Abstract
Non-small cell lung cancer (NSCLC), caused by various mutations in a spectrum of cancer driver genes, may have distinct pathological characteristics and drug responses. Extensive genetic screening and pathological characterization is required for the design of customized therapies to improve patient outcomes. Notably, NSCLC in never-smokers exhibits distinctive clinicopathological features, which are frequently associated with tumorigenic mutations, and thus may be treated as a unique disease entity. However, to the best of our knowledge, these mutations have not been extensively and accurately characterized in an NSCLC study with a large sample size. Therefore, the present study enrolled a large cohort of NSCLC patients, which consisted of 358 never-smokers, for the screening of genetic alterations in the epidermal growth factor receptor (EGFR), ret proto-oncogene (RET), anaplastic lymphoma kinase (ALK), Kirsten rat sarcoma viral oncogene homolog (KRAS) and B-Raf proto-oncogene serine/threonine kinase (BRAF) tumorigenic genes. It was identified that the mutation rate was 47.8, 7.5, 3.6, 1.4 and 0.3% for EGFR, ALK, KRAS, RET and BRAF, respectively. In addition, clinicopathological features associated with these mutations were characterized. EGFR mutations were more frequently observed in female and older patients. By contrast, KRAS mutations were more frequently detected in male patients, and ALK and RET translocations in younger patients. The cancer cells were frequently well-differentiated in carcinoma cases exhibiting EGFR mutations, however, were less differentiated in those with ALK translocations. In conclusion, the present study determined the frequency of oncogenic alterations and associated clinicopathological features in NSCLC exhibited by never-smokers using a large sample size. The results of the present study may enrich our knowledge of NSCLC in never-smokers and provide useful insights for improvement of the outcome of molecularly targeted therapies for the treatment of NSCLC.
Keywords: lung cancer, EGFR, ALK, RET, KRAS, BRAF, never-smokers
Introduction
Lung cancer is the number one cause of cancer-associated mortality (1). The high mortality rates associated with lung cancer are largely due to the poor outcomes of conventional treatments, including the use of surgical removal combined with adjuvant radiation and chemotherapy (2). Significant improvements have been achieved due to increased efforts to determine the molecular mechanisms underlying tumorigenesis, which has led to the identification of multiple oncogenic alterations, including those observed in epidermal growth factor receptor (EGFR) (3), Kirsten rat sarcoma viral oncogene homolog (KRAS) (4), B-Raf proto-oncogene, serine/threonine kinase (BRAF) (5), anaplastic lymphoma kinase (ALK) (6), ROS proto-oncogene 1 (7) and ret proto-oncogene (RET) (7–9). It was previously demonstrated that patients carrying EGFR mutations exhibited a significant response to the EGFR tyrosine kinase inhibitors (TKIs) gefitinib and erlotinib as a first-line therapy (3,10). By contrast, patients carrying ALK fusions exhibited a poor response to these drugs (11), but responded well to the ALK TKI crizotinib (12). Thus, targeted treatment based on the results of molecular and pathological diagnosis has become a new standard for the treatment of lung cancer (13).
Although the majority of lung cancer cases are associated with an extensive history of cigarette smoking, the prevalence of lung cancer death in non-smokers remains high (14). In the United States, 10–15% of lung cancer cases are diagnosed in patients who are considered never-smokers (15). If listed as a separate category, lung cancer in never-smokers would rank among the top 10 most commonly observed fatal cancer cases in the United States (14,16). This ranking in never-smokers is likely to rise due to increased public awareness of the life-threatening hazards caused by cigarette smoking, resulting in a drop in the population of smokers and thus an increase in the population of never-smokers (17).
A previous clinical study demonstrated that targeted therapy in never-smoker lung cancer patients typically produces an improved response compared with that in smokers (18). It has been suggested that the molecular profiles of lung cancer cases are likely to vary between heavy smokers and never-smokers. Accumulating evidence based on molecular and clinicopathological studies has suggested that non-small cell lung cancer (NSCLC) in never-smokers should be considered as a distinct entity (19). Thus, it is critical to determine the mutation state of NSCLC in never-smokers as a unique type of cancer, for the purpose of cancer research and clinical translation. With this aim in mind, the present study performed a large-scale screen for tumorigenic alterations in the oncogenes EGFR, KRAS, BRAF ALK and RET in 358 Chinese NSCLC adenocarcinoma patients who were exclusively never-smokers. The clinicopathological characteristics associated with these genetic alterations were additionally determined. The present study may yield a clear picture concerning the molecular profile of NSCLC in never-smokers, thus providing valuable information for cancer research and the improvement of targeted therapies for the treatment of NSCLC.
Materials and methods
Specimen collection
The present study was approved by the Institutional Review Boards of Shanghai Chest Hospital, Shanghai Jiao Tong University (Shanghai, China), and Chongqing Cancer Institute (Chongqing, China). All participants underwent lung resection and needle aspiration, and provided written informed consent. Samples were snap-frozen with liquid nitrogen at the time of resection and stored at −80°C until required. All cases were independently reviewed by two pathologists during disease diagnosis. Patients were considered never-smokers if they had never smoked or had smoked <100 cigarettes in their lifetime (15).
Detection of mutations in EGFR, KRAS and BRAF
Genomic DNA was extracted with the QIAamp DNA formalin-fixed paraffin-embedded (FFPE) kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's protocols. EGFR, KRAS and BRAF mutations were detected by amplification refractory mutation system in multiple quantitative polymerase chain reaction (ARMS-multi-qPCR) analysis with the Human EGFR Mutation Detection kit (YuanQi Bio-Pharmaceutical Co., Ltd., Shanghai, China) and the Human KRAS and BRAF Mutation Detection kit (YuanQi Bio-Pharmaceutical Co., Ltd.), respectively. The PCR conditions used were as follows: 42°C for 5 min, 94°C for 3 min, followed by 40 cycles at 94°C for 15 sec and 60°C for 1 min on the 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The following primers were used: EGFR-exon (E)18 forward, 5′-CAAGTGCCGTGTCCTGG-3′ and reverse, 5′-CCTTACCTTATACACCGTGCC-3′; EGFR-E19 forward, 5′-CGGTGCATCGCTGGTAACAT-3′ and reverse, 5′-ATGGACCCCCACACAGC-3′; EGFR-E20 forward, 5′-CTGGCCACCATGCGAAG-3′ and reverse, 5′-TCCTGGCTCCTTATCTCCC-3′; EGFR-E21 forward, 5′-GCTTCTTCCCATGATGATCTG-3′ and reverse, 5′-CTGGTCCCTGGTGTCAGG-3′; KRAS forward, 5′-TTTGTATTAAAAGGTACTGGTGG-3′ and reverse, 5′-CCTCTATTGTTGGATCATATTCG-3′; and BRAF forward, 5′-ACTCTTCATAATGCTTGCTCTG-3′ and reverse, 5′-TGAATACTGGGAACTATGAAAATAC-3′. All PCR products were subjected to direct sequencing to verify mutations in EGFR, KRAS and BRAF. The following probes were used: for EGFR-E18, 5′-GGTGACCCTTGTCTCTGTGTTC-3′; EGFR-E19, 5′-ATCACTGGGCAGCATGTG-3′; EGFR-E20, 5′-CCCTGATTACCTTTGCGAT-3′; EGFR-E21, 5′-TGATCTGTCCCTCACAGCAG-3′; KRAS, 5′-TGTATTAAAAGGTACTGGTGGAG-3′; and BRAF 5′-TGAGACCTTCAATGACTTTCTAG-3′. All primers and probes were purchased from Sangon Biotech Co., Ltd., Shanghai, China.
Detection of ALK and RET fusion variants
Multiplex one-step reverse transcription (RT)-PCR was performed to detect ALK fusion gene variants. The Human Lung Cancer Related Fusion Gene Detection kit (YuanQi Bio-Pharmaceutical Co., Ltd.) was used according to the manufacturer's protocols. In brief, the mixture for each reaction contained 3 µl total RNA extracted from the tumor specimen, 12 µl Multiplex RT-PCR buffer, 2.5 µl Multiplex Enzyme mix and 300 nmol/l primers in a total volume of 25 µl. The PCR conditions used were as follows: 42°C for 30 min, 94°C for 5 min, followed by 40 cycles at 94°C for 15 sec and 60°C for 1 min on the 7500 Real Time PCR System. A total of two experiments were performed separately to detect echinoderm microtubule-associated protein-like 4 (EML4)-ALK fusion or alternative ALK fusions [transforming growth factor (TGF)-ALK, kinesin light chain 1 (KLC1)-ALK and kinesin family member 5B (KIF5B)-ALK]. The following forward primers were used for detecting EML4-ALK variants: EML4-E2 (V5a and 5b) forward, 5′-GTGGCCTCAGTGAAAAAATC-3′; EML4-E6 (V3a and 3b) forward, 5′-TAAAGATGTCATCATCAACCAAG-3′; EML4-E13 (V1 and 6) forward, 5′-CCTGGGAAAGGACCTAAAG-3′; EML4-E14 (V4b and 7) forward, 5′-GGGAAAGGACCTAAAGGTG-3′; EML4-E15 (V4a) forward, 5′-TGATGGCTTCCAAATAGAAGTAC-3′; EML4-E17 (V9) forward, 5′-ACGGGAATGAACAGCTCTCT-3′; and EML4-E20 (V2) forward, 5′-CGGGAGACTATGAAATATTGTACT-3′. The primers for alternative ALK fusion variants included: TGF-E3 forward, 5′-GAGAACCAGGACCTTCCACC-3′; KLC1-E9 forward, 5′-ATTCTCACTCGTGCACATGAAA-3′; KIF5B-E15 forward, 5′-AAAAGACCTTGCAGAAATAGGAA-3′; KIF5B-E17 forward, 5′-TCTGTCGATGCCCTCAGTG-3′; and KIF5B-E24 forward, 5′-TCAGGTCAAAGAATATGGCCA-3′. The common reverse primer for all ALK fusion variants was 5′-GCTTGTACTCAGGGCTCTGC-3′. Multiplex One-step RT-PCR was additionally used to detect RET fusion variants, including KIF5B-RET and coiled-coil domain containing 6 (CCDC6)-RET. RET Fusion Gene Detection kit (YuanQi Bio-Pharmaceutical Co., Ltd.) was used according to the manufacturer's protocols. All PCR products were subjected to DNA sequencing with the probe 5′-AGCTCCTGGTGCTTCCGGCG-3′ for all ALK fusion products. The expression of ALK tyrosine kinase was examined by immunohistochemistry using the ALK (D5F3) CDx Assay kit (Ventana Medical Systems, Inc., Tucson, AZ, USA) containing the rabbit monoclonal antibody against ALK (clone D5F3; catalog no., 790–4796; dilution, 1:250), which detected endogenous levels of total ALK protein, as well as ALK fusion proteins. The experiments were performed on FFPE sections, as described previously (20).
Statistical analysis
P-values were determined by Fisher's exact test or χ2 test using Prism 6 analysis software (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 and P<0.01 were considered to indicate a statistically significant and highly significant difference, respectively.
Results
Clinical characteristics of enrolled patients
Between January 2012 and June 2013, resected lung adenocarcinoma samples were collected from a total of 358 patients who had been diagnosed with NSCLC at Shanghai Chest Hospital or Chongqing Cancer Institute. The cohort consisted of 274 female patients (76.5%) and 84 male patients (23.5%) (Table I). The median age of the patient cohort was 57.1 years. While ~36.3% of patients were 50–59 years old, and 41.6% were ≥60 years, only 22.1% of patients were <50 years old (Table I). All specimens were selected based on the following criteria: i) Re-review confirmed a pathological diagnosis of lung adenocarcinoma; ii) the tumor specimen contained a minimum of 70% tumor cells; iii) sufficient tissue was available for comprehensive analysis; iv) the patient was a never-smoker; and v) the patient did not receive any neoadjuvant treatment. Based on the differentiation level of cancer cells, the specimens could be divided into three groups, which comprised poorly-, moderately- or well-differentiated carcinoma. The number of specimens was similar among these groups, ranging from 32.1 to 35.8% (Table I).
Table I.
Clinical characteristics of 358 never-smokers with non-small cell lung cancer.
| Variable | Patients, n (%) |
|---|---|
| Gender | |
| Male | 84 (23.5) |
| Female | 274 (76.5) |
| Age, years | |
| <40 | 5 (1.4) |
| 40–49 | 74 (20.7) |
| 50–59 | 130 (36.3) |
| ≥60 | 149 (41.6) |
| Differentiation | |
| Poorly | 115 (32.1) |
| Moderately | 128 (35.8) |
| Well | 115 (32.1) |
Mutations were detected in EGFR, KRAS, BRAF, EML4-ALK and KIF5B-RET
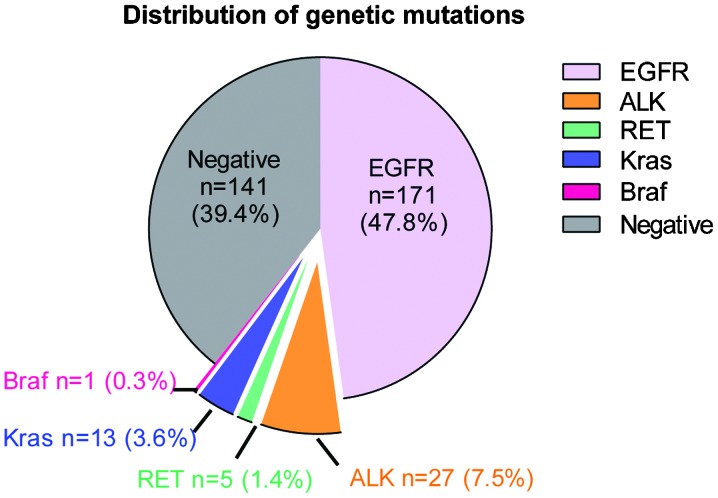
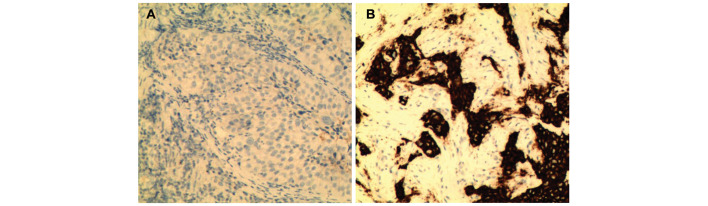
Out of a total of 358 NSCLC patents, genetic alterations were detected in 217 carcinoma specimens. Among these positive cases, there were 171 patients carrying EGFR mutations, accounting for 47.8% of all patients (Fig. 1). A total of 27 patients were detected as exhibiting EML4-ALK fusion genes, 13 with KRAS mutations, 5 with KIF5B-RET fusion variants and 1 with BRAF mutations, accounting for 7.5, 3.6, 1.4 and 0.3%, respectively (Fig. 1). No KIF5B-ALK or TFG-ALK fusion variants were detected in the samples. These results were confirmed by immunohistochemistry. Representative images revealed that ALK was absent in EML4-ALK-negative carcinoma cases (Fig. 2A), but was highly expressed in EML4-ALK-positive samples (Fig. 2B). None of the specimens carried mutations in >1 gene. However, there was one patient (>60 years old) who carried two EGFR mutations (E19 and E21) in their tumor specimen. These results supported the observation that mutations in the investigated tumorigenic genes are typically mutually exclusive (21).
Figure 1.
Pie graph showing the frequency of genetic alterations in never-smokers with NSCLC. The frequency of genetic mutations or alterations in EGFR, ALK, RET, KRAS and BRAF was determined in 358 never-smokers with NSCLC. The number and percentage of carcinoma cases harboring each of these genetic mutations is indicated in the graph. EGFR, epidermal growth factor receptor; ALK, anaplastic lymphoma kinase; RET, ret proto-oncogene; KRAS, Kirsten rat sarcoma viral oncogene homolog; BRAF, B-Raf proto-oncogene, serine/threonine kinase.
Figure 2.
Representative images showing ALK expression in lung cancer. Immunohistochemistry was performed using an antibody against ALK on formalin-fixed paraffin-embedded tissue sections collected from non-small cell lung cancer patients who were never-smokers. Hematoxylin and eosin staining was additionally performed on the sections. (A) ALK protein was absent in EML4-ALK-negative carcinoma. (B) ALK protein was aberrantly expressed in EML4-ALK-positive carcinoma. ALK, anaplastic lymphoma kinase; EML4, echinoderm microtubule-associated protein-like 4.
EGFR mutations were most prevalent in E19, with 102 cases, accounting for 28.5% of all patients and 59.6% of patients exhibiting EGFR mutations (Table II). EGFR mutations were additionally identified frequently in E21, with 63 cases, accounting for 17.6% of all patients and 36.8% of patients with EGFR mutations (Table II). Markedly fewer cases were detected with mutations in E18 (3 cases; 0.8% of all patients) or E20 (4 cases; 1.1% of all patients), accounting for <5% of combined patients exhibiting EGFR mutations (Table II). Detailed data is listed in Table II according to gender, age, differentiation and histology.
Table II.
Characterization of EGFR mutations in 358 Chinese never-smokers exhibiting non-small cell lung cancer.
| All EGFR | E18 | E19 | E20 | E21 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variable | n | % | n | % | n | % | n | % | n | % |
| Gender | ||||||||||
| Male | 30 | 35.7 | 0 | 0.0 | 18 | 21.4 | 0 | 0.0 | 12 | 14.3 |
| Female | 141 | 51.5 | 3 | 1.1 | 84a | 30.7 | 4 | 1.5 | 51a | 18.6 |
| Age, years | ||||||||||
| <40 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| 40–49 | 32 | 43.2 | 1 | 1.4 | 19 | 25.7 | 1 | 1.4 | 11 | 14.9 |
| 50–59 | 60 | 46.2 | 1 | 0.8 | 38 | 29.2 | 2 | 1.5 | 19 | 14.6 |
| ≥60 | 79 | 53.0 | 1 | 0.7 | 45a | 30.2 | 1 | 0.7 | 33a | 22.1 |
| Diff | ||||||||||
| Poorly | 56 | 48.7 | 2 | 1.7 | 36 | 31.3 | 1 | 0.9 | 17 | 14.8 |
| Mod | 51 | 39.8 | 1 | 0.8 | 28a | 21.9 | 2 | 1.6 | 21a | 16.4 |
| Well | 64 | 55.7 | 0 | 0.0 | 38 | 33.0 | 1 | 0.9 | 25 | 21.7 |
| Total | 171 | 47.8 | 3 | 0.8 | 102a | 28.5 | 4 | 1.1 | 63a | 17.6 |
One patient with double mutations (E19/E21). Diff, differentiation; Mod, moderately; EGFR, epidermal growth factor receptor; E, exon.
Gender may affect the occurrence of oncogenic mutations
Subgroup analysis was performed in order to uncover clinical features that were associated with the identified genetic mutations in the present study (Table III). In order to determine whether gender affected the frequency of investigated tumorigenic mutations in the present patient cohort, subtype analysis was performed according to patient gender, as shown in Table III. The percentage of patients who possessed EGFR mutations was 35.7% (30/84) of male patients and 51.5% (141/274) of female patients. The difference in the number of patients with or without EGFR mutations between male (30 vs. 54) and female (141 vs. 133) patients was markedly significant (P<0.01), suggesting that gender is an important factor that may affect EGFR mutations in NSCLC in never-smokers. Thus, EGFR mutations were more likely to be detected in female patients compared with male patients in the subset of NSCLC exhibited by never-smokers. Using an identical analysis method, a significant difference was observed (P<0.05) in the prevalence of KRAS mutations between male and female patients. The frequency of KRAS mutations was 7.1% in male patients and 2.6% in female patients. No significant difference was observed in the prevalence of mutations for ALK, RET and BRAF genes between male and female patients, suggesting that gender may have no significant effect on these mutations in NSCLC in never-smokers.
Table III.
Characteristics of genetic mutations in 358 never-smokers with non-small cell lung cancer.
| EGFR | ALK | KRAS | RET | BRAF | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | −, n (%) | +, n (%) | P-value | −, n (%) | +, n (%) | P-value | −, n (%) | +, n (%) | P-value | −, n (%) | +, n (%) | P-value | −, n (%) | +, n (%) | P-value |
| Frequency | 187 (52.5) | 171 (47.8) | 331 (92.5) | 27 (7.5) | 345 (96.4) | 13 (3.6) | 353 (98.6) | 5 (1.4) | 357 (99.7) | 1 (0.3) | |||||
| Gender | |||||||||||||||
| Male | 54 (64.3) | 30 (35.7) | 0.01 | 78 (92.9) | 6 (7.1) | 0.87 | 78 (92.9) | 6 (7.1) | 0.05 | 84 (100.0) | 0 (0.0) | 0.21 | 84 (100.0) | 0 (0.0) | 0.58 |
| Female | 133 (48.5) | 141 (51.5) | 253 (92.3) | 21 (7.7) | 267 (97.4) | 7 (2.6) | 269 (98.2) | 5 (1.8) | 273 (99.6) | 1 (0.4) | |||||
| Age, years | |||||||||||||||
| <40 | 5 (100.0) | 0 (0.0) | 0.04 | 3 (60.0) | 2 (40.0) | 0.0002 | 5 (100.0) | 0 (0.0) | 0.35 | 4 (80.0) | 1 (20.0) | 0.0001 | 5 (100.0) | 0 (0.0) | 0.82 |
| 40–49 | 42 (56.8) | 32 (43.2) | 63 (85.1) | 11 (14.9) | 73 (98.6) | 1 (1.4) | 70 (94.6) | 4 (5.4) | 74 (100.0) | 0 (0.0) | |||||
| 50–59 | 70 (53.8) | 60 (46.2) | 121 (93.1) | 9 (6.9) | 124 (95.4) | 6 (4.6) | 130 (100.0) | 0 (0.0) | 129 (99.2) | 1 (0.8) | |||||
| ≥60 | 70 (47.0) | 79 (53.0) | 144 (96.6) | 5 (3.4) | 143 (96.0) | 6 (4.6) | 149 (100.0) | 0 (0.0) | 149 (100.0) | 0 (0.0) | |||||
| Diff | |||||||||||||||
| Poorly | 59 (51.3) | 56 (48.7) | 0.05 | 105 (89.7) | 12 (10.3) | 0.004 | 112 (95.7) | 5 (4.3) | 0.75 | 114 (97.4) | 3 (2.6) | 0.24 | 116 (99.1) | 1 (0.9) | 0.36 |
| Mod | 77 (60.2) | 51 (39.8) | 110 (88.7) | 14 (11.3) | 119 (96.0) | 5 (4.0) | 122 (98.4) | 2 (1.6) | 124 (100.0) | 0 (0.0) | |||||
| Well | 51 (44.3) | 64 (55.7) | 116 (99.1) | 1 (0.9) | 114 (97.4) | 3 (2.6) | 117 (100.0) | 0 (0.0) | 117 (100.0) | 0 (0.0) | |||||
| Avg. age, years | 57.7 | 54.4 | 59.3 | 42.8 | 58.0 | ||||||||||
Diff, differentiation; Mod, moderate; Avg., average; EGFR, epidermal growth factor receptor; ALK, anaplastic lymphoma kinase; KRAS, Kirsten rat sarcoma viral oncogene homolog; RET, ret proto-oncogene; BRAF, B-Raf proto-oncogene, serine/threonine kinase.
Genetic mutations may be affected by ageing
Subsequently, the effect of ageing on the occurrence of oncogenic mutations was determined. Consistent with previous reports (22,23), in the present study, EGFR mutations were more likely to be identified in older patients compared with younger patients. This was determined by comparing the distribution in four age subgroups between patients with and without EGFR mutations (P<0.04; Table III). By contrast, mutations in KIF5B-RET and EML4-ALK were more likely to be detected in younger patients compared with older patients, as there was a significant difference in the distribution of patients in four distinct age groups between wild-type and mutated KIF5B-RET or EML4-ALK, respectively (P<0.001; Table III). The median age was 42.8±1.6 years in KIF5B-RET-positive patients or 54.4±0.6 years in EML4-ALK-positive patients, compared with 57.9±0.8 years in mutation-negative patients (Table III). These results suggested a potential early onset of the disease in the patients exhibiting KIF5B-RET or EML4-ALK mutations. No evidence suggested that ageing was a significant factor in the occurrence of mutations in KRAS and BRAF, as determined by Fisher's exact test (Table III).
An association is present between genetic mutations and the level of differentiation in cancer
The differentiation level of carcinoma cases was examined, and it was identified that those expressing EML4-ALK were more likely to be poorly- or moderately-differentiated, but not well-differentiated, compared with mutation-negative patients (P<0.01; Table III). By comparing the differentiation level between tumors with and without EGFR mutations, the cancer cells were more likely to be well-differentiated in carcinoma exhibiting EGFR mutations compared with wild-type carcinoma (P<0.05; Table III). The differentiation level of carcinoma carrying alternative types of gene mutation was not significantly different from the remaining patients. These results suggested that ALK fusions activated cancer types that were less differentiated, but that possessed a more rapid rate of growth and were more resistant to conventional treatment. By contrast, EGFR mutations demonstrated the opposite properties for these aspects. The tumors were well-differentiated, with a slower rate of growth and possessed an improved response to conventional treatment (3,10). Therefore, screening for genetic mutations and determining cell differentiation levels may be important steps for improving the efficiency of targeted treatments in NSCLC.
Discussion
Novel discoveries in the molecular genetics of cancer have revolutionized the treatment of the disease by replacing traditional methods with customized therapies based on the clinical pathology and molecular diagnosis of genetic mutations (13). Thus, it is crucial to accurately determine tumorigenic alterations for the success of subsequent treatments. NSCLC in never-smokers has been proposed to be a distinct disease entity due to its unique molecular and clinical properties (19). Although the frequency of oncogenic mutations in never-smokers has been investigated in a large number of studies, the results have been varied and unclear, as never-smoker patients are typically included as a small proportion of the investigated subjects together with a relatively larger amount of smoker patients. In addition, only one mutation at a time has been traditionally investigated in the majority of these previous studies, thus avoiding a direct comparison between various mutations among the same group of patients, which may vary compared with other groups of patients in molecular and clinical characteristics due to differences in race, region, economy and environment. In order to overcome these problems, a comprehensive study was performed to determine the frequency of genetic alterations in five known oncogenes and their associated clinical features in 358 NSCLC patients who were exclusively never-smokers. Using this large-scale screen, a precise molecular profile was generated concerning tumorigenic alterations in NSCLC adenocarcinoma in never-smokers as a distinct disease entity.
EGFR and KRAS represent the two most frequently mutated genes in lung cancer, with a frequency of >10% in each case (21). However, the results of the present study indicated that only EGFR was frequently mutated in never-smoker patients, while KRAS and all other investigated genes were infrequently (<10%) altered in this patient cohort. This supported the idea that NSCLC in never-smokers is a distinct entity in NSCLC, at least with regard to the state of oncogenic mutations. While the causes of these de novo mutations remain to be elucidated, it is agreed that various mutations may have specific impacts on signaling pathways that regulate cellular proliferation and survival (24). For research and clinical purposes, types of cancer with infrequent tumorigenic mutations, particularly those demonstrating poor responses to EGFR TKIs and other targeted therapies, may be classified as rare diseases requiring different attention and treatments. An improved classification and diagnosis of lung cancer should consider molecular profiling and pathological characteristics in patients.
The present study reported that 47.8% of patients (171/358) in the present cohort exhibited EGFR mutation(s), which was similar to the results of a previous study reporting a frequency of 49.8% in never-smoker patients (25). In the present study, there were 102 patients exhibiting EGFR E19 microdeletions and 63 patients demonstrating E21 mutations, including 58 cases with L858R point mutations. These numbers were consistent with previous studies demonstrating that the two most common mutations were located in E19 and E21 (3,10). Notably, one patient exhibited two EGFR mutations simultaneously, E19 (E746-A750 DEL) and E21 (L858R). This patient was a female >60 years old who exhibited adenocarcinoma with moderately-differentiated cells. Subgroup analyses suggested that the incidence of EGFR mutations was significantly affected by gender and age. In particular, mutations were more likely to occur in female patients compared with male patients and in older patients compared with younger patients. In addition, it was observed that EGFR mutations were identified more frequently in well-differentiated tumor cells, which were typically less aggressive and grew more slowly compared with less-differentiated cancer cells.
Mutations in KRAS are identified in a wide range of types of cancer, including cancer of the pancreas, large intestine and lungs (26). Although 15–25% of NSCLC patients have been observed to exhibit KRAS mutations in previous studies, only 3.6% of patients in the present study cohort possessed KRAS mutations, which was consistent with the idea that the occurrence of this mutation may be associated with smoking (27–29). In the present study, it was additionally identified that KRAS mutations were more frequently detected in male patients compared with female patients, suggesting the effect of gender on the incidence of KRAS mutations. The majority of the mutations have been identified in E12 and less frequently in E13 (30), and mutations in these exons are able to cause sustained activation of RAS signaling leading to tumorigenesis (21,31). However, despite a large sample size in the present study, mutations were only identified in E12 at positions 12 and 13, and not in E13, suggesting that the mutation rate in E13 may be low or undetectable in NSCLC in never-smokers. It was additionally identified that EGFR and KRAS mutations were mutually exclusive, as has been reported previously (21). As patients exhibiting KRAS mutations typically demonstrate a worse response to EGFR TKIs, including gefitinib or erlotinib, compared to those with wild-type KRAS (28), KRAS mutations may be utilized as a negative predictive marker for responses to EGFR TKI-based therapy (28).
Somatic point mutations in BRAF occur more frequently in melanoma and thyroid, colon and ovarian cancer (5). The V600E substitution, which disrupts an inhibitory interaction between the P-loop and the activation segment, and leads to constitutive kinase activation, accounts for ~90% of BRAF missense mutations identified in human tumors (32,33). Previous studies have reported that the frequency of BRAF mutation in lung cancer is typically low, normally below or around 1% (34,35). Similarly, the present study detected a V600E point mutation in one patient, accounting for 0.3% of NSCLC in never-smokers in the present patient cohort.
The ALK gene encodes a receptor tyrosine kinase, which has been identified in a number of fusion proteins in cancer (6,36–39). Adenocarcinoma appears to be the major NSCLC cell type to exhibit EML4-ALK fusions. Previous studies, primarily involving East Asian patients, have reported that 3–7% of lung tumors exhibit EML4-ALK fusions (6,40–43). A total of 27 cases (7.5%) in the present study demonstrated EML4-ALK fusions. No other types of ALK fusions were detected in the current patient cohort. The results of the present study suggested that the onset age of lung cancer is likely to be younger in patients possessing ALK fusions compared to those without. Furthermore, the carcinoma cases were less differentiated in patients exhibiting ALK fusions compared to those without ALK fusions. These features may be at least in part responsible for an observed poor response to EGFR TKIs associated with ALK fusion (11), but an improved response to ALK TKI crizotinib (12). In addition to ALK fusion, KIF5B-RET fusion was detected in 1.4% of patients; however, no CCDC6-RET fusions were observed in the present study. Similar to EML4-ALK fusion, KIF5B-RET fusion is likely to be associated with early disease onset, as it was identified more frequently in younger patients. No other clinical features were identified as being significantly associated with RET mutations. However, this may have been due to the low frequency of this genetic alteration, which may have required a larger sample size to reach a statistically significant level.
In conclusion, the present study screened the genetic mutations in multiple oncogenes and determined clinical features associated with these mutations in a large cohort of NSCLC patients who were exclusively never-smokers. It was identified that EGFR mutations, but not other mutations, frequently occurred in NSCLC in never-smokers. It was additionally identified that gender may be associated with mutations in EGFR and KRAS, differentiation level with EGFR and ALK, and ageing with EGFR, ALK and RET. The results of the present study may provide valuable insights for the enhancement of our knowledge of lung cancer and facilitate the advancement of tailored therapies that are targeted to tumorigenic mutations.
Glossary
Abbreviations
- EGFR
epidermal growth factor receptor
- ALK
anaplastic lymphoma kinase
- RET
ret proto-oncogene
- EML4
echinoderm microtubule-associated protein-like 4
- KRAS
Kirsten rat sarcoma viral oncogene homolog
- BRAF
B-Raf proto-oncogene, serine/threonine kinase
- NSCLC
non-small cell lung cancer
- TKI
tyrosine kinase inhibitor
- RT-PCR
reverse transcription-polymerase chain reaction
- FFPE
formalin-fixed paraffin-embedded
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH. Eastern Cooperative Oncology Group: Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Santos E, Martin-Zanca D, Reddy EP, Pierotti MA, Della Porta G, Barbacid M. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science. 1984;223:661–664. doi: 10.1126/science.6695174. [DOI] [PubMed] [Google Scholar]
- 5.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 6.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 7.Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 8.Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, Sakamoto H, Tsuta K, Furuta K, Shimada Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, Curran JA, Balasubramanian S, Bloom T, Brennan KW, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 11.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, Solomon B, Stubbs H, Admane S, McDermott U, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thunnissen E, Bubendorf L, Dietel M, Elmberger G, Kerr K, Lopez-Rios F, Moch H, Olszewski W, Pauwels P, Penault-Llorca F, Rossi G. EML4-ALK testing in non-small cell carcinomas of the lung: A review with recommendations. Virchows Arch. 2012;461:245–257. doi: 10.1007/s00428-012-1281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buettner R, Wolf J, Thomas RK. Lessons learned from lung cancer genomics: The emerging concept of individualized diagnostics and treatment. J Clin Oncol. 2013;31:1858–1865. doi: 10.1200/JCO.2012.45.9867. [DOI] [PubMed] [Google Scholar]
- 14.Thun MJ, Hannan LM, Adams-Campbell LL, Boffetta P, Buring JE, Feskanich D, Flanders WD, Jee SH, Katanoda K, Kolonel LN, et al. Lung cancer occurrence in never-smokers: An analysis of 13 cohorts and 22 cancer registry studies. PLoS Med. 2008;5:e185. doi: 10.1371/journal.pmed.0050185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pomerleau CS, Pomerleau OF, Snedecor SM, Mehringer AM. Defining a never-smoker: Results from the nonsmokers survey. Addict Behav. 2004;29:1149–1154. doi: 10.1016/j.addbeh.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 17.Office of the Surgeon General. Rockville, MD: 2014. U.S. Department of Health and Human Services: The Health Consequences of Smoking-50 Years of Progress: A Report of the Surgeon General; pp. 139–351. [Google Scholar]
- 18.Gazdar AF, Thun MJ. Lung cancer, smoke exposure, and sex. J Clin Oncol. 2007;25:469–471. doi: 10.1200/JCO.2006.09.4623. [DOI] [PubMed] [Google Scholar]
- 19.Scagliotti GV, Longo M, Novello S. Nonsmall cell lung cancer in never smokers. Curr Opin Oncol. 2009;21:99–104. doi: 10.1097/CCO.0b013e328321049e. [DOI] [PubMed] [Google Scholar]
- 20.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Jänne PA, Costa DB, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karachaliou N, Mayo C, Costa C, Magrí I, Gimenez-Capitan A, Molina-Vila MA, Rosell R. KRAS mutations in lung cancer. Clin Lung Cancer. 2013;14:205–214. doi: 10.1016/j.cllc.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 22.Choi YH, Lee JK, Kang HJ, Lee TS, Kim HR, Kim CH, Koh JS, Baek HJ, Lee JC, Na II. Association between age at diagnosis and the presence of EGFR mutations in female patients with resected non-small cell lung cancer. J Thorac Oncol. 2010;5:1949–1952. doi: 10.1097/JTO.0b013e3181f38816. [DOI] [PubMed] [Google Scholar]
- 23.Ueno T, Toyooka S, Suda K, Soh J, Yatabe Y, Miyoshi S, Matsuo K, Mitsudomi T. Impact of age on epidermal growth factor receptor mutation in lung cancer. Lung Cancer. 2012;78:207–211. doi: 10.1016/j.lungcan.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Mitsudomi T. Advances in target therapy for lung cancer. Jpn J Clin Oncol. 2010;40:101–106. doi: 10.1093/jjco/hyp174. [DOI] [PubMed] [Google Scholar]
- 25.An SJ, Chen ZH, Su J, Zhang XC, Zhong WZ, Yang JJ, Zhou Q, Yang XN, Huang L, Guan JL, et al. Identification of enriched driver gene alterations in subgroups of non-small cell lung cancer patients based on histology and smoking status. PLoS One. 2012;7:e40109. doi: 10.1371/journal.pone.0040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bos JL. ras oncogenes in human cancer: A review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 27.Thu KL, Vucic EA, Chari R, Zhang W, Lockwood WW, English JC, Fu R, Wang P, Feng Z, MacAulay CE, et al. Lung adenocarcinoma of never smokers and smokers harbor differential regions of genetic alteration and exhibit different levels of genomic instability. PLoS One. 2012;7:e33003. doi: 10.1371/journal.pone.0033003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao C, Qiu LX, Liao RY, Du FB, Ding H, Yang WC, Li J, Chen Q. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: A meta-analysis of 22 studies. Lung Cancer. 2010;69:272–278. doi: 10.1016/j.lungcan.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 29.Le Calvez F, Mukeria A, Hunt JD, Kelm O, Hung RJ, Tanière P, Brennan P, Boffetta P, Zaridze DG, Hainaut P. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: Distinct patterns in never, former, and current smokers. Cancer Res. 2005;65:5076–5083. doi: 10.1158/0008-5472.CAN-05-0551. [DOI] [PubMed] [Google Scholar]
- 30.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 31.Cataldo VD, Gibbons DL, Pérez-Soler R, Quintás-Cardama A. Treatment of non-small-cell lung cancer with erlotinib or gefitinib. N Engl J Med. 2011;364:947–955. doi: 10.1056/NEJMct0807960. [DOI] [PubMed] [Google Scholar]
- 32.Vakiani E, Solit DB. KRAS and BRAF: Drug targets and predictive biomarkers. J Pathol. 2011;223:219–229. doi: 10.1002/path.2796. [DOI] [PubMed] [Google Scholar]
- 33.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R. Cancer Genome Project: Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 34.Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382:720–731. doi: 10.1016/S0140-6736(13)61715-8. [DOI] [PubMed] [Google Scholar]
- 35.Brustugun OT, Khattak AM, Trømborg AK, Beigi M, Beiske K, Lund-Iversen M, Helland Å. BRAF-mutations in non-small cell lung cancer. Lung Cancer. 2014;84:36–38. doi: 10.1016/j.lungcan.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 36.Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 37.Palmer RH, Vernersson E, Grabbe C, Hallberg B. Anaplastic lymphoma kinase: Signalling in development and disease. Biochem J. 2009;420:345–361. doi: 10.1042/BJ20090387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coffin CM, Hornick JL, Fletcher CD. Inflammatory myofibroblastic tumor: Comparison of clinicopathologic, histologic, and immunohistochemical features including ALK expression in atypical and aggressive cases. Am J Surg Pathol. 2007;31:509–520. doi: 10.1097/01.pas.0000213393.57322.c7. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, Wang L, Soda M, Kikuchi A, Igarashi T, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 40.Inamura K, Takeuchi K, Togashi Y, Nomura K, Ninomiya H, Okui M, Satoh Y, Okumura S, Nakagawa K, Soda M, et al. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol. 2008;3:13–17. doi: 10.1097/JTO.0b013e31815e8b60. [DOI] [PubMed] [Google Scholar]
- 41.Takeuchi K, Choi YL, Soda M, Inamura K, Togashi Y, Hatano S, Enomoto M, Takada S, Yamashita Y, Satoh Y, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res. 2008;14:6618–6624. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- 42.Shinmura K, Kageyama S, Tao H, Bunai T, Suzuki M, Kamo T, Takamochi K, Suzuki K, Tanahashi M, Niwa H, et al. EML4-ALK fusion transcripts, but no NPM-, TPM3-, CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non-small cell lung carcinomas. Lung Cancer. 2008;61:163–169. doi: 10.1016/j.lungcan.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 43.Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, Cheng LC, Ho KK, Au JS, Chung LP, Wong Pik M. University of Hong Kong Lung Cancer Study Group: The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]