Abstract
Previously, we reported an electron spin echo envelope modulation (ESEEM) spectroscopic approach for probing the local secondary structure of membrane proteins and peptides utilizing 2H isotopic labeling and site-directed spin-labeling (SDSL). In order to probe the secondary structure of a peptide sequence, an amino acid residue (i) side chain was 2H-labeled, such as 2H-labeled d10-Leucine, and a cysteine residue was strategically placed at a subsequent nearby position (denoted as i + 1 to i + 4) to which a nitroxide spin label was attached. In order to fully access and demonstrate the feasibility of this new ESEEM approach with 2H-labeled d10-Leu, four Leu residues within the AChR M2δ peptide were fully mapped out using this ESEEM method. Unique 2H-ESEEM patterns were observed with the 2H-labeled d10-Leu for the AChR M2δ α-helical model peptide. For proteins and peptides with an α-helical secondary structure, deuterium modulation can be clearly observed for i ± 3 and i ± 4 samples, but not for i ± 2 samples. Also, a deuterium peak centered at the 2H Larmor frequency of each i ± 4 sample always had a significantly higher intensity than the corresponding i + 3 sample. This unique feature can be potentially used to distinguish an α-helix from a π-helix or 310-helix. Moreover, 2H modulation depth for ESEEM samples on Leu10 were significantly enhanced which was consistent with a kinked or curved structural model of the AChR M2δ peptide as suggested by previous MD simulations and NMR experiments.
Graphical abstract

INTRODUCTION
A majority of membrane protein structural motifs fall into two categories: membrane-spanning or surface-associated α-helix or α-helix bundles and β-barrels.1,2 More than 70% of membrane proteins with solved 3-D structures are proteins comprised of α-helices.1 As a result of the abundance of secondary structures in membrane proteins, assembly, packing, and interaction of membrane proteins are largely affected, if not dictated by the secondary structure of membrane proteins.3 Generally, better knowledge about the secondary structure, particularly the site-specific secondary structure, is useful toward a better understanding of membrane proteins function, dynamics, and protein—lipid interactions.4 Also, the formation and transition of secondary structural components are crucial for a variety of cellular processes ranging from protein folding and refolding to the amyloid deposits in various neurodegenerative disorders such as Alzheimer’s, Huntington’s, and Parkinson’s disease.5
There are several established biophysical techniques that are used to study secondary structures of membrane proteins such as circular dichroism, solid-state nuclear magnetic resonance (NMR), FT-Raman, and ATR FT-IR.6–10 The Lorigan lab is developing a powerful novel ESEEM approach to probe the local secondary structure of membrane proteins that is advantageous when compared to other structural biological techniques.11–13ESEEM spectroscopy coupled with site-directed spin-labeling (SDSL) can provide valuable local secondary structural information (α-helix and β-strand) of membrane proteins and peptides in lipid bilayers.13 Moreover, the high sensitivity of this ESEEM approach only requires a small amount of sample and a short amount of data acquisition time.11 Those features make this approach extremely suitable for studying inherently difficult systems such as membrane protein systems.14
Figure 1 shows the SDSL and isotopic labeling scheme for this ESEEM approach on a model α-helical peptide (AChR M2δ). For this ESEEM approach, the side chain of one amino acid (such as Leu) in a model peptide at position i was selectively labeled with 2H (blue in Figure 1). A nitroxide spin label was attached to a mutated cysteine residue on a subsequent position on each sample (denoted as i + 1 to i + 4, yellow in Figure 1) which is one, two, three, or four amino acids away from the 2H-labeled Leu.12 ESEEM spectroscopy can detect the weak dipolar coupling between the spin label and 2H atoms up to 8 Å. When the 2H-labeled amino acid and spin-labeled cysteine are three or four amino acids away (i + 3 or i + 4), the 2H-labeled amino acid and the spin label point to the same side of the helix (Figure 1A). Thus, weak dipolar couplings between 2H nuclei and the nitroxide can be detected for i + 3 and i + 4 samples. Due to the fact that a typical α-helix has 3.6 amino acids per turn and a 5.4 Å pitch, the 2H-labeled amino acid side chain and the nitroxide spin label point to opposite sides of the helix when they are one or two amino acids away (i + 1 or i + 2). As shown in Figure 1B, the distance between the 2H on the amino acid side chain and the nitroxide spin label is larger than the ESEEM detection limitation. Thus, deuterium modulation would not be detected in the ESEEM time domain data or in the frequency domain data.11–13
Figure 1.
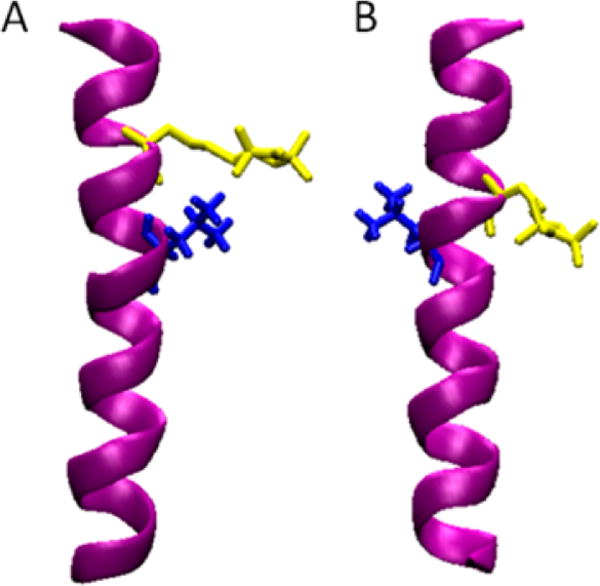
ESEEM experiment SDSL and isotopic label paradigm with a model α-helix (AChR M2δ peptide in purple) for (A) the i ± 3 sample and (B) the i ± 2 sample. 2H-labeled d10-Leu residue is in blue at the 10 position. The Cys residue attached with MTSL is in yellow.
Previously, we demonstrated the feasibility of this ESEEM approach using 2H-labeled d10-Leu and 2H-labeled d8-Val as probes.11,12 Since 2H-labeled d10-Leu has been shown as a very efficient 2H-labeled probe for this ESEEM approach, a more indepth understanding about its ESEEM pattern and variations at different positions could be extremely helpful for its future application in biological systems.11 Here, we further explore the ESEEM signal pattern of an α-helix with 2H-labeled d10-Leu residues and provide a library of valuable data for this 2H-labeled probe for the first time. Multiple 2H-labeled d10-Leu residues on AChR M2δ peptides were mapped out on both sides with SDSL to provide a more detailed description of the ESEEM pattern. All of the ESEEM data sets observed at different sites showed a similar distinguishing α-helical ESEEM spectra pattern. Also, modulation depth of the i ± 4 sample for each set of data was larger than the corresponding i ± 3 sample for 2H-labeled d10. This regularity can potentially be used to distinguish an α-helical structure from other less common helical structures such as 310-helix or π-helix.
EXPERIMENTAL METHODS
The M2δ peptide of the nicotinic acetylcholine receptor (AChR) with 23 amino acid residues was used as an α-helical model for transmembrane peptides and proteins (denoted as AChR M2δ).15,16 Table 1 shows the amino acid sequences of the wild type and all experimental constructs of the M2δ peptides. For this study, four Leu residues at positions 10,11, 17, and 18 were mapped out with this ESEEM approach. Four different peptides were designed on the left (−) and the right (+) side for each Leu residue. The 2H-labeled d10-Leu was at position i with the cysteine (denoted as X) at four successive positions (denoted as i + 1 to i + 4).
Table 1.
Peptide Sequences of Wild Type AChR M2δ and ESEEM Experimental Constructsa
| N-terminal (−) | C-terminal (+) | |
|---|---|---|
| wild type | NH2-EKMSTAISVLLAQAVFLLLTSQR-COOH | |
| Leu10 | NH2-EKMSTAISXiLAQAVFLLLTSQR-COOH | NH2-EKMSTAISViXAQAVFLLLTSQR-COOH |
| NH2-EKMSTAIXViLAQAVFLLLTSQR-COOH | NH2-EKMSTAISViLXQAVFLLLTSQR-COOH | |
| NH2-EKMSTAXSViLAQAVFLLLTSQR-COOH | NH2-EKMSTAISViLAXAVFLLLTSQR-COOH | |
| NH2-EKMSTXISViLAQAVFLLLTSQR-COOH | NH2-EKMSTAISViLAQXVFLLLTSQR-COOH | |
| Leu11 | NH2-EKMSTAISVXiAQAVFLLLTSQR-COOH | NH2-EKMSTAISVLiXQAVFLLLTSQR-COOH |
| NH2-EKMSTAISXLiAQAVFLLLTSQR-COOH | NH2-EKMSTAISVLiAXAVFLLLTSQR-COOH | |
| NH2-EKMSTAIXVLiAQAVFLLLTSQR-COOH | NH2-EKMSTAISVLiAQXVFLLLTSQR-COOH | |
| NH2-EKMSTAXSVLiAQAVFLLLTSQR-COOH | NH2-EKMSTAISVLiAQAXFLLLTSQR-COOH | |
| Leu17 | NH2-EKMSTAISVLLAQAVXiLLTSQR-COOH | NH2-EKMSTAISVLLAQAVFiXLTSQR-COOH |
| NH2-EKMSTAISVLLAQAXFiLLTSQR-COOH | NH2-EKMSTAISVLLAQAVFiLXTSQR-COOH | |
| NH2-EKMSTAISVLLAQXVFiLLTSQR-COOH | NH2-EKMSTAISVLLAQAVFiLLXSQR-COOH | |
| NH2-EKMSTAISVLLAXAVFiLLTSQR-COOH | NH2-EKMSTAISVLLAQAVFiLLTXQR-COOH | |
| Leu18 | NH2-EKMSTAISVLLAQAVFXiLTSQR-COOH | NH2-EKMSTAISVLLAQAVFLiXTSQR-COOH |
| NH2-EKMSTAISVLLAQAVXLiLTSQR-COOH | NH2-EKMSTAISVLLAQAVFLiLXSQR-COOH | |
| NH2-EKMSTAISVLLAQAXFLiLTSQR-COOH | NH2-EKMSTAISVLLAQAVFLiLTXQR-COOH | |
| NH2-EKMSTAISVLLAQXVFLiLTSQR-COOH | NH2-EKMSTAISVLLAQAVFLiLTSXR-COOH | |
Wild type and experimental constructs of AChR M2δ (α-helix) are listed in this table. i is the positions where 2H-labeled d10-Leu was placed. X is the position for MTSL incorporation.
All peptides were synthesized using Fmoc solid phase peptide synthesize chemistry on a CEM microwave solid phase peptide synthesizer.17 A resin with a low loading (0.2 mmol/g) and a high swallow rate was chosen to increase the yield of this relatively hydrophobic peptide sequence. 2H-labeled d10-Leu (Isotec) was dissolved in dimethylformamide and used as the 2H probe and incorporated into each peptide at a designated position (i). Those peptides were cleaved from their resin supports in a cleavage cocktail with trifluoroacetic acid/anisole/triisopropylsilane/H2O (85%/5%/5%/5%) for 3 h. The cleavage cocktail was evaporated by N2 gas flow until peptide precipitation started to appear. Methyl tert-butyl ether was added to assist the precipitation of peptide and wash off any possible residual trifluoroacetic acid. The crude peptides were dried under a vacuum overnight. Reverse-phase HPLC was used for purification with a C4 preparation column and a gradient of 5%–95% solvent B (90% acetonitrile).18 Purified peptides were labeled with a fivefold excess of MTSL (Toronto Research Chemicals) in DMSO for 20 h, and excess MTSL was removed by HPLC. MALDI-TOF was utilized to confirm the molecular weight and the purity of target peptides. HPLC fractions for pure and labeled peptides were lyophilized to a powder form for further usage and storage.
For these experiments, bicelles were used as a membrane mimic system to yield high-quality ESEEM data. MTSL-labeled M2δ peptides were integrated into DMPC/DHPC (3.5/1) bicelles at a 1:1000 molar ratio. X-band CW-EPR (~9 GHz) spectroscopy was used to measure spin concentrations (~150 μM) of all bicelle samples. Three-pulse ESEEM measurements were performed on a Bruker ELEXSYS E580 with an ER 4118X MS3 resonator using a 200 ns tau value with a microwave frequency of ~9.269 GHz at 80 K.11–13 For all samples, a starting T of 386 ns and 512 points in 12 ns increments were used to collect the spectra. All ESEEM data were obtained with 40 μL of bicelle samples and 40 scans.11,12
The original ESEEM time domain data were fit to a two-component exponential decay.11,12 The maximum value of the exponential fit was scaled to 1, and the same factor was applied to the time domain data. The exponential fit was then subtracted from the time domain data and yielded a scaled ESEEM spectrum with modulation about zero. A cross-term averaged Fourier transformation (FT) was performed to the resulting spectrum to generate the corresponding frequency domain with minimized dead time artifacts.11,12 Maximum deuterium peaks intensities at 2.3 MHz were measured and recorded for further analysis.
RESULTS
Figure 2 shows three-pulse ESEEM data for 2H-labeled d10-Leu18 (i−1 through i−4) M2δ peptides incorporated into DMPC/DHPC (3.5/1) bicelles. In the time domain data (Figure 2 left), 2H modulation is clearly observed for i − 3 and i − 4 samples of 2H-labeled d10-Leu18 M2δ peptides. Also, a corresponding 2H peak is clearly observed for those samples centered at the 2H Larmor frequency of 2.3 MHz in the frequency domain data (Figure 2 right). However, there was no 2H modulation observed for the 2H-labeled d10-Leu11 i − 2 or i − 1 M2δ samples. These results reveal a unique ESEEM pattern for an α-helix which is consistent with previous ESEEM results.11–13 Despite the longer side chain with more flexibility of the Leu amino acid, ESEEM spectra still revealed a similar pattern for this α-helix. At the same time, the modulation depth in the time domain data and the FT peak intensity in the frequency domain data of i − 3 and i − 4 positions were comparable to previous results.11 The high signal-to-noise ratio of 2H-labeled d10-Leu makes it a very efficient side chain probe for this ESEEM technique.
Figure 2.
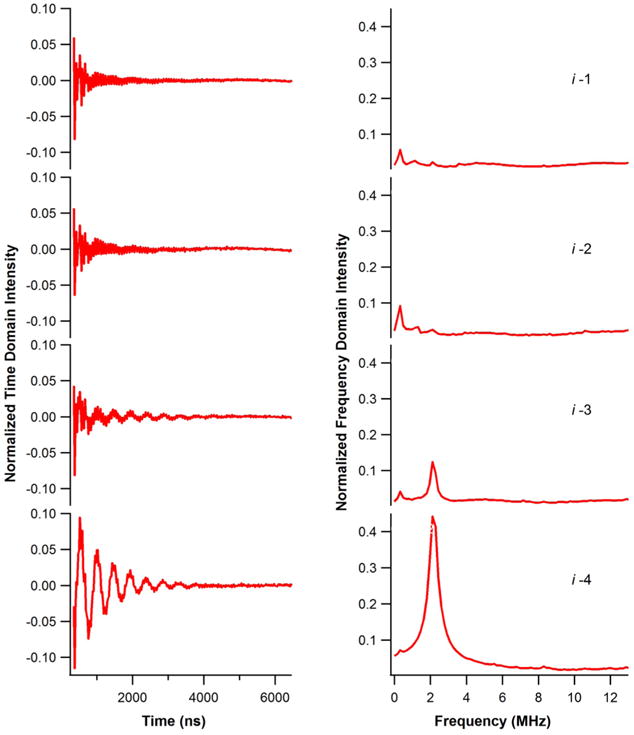
Three-pulse ESEEM experimental data of AChR M2δ with 2H-labeled d10-Leu18 at the N-terminal (−) side in DMPC/DHPC (3.5:1) bicelles at τ = 200 ns for i + 1 to i + 4 in the time domain (left) and the frequency domain (right).
ESEEM data for all eight sets of AChR M2δ samples were collected under the same sample and experimental conditions. The original time domain and frequency domain data are shown in the Supporting Information (Figures S1–S4). Normalized 2H frequency domain FT peak intensities for all data sets were measured and plotted in Figure 3. Several differences were noticed depending upon the location of the 2H-labeled d10-Leu and the spin label. 2H peak intensities for i ± 4 positions varied from 0.1 to 0.6, while for i ± 3 positions it varied from 0.03 to 0.3. Any frequency domain spectra with an obvious 2H peak had a normalized intensity larger than 0.02 (indicted by the red line). Despise the variation of peak intensities between different data sets; it is obvious that all of them have the same pattern within each set of i ± 1 through i ± 4 data as demonstrated in Figure 3A. No 2H modulation was observed for any of the i ± 2 samples. Most i ± 1 positions did not show any modulation above the noise level. Leu11 minus 1 and Leu17 minus 1 position showed a minor 2H peak near the noise level but were several folds lower than its corresponding i ± 3 and i ± 4 positions. Clearly, ESEEM data from all i ± 3 and i ± 4 positions showed significant 2H modulation in the time domain and a strong 2H peak in the frequency domain (see Figures S1–S4). Also, all of the data sets demonstrated high sensitivity with excellent signal-to-noise ratios with less than 2 h of total data acquisition time.
Figure 3.
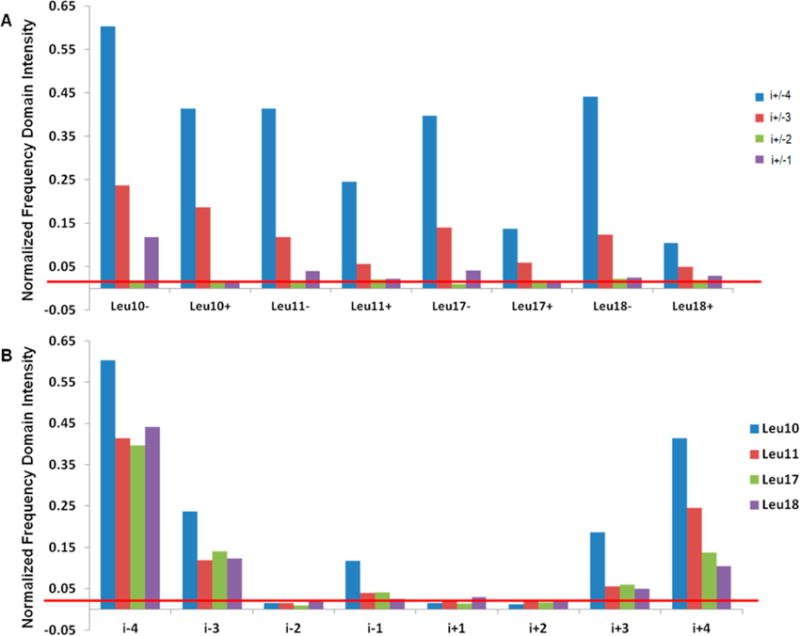
Normalized ESEEM FT domain intensity from all four 2H-labeled d10-Leu data sets. (A) ESEEM data for each position was grouped together to demonstrate the ESEEM pattern observed for different 2H-labeled residues of an α-helix. (B) The same data was rearranged for visualizing the 2H peak intensity variation from i − 4 to i + 4.
In Figure 3B, the ESEEM data are reorganized according to different positions (i ± x) for comparison. The results clearly indicate that most 2H peak amplitudes of i ± 3 and i ± 4 samples on the N-terminal side (−) were higher than the corresponding C-terminal side (+). However, the ESEEM data of the Leu10 position showed significantly larger 2H FT peak amplitudes for both the N-terminal and C-terminal sides. Also, a 2H FT peak for the i − 1 sample is observed for this Leu position.
Figure 4 compares the normalized frequency domain 2H FT peak intensities of i ± 4 and i ± 3 positions for a particular 2H-labeled d10-Leu. The peak intensities of the i ± 4 sample were plotted against the corresponding i ± 3 sample. The red line in Figure 4 represents equal 2H FT peak intensities at i ± 4 and i ± 3 positions, whereas the blue line is indicative of the i ± 4 peak twice as large as the corresponding i ± 3 peak. The graph clearly indicates that all ESEEM data from the 2H-labeled d10-Leu AChR M2δ peptides fell in this region, which indicated that i ± 4 samples always showed a peak with at least a two-fold increase in the 2H FT peak intensity rather than the corresponding i ± 3 with d10-Leu isotopic probe for an α-helical structure.
Figure 4.
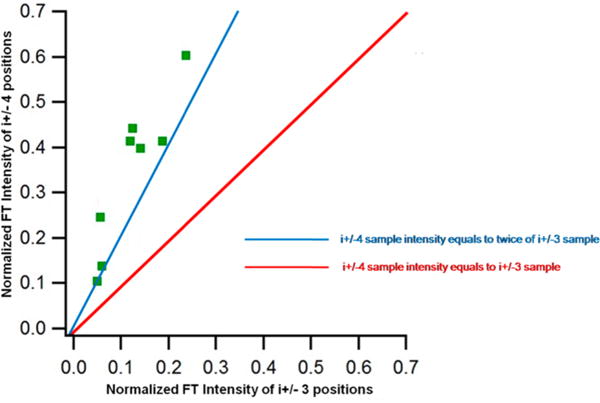
Frequency domain 2H peak intensity comparison between i ± 4 positions and i ± 3 positions for all ESEEM data. Red line represents that the ESEEM 2H FT peak intensity of the i ± 4 is equal to the corresponding i ± 3 sample. Blue line represents that the ESEEM 2H FT peak intensity of the i ± 4 sample is as twice that of the corresponding i ± 3 sample.
DISCUSSION
The nicotinic acetylcholine receptor is a ligand-gated ion channel receptor which is important for signal transduction across plasma membranes.19 It consists of five protein subunits with each of them containing four transmembrane helices, known as M1–M4. The M2 segment is a membrane-spanning α-helix with 23 amino acid residues that is highly conserved and responsible for assembly of the channel pore. High-resolution structures of both the AChR protein and the isolated M2 segment peptide have been obtained.16,20–22 In addition, it has been shown that the AchR M2δ peptide has a 14° tilt angle with respect to the membrane normal upon insertion into DMPC bilayers.18,23 In previous ESEEM studies utilizing 2Hlabeled d8 and 2H-labeled d10-Leu as the 2H probe, ESEEM data indicated that the distance between 2H atoms on the amino acid side chain and the spin label are within 8 Å for i + 3 and i + 4 positions, but not for i + 1 or i + 2 positions.11,12
General ESEEM Pattern for α-Helix
In this research, all ESEEM data showed a similar pattern, which is indicative of an α-helical structure. Weak dipolar coupling can be detected between 2H nuclei on the Leu side chain and a nitroxide spin label for i ± 3 and i ± 4 positions for all Leu residues on both the N-terminal (−) and the C-terminal (+) sides. Those results indicated that side chain distances between 2H-labeled Leu residues and spin labels are within the 8 Å detection limit for i ± 3 and i ± 4 positions due to the unique 3.6 amino acids per turn feature of the α-helical structure. Only minor 2H peaks around the noise level, if any, were detected for i ± 1 positions. For all the constructs that had been tested with this ESEEM approach, none of them showed any 2H modulation for i ± 2 positions. Different conformations of MTSL and Leu might be favored due to unique side chain or tertiary interactions depending upon their environment. All of these factors play a role in the 2H modulation depth and can affect the corresponding FT intensity. However, all of the ESEEM results obtained so far demonstrated that the ESEEM spectra pattern for an α-helix (i ± 1 to i ± 4) was not affected by the flexibility of the MTSL or the Leu side chain, which verify the reliability of this ESEEM approach on identifying secondary structural motifs.
Relative 2H Peak Intensity for i ± 3 and i ± 4 Positions
Previous ESEEM studies have revealed a distinguishing pattern for an α-helical secondary structure with 2H-labeled d10-Leu and 2H-labeled d8-Val.11,12 2H modulation can be detected for i ± 3 and i ± 4 positions, but not i ± 1 or i ± 2 positions. Beside the similar pattern that 2H-labeled d10-Leu and 2H-labeled d8-Val share for an α-helix, Leu demonstrated some unique features due to the longer and more flexible side chain. The i ± 4 to i ± 3 ratio shown in Figure 4 reveals a unique pattern for Leu in which the i ± 4 positions have much larger 2H peaks when compared to the corresponding i ± 3 positions. Since a standard α-helix has a 3.6 amino acid per turn regularity, the angle between the side chain of the amino acid and the MTSL with respect to the helical axis was smaller in i ± 4 positions than i ± 3 positions.24–26 As the side chain gets longer, the distance between the 2H atoms on the Leu side chain and the nitroxide spin label reflect this angle difference more significantly. Thus, the ESSEM results always showed a larger 2H FT peak when utilizing 2H-labeled d10-Leu as a probe. The ESEEM results indicate that the small angular difference between i ± 4 and i ± 3 positions of an α-helix can be detected with this ESEEM approach.
With this unique pattern of 2H-labeled d10-Leu, this new approach could potentially identify less abundant helical structures such as a 310-helix or a π-helix. In the case of the 310-helix, the i ± 3 position should have a larger 2H FT ESEEM peak than the corresponding i ± 4 position due to the 3.1 amino acid per turn regularity while i ± 1 and i ± 2 positions should not show any 2H modulation.27 As for the π-helix, it has four amino acids per turn. Thus, the i ± 3 and the i ± 1 should have similar 2H modulation depths, while the i ± 2 would not show any modulation as a normal α-helical structure. Also, the i ± 4 position should have the largest peak when compared to the corresponding i ± 1 and i ± 3 positions.
ESEEM 2H Peak Intensity and 2H-Labeled Side Chain Orientations
The ESEEM modulation depth is related to 1/r6, where r is the distance between nuclei on the 2H-labeled side chain and the spin label. This distance varies because of multiple 2H nuclei and the different conformations of both the spin label and the side chain. The MTSL spin label has three torsion angle rotations about χ1, χ2, and χ3 and two additional free torsion angle rotations about χ4 and χ5.28 However, it can be seen in Figure 3B that all ESEEM 2H FT peak amplitudes of the i ± 3 and i ± 4 positions on the N-terminal side (−) were larger than the corresponding C-terminal side (+), which indicated that the spin label and 2H-labeled side chain were generally closer together on the N-terminal side consistently regardless of the MTSL position (i ± 3 or i ± 4). Thus, it is more likely that those distances were dominated by the relative orientation of the 2H-labeled Leu side chain, which was fixed for each position probed rather than the spin-labeled Cys side chain in these cases. Leu side chains have two torsion angle rotations about χ1 and χ2 and two free rotation modes about the Cγ and Cδ bonds, which correspond to two (CD3) methyl groups.24 Thus, different conformations of the Leu side chain might be favored due to dynamic and tertiary interactions that can affect the observed 2H modulation depth.24,26,29 The ESEEM data suggest that 2H-labeled Leu side chains were orientated more toward the N-terminal side of the peptide on 10, 11, 17, and 18 positions. Previous computational simulation studies have indicated that the AChR M2δ peptide has more polar amino acids and is more flexible on the N-terminal end.30 Thus, interactions of those polar side chains with the membrane surface and water environment outside the membrane could cause the amino acid side chains on the N-terminal end to tilt slightly toward the surface of the membrane bilayer.31 As a consequence, it is more favorable for the Leu side chains to tilt toward the N-terminal side. Also, the kink in the peptide may play a role in this observation (see below). Additional membrane peptides will be probed to study this.
ESEEM Pattern Deviation at the Leu10 Position Is Consistent with the Kinked Model of the M2δ Peptide
The structure of the AChR M2δ peptide has been characterized via solution NMR in dodecylphosphocholine (DPC) micelles (PDB: 1A11) and by solid-state NMR in mechanically oriented 1, 2-dimyristoyl-sn-glycerophosphcholine (DMPC) bilayers (PDB: 1EQ8).16 The results indicate that the M2δ peptide is a transmembrane α-helix with no obvious kink. However, it should be noted the solution NMR structure was conducted in a DPC micelle complex and not a lipid bilayer.32 Also, mechanically aligned solid-state NMR structural studies require samples with a highly oriented lipid bilayer, which is difficult to achieve and highly lipid- or peptide-dependent.33 In contrast to those early NMR structures, cryo-EM, molecular modeling, and magical angle spinning (MAS) solid-state NMR studies of the AChR M2δ peptide suggested the helix is kinked in the vicinity of Leu11.22,34,35 Early mutagenesis studies and sequence comparisons suggested that Leu11 plays a key role in the gating mechanism of the AChR channel.36Cryo-EM and molecular modeling studies proposed the open and closed states of the AChR channel with a bending motion at this position.22 In the closed state, the AChR M2δ segment is kinked so that the Leu11 side chain adapts a conformation to prevent the ion conduction.37 In addition, the MAS solid-state NMR results showed peptide backbone torsion angles at positions Leu10, Leu11, and Ala12 which deviate from a classic α-helical conformation.20
As mentioned above, 2H ESEEM peak amplitudes for the Leu10 position of M2δ peptide were enhanced on both the N-terminal (−) and the C-terminal (+) side at i ± 3 and i ± 4 positions when compared to all other Leu residues in this study. Also, both sides of the Leu10 i ± 1 position showed 2H modulation larger than the other i ± 1 position on the same side. The 2H FT peak intensity at the i − 1 position is especially significant when compared to the other i ± 1 position (Figure 3B). The larger ESEEM FT intensity at the Leu10 position clearly indicates that 2H atoms on the Leu side chain and spin-label are closer to each other around Leu10 when compared to other positions. This can be explained by the structure irregularity such as a kink or curve at this site, which have been suggested by previous NMR, electron microscopy (EM), and computational simulation studies.20,34,38 The kinked model of the AChR M2δ segment shows a slight curve around residue Leu11 (Figure 5B, right). Figure 5C illustrates the effects of the kink at Leu11 on the side chain proximities on both the inner and outer sides of the channel. The side chain of Leu11 (red dots) points toward the center of the channel, while the helix bends away from the center of the channel due to the kink. Side chains of residues such as Leu10, Ala6, and Ala14 locate in the outer side of the channel and point outward from the center of the channel (blue dots). As shown in Figure 5C, the outer side of the helix would be more crowded with side chains (right) when compared to that of the straight peptide (left). Thus, side chains of those residues located in the outer side of the channel would be closer to each other due to the kink. As a result, closer distances between Leu10 side chain and MTSL at Ala6 (Leu10 i − 4) and Ala14 (Leu10 i + 4) positions, which are indicated by enhanced ESEEM 2H FT peaks, were observed. Larger ESEEM 2H FT peaks observed for Leu10 position samples are consistent with previously reported kinked model of AChR M2δ peptide.20,34,38
Figure 5.
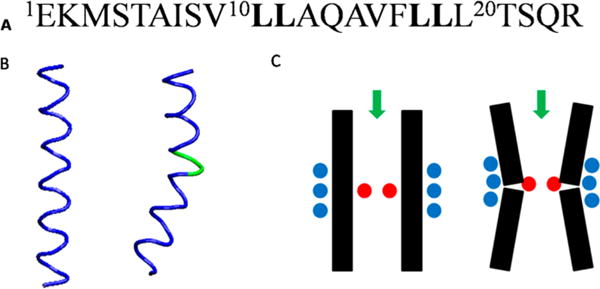
(A) AChR M2δ peptide sequence with Leu residues highlighted in bold. (B) Structural representations of the straight (left) and the kinked (right) M2δ peptides. (C) Structural representations of the AChR channel with straight (left) and kinked (right) M2δ peptides. Green arrows point toward the inside of the channel. Leu11 is represented by red dots, while Ala6, Leu10, and Ala14 are shown as blue dots.
CONCLUSIONS
In this study, 2H-labeled d10-Leu has been shown to be a very powerful secondary structural probe with high sensitivity and an excellent signal-to-noise ratio to study the local α-helical secondary structure. The ESEEM data from four different Leu residues on the AChR M2δ peptide further validates this structural biology approach and provides researchers with a reference to probe α-helical secondary structural components for proteins and peptides. Moreover, the ratio of 2H FT peak intensities between the i ± 4 and the i ± 3 samples can be potentially utilized to determine less predominant helical structures such as a 310-helix and a π-helix. Further studies need to be conducted to explore the application of this ESEEM approach to identify and distinguish more secondary structures and structural motifs. Also, different 2H-labeled amino acids with different numbers of 2H atoms, side chain length, and rigidity should be studied with this ESEEM approach to establish ESEEM patterns for different secondary structures. Due to the presence of multiple 2H atoms on the probe and side chain flexibility, it is still difficult to obtain quantitative distance information. However, with 2H-labeled side chains with less 2H atoms and more rigid spin labels such as tetrathiatriarylmethyl (TAM), 2,2,6,6-tetramethylpiperidine-1-oxyl-4-amino-4-carboxylic acid (TOAC) and bifunctional spin label (BSL), more quantitative distance information can be obtained.39,40
This ESEEM method uses SDSL and selective deuterium labels, both of which can be incorporated into standard expression systems using site-directed mutagenesis and selective isotopic labeling techniques for applications to larger protein systems. Thus, this new ESEEM secondary structure approach can be applied to a wide variety of different protein systems that are not amiable to other biophysical techniques.
Supplementary Material
Acknowledgments
This work was generously supported by National Institutes of Health Grant R01 GM108026 and by the National Science Foundation Grant CHE-1305664. The pulsed EPR spectrometer was purchased through the NSF and the Ohio Board of Regents grants (MRI-0722403).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpcb.5b09040.
Three-pulse ESEEM experimental data of AChR M2δ with 2H-labeled d10-Leu at the N-terminal (−) and C-terminal (+) sides in DMPC/DHPC (3.5:1) bicelles at τ = 200 ns for i + 1 to i + 4 in time domain and frequency domain (PDF)
The authors declare no competing financial interest.
References
- 1.McLuskey K, Roszak AW, Zhu Y, Isaacs NW. Crystal Structures of All-Alpha Type Membrane Proteins. Eur Biophys J. 2010;39:723–755. doi: 10.1007/s00249-009-0546-6. [DOI] [PubMed] [Google Scholar]
- 2.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of Proteins in Membranes Database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 3.Kurochkina N. Helix-Helix Interactions and Their Impact on Protein Motifs and Assemblies. J Theor Biol. 2010;264:585–592. doi: 10.1016/j.jtbi.2010.02.026. [DOI] [PubMed] [Google Scholar]
- 4.Bordag N, Keller S. Alpha-Helical Transmembrane Peptides: A “Divide and Conquer″” Approach to Membrane Proteins. Chem Phys Lipids. 2010;163:1–26. doi: 10.1016/j.chemphyslip.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Gross M. Proteins That Convert from Alpha Helix to Beta Sheet: Implications for Folding and Disease. Curr Protein Pept Sci. 2000;1:339–347. doi: 10.2174/1389203003381289. [DOI] [PubMed] [Google Scholar]
- 6.Whitmore L, Wallace BA. Protein Secondary Structure Snalyses from Circular Dichroism Spectroscopy: Methods and Reference Databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 7.Greenfield NJ. Using Circular Dichroism Spectra To Estimate Protein Secondary Structure. Nat Protoc. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu X, Lorigan GA. Secondary Structure, Backbone Dynamics, and Structural Topology of Phospholamban and Its Phosphorylated and Arg9cys-Mutated Forms in Phospholipid Bilayers Utilizing 13C and 15N Solid-State NNR Spectroscopy. J Phys Chem B. 2014;118:2124–2133. doi: 10.1021/jp500316s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roach CA, Simpson JV, JiJi RD. Evolution of Quantitative Methods in Protein Secondary Structure Determination via Deep-Ultraviolet Resonance Raman Spectroscopy. Analyst. 2012;137:555–562. doi: 10.1039/c1an15755h. [DOI] [PubMed] [Google Scholar]
- 10.Carbonaro M, Nucara A. Secondary Structure of Food Proteins by Fourier Transform Spectroscopy in the Mid-Infrared Region. Amino Acids. 2010;38:679–690. doi: 10.1007/s00726-009-0274-3. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Sahu ID, Mayo DJ, McCarrick RM, Troxel K, Zhou A, Shockley E, Lorigan GA. Enhancement of Electron Spin Echo Envelope Modulation Spectroscopic Methods To Investigate the Secondary Structure of Membrane Proteins. J Phys Chem B. 2012;116:11041–11045. doi: 10.1021/jp304669b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayo D, Zhou A, Sahu I, McCarrick R, Walton P, Ring A, Troxel K, Coey A, Hawn J, Emwas AH, Lorigan GA. Probing the Structure of Membrane Proteins with Electron Spin Echo Envelope Modulation Spectroscopy. Protein Sci. 2011;20:1100–1104. doi: 10.1002/pro.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou A, Abu-Baker S, Sahu ID, Liu L, McCarrick RM, Dabney-Smith C, Lorigan GA. Determining α-Helical and β-Sheet Secondary Structures via Pulsed Electron Spin Resonance Spectroscopy. Biochemistry. 2012;51:7417–7419. doi: 10.1021/bi3010736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klare JP, Steinhoff HJ. Spin Labeling EPR. Photosynth Res. 2009;102:377–390. doi: 10.1007/s11120-009-9490-7. [DOI] [PubMed] [Google Scholar]
- 15.Oblatt-Montal M, Bühler LK, Iwamoto T, Tomich JM, Montal M. Synthetic Peptides and Four-Helix Bundle Proteins as Model Systems for the Pore-Forming Structure of Channel Proteins. I. Transmembrane Segment M2 of the Nicotinic Cholinergic Receptor Channel Is a Key Pore-Lining Structure. J Biol Chem. 1993;268:14601–14607. [PubMed] [Google Scholar]
- 16.Opella SJ, Marassi FM, Gesell JJ, Valente AP, Kim Y, Oblatt-Montal M, Montal M. Structures of the M2 Channel-Lining Segments from Nicotinic Acetylcholine and NMDA Receptors by NMR Spectroscopy. Nat Struct Biol. 1999;6:374–379. doi: 10.1038/7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandrudu S, Simerska P, Toth I. Chemical Methods for Peptide and Protein Production. Molecules. 2013;18:4373–4388. doi: 10.3390/molecules18044373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayo DJ, Inbaraj JJ, Subbaraman N, Grosser SM, Chan CA, Lorigan GA. Comparing the Structural Topology of Integral and Peripheral Membrane Proteins Utilizing Electron Paramagnetic Resonance Spectroscopy. J Am Chem Soc. 2008;130:9656–9657. doi: 10.1021/ja803590w. [DOI] [PubMed] [Google Scholar]
- 19.Itier V, Bertrand D. Neuronal Nicotinic Receptors: from Protein Structure to Function. FEBS Lett. 2001;504:118–125. doi: 10.1016/s0014-5793(01)02702-8. [DOI] [PubMed] [Google Scholar]
- 20.Long JR, Mills FD, Raucci F. A High Resolution Structure of the Putative Hinge Region in M2 Channel-Lining Segments of the Nicotinic Acetylcholine Receptor. Biochim Biophys Acta Biomembr. 2007;1768:2961–2970. doi: 10.1016/j.bbamem.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Sankararamakrishnan R, Sansom MS. Structural Features of Isolated M2 Helices of Nicotinic Receptors: Simulated Annealing via Molecular Dynamics Studies. Biophys Chem. 1995;55:215–230. doi: 10.1016/0301-4622(95)00006-j. [DOI] [PubMed] [Google Scholar]
- 22.Unwin N. Acetylcholine Receptor Channel Imaged in the Open State. Nature. 1995;373:37–43. doi: 10.1038/373037a0. [DOI] [PubMed] [Google Scholar]
- 23.Newstadt JP, Mayo DJ, Inbaraj JJ, Subbaraman N, Lorigan GA. Determining the Helical Tilt of Membrane Peptides Using Electron Paramagnetic Resonance Spectroscopy. J Magn Reson. 2009;198:1–7. doi: 10.1016/j.jmr.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batchelder LS, Sullivan CE, Jelinski LW, Torchia DA. Characterization of Leucine Side-Chain Reorientation in Collagen-Fibrils by Solid-State 2H NMR. Proc Natl Acad Sci USA. 1982;79:386–389. doi: 10.1073/pnas.79.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Columbus L, Kalái T, Jekö J, Hideg K, Hubbell WL. Molecular Motion of Spin Labeled Side Chains in Alpha-Helices: Analysis by Variation of Side Chain Structure. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 26.Mulder FA. Leucine Side-Chain Conformation and Dynamics in Proteins from 13C NMR Chemical Shifts. ChemBioChem. 2009;10:1477–1479. doi: 10.1002/cbic.200900086. [DOI] [PubMed] [Google Scholar]
- 27.Kubota T, Lacroix JJ, Bezanilla F, Correa AM. Probing α-3(10) Transitions in a Voltage-Sensing S4 Helix. Biophys J. 2014;107:1117–1128. doi: 10.1016/j.bpj.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beier C, Steinhoff HJ. A Structure-Based Simulation Approach for Electron Paramagnetic Resonance Spectra Using Molecular and Stochastic Dynamics Simulations. Biophys J. 2006;91:2647–2664. doi: 10.1529/biophysj.105.080051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wand AJ. Dynamic Activation of Protein Function: A View Emerging from NMR Spectroscopy. Nat Struct Biol. 2001;8:926–931. doi: 10.1038/nsb1101-926. [DOI] [PubMed] [Google Scholar]
- 30.Kessel A, Shental-Bechor D, Haliloglu T, Ben-Tal N. Interactions of Hydrophobic Peptides with Lipid Bilayers: Monte Carlo Simulations with M2delta. Biophys J. 2003;85:3431–3444. doi: 10.1016/S0006-3495(03)74765-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kessel A, Haliloglu T, Ben-Tal N. Interactions of the M2delta Segment of the Acetylcholine Receptor with Lipid Bilayers: A Continuum-Solvent Model Study. Biophys J. 2003;85:3687–3695. doi: 10.1016/S0006-3495(03)74785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A Collision Gradient Method To Determine the Immersion Depth of Nitroxides in Lipid bilayers: Application to Spin-Labeled Mutants of Bacteriorhodopsin. Proc Natl Acad Sci USA. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Byström T, Strandberg E, Kovacs FA, Cross TA, Lindblom G. Influence of Transmembrane Peptides on Bilayers of Phosphatidylcholines with Different Acyl Chain Lengths Studied by Solid-State NMR. Biochim Biophys Acta Biomembr. 2000;1509:335–345. doi: 10.1016/s0005-2736(00)00316-3. [DOI] [PubMed] [Google Scholar]
- 34.Sankararamakrishnan R, Adcock C, Sansom MS. The Pore Domain of the Nicotinic Acetylcholine Receptor: Molecular Modeling, Pore Dimensions, and Electrostatics. Biophys J. 1996;71:1659–1671. doi: 10.1016/S0006-3495(96)79370-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hung A, Tai K, Sansom MS. Molecular Dynamics Simulation of the M2 Helices within the Nicotinic Acetylcholine Receptor Transmembrane Domain: Structure and Collective Motions. Biophys J. 2005;88:3321–3333. doi: 10.1529/biophysj.104.052878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galzi JL, Devillers-Thiéry A, Hussy N, Bertrand S, Changeux JP, Bertrand D. Mutations in the Channel Domain of a Neuronal Nicotinic Receptor Convert Ion Selectivity from Cationic to Anionic. Nature. 1992;359:500–505. doi: 10.1038/359500a0. [DOI] [PubMed] [Google Scholar]
- 37.Unwin N. Refined Structure of the Nicotinic Acetylcholine Receptor at 4A Resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 38.Tikhonov DB, Zhorov BS. Kinked-Helices Model of the Nicotinic Acetylcholine Receptor Ion Channel and Its Complexes with Blockers: Simulation by the Monte Carlo Minimization Method. Biophys J. 1998;74:242–255. doi: 10.1016/S0006-3495(98)77783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fielding JA, Concilio MG, Heaven G, Hollas MA. New Developments in Spin Labels for Pulsed Dipolar EPR. Molecules. 2014;19:16998–17025. doi: 10.3390/molecules191016998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahu ID, McCarrick RM, Troxel KR, Zhang R, Smith JH, Dunagan MM, Swartz MS, Rajan PV, Kroncke BM, Sanders CR, Lorigan GA. DEER EPR Measurement for Membrane Protein Structures via Bifunctional Spin Labels and Lipodisq Nanoparticles. Biochemistry. 2013;52:6627–6632. doi: 10.1021/bi4009984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


