untreated cells. The results demonstrated that all the GSTP1 genes in the untreated QGY7703 cells were methylated. However, after treatment with elemene, unmethylated GSTP1 genes were found in the QGY7703 cells. The GSTP1 gene is located at q13 of human chromosome 11 and encoded an enzyme with detoxicating and protein-binding effects (37). The GSTP1 protein's main function is to catalyze the reactions between glutathione and electron-containing compounds, which could help metabolize carcinogens and exogenous drugs into low- or non-toxic metabolites and thus exert anti-cytotoxic and anti-carcinogen effects (37). Several previous studies showed that GSTP1 inactivation induced by hypermethylation is mainly found in several human tumors, including prostate, renal, breast, and liver cancers (10–12). Tchou et al (38) found that GSTP1 in HCC tissues and cell lines were hypermethylated, with the rate of methylation of 85%. Additionally, GSTP1 protein levels reduced significantly, and its absence was found in 90% of the tissues or cells. In our previous studies, the GSTP1 gene methylation was investigated in 35 liver cancer tissues and adjacent tissues, as well as in 20 normal liver tissues (Wu et al, unpublished data). In that study, the positive methylation rate was 57.1% in liver cancer tissues, which was significantly higher than in the adjacent tissues (25.7%, P<0.01). However, no methylation was observed in normal liver tissues, suggesting that GSTP1 expression is highest in normal liver tissues and lowest in liver cancer tissues. In the present study, GSTP1 in the HCC cell line was completely methylated (100%), which further confirmed that GSTP1 methylation could be involved in HCC's development and progression. GSTP1 could also be used as a promising molecular biomarker of great clinical significance in helping early HCC screening and diagnosis. Unmethylated GSTP1 were demonstrated in elemene-treated QGY7703 cells, suggesting that elemene could reverse tumor suppressor gene methylation. However, further studies are needed to investigate the exact mechanisms involved. In summary, the present study provides insight into elemene's anti-tumor mechanisms and provided a novel method to identify novel demethylation drugs in TCM to treat tumors.
References
- 1.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 2.Bharadwaj M, Roy G, Dutta K, Misbah M, Husain M, Hussain S. Tackling hepatitis B virus-associated hepatocellular carcinoma-the future is now. Cancer Metastasis Rev. 2013;32:229–268. doi: 10.1007/s10555-012-9412-6. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;65:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 4.Bruix J, Llovet JM. Major achievements in hepatocellular carcinoma. Lancet. 2009;373:614–616. doi: 10.1016/S0140-6736(09)60381-0. [DOI] [PubMed] [Google Scholar]
- 5.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 6.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, Feng J, Ye S, Yang TS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: A phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 7.Choi JD, Lee JS. Interplay between epigenetics and genetics in cancer. Genomics Inform. 2013;11:164–173. doi: 10.5808/GI.2013.11.4.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rongrui L, Na H, Zongfang L, Fanpu J, Shiwen J. Epigenetic mechanism involved in the HBV/HCV-related hepatocellular carcinoma tumorigenesis. Curr Pharm Des. 2014;20:1715–1725. doi: 10.2174/13816128113199990533. [DOI] [PubMed] [Google Scholar]
- 9.Sato K, Satoh K, Tsuchida S, Hatayama I, Shen H, Yokoyama Y, Yamada Y, Tamai K. Specific expression of glutathione S-transferase Pi forms in (pre) neoplastic tissues: Their properties and functions. Tohoku J Exp Med. 1992;168:97–103. doi: 10.1620/tjem.168.97. [DOI] [PubMed] [Google Scholar]
- 10.Fukushige S, Horii A. DNA methylation in cancer: A gene silencing mechanism and the clinical potential of its biomarkers. Tohoku J Exp Med. 2013;229:173–185. doi: 10.1620/tjem.229.173. [DOI] [PubMed] [Google Scholar]
- 11.Hessels D, Schalken JA. Urinary biomarkers for prostate cancer: A review. Asian J Androl. 2013;15:333–339. doi: 10.1038/aja.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White DL, Li D, Nurgalieva Z, El-Serag HB. Genetic variants of glutathione S-transferase as possible risk factors for hepatocellular carcinoma: A HuGE systematic review and meta-analysis. Am J Epidemiol. 2008;167:377–389. doi: 10.1093/aje/kwm315. [DOI] [PubMed] [Google Scholar]
- 13.Yoo CB, Jones PA. Epigenetic therapy of cancer: Past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 14.Saito Y, Hibino S, Saito H. Alterations of epigenetics and microRNA in hepatocellular carcinoma. Hepatol Res. 2014;44:31–42. doi: 10.1111/hepr.12147. [DOI] [PubMed] [Google Scholar]
- 15.Borthakur G, El Ahdab SE, Ravandi F, Faderl S, Ferrajoli A, Newman B, Issa JP, Kantarjian H. Activity of decitabine in patients with myelodysplastic syndrome previously treated with azacitidine. Leuk Lymphoma. 2008;49:690–695. doi: 10.1080/10428190701882146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karahoca M, Momparler RL. Pharmacokinetic and pharmacodynamic analysis of 5-aza-2′-deoxycytidine (decitabine) in the design of its dose-schedule for cancer therapy. Clin Epigenetics. 2013;5:3. doi: 10.1186/1868-7083-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Mao Y, Hou L, Cui X. The effect of beta-elemene on alpha-tubulin polymerization in human hepatoma HepG2 cells. Chin J Cancer Res. 2013;25:770. doi: 10.3978/j.issn.1000-9604.2013.12.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng X, Zhao Y, Liang X, Wu L, Cui S, Guo A, Wang W. Assessing the quality of RCTs on the effect of beta-elemene, one ingredient of a Chinese herb, against malignant tumors. Contemp Clin Trials. 2006;27:70–82. doi: 10.1016/j.cct.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y, Liu G, Zhang Y, Zhu H, Ren Y, Shen YM. Synthesis and in vitro anti-proliferative activity of beta-elemene monosubstituted derivatives in HeLa cells mediated through arrest of cell cycle at the G1 phase. Bioorg Med Chem. 2009;17:1118–1124. doi: 10.1016/j.bmc.2008.12.040. [DOI] [PubMed] [Google Scholar]
- 20.Bohari SP, Hukins DW, Grover LM. Effect of calcium alginate concentration on viability and proliferation of encapsulated fibroblasts. Biomed Mater Eng. 2011;21:159–170. doi: 10.3233/BME-2011-0665. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Zhang H, Sun Y. Phase III clinical trial of elemenum emulsion in the management of malignant pleural and peritoneal effusions. Zhonghua Zhong Liu Za Zhi. 1996;18:464–467. (In Chinese) [PubMed] [Google Scholar]
- 22.Mao Y, Zhang J, Hou L, Cui X. The effect of beta-elemene on alpha-tubulin polymerization in human hepatoma HepG2 cells. Chin J Cancer Res. 2013;25:770–776. doi: 10.3978/j.issn.1000-9604.2013.12.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee RX, Li QQ, Reed E. β-elemene effectively suppresses the growth and survival of both platinum-sensitive and -resistant ovarian tumor cells. Anticancer Res. 2012;32:3103–3113. [PMC free article] [PubMed] [Google Scholar]
- 24.Liu JS, He SC, Zhang ZL, Chen R, Fan L, Qiu GL, Chang S, Li L, Che XM. Anticancer effects of β-elemene in gastric cancer cells and its potential underlying proteins: A proteomic study. Oncol Rep. 2014;32:2635–2647. doi: 10.3892/or.2014.3490. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Chen YJ, Wu MD. Herbal extract elemene intrathoracic injection in the treatment of lung cancer patients with malignant pleural effusion: A meta-anaylsis. J Cancer Res Ther. 2014;10(Suppl 1):56–59. doi: 10.4103/0973-1482.139761. [DOI] [PubMed] [Google Scholar]
- 26.Guan C, Liu W, Yue Y, Jin H, Wang X, Wang XJ. Inhibitory effect of β-elemene on human breast cancer cells. Int J Clin Exp Pathol. 2014;7:3948–3956. [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang ZY, Qin SK, Yin XJ, Chen YL, Zhu L. Synergistic effects of Endostar combined with β-elemene on malignant ascites in a mouse model. Exp Ther Med. 2012;4:277–284. doi: 10.3892/etm.2012.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi H, Liu L, Liu L, Geng J, Zhou Y, Chen L. β-Elemene inhibits the metastasis of B16F10 melanoma cells by downregulation of the expression of uPA, uPAR, MMP-2 and MMP-9. Melanoma Res. 2014;24:99–107. doi: 10.1097/CMR.0000000000000043. [DOI] [PubMed] [Google Scholar]
- 29.Guo HQ, Zhang GN, Wang YJ, Zhang YK, Sodani K, Talele TT, Ashby CR, Jr, Chen ZS. β-Elemene, a compound derived from Rhizoma zedoariae, reverses multidrug resistance mediated by the ABCB1 transporter. Oncol Rep. 2014;31:858–866. doi: 10.3892/or.2013.2870. [DOI] [PubMed] [Google Scholar]
- 30.Yang H, Wang X, Yu L. The antitumor activity of elemene is associated with apoptosis. Zhonghua Zhong Liu Za Zhi. 1996;18:169–172. (In Chinese) [PubMed] [Google Scholar]
- 31.Zhu T, Zhao Y, Zhang J, Li L, Zou L, Yao Y, Xu Y. β-Elemene inhibits proliferation of human glioblastoma cells and causes cell-cycle G0/G1 arrest via mutually compensatory activation of MKK3 and MKK6. Int J Oncol. 2011;38:419–426. doi: 10.3892/ijo.2010.855. [DOI] [PubMed] [Google Scholar]
- 32.Dai ZJ, Tang W, Lu WF, Gao J, Kang HF, Ma XB, Min WL, Wang XJ, Wu WY. Antiproliferative and apoptotic effects of β-elemene on human hepatoma HepG2 cells. Cancer Cell Int. 2013;13:27. doi: 10.1186/1475-2867-13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang XS, Yang W, Tao SJ, Li K, Li M, Dong JH, Wang MW. Effect of delta-elemene on Hela cell lines by apoptosis induction. Yakugaku Zasshi. 2006;126:979–990. doi: 10.1248/yakushi.126.979. [DOI] [PubMed] [Google Scholar]
- 34.Zheng CP, Tong XM, Yao HP, Yang J, Xu J, Cai XP, Liu Z. Beta-elemene enhances aclarubicin-induced apoptotic effect in HL-60 cells and its mechanism. Zhonghua Xue Ye Xue Za Zhi. 2009;30:821–824. (In Chinese) [PubMed] [Google Scholar]
- 35.Zhao YS, Zhu TZ, Chen YW, Yao YQ, Wu CM, Wei ZQ, Wang W, Xu YH. B-elemene inhibits Hsp90/Raf-1 molecular complex inducing apoptosis of glioblastoma cells. J Neurooncol. 2012;107:307–314. doi: 10.1007/s11060-011-0770-7. [DOI] [PubMed] [Google Scholar]
- 36.Yao CC, Tu YR, Jiang J, Ye SF, Du HX, Zhang Y. β-elemene reverses the drug resistance of lung cancer A549/DDP cells via the mitochondrial apoptosis pathway. Oncol Rep. 2014;31:2131–2138. doi: 10.3892/or.2014.3083. [DOI] [PubMed] [Google Scholar]
- 37.Rezaei MK, Shobbar ZS, Shahbazi M, Abedini R, Zare S. Glutathione S-transferase (GST) family in barley: identification of members, enzyme activity, and gene expression pattern. J Plant Physiol. 2013;170:1277–1284. doi: 10.1016/j.jplph.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Tchou JC, Lin X, Freije D, Isaacs WB, Brooks JD, Rashid A, De Marzo AM, Kanai Y, Hirohashi S, Nelson WG. GSTP1 CpG island DNA hypermethylation in hepatocellular carcinomas. Int J Oncol. 2000;16:663–676. doi: 10.3892/ijo.16.4.663. [DOI] [PubMed] [Google Scholar]