Abstract
Vip proteins, a new group of insecticidal toxins produced by Bacillus thuringiensis, are effective against specific pests including Spodoptera litura. Here, we report construction of a transcriptome database of S. litura by de novo assembly along with detection of the transcriptional response of S. litura larvae to Vip3Aa toxin. In total, 56,498 unigenes with an N50 value of 1,853 bp were obtained. Results of transcriptome abundance showed that Vip3Aa toxin provoked a wide transcriptional response of the S. litura midgut. The differentially expressed genes were enriched for immunity-related, metabolic-related and Bt-related genes. Twenty-nine immunity-related genes, 102 metabolic-related genes and 62 Bt-related genes with differential expression were found. On the basis of transcriptional profiling analysis, we focus on the functional validation of trypsin which potentially participated in the activation of Vip3Aa protoxin. Zymogram analysis indicated that the presence of many proteases, including trypsin, in S. litura larvae midgut. Results of enzymolysis in vitro of Vip3Aa by trypsin, and bioassay and histopathology of the trypsin-digested Vip3Aa toxin showed that trypsin was possibly involved in the Vip3Aa activation. This study provides a transcriptome foundation for the identification and functional validation of the differentially expressed genes in an agricultural important pest, S. litura.
Spodoptera litura (Fab.) (Noctuidae: Lepidoptera) is notorious as one of the most destructive pests of agricultural crops, including more than 120 host plants globally1,2. It is a serious pest of soybean, cotton, groundnut, taro, tobacco, flax and some important vegetables, including potato, eggplants, cabbage, capsicum, brinjal, okra, brassica and cucurbits3,4. The species is responsible for huge yield losses annually in many crops due to its vigorous defoliation5. It has shown resistance to numerous synthetic insecticides, which has led to frequent control failure in crops6,7,8. As a consequence, considerable attention has been paid to biological insecticides as alternatives for controlling pests.
Bacillus thuringiensis is the most widely used commercial biopesticide with wide pathogenicity against numerous species9. At present, the Cry toxins produced by this bacterium are the most well-known biocontrol agents10. In addition to Cry toxins, some B. thuringiensis strains produce other insecticidal proteins during the vegetative growth phase, named vegetative insecticidal proteins (Vip)11,12. Vip toxins share no sequence homology with Cry toxins13 and have been classified into four subfamilies, Vip1, Vip2, Vip3 and Vip4, according to their sequence homology (http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/Bt/vip.html). To date, most studies on Vip toxins have been carried out with Vip3A. Since the discovery of the first Vip toxin, a great number of B. thuringiensis collections have been screened for new vip genes14,15,16,17 and the insecticidal activities of this kind of toxins have also been tested10,18,19,20. Vip3A toxins have already been incorporated into crops for the control of Lepidopteran pests21, but the mode of action of the toxin is still unclear. It is commonly accepted that, similar to Cry toxins, Vip3A toxins need to be activated by proteolysis and then the active forms bind to specific receptors on the midgut brush border membrane which eventually lead to pore formation and cell lysis22,23,24. Vip3A toxins, however, differ in their mode of action from Cry toxins in several key steps, especially in the binding step23,24,25. The binding sites of Vip toxins and Cry toxins in Helicoverpa armigera, Spodoptera frugiperda and Agrotis segetum have been shown to be different23,26,27,28. This contribute to the insecticidal activity of B. thuringiensis against insects, and is especially useful for the management of insect resistance to Cry toxins.
Recently, proteomic analysis and transcriptional profiling analysis have been used to investigate the defense response of some insects to Cry toxins29,30,31,32,33, and to gain insight into the mode of action of Cry toxins34,35,36,37. However, there is no report about the defense response of S. litura to Vip toxins by transcriptional profiling analysis. In the present study, the complete transcriptome information of S. litura was obtained and a genome-wide high-throughput transcriptome sequencing was used for comparison of Vip3Aa-treated and non-treated larvae of S. litura. The obtained transcriptome sequences provided genetic information to investigate the complex response of S. litura larva to Vip3Aa toxin by monitoring gene expression levels. Selected genes with different expression patterns were further validated by qRT-PCR. On the basis of transcriptional profiling analysis, the function of trypsin was validated by enzymolysis in vitro, bioassay and histopathology. Transcriptional profiling could provide a better understanding of the complex response of the S. litura midgut to Vip3Aa toxin and laid a foundation for further verifying the functions of the genes with different expression of S. litura, which are useful for future biocontrol strategies.
Results
Sequence Analysis and De Novo Assembly
A complete transcriptome was generated to obtain as many functional gene transcripts as possible. After removing low-quality sequences and adaptor sequences, a total of 56,108,398 clean reads were generated for control S. litura larvae midgut and 56,586,122 for Vip3Aa-treated larvae midgut. The quality value ≥30 with more than 89.79% bases for each sample suggested the sequencing was highly accurate. The GC content was about 48%, and 83.66–89.66% of the reads were mapped to unique sequences. After assembly using the Trinity method, 56,498 unigenes with an N50 value of 1,853 bp and a mean length of 827.6 bp were obtained (Table 1). Unigenes with a length between 200 and 300 bp represented the highest proportion.
Table 1. Unigene assembly of S. litura transcriptome.
| Length range (nt) | Unigene count | Percentage |
|---|---|---|
| 200–300 | 23,039 | 40.78% |
| 300–500 | 13,460 | 23.82% |
| 500–1000 | 8,472 | 15.00% |
| 1000–2000 | 5,580 | 9.88% |
| 2000+ | 5,945 | 10.52% |
| Total Number | 56,498 | |
| Total Length | 46,757,968 | |
| N50 Length | 1,853 | |
| Mean Length | 827.604 |
Functional Annotation
The unigenes were annotated against the following databases: NCBI nonredundant protein database (Nr), SwissProt database, Protein family (Pfam), euKaryotic Orthologous Groups of proteins (KOG), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) database. A total of 21,701 unigenes (38.41%) were successfully annotated, including 20,541 (36.35%) present in the Nr database, 13,126 (23.23%) in the SwissProt database and 6,296 (11.14%) in the KEGG database. Nr database queries also revealed the distribution and frequency of the species in the homology search (Fig. 1A). A high percentage of S. litura sequences closely matched sequences of silkworm (Bombyx mori, 38.38%) and monarch butterfly (Danaus plexippus, 22.48%).
Figure 1. Overview of S. litura annotations.
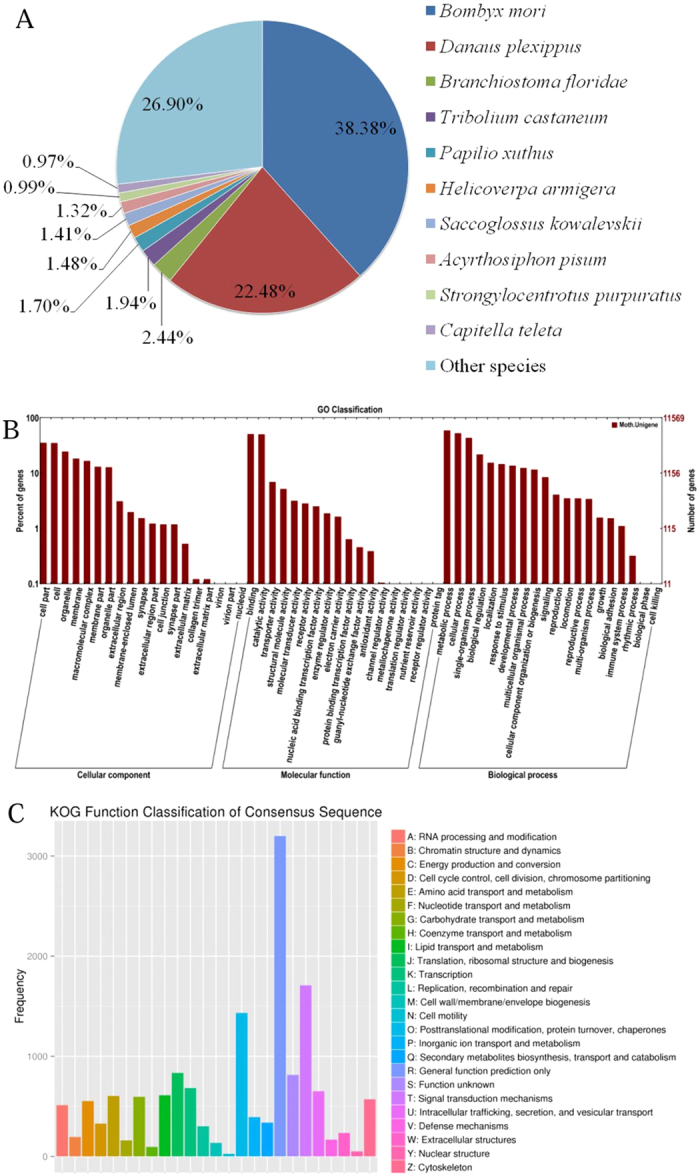
(A) Species distribution of the BLASTX results. (B) GO categories of all unigenes and DEGs. (C) euKaryotic orthologous Groups (KOG) classification.
The GO distributions of the unigenes were classified into three categories (57 subcategories): cellular components (19 subcategories), molecular functions (18 subcategories), and biological processes (20 subcategories) (Fig. 1B). In the category of cellular components, the clusters relating to “cell part” with 4,096 genes and “cell” with 4,068 genes were enriched. In the category of molecular functions, the clusters of “binding” and “catalytic activity” were highly represented (5,091 and 5,841 genes, respectively). In the category of biological processes, “metabolic processes”, “cellular processes” and “single-organism processes” were large compared to other subcategories with 6,855, 6,120 and 5,041 genes, respectively. In addition, the orthologous functions of the identified unigenes were defined using the KOG database (Fig. 1C). The “general function prediction” cluster (3,198, 21.07%) occupied the highest proportion, followed by “Signal transduction mechanisms” (1,708, 11.25%) and “Posttranslational modification, protein turnover, chaperones” (1,432, 9.43%). The two smallest groups were “Cell motility” and “Nuclear structure”.
Complex response of S. litura midgut to Vip3Aa toxin
The hierarchical cluster generated from transcriptome data was used to determine the profiles of the differentially expressed unigenes between control and Vip3Aa-treated S. litura (Figure S1A in Supplementary Material). The overall pattern of transcript changes after Vip3Aa ingestion were shown by gathering the same or similar expression unigenes in the same cluster. A total of 1,253 significantly altered unigenes was detected between control and Vip3Aa-treated S. litura samples (Figure S1B in Supplementary Material). The number of up-regulated genes (742) was greater than the number of down-regulated genes (511), which is similar to the report for S. exigua38.
In order to detect the complex response of the S. litura midgut to Vip3Aa toxin, GO annotation, KEGG database and BLAST searches were combined to identify differentially expressed in immunity-related, metabolic-related and Bt-related genes. In total, 29 differentially expressed immunity-related genes were identified (Table S1 in Supplementary Material), including 12 genes for recognition, 11 genes for signal transduction and melanization processes, 3 genes for antimicrobial effector and 3 genes for other immune-related proteins. The majority of these genes were up-regulated after Vip3Aa feeding except PGRP (peptidoglycan recognition protein) and superoxide dismutase. Differentially expressed metabolic genes from the transcriptome data were identified and listed in Figure S2 in Supplementary Material. In total, 102 metabolic genes were differentially expressed between control and Vip3Aa-treated S. litura, and participated in one or more pathways. Five types of mainly basal metabolic systems were identified, including amino acid metabolism, metabolism of cofactors and vitamins, lipid metabolism, xenobiotics biodegradation and metabolism, and carbohydrate metabolism. Bt-related genes mainly included genes encode proteases (51 genes with differential expression, part of them was shown in Table 2) participated in the activation or degradation of Vip protoxin and unigenes homologous to the potentially Bt toxin receptors (11 genes with differential expression, shown in Table S2 in Supplementary Material). Other genes with high level regulation after Vip3Aa feeding were shown in Table S3 in Supplementary Material.
Table 2. Potential serine protease genes of S. litura midgut in response to the ingestion of Vip3Aa toxin.
| WL-vs-M1 |
Unigene ID | nr_annotation | MC-vs-MVT2 |
||||
|---|---|---|---|---|---|---|---|
| Regulated | log2FC | FDR | FDR | log2FC | Regulated | ||
| up | 4.7040 | 4.46E-09 | c19837 | serine protease 36 [Mamestra configurata] | 0 | 5.8649 | up |
| up | 3.8193 | 1.89E-08 | c19623 | serine protease 62 [Mamestra configurata] | 0 | 3.9953 | up |
| up | 3.2438 | 1.15E-06 | c17572 | serine protease 54 [Mamestra configurata] | – | – | – |
| up | 2.9806 | 2.71E-05 | c18669 | serine protease 37 [Mamestra configurata] | 0 | 2.7062 | up |
| up | 2.7414 | 5.32E-05 | c14961 | serine protease 1 [Mamestra configurata] | – | – | – |
| up | 2.6627 | 0.00042 | c23985 | serine protease 37 [Mamestra configurata] | – | – | – |
| up | 2.4466 | 0.000986 | c11259 | serine protease 40 [Mamestra configurata] | 0 | −2.5298 | down |
| up | 2.4116 | 0.000661 | c21832 | serine protease 1 [Mamestra configurata] | 0 | −3.7417 | down |
| up | 2.4108 | 0.000949 | c17613 | serine protease 37 [Mamestra configurata] | 2.73E-07 | 1.2901 | up |
| up | 2.2285 | 0.006531 | c19310 | serine protease 3 [Lonomia obliqua] | 0 | −2.5281 | down |
| up | 2.1944 | 0.004283 | c19576 | serine protease 1 [Mamestra configurata] | 0.000445 | 1.0116 | up |
| up | 4.5718 | 2.00E-11 | c20161 | trypsinogen [Helicoverpa punctigera] | 0 | 2.3939 | up |
| up | 3.9148 | 2.79E-09 | c19094 | trypsin-like serine protease [Spodoptera litura] | 0 | 2.4131 | up |
| up | 3.5502 | 2.94E-06 | c6150 | trypsin-like serine protease [Spodoptera litura] | – | – | – |
| up | 3.5361 | 1.07E-06 | c11399 | trypsin-like protease [Helicoverpa armigera] | – | – | – |
| up | 3.0159 | 6.21E-06 | c19675 | trypsin precursor Hz19 [Helicoverpa zea] | – | – | – |
| up | 2.9418 | 1.32E-05 | c19527 | PREDICTED: trypsin, alkaline C-like [Bombyx mori] | – | – | – |
| up | 2.8859 | 3.59E-05 | c19330 | trypsin-like serine protease 9 [Ostrinia nubilalis] | 2.48E-10 | −1.4885 | down |
| up | 2.8332 | 0.00013 | c18929 | trypsin [Helicoverpa armigera] | 0 | −2.5266 | down |
| up | 2.7896 | 3.64E-05 | c21150 | PREDICTED: trypsin-1-like [Bombyx mori] | – | – | – |
| up | 2.7312 | 0.000566 | c5682 | trypsin-like protease [Helicoverpa armigera] | 0 | 2.5265 | up |
| up | 2.6323 | 0.001288 | c19277 | trypsin-like protease [Helicoverpa armigera] | 0 | 2.7106 | up |
| up | 2.6305 | 0.000135 | c20871 | trypsin-like serine protease [Spodoptera litura] | – | – | – |
| up | 2.6298 | 0.000217 | c11401 | PREDICTED: LOW QUALITY PROTEIN: trypsin CFT-1-like [Bombyx mori] | – | – | – |
| up | 2.5242 | 0.00156 | c18046 | trypsin-like protease [Helicoverpa armigera] | 1.50E-05 | 1.2165 | up |
| up | 2.1932 | 0.005754 | c19037 | trypsin [Heliothis virescens] | 3.73E-07 | 1.2887 | up |
| up | 4.6265 | 1.74E-09 | c25196 | chymotrypsin-like protein 2 [Spodoptera litura] | – | – | – |
| up | 3.7952 | 8.75E-06 | c25963 | chymotrypsin-like protein 2 [Spodoptera litura] | 1.33E-09 | −2.9884 | down |
| up | 3.7516 | 0.000108 | c20707 | chymotrypsin-like protein 2 [Spodoptera litura] | 0 | 5.0745 | up |
| up | 3.1787 | 6.98E-05 | c20135 | chymotrypsin, partial [Heliothis virescens] | 0 | 5.0409 | up |
| up | 2.5806 | 0.00031 | c19121 | chymotrypsin-like protein 2 [Spodoptera litura] | 1.73E-05 | −1.1248 | down |
| up | 2.5024 | 0.000657 | c19149 | chymotrypsin-like protease [Helicoverpa armigera] | – | – | – |
| up | 2.1547 | 0.003726 | c19321 | chymotrypsin-like protein precursor [Spodoptera litura] | 0 | 2.7072 | up |
| up | 2.0730 | 0.00687 | c20751 | chymotrypsin-like serine protease 14 [Ostrinia nubilalis] | – | – | – |
1WL-vs-M: whole larvea vs. midgut.
2MC-vs-MVT: midgut control vs. midgut of larvea with vip3Aa treatment.
To validate the transcriptome data, qRT-PCR analysis was performed for 12 differentially expressed genes including five up-regulated and seven down-regulated unigenes (underlined unigenes in Table 2, Tables S1–S3 in Supplementary Material). The regulated genes selected for confirmation included immune-related unigenes, Bt-related unigenes and other significantly differentially expressed unigenes. The qRT-PCR validation results were presented as fold-changes normalized to the β-actin gene. A comparison of the mean values of qRT-PCR results and the DEGs is summarized in Fig. 2. The consistent expression trend in qRT-PCR results and the original DEGs indicates the reliability of gene expression profiling from transcriptome analysis.
Figure 2. Validation of transcriptome data by qRT-PCR.
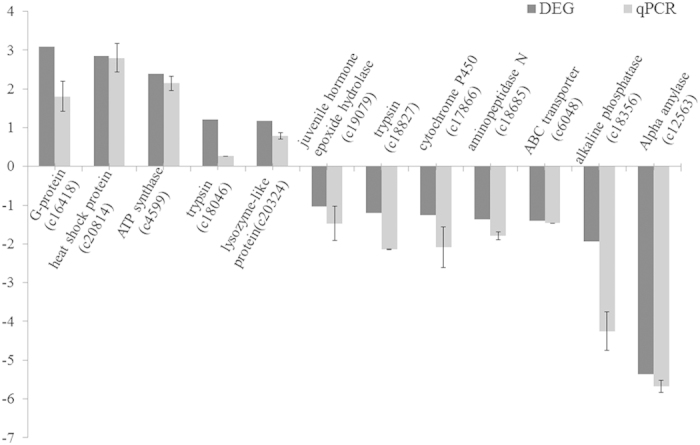
The means of at least three biological replicates are presented as log2FC ± SE.
Identification of serine protease genes of S. litura midgut potentially in response to the ingestion of Vip3Aa toxin
Activation or degradation of B. thuringiensis protoxin is a primary factor influencing its toxicity after ingestion. Unigenes encode proteins participated in the activation or degradation of Vip protoxin were extracted from transcriptome data for the identification of the differentially expressed genes. Serine proteases were found to be constituted the most abundant differentially expressed group of transcripts when S. litura larvae feeding with Vip3Aa toxin. Most of the serine proteases genes in midgut, which were up-regulated relative to whole larvae (Table 2, left), were found differentially expressed when S. litura larvae feeding with Vip3Aa toxin (Table 2, right). This result provided a preliminary proof that serine protease maybe involved in the solubilization of Vip3Aa toxin. Due to the complexity of serine protease family, we select one of serine protease, trypsin, for the further validation.
Validation of trypsin possibly involved in the Vip3Aa toxin activation
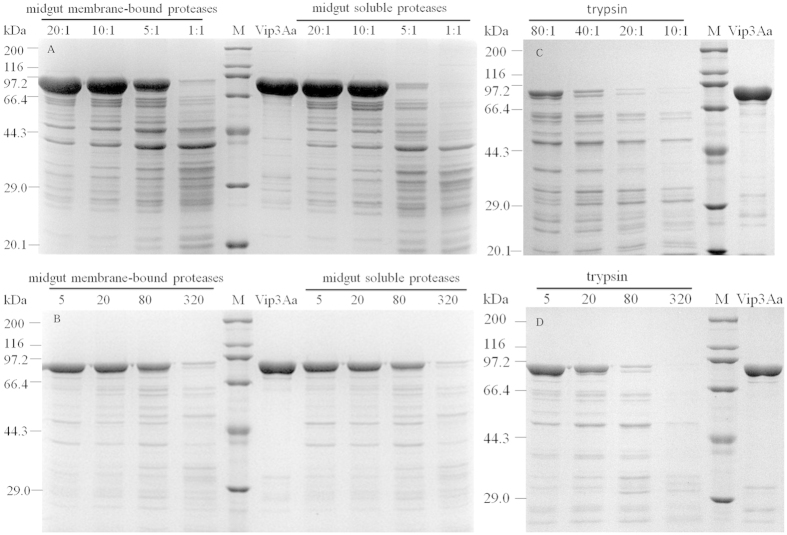
Using a general substrate, casein, as substrate, zymogram analysis was performed with the soluble proteases and membrane-bound proteases prepared from S. litura larval midgut and commercial trypsin. Trypsin can degrade casein into a band of 25.7 kDa (Fig. 3, lane 3). Zymogram of the soluble proteases and membrane-bound proteases showed the presence of many active proteases including trypsin (Fig. 3, lane 1 and 2).
Figure 3. Zymogram analysis of S. litura larvae midgut proteases.
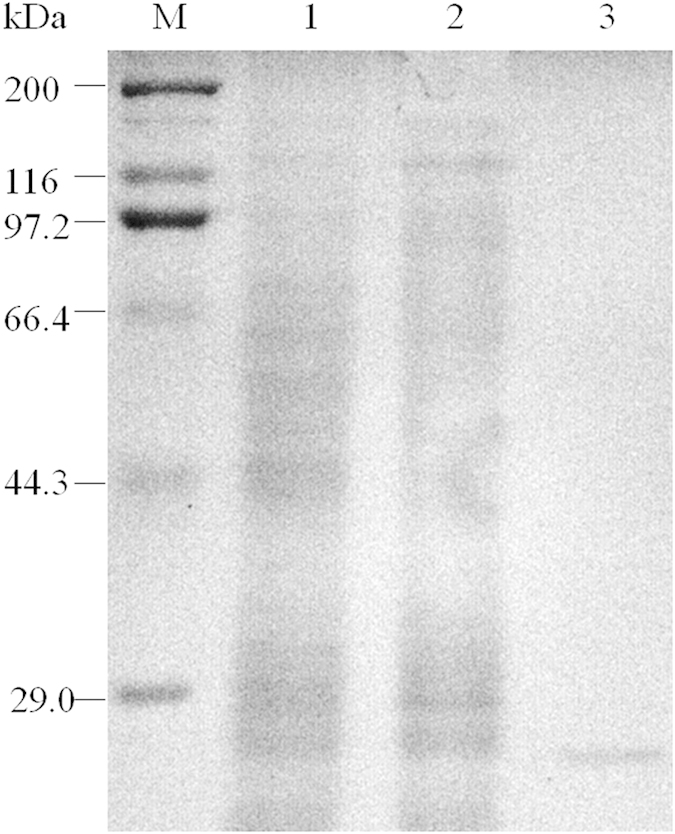
Midgut soluble proteases (lane 1), midgut membrane-bound proteases (lane 2) and trypsin (lane 3) were separated by 12% SDS-PAGE and their activity revealed using casein as substrates.
After confirmation the presence of trypsin in S. litura larval midgut, We further investigated enzymolysis in vitro of Vip3Aa by the soluble proteases and membrane-bound proteases prepared from S. litura larval midgut and trypsin with different concentrations and incubation times. Although their SDS-PAGE profiles were slightly different, the bands between 66.4 kDa and 29 kDa are similar especially in five major bands about 64, 62, 59, 45 and 39 kDa (Fig. 4A,C). The SDS-PAGE profiles obtained with different incubation times indicated that Vip3Aa proteolysis occurred rapidly as detectable small fragments was found after 5 min (Fig. 4B,D).
Figure 4. Proteolysis processing of Vip3Aa toxin by S. litura midgut proteases and commercial trypsin.
(A,C) Digestion of Vip3Aa toxin by different concentrations of proteases. Vip3Aa/proteases (midgut soluble, membrane-bound proteases or trypsin) ratio was showed in the figure (μg/μg). The toxins digestion with different incubation times were conducted for 5, 20, 80 and 320 min at the optimal activation concentration (B,D). Molecular mass markers (M) in kDa are indicated in the middle of the figure.
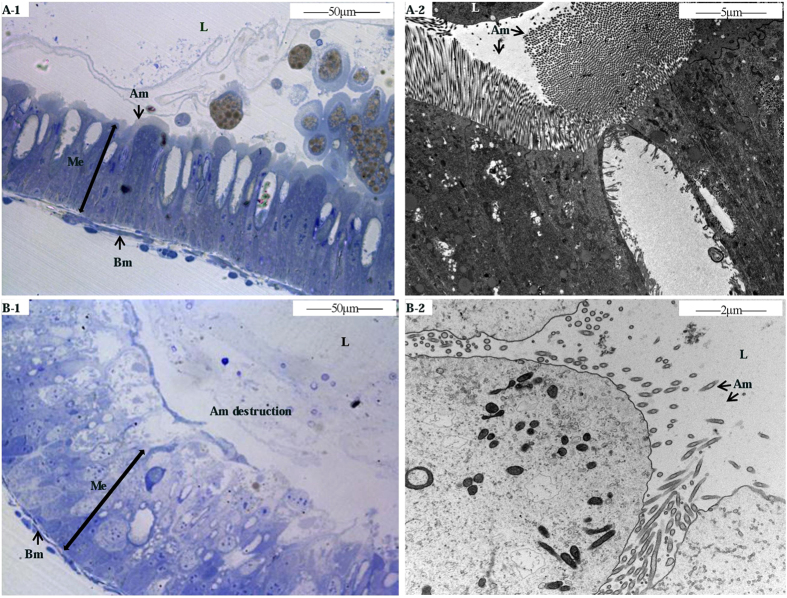
The insecticidal activity and histopathology of the trypsin-digested Vip3Aa toxin were also assayed. The digested products show high toxicity to first instar S. litura larvae with an LC50 of 77.052 ng/cm2 artificial diet with 95% confidence limits of (66.304–90.649 ng/cm2). Compared to control untreated-midgut, the observations of midgut cross sections showed wide damage to S. litura larvae midgut epithelium induced by the digested products. The histopathological changes of larvae fed with the trypsin-digested Vip3Aa toxin mainly included vacuolization of the cytoplasm, cells swelling and brush border membrane destruction (Fig. 5B), while control untreated-midgut showed a well-preserved layer of epithelial cells with unaffected apical microvilli membrane (Fig. 5A). These results demonstrated that trypsin is possibly involved in the activation of Vip3Aa toxin.
Figure 5. Histopathological effects of the trypsin-activated Vip3Aa toxin on S. litura larvae midgut.
general aspects of the midgut (A) and midgut of larvae fed with trypsin-activated Vip3Aa toxin (B). Me midgut epithelium, Am apical membrane, Bm basement membrane, L lumen.
Discussion
Recently, transcriptome sequencing has become a crucial research method on account of its peculiarity of low cost and high throughput39. We presented here the complete transcriptome information of S. litura, and a complex response of the S. litura larval midgut to Vip3Aa toxin. Due to the lack of a reference genome of S. litura, the unigenes were identified and annotated using public protein databases including the Nr, GO, KEGG, KOG, Swiss-Prot and Pfam databases. Over the past few years, GO terms have been utilized for the functional annotation and visualization of unigenes. A framework for categorizing genes was provided, namely biological process, molecular function, and cellular compartment. In the present study, 11,569 unigenes were assigned to one or more ontologies. KEGG analysis was utilized for demonstrating the various biological pathways40, and 6,296 unigenes were assigned to KEGG pathways. Collectively, the data portrayed a perspective of S. litura transcriptome information and also the differential expression of S. litura genes induced by Vip3Aa toxin treatment.
Previous studies have demonstrated that Cry intoxication of insects induced differential gene expression related to immunity and metabolism31,41. As mentioned above, the majority of immune-related genes we observed were up-regulated after Vip3A intoxication. In the insect innate immune response, successful recognition of a potential pathogen is the initial process. It is currently known that this recognition is mediated by pattern recognition proteins (PRPs)42,43. However, after Vip3Aa feeding, we found most of PRPs such as hemolin, β-1,3-glucan recognition proteins (βGRPs), C-type lectins(CTLs), scavenger receptors (SCRs) were all over-expressed, but peptidoglycan recognition proteins (PGRPs) were down-regulated. The silimar result had also been found in S. exigua after Vip3A feeding38. It can be speculated that PRPs probably have other functions, besides the mediation of pathogen recognition. In addition, melanization occurs regularly in the insect midgut and two up-regulated genes of the melanization cascade were observed after Vip3A intoxication. After recognition and melanization of the invasive signals, the immune system of insects mainly relies on signaling pathways to produce the effector molecules. The main signal transduction pathways involved in insect immunity are the janus kinase/signal transduction and activator of transcription (JAK/STAT), Mitogen Activated Protein Kinase (MAPK), Toll, immune deficiency (IMD), and c-jun Nterminal kinase (JNK) pathways38,44. We identified signal transduction-related genes related to JAK/STAT, Toll and MAPK pathways. The final effectors insects produce upon infection are induced following successive signal recognition, melanization, and transduction. Part of the synthesized effector molecules play direct roles in elimination of infections, phenol oxidase-dependent melanization and other immune-related mechanism48. Vip3Aa feeding also revealed up-regulation of other associated immune-related genes, such as Hdd family.
The basic metabolism response is very important for maintaining the normal physiological activities of organisms. Analysis of the differential expression of genes related to basal metabolic pathways between control and Vip3Aa-treated S. litura contributes to understanding the interaction between insect and toxin. In this study, we found that the number of unigenes and transcripts related to metabolism was significantly changed after infection. The results shown in Figure S2 (Supplementary Material) indicated that Vip3Aa affected the basal metabolic system-related genes of S. litura in many pathways, including glycandegradation, oxidative phosphorylation, and fatty acid biosynthesis, leading to over or reduced expression. However, the mechanisms of the basal metabolic pathways modulation are not exactly known.
After Vip protoxin is ingested by insect, it is processed to an active toxin which can bind with insect gut receptors45. Previous studies in some lepidopteran insects had reported that a 62 kDa fragment produced by the Vip3A protoxin enzymolysis is the active form of this protein20,22,24, and that trypsin are responsible for the activation of Cry protoxin46. However, for S. litura, there is so far no report about the activation process of Vip3Aa toxin and the role of trypsin in the Vip3Aa activation. By means of transcriptome analysis, We found that 34 serine proteases genes in midgut were up-regulated relative to whole larvae (Table 2, left), and that 14 genes were up-regulated and 7 genes were down-regulated in Vip3Aa-treared midgut compared to non-treated midgut (Table 2, right). The significant changes in the gene expression after Vip3Aa toxin ingestion suggested that the candidate genes may involved in the process of toxin activation or degradation. Zymogram of the soluble proteases and membrane-bound proteases showed the presence of many active proteases including trypsin (Fig. 3). In enzymolysis in vitro of Vip3Aa toxin by trypsin, a 62 kDa fragment which is considered as the active form in some other lepidopteran insects also appeared. The trypsin-digested Vip3Aa toxin is high toxicity to first instar S. litura larvae and can cause wide damage to midgut epithelium. These result demonstrated that trypsin is possibly involved in the activation of Vip3Aa toxin, which is the same as the report in Cry toxin.
Although previous studies about some Lepidopteran pests have shown that the binding sites of Vip toxins and Cry toxins are different, some general characterized receptors genes for Cry toxin were found to be differentially expressed after Vip3Aa ingestion (Table S2). In the Cry toxins pore-formation model of Lepidoptera, the activated toxin first binds to cadherin to form toxin oligomers, which may involved in alkaline phosphatases (ALPs) and N-aminopeptidases (APNs). Two differentially expressed cadherin-like transcript were found after Vip3Aa ingestion, but it is unclear why the pattern of transcript changes were different with one up-regulated and one down-regulated. Unigenes homologous to ALPs and APNs were found down-regulated, whereas G-proteins were up-regulated. The changes of ALPs and G-proteins are the same as the report in S. exigua38. Further studies are needed to know their functions.
In summary, in this study, the complete transcriptome information of S. litura has been obtained, and the overall response of the S. litura midgut to Vip3Aa toxin has been described for the first time. On the basis of transcriptional profiling analysis, we further investigated the in vitro activation process of Vip3Aa toxin, and demonstrated that trypsin is possibly involved in the Vip3Aa toxin activation. The obtained transcriptome data provides a foundation for the identification and functional validation of the differentially expressed genes of S. litura after Vip3Aa toxin feeding.
Materials and Methods
Insects and toxin
The colony of S. litura used in the experiments was purchased from Ke Yun Biology co. (China) and reared on artificial diet under the following conditions: temperature 27 ± 2 °C, relative humidity 55 ± 5% and photoperiod 16:8 h (light/dark).
The vip3Aa gene (GenBank Accession No. AF500478) was from B. thuringiensis WB5, a native strain isolated from soil collected in Wuyi mountain (China). The pCzn1-vip3Aa, a recombinant plasmid constructed in our previous study, was transformed into E. coli BL21 and the transformants were induced with 0.2 mM isopropyl-D-thiogalactopyranoside (IPTG) at 16 °C for 4 h. After centrifugation at 1,2000 g at 4 °C for 10 min, the pellet was resuspended and sonicated on ice, then centrifuged at 1,2000 g at 4 °C for 10 min. The supernatant was purified using Ni-IDA-Sepharose affinity column, dialyzed with PBS buffer and stored at −20 °C for the feeding experiments. The concentrations of Vip3Aa toxin were measured according to the Bradford method47.
Treatment of S. litura larvae with Vip3Aa
Third instar larvae of S. litura were selected for the feeding experiments. The larvae were reared on artificial diet containing Vip3Aa toxin with an LC50 of 200 ng/cm2, which resulted in a growth inhibition but did not cause visible death of the larvae for 24 hours. Larvae treated with PBS buffer were used as negative control. All treated larvae were incubated for 24 h at 27 ± 2 °C, 55 ± 5% RH, and L16:D8 h. Ten larvae were used for each treatment, and at least five larvae that had fed (as determined by observing the food bites) were dissected to obtain midgut tissues for further processing. Both treatments were performed in triplicate.
RNA isolation, cDNA library construction, and Illumina sequencing
Total RNA was extracted from the whole body and midgut of S. litura respectively, using the TRIzol Reagent (Invitrogen) following the protocols provided by the manufacturers. The integrity, quantity and quality of total RNA were determined using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA), the Qubit® RNA Assay Kit in Qubit®2.0 Flurometer (Life Technologies, CA, USA) and the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA).
The mRNA was enriched and isolated from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using a RNA fragmentation kit, followed by synthesis of first and second strand cDNA using M-MuLV Reverse Transcriptase (RNase H-), DNA polymerase I and RNaseH. The cDNA fragments were end-repaired and ligated to a NEBNext Adaptor after adenylation of the 3′ ends, then purified with the AMPure XP system (Beckman Coulter, Beverly, USA) to create a cDNA library. Library quality was assessed on the Agilent Bioanalyzer 2100 system. After clustering, the products were sequenced on an Illumina Hiseq 2500 platform.
De Novo Assembly and Data analysis
The raw data were first filtered by removing the adaptor sequences, reads containing ploy-N and low quality reads. Then the quality value and base distribution of the raw data were examined by calculating Q20, Q30, GC-content and sequence duplication level of the clean data. De novo assembly of the mRNA-seq reads was accomplished using Trinity48 with K-mer = 25. Annotations of the unigenes were performed by a BLASTx search against the Nr, Swiss-Prot, KOG, KEGG databases and Pfam, and by a Blast2GO search against the GO database. Gene expression levels were calculated by RSEM49. Differential expression of the unigenes was performed using the DEGseq50 R package. False discovery rate (FDR) <0.01 and log2 ratio ≥1 were set as thresholds for identifying significant differential expression between control and Vip3Aa treated larvae.
Quantitative real-time PCR (qRT-PCR) validation
To confirm the data, a subset of differentially expressed genes was validated by quantitative real time PCR (qRT-PCR). Total RNA from each sample was extracted as described above. Reverse transcription was performed using a PrimeScript RT reagent kit with the gDNA Eraser (TOYOBO). Quantitative real-time PCR was conducted on the CFX96 Real-Time System (Bio-Rad, Hercules, CA, USA) using the SybrGreen method with Premix Ex Taq II (Takara, Kyoto, Japan). The selected genes were verified with the following cycling conditions: 95 °C for 30 s, followed by 40 cycles of 95 °C for 30 s, 60 °C for 35 s. The melting curve analysis was used to analyze the specificity of the qPCR product. The sequences of the primers used are listed in Table S4 in Supplementary Material. A β-actin served as an internal control. The relative gene expression values were calculated using the 2−△△Ct method51.
Preparation of S. litura larvae midgut proteases and Zymogram analysis
Third instar S. litura larvae were collected and anesthetized on ice for 30 min, then dissected the midgut. The soluble fraction and membrane fraction extracts of the midgut were obtained by centrifugation after homogenating52, then separated by SDS-PAGE. After washed in turns with Triton X-100 (2.5%) and water, the gel was incubated with 2% casein in 50 mM Na2CO3 buffer (pH 9.6) at 37 °C for 3 h. Then the protease activity was visible after Coomassie staining.
Enzymolysis in vitro of Vip3Aa toxin by commercial trypsin and S. litura midgut proteases
The purified Vip3Aa toxin was mixed with each of different concentrations of S. litura larvae midgut proteases and commercial trypsin (Sigma) in 100 mM PBS buffer (pH 7.2, similar to S. litura midgut) and incubated for 1 h at 30 °C. To investigate the enzymolysis kinetic of the purified Vip3Aa toxin, it was mixed with each protease as mentioned above and incubated for diverse incubation times (between 5 and 320 min) at 30 °C. The Vip3Aa/protease ratio was the optimal activation concentration. The proteolytic reaction was stopped by the addition of Laemmli sample buffer (5×) followed by heat denaturation. Samples were separated by 12% SDS–PAGE and stained with Coomassie blue.
Bioassay and histopathology observation of the trypsin-digested Vip3Aa toxin
The toxicity to first instar S. litura larvae was tested by feeding with five different concentrations of the trypsin-digested Vip3Aa toxin in artificial diet. PBS buffer was used as a negative control. All treatments were in sextuplicate with ten larvae per replicate. The bioassays were performed in a growth chamber under the following conditions: temperature 27 ± 2 °C, relative humidity 55 ± 5% and photoperiod 16:8 h (light/dark). The mortality of each treatment was recorded every other day. Median lethal concentrations (LC50) and ninety percent lethal concentrations (LC90) were estimated from mortality data by probit analysis (DPS software).
Third instar S. litura larvae were starved for 2 h, and then fed with artificial diet supplemented with the trypsin-digested Vip3Aa toxin (200 ng/cm2 of diet) for 24 h. Artificial diet supplemented with PBS buffer was used as a negative control. After chilled on ice for 30 min, the midguts were excised and fixed in 2.5% glutaraldehyde and 1% osmic acid stationary liquid. Tissue were dehydrated using increasing ethanol concentrations, then rinsed in 100% acetone and fixed by embedding and curing. Images were captured with transmission electron microscopy (TEM, JEM-2010(HR)).
Additional Information
How to cite this article: Song, F. et al. Transcriptional profiling analysis of Spodoptera litura larvae challenged with Vip3Aa toxin and possible involvement of trypsin in the toxin activation. Sci. Rep. 6, 23861; doi: 10.1038/srep23861 (2016).
Supplementary Material
Acknowledgments
We thanks Dr. Brian McGarvey for critical reading and revising of the manuscript. This study is supported by Project of Fujian-Taiwan Joint Center for Ecological Control of Crop Pests (No. Minjiaoke [2013] 51), National Natural Science Foundation of China (No. 31401802) and State Administration of Grain (No. 201313002-3).
Footnotes
Author Contributions F.F.S. and S.Q.W. contributed to the design of the research. F.F.S., C.C. and M.N.L. participated in the materials preparation and experiments performing. F.F.S., E.S.S. and S.Q.W. performed the transcriptome analysis. F.F.S. drafted the manuscript. Z.P.H. and X.G. participated in revision of the text. All authors reviewed the manuscript.
References
- Knipling E. F. Regional management of the fall armyworm: A realistic approach? Fla. Entomol. 63, 468–480 (1980). [Google Scholar]
- Zhou J., Zhang G. & Zhou Q. Molecular characterization of cytochrome P450 CYP6B47 cDNAs and 5′-flanking sequence from Spodoptera litura (Lepidoptera: Noctuidae): its response to lead stress. J. Insect Physiol. 58, 726–736 (2012). [DOI] [PubMed] [Google Scholar]
- Baskar K. & Ignacimuthu S. Antifeedant, larvicidal and growth inhibitory effects of ononitol monohydrate isolated from Cassia tora L. against Helicoverpa armigera (Hub.) and Spodoptera litura (Fab.) (Lepidoptera: Noctuidae). Chemosphere 88, 384–388 (2012). [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. Genomic sequence analysis of granulovirus isolated from the tobacco cutworm, Spodoptera litura. Plos One 6, e28163 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Ye Z., Huang S., Ding J. & Luo R. The correlations of the different host plants with preference level, life duration and survival rate of Spodoptera litura. Fabricius. Chin. J. Eco-Agric. 12, 40–42 (2004). [Google Scholar]
- Saleem M. A., Ahmad M., Ahmad M., Aslam M. & Sayyed A. H. Resistance to selected organochlorin, organophosphate, carbamate and pyrethroid, in Spodoptera litura (Lepidoptera: Noctuidae) from Pakistan. J. Econ. Entomol. 101, 1667–1675 (2008). [DOI] [PubMed] [Google Scholar]
- Shad S. A., Sayyed A. H. & Saleem M. A. Cross-resistance, mode of inheritance and stability of resistance to emamectin in Spodoptera litura (Lepidoptera: Noctuidae). Pest Manag. Sci. 66, 839–846 (2010). [DOI] [PubMed] [Google Scholar]
- Shad S. A. et al. Field evolved resistance to carbamates, organophosphates, pyrethroids, and new chemistry insecticides in Spodoptera litura Fab. (Lepidoptera: Noctuidae). J. Pest Sci. 85, 153–162 (2012). [Google Scholar]
- Schnepf E. et al. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62, 775–806 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Martinez P., Hernandez-Rodriguez C. S., Rie J. V., Escriche B. & Ferre J. Insecticidal activity of Vip3Aa, Vip3Ad, Vip3Ae, and Vip3Af from Bacillus thuringiensis against lepidopteran corn pests. J. Invertebr. Pathol. 113, 78–81 (2013). [DOI] [PubMed] [Google Scholar]
- Zhu C., Ruan L., Peng D., Yu Z. & Sun M. Vegetative insecticidal protein enhancing the toxicity of Bacillus thuringiensis subsp kurstaki against Spodoptera exigua. Lett. Appl. Microbiol. 42, 109–114 (2006). [DOI] [PubMed] [Google Scholar]
- Donovan W. P., Donovan J. C. & Engleman J. T. Gene knockout demonstrates that vip3A contributes to the pathogenesis of Bacillus thuringiensis toward Agrotis ipsilon and Spodoptera exigua. J. Invertebr. Pathol. 78, 45–51 (2001). [DOI] [PubMed] [Google Scholar]
- Caccia S., Chakroun M., Vinokurov K. & Ferre J. Proteolytic processing of Bacillus thuringiensis Vip3A proteins by two Spodoptera species. J. Insect Physiol. 67, 76–84 (2014). [DOI] [PubMed] [Google Scholar]
- Abdelkefi-Mesrati L., Tounsi S. & Jaoua S. Characterization of a novel vip3-type gene from Bacillus thuringiensis and evidence of its presence on a large plasmid. FEMS microbiol. lett. 244, 353–358 (2005). [DOI] [PubMed] [Google Scholar]
- Palma L., De Escudero I. R., Maeztu M., Caballero P. & Muñoz D. Screening of vip genes from a Spanish Bacillus thuringiensis collection and characterization of two Vip3 proteins highly toxic to five lepidopteran crop pests. Biol. Control 66, 141–149 (2013). [Google Scholar]
- Palma L. et al. Vip3C, a novel class of vegetative insecticidal proteins from Bacillus thuringiensis. Appl. Environ. Microb. 78, 7163–7165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattar S. & Maiti M. K. Molecular characterization of a novel vegetative insecticidal protein from Bacillus thuringiensis effective against sap-sucking insect pest. J. Microbiol. Biotechn. 21, 937–946 (2011). [DOI] [PubMed] [Google Scholar]
- De Escudero I. R. et al. A screening of five Bacillus thuringiensis Vip3A proteins for their activity against lepidopteran pests. J. Invertebr. Pathol. 117, 51–55 (2014). [DOI] [PubMed] [Google Scholar]
- Bergamasco V. B., Mendes D. R., Fernandes O. A., Desiderio J. A. & Lemos M. V. Bacillus thuringiensis Cry1Ia10 and Vip3Aa protein interactions and their toxicity in Spodoptera spp. (Lepidoptera). J. Invertebr. Pathol. 112, 152–158 (2013). [DOI] [PubMed] [Google Scholar]
- Chakroun M. et al. Susceptibility of Spodoptera frugiperda and S. exigua to Bacillus thuringiensis Vip3Aa insecticidal protein. J. Invertebr. Pathol. 110, 334–339 (2012). [DOI] [PubMed] [Google Scholar]
- Raybould A. & Quemada H. Bt crops and food security in developing countries: realised benefits, sustainable use and lowering barriers to adoption. Food Secur. 2, 247–259 (2010). [Google Scholar]
- Lee M. K., Miles P. & Chen J. S. Brush border membrane binding properties of Bacillus thuringiensis Vip3A toxin to Heliothis virescens and Helicoverpa zea midguts. Biochem. Biophys. Res. Commun. 339, 1043–1047 (2006). [DOI] [PubMed] [Google Scholar]
- Liu J. G., Yang A. Z., Shen X. H., Hua B. G. & Shi G. L. Specific binding of activated Vip3Aa10 to Helicoverpa armigera brush border membrane vesicles results in pore formation. J. Invertebr. Pathol. 108, 92–97 (2011). [DOI] [PubMed] [Google Scholar]
- Lee M. K., Walters F. S., Hart H., Palekar N. & Chen J. S. The Mode of action of the Bacillus thuringiensis vegetative insecticidal protein Vip3A differs from that of Cry1Ab-endotoxin. Appl. Environ. Microb. 69, 4648–4657 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz R. W. A review of Vip3A mode of action and effects on Bt Cry protein-resistant colonies of lepidopteran larvae. Southwest. Entomol. 35, 391–394 (2010). [Google Scholar]
- Gouffon C., Van Vliet A., Van Rie J., Jansens S. & Jurat-Fuentes J. L. Binding sites for Bacillus thuringiensis Cry2Ae toxin on heliothine brush border membrane vesicles are not shared with Cry1A, Cry1F, or Vip3A toxin. Appl. Environ. Microb. 77, 3182–3188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena J. A., Hernandez-Rodriguez C. S. & Ferre J. Interaction of Bacillus thuringiensis Cry1 and Vip3A proteins with Spodoptera frugiperda midgut binding sites. Appl. Environ. Microb. 75, 2236–2237 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Hamadou-Charfi D., Boukedi H., Abdelkefi-Mesrati L., Tounsi S. & Jaoua S. Agrotis segetum midgut putative receptor of Bacillus thuringiensis vegetative insecticidal protein Vip3Aa16 differs from that of Cry1Ac toxin. J. Invertebr. Pathol. 114, 139–143 (2013). [DOI] [PubMed] [Google Scholar]
- Rodezno A. C. et al. Comparative Proteomic Analysis of Aedes aegypti larval midgut after intoxication with Cry11Aa toxin from Bacillus thuringiensis. Plos One 7, e37034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras E., Rausell C. & Real M. D. Proteome response of Tribolium castaneum larvae to Bacillus thuringiensis toxin producing strains. Plos One 8, e55330 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppert B. et al. Transcriptome profiling of the intoxication response of Tenebrio molitor larvae to Bacillus thuringiensis Cry3Aa protoxin. Plos One 7, e34624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y. et al. Midgut transcriptome response to a Cry toxin in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Gene 533, 180–187 (2014). [DOI] [PubMed] [Google Scholar]
- Yao J., Buschman L. L., Lu N., Khajuria C. & Zhu K. Y. Changes in gene expression in the larval gut of Ostrinia nubilalis in Response to Bacillus thuringiensis Cry1Ab protoxin ingestion. Toxins 6, 1274–1294 (2014). [DOI] [PMC free article] [PubMed]
- Guo Z., Zhu Y., Huang F., Luttrell R. & Leonard R. Microarray analysis of global gene regulation in the Cry1Ab-resistant and Cry1Ab-susceptible strains of Diatraea saccharalis. Pest Manag. Sci. 68, 718–730 (2012). [DOI] [PubMed] [Google Scholar]
- Paris M. et al. Transcription profiling of resistance to Bti toxins in the mosquito Aedes aegypti using next-generation sequencing. J. Invertebr. Pathol. 109, 201–208 (2012). [DOI] [PubMed] [Google Scholar]
- Tetreau G. et al. Larval midgut modifications associated with Bti resistance in the yellow fever mosquito using proteomic and transcriptomic approaches. BMC Genomics 13, 248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual L. et al. The transcriptome of Spodoptera exigua larvae exposed to different types of microbes. Insect Biochem. Molec. 42, 557–570 (2012). [DOI] [PubMed] [Google Scholar]
- Bel Y., Jakubowska A. K., Costa J., Herrero S. & Escriche B. Comprehensive analysis of gene expression profiles of the beet armyworm Spodoptera exigua larvae challenged with Bacillus thuringiensis Vip3Aa toxin. Plos One 8, e81927 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C. et al. Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. The Plant cell 25, 1960–1978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzodek C., Buchel F., Ruff M., Drager A. & Zell A. Precise generation of systems biology models from KEGG pathways. BMC Syst. Biol. 7, 15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V. M. et al. Altered gene expression in Choristoneura fumiferana and Manduca sexta in response to sublethal intoxication by Bacillus thuringiensis Cry1Ab toxin. Insect Mol. Biol. 16, 25–35 (2007). [DOI] [PubMed] [Google Scholar]
- Kim J. H. et al. Comparison of the humoral and cellular immune responses between body and head lice following bacterial challenge. Insect Biochem. Molec. 41, 332–339 (2011). [DOI] [PubMed] [Google Scholar]
- Wang Y., Sumathipala N., Rayaprolu S. & Jiang H. Recognition of microbial molecular patterns and stimulation of prophenoloxidase activation by a beta-1,3-glucanase-related protein in Manduca sexta larval plasma. Insect Biochem. Molec. 41, 322–331 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J. D. et al. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 15, 645–656 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelkefi-Mesrati L. et al. Investigation of the steps involved in the difference of susceptibility of Ephestia kuehniella and Spodoptera littoralis to the Bacillus thuringiensis Vip3Aa16 toxin. J. Invertebr. Pathol. 107, 198–201 (2011). [DOI] [PubMed] [Google Scholar]
- Oppert B. Protease interactions with bacillus thuringiensis insecticidal toxins. Arch. Insect biochem. 42, 1–12 (1999). [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- Grabherr M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. & Dewey C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Wang X. & Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138 (2010). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Li H., Chougule N. P. & Bonning B. C. Interaction of the Bacillus thuringiensis delta endotoxins Cry1Ac and Cry3Aa with the gut of the pea aphid, Acyrthosiphon pisum (Harris). J. Invertebr. Pathol. 107, 69–78 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.