Abstract
The current study aimed to develop a method to rapidly, sensitively and practically screen for the epidermal growth factor receptor (EGFR) T790M mutation. This method combines an allele-specific competitive blocker (ACB) with a TaqMan quantitative polymerase chain reaction (PCR) amplification refractory mutation system (ARMS) in a one-step reaction. Using a mimic of a human genomic DNA panel containing serially diluted mutant alleles, the performance efficacy of this method was assessed. Using this method, the EGFR T790M mutation was detected in tyrosine kinase inhibitor (TKI)-naïve samples obtained from 27 non-small cell lung cancer (NSCLC) patients with EGFR-activating mutations. The association between de novo T790M mutations and the clinical benefit of EGFR-TKI treatment was also analysed. The sensitivity of this method was as low as 0.01%. In the samples from the 27 NSCLC patients, this method identified 6 mutant patients (22.2%), which was higher than the detection rate with scorpion ARMS (0.0%). No clinical variables were associated with the occurrence of a de novo T790M mutation. The median progression-free survival time in the TKI-naïve patients with a T790M mutation was shorter that that of patients without the mutation, but the difference was not significant (3.2 vs. 19.5 months, respectively; P=0.256). The median overall survival time in the groups with or without T790M mutation also did not significantly differ (10 vs. 20 months, respectively; P=0.689). Overall, the ACB-ARMS PCR method could be useful for detecting the EGFR T790M mutation in clinical samples that contain only a small number of mutant alleles. The clinical significance of a de novo T790M mutation should be further investigated.
Keywords: epidermal growth factor receptor, T790M mutation, non-small cell lung cancer, allele-specific competitive blocker-amplification refractory mutation system TaqMan quantitative polymerase chain reaction
Introduction
The epidermal growth factor receptor (EGFR) T790M mutation has gradually become a research hotspot in the field of targeted non-small cell lung cancer (NSCLC) therapy in recent years. Secondary T790M mutation accounts for ~50% of acquired EGFR tyrosine kinase inhibitor (TKI) resistance in originally highly responsive NSCLC patients with EGFR activating mutations, including exon 19 small in-frame deletions and exon 21 L858R mutations (1). De novo T790M EGFR mutations have been observed in TKI-naïve patients and could predict a shorter response duration following EGFR-TKI treatment (2,3). Furthermore, the incidence of de novo T790M mutation may be more prevalent than expected due to the limited sensitivity of detection methods, such as direct sequencing (2–5). Next generation EGFR-TKIs, including AZD9291, CO-1686 and HM61713, could inhibit EGFR T790M and activating mutations, and have been proven beneficial to NSCLC patients with EGFR-TKI (gefitinib or erlotinib) resistance caused by secondary T790M mutation (6–8). Thus, detecting the EGFR T790M mutation in NSCLC patients is important for monitoring the presence of acquired resistance and for selecting patients for treatment with next generation EGFR-TKIs (1,6–8). However, reliable methods for EGFR T790M mutation detection have not yet been established and widely accepted. The major difficulty in establishing such a method is that EGFR T790M mutant cells are mixed with a large amount of wild-type cells derived from the site of tissue sampling.
A number of methods to overcome this obstacle have been reported, including mutant-enriched polymerase chain reaction (PCR) (9,10), the amplification refractory mutation system (ARMS) (4,11), peptide nucleic acid (PNA)-clamping PCR (12–15), combining scorpion ARMS with whole genome amplification (16), combining co-amplification at lower denaturation temperature-PCR with TaqMan technology (17), molecular beacon-based quantitative PCR (18), the beads, emulsion, amplification and magnetics (BEAMing) assay (19), PCR-clone hybridisation (5) and matrix-assisted laser desorption/ionisation-time of flight mass spectrometry (2). Although these methods have significantly improved the detection limit ranging from 1 to 0.01%, the majority of these methods are unavailable and cannot be used easily in a clinical setting. Additionally, certain methods involve a number of steps of post-PCR processing, which may increase PCR-product contamination.
In the present study, a sensitive and practical method was developed that utilises an allele-specific competitive blocker (ACB) coupled with ARMS TaqMan quantitative PCR in a one-step reaction tube. This method allowed the preferential amplification of the mutant DNA through use of an ACB to prohibit wild-type allele elongation and was thus used to screen for the T790M mutation in 27 TKI-naïve NSCLC specimens.
Patients and methods
Plasmid construction
Recombinant plasmids encoding wild-type and T790M (2169 C>T) mutant EGFR exon 20 were constructed according to the method previously reported by Board et al (20). Briefly, corresponding outer and mutant primers (Sangon Biotech Co., Ltd., Shanghai, China) were used to yield half fragments that each had complimentary ends and contained a mutant base. The sequences of the outer primers were as follows: Primer a, 5′-TTCACAGCCCTGCGTAAAC-3′; primer d, 5′-TTTCCACATGCAGATGGGAC-3′. The sequences of the mutant primers as follows: Primer b, 5′-CGAAGGGCATGAGCTGCATGATGAGCTGCACGGTGG-3′; primer c, 5′-CCACCGTGCAGCTCATCATGCAGCTCATGCCCTTC-3′. The PCR assay was performed in a 20-µl mixture containing 2 µl of 10X PCR buffer, 0.5 units of HotStarTaq DNA polymerase (Qiagen China Co., Ltd.), 0.25 µM of each primer (Sangon China Co., Ltd.) and 2 µl of DNA in a PCR instrument (ABI 2720; Applied Biosystems, Beijing, China). The PCR conditions were as follows: Initial denaturation at 95°C for 5 min, followed by 35 cycles at 95°C for 15 sec, 54°C for 30 sec and 72°C for 30 sec, then a hold at 72°C for 7 min and a final permanent hold at 4°C. The PCR products were mixed and amplified with outer primers and the PCR process was same as the aforementioned method. The self-priming of the complementary half fragments and subsequent amplification created a final product with a mutant base. The product was ligated into the pMD19-T plasmid (Tiangen Biotech Co., Ltd., Beijing, China) to generate recombinant containing mutant alleles, which were confirmed by sequencing (Sangon, Biotech Co., Ltd.). The plasmid DNA was extracted using Tiangen Plasmids DNA kits (Tiangen Biotech Co., Ltd.).
Study population, sample collection and processing
In total, 27 NSCLC patients with activating mutations (19 Del or 21 L858R) were recruited for this study between January 2008 and December 2012 at Peking Union Medical College Hospital (PUMCH; Beijing, China). All patients received gefitinib as a first-line or multiple-line therapy during the course of cancer treatment. The clinical data of the patients, including the demographics, pathological type, stage, smoking status and treatment information, were obtained from each patient's medical records and were reviewed by the physicians. The progression-free survival (PFS) time was calculated based on the time of gefitinib treatment initiation until disease progression or mortality. Overall survival (OS) time was defined as the period following the date of diagnosis, not the date of informed consent, until mortality.
A tumour biopsy sample was obtained from each patient (n=27) prior to gefitinib treatment. The samples were formalin-fixed, paraffin-embedded (FFPE) and retrieved from the Department of Pathology, PUMCH. The tumour tissues were isolated by manual microdissection. The tumour content in each microdissected sample was ≥75%. The DNA was extracted using a QIAamp DNA FFPE Tissue kit (Qiagen China Co., Ltd.) according to the manufacturer's protocols, and then stored at −20°C for further testing.
The study protocol was approved by the Institutional Review Boards of PUMCH. All patients provided written informed consent for the procurement of tumour specimens.
Mimic human genomic DNA panel of different concentrations of T790M mutant EGFR
Serially diluted plasmids (10 µl) containing 10,000, 1,000, 100 or 10 copies/µl of the T790M mutation were each added to 90 µl of human genomic DNA (30 ng/µl; Sigma-Aldrich China Inc., Shanghai, China). As one copy of genomic DNA is equivalent to 3 pg of DNA on average, this mimic human genomic DNA panel consisted of 10, 1, 0.1 and 0.01% mutants.
Genotyping by ACB-ARMS TaqMan quantitative PCR (ACB-ARMS PCR) assay
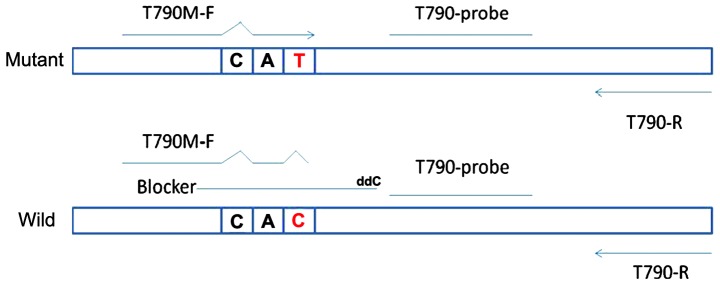
The ACB-ARMS PCR assay for the detection of the EGFR T790M mutation was established in the molecular laboratories of the Beijing BGI-GBI Biotech Co., Ltd. (Beijing, China). In the ACB-ARMS PCR assay, dideoxynucleotide-labelled oligonucleotides were used as a competitive blocker to suppress the amplification of the wild-type allele via subtle differences in the melting temperature between the blocker-wild-type and blocker-mutant DNA hybrids. The principle of this assay is depicted in Fig. 1, and the sequences are shown in Table I.
Figure 1.
A schematic presentation of allele-specific competitive blocker-PCR. The forward primer of T790M mutation harbours another mismatched site (C>T) at the third base of 3′-ends to decrease the non-specific combination of primer with wild-type template alleles. The non-extendable blocker used here consisted of 23 bp of normal base sequences of the EGFR exon 20 gene, overlapped with the forward primer by a few bases, with the variant base position approximately in the middle of the oligonucleotide, and was labelled with dideoxynucleotides in the 3′-end to prohibit wild-type allele elongation with DNA polymerase. The inhibition of wild-type template amplification is temperature-independent, and its value for the blocker-wild type hybrids is higher than that for the blocker-T790M mutation. With appropriate annealing and extension conditions, the blocker preferentially hybridises to wide-type alleles rather than to mutant alleles. Consequently, DNA replication proceeds less smoothly in wild-type DNA than in mutant DNA. Finally, the PCR selectively amplifies mutant alleles. PCR, polymerase chain reaction.
Table I.
Sequences of primers, blocker and probes.
| Detection | Primers (5′-3′) | Probes (5′-3′) |
|---|---|---|
| T790M | ||
| ARMS | F: 5′-CACCGTGCAGCTCATTAT-3′ | 5′FAM-CCTTCCCTGGACTATGT-BHQ3′ |
| R: 5′-CACACACCAGTTGAGCAGGTACT-3′ | ||
| Blocker | 5′-TGCAGCTCATCACGCAGCTCATG-ddC3′ | |
| Internal control | F: 5′-TGCCAAGGCACGAGTAACAAG-3′ | 5′FAM-TCTCAGCCTCCAGAGGATGTTCAA |
| R: 5′TCCAAATTCCCAAGGACCAC-3′ | TAACT-BHQ3′ |
ARMS, amplification refractory mutation system.
The ACB-ARMS PCR assay was performed in a 20-µl mixture containing 2 µl of 10X PCR buffer, 0.5 units of HotStarTaq DNA polymerase (Qiagen China Co., Ltd.), 0.15 µM of probe, 0.15 µM of blocker, 0.25 µM of each primer (Sangon China Co., Ltd.) and 2 µl of DNA in a fluorometric PCR instrument (ABI 7300 and StepOne; Applied Biosystems). The detection includes two phases: i) Step one enriches the T790M mutation with 5 cycles of 95°C for 15 sec, 68°C for 20 sec, 60°C for 30 sec and 72°C for 20 sec after denaturing at 95°C for 5 min; ii) step two entails normal amplification with 40 cycles of 95°C for 15 sec, 68°C for 20 sec and 60°C for 45 sec (fluorescence collection). The mimic human genomic DNA was used as the positive control, RNase and DNase free water was used as the negative control and β-actin was used as the reference control. The assay was repeated three times.
Validation of T790M mutation by clone sequencing
The T790M-positive samples detected by ACB-ARMS PCR were confirmed with clone sequencing. The wild human genomic DNA was used as the controls. The forward primer used here was similar to T790M ARMS-F, but the mutated base was reduced and lacked the third mismatched base. The sequences of the primers for PCR were as follows: Forward, 5′-CACCGTGCAGCTCATCA-3′ and reverse, 5′-GATGGGACAGGCACTGATTT-3′. The length of the PCR product was 307 bp. The blocker remained unchanged. The PCR was performed in a 20-µl mixture containing 2 µl of 10X PCR buffer, 0.15 µM of probe, 0.15 µM of blocker, 0.25 µM of each primer, 0.5 units of HotStarTaq DNA polymerase and 2 µl of DNA in a fluorometric PCR instrument (ABI 7300 and StepOne; Applied Biosystems). The PCR procedure included an initial denaturation at 95°C for 5 min followed by 35 cycles at 95°C for 30 sec, 68°C for 20 sec for blocker binding, 60°C for 30 sec and 72°C for 30 sec, then a hold at 72°C for 7 min and a final permanent hold at 4°C. The product was ligated into the pMD19-T plasmid (Tiangen Biotech Co., Ltd.). The positive monoclonal colonies identified by colony PCR were sequenced (Sangon Biotech Co., Ltd.).
Statistical analysis
For categorical data, Fisher's exact test was performed to compare differences between groups. The Kaplan-Meier method was used to estimate survival curves for PFS and OS. Log-rank tests were used to compare survival curves between different EGFR mutations. A two-sided P-value of <0.05 was considered to indicate a statistically significant difference. All analyses were performed using SPSS software, version 13.0 (SPSS Inc., Chicago, IL, USA).
Results
Sensitivity of ACB-ARMS PCR for detecting EGFR T790M mutation
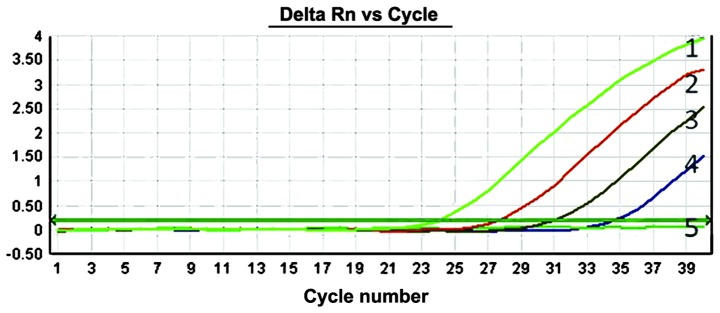
To evaluate the sensitivity of the ACB-ARMS PCR assay, mimic human genomic DNA panels consisting of 10, 1, 0.1, 0.01 and 0% T790M mutant plasmids were used. The results of detecting T790M mutation by this method showed that can ACB-ARMS PCR definitely detect 10, 1, 0.1 and 0.01% mutant plasmids in mimic samples (Fig. 2), which indicated that the lower limit (sensitivity) of this method was 0.01% (one mutant in the presence of 10,000 wild type genes), corresponding to <10 copies.
Figure 2.
Detection of serially diluted T790M mutants in mimic samples by the allele-specific competitive blocker-amplification refractory mutation system TaqMan quantitative polymerase chain reaction array. The amplification curves marked as numbers 1, 2, 3, 4 and 5 correspond to the 10, 1, 0.1, 0.01 and 0% T790M mutant proportions, respectively, in a background of 30 ng/µl human genomic DNA.
EGFR T790M mutation detected by ACB-ARMS PCR in clinical samples
Patient characteristics
Of the 27 patients with NSCLC, 15 were female (55.6%), and adenocarcinoma was the most common pathological type (26 patients; 96.3%). The majority of patients were non-smokers (20 patients; 74.1%). According to the TNM staging system (21), advanced disease at stages IIIB or IV was identified in 81.5% of all enrolled patients. All patients had EGFR activating mutations, including 19 Del in 13 patients (48.1%) and 21 L858R in 14 patients (51.9%), and no T790M mutation was detected by scorpion ARMS PCR (catalogue no. EG-04; Qiagen China Co., Ltd.). The patient data are summarised in Table II.
Table II.
Demographics and clinical characteristics of enrolled patients.
| T790M detected by ACB-ARMS PCR | ||||
|---|---|---|---|---|
| Characteristic | Total, n (%) | Positive, n (%) | Negative, n (%) | P-value |
| Age, years | 0.363 | |||
| ≤60 | 17 (63.0) | 5 (18.5) | 12 (44.4) | |
| >60 | 10 (37.0) | 1 (3.7) | 9 (33.3) | |
| Gender | 1.000 | |||
| Female | 15 (55.6) | 3 (11.1) | 12 (44.4) | |
| Male | 12 (44.4) | 3 (11.1) | 9 (33.3) | |
| Smoking status | 0.290 | |||
| Never-smoker | 20 (74.1) | 3 (11.1) | 17 (63.0) | |
| Smoker | 7 (25.9) | 3 (11.1) | 4 (14.8) | |
| Pathology | 0.222 | |||
| Squamous | 1 (3.7) | 1 (3.7) | 0 (0.0) | |
| Adenocarcinoma | 26 (96.3) | 5 (18.5) | 21 (77.8) | |
| EGFR activating mutation | 0.648 | |||
| 19 Del | 13 (48.1) | 2 (7.4) | 11 (40.7) | |
| 21 L858R | 14 (51.9) | 4 (14.8) | 10 (37.0) | |
| Stage | 1.000 | |||
| IA-IIIA | 5 (18.5) | 1 (3.7) | 4 (14.8) | |
| IIIB-IV | 22 (81.5) | 5 (18.5) | 17 (63.0) | |
| Line of TKI therapy | 0.628 | |||
| First | 9 (33.3) | 1 (3.7) | 8 (29.6) | |
| Second or multiple | 18 (66.7) | 5 (18.5) | 13 (48.1) | |
ACB-ARMS PCR, allele-specific competitive blocker-amplification refractory mutation system TaqMan quantitative polymerase chain reaction; TKI, tyrosine kinase inhibitor.
T790M mutation is not rare in NSCLC patients with activating mutations prior to TKI treatment
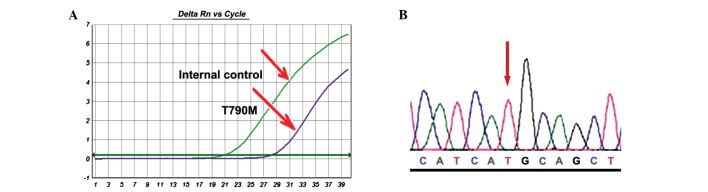
The sensitive ACB-ARMS PCR assay was used to analyse the T790M mutation. Of the 27 specimens pre-treated with EGFR-TKIs, 6 patients (22.2%) with T790M were identified (Fig. 3A). These results were confirmed by performing the ACB-PCR assay in triplicate and employing clone sequencing (Fig. 3B). The clone sequencing showed that all mutant cases harboured T790M mutant colonies in different proportions, which also suggested that the ACB-PCR assay is highly specific.
Figure 3.
Detection of T790M mutation in exon 20 by (A) allele-specific competitive blocker-amplification refractory mutation system TaqMan quantitative polymerase chain reaction and (B) clone sequencing (patient 12).
Fisher's exact test was used to evaluate the association between the T790M mutation and clinical variables, such as gender, age, smoking status, tumour stage, EGFR activating mutation and pathological type. However, none of these clinical factors were found to be associated with the occurrence of T790M in the NSCLC patients with EGFR-activating mutations.
PFS time, OS time and T790M mutation in NSCLC patients with activating mutations
Among the present cohort of 27 patients with EGFR-activating mutations, the median PFS time in the TKI-naïve patients with a T790M mutation (n=6) was shorter than in those without a T790M mutation (n=21), but the difference was not significant (3.2 vs. 19.5 months, respectively; P=0.256; Fig. 4A). Furthermore, the median OS time in the groups with or without T790M mutation did not significantly differ (10 vs. 20 months, respectively; P=0.689; Fig. 4B).
Figure 4.
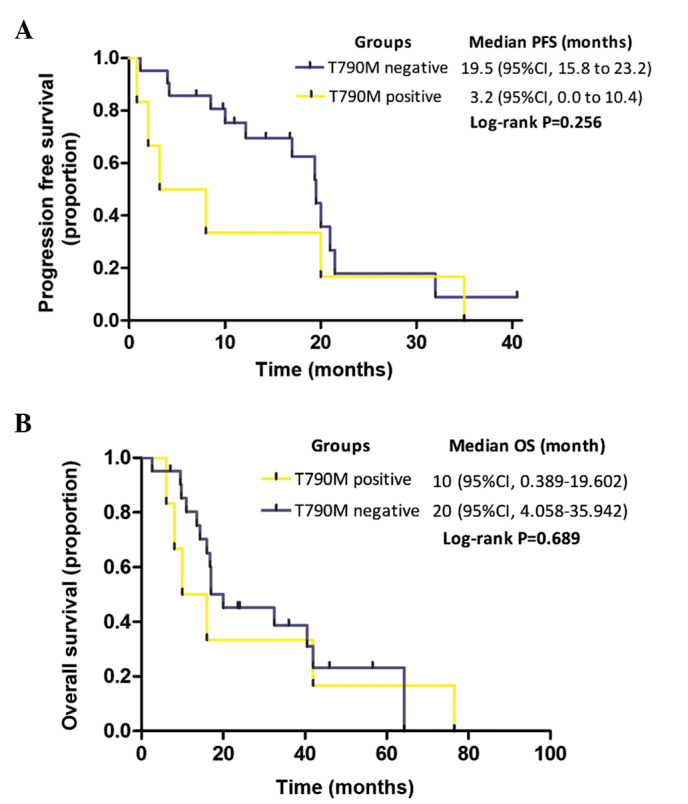
Kaplan-Meier curves for (A) PFS and (B) OS are shown for patients according to their T790M mutation status, as detected by allele-specific competitive blocker-amplification refractory mutation system TaqMan quantitative polymerase chain reaction method in pre-tyrosine kinase inhibitor treatment samples. PFS, progression-free survival; OS, overall survival; CI, confidence interval.
Discussion
The detection of the EGFR T790M mutation has become increasingly important, as it could be used to monitor acquired resistance to EGFR-TKIs (1), predict EGFR-TKI response duration and patient prognosis (2,3,5), and identify appropriate patients for treatment with next-generation EGFR-TKIs (6–8). Direct sequencing is a straightforward method that is widely used to detect the EGFR T790M mutation. However, the detection limit of direct sequencing is generally 25–30%, and homogeneous tumour samples may not be available in a clinical setting. To date, several strategies to improve the detection limit of the EGFR T790M mutation have been proposed (as summarised in Table III). Although the sensitivities of these methods are greatly improved, each method features unique advantages and disadvantages or difficulties in practical situations, such as the requirement for special equipment and expensive reagents or the involvement of complicated procedures. Of these strategies, the various PCR-based methods are the most suitable and easy to use in order to detect this known mutation in clinical practice. Hence, a PCR-based assay was developed in the present study. This method was sensitive, cost-effective and clinically applicable for the detection of the EGFR T790M mutation.
Table III.
Documented methods for T790M mutation detection.
| T790M mutation detection rate, % (sample type) | ||||||
|---|---|---|---|---|---|---|
| First author, year | Methods | Detection range, % | Disadvantages | Pre-treatment | TKI-resistance | (Ref.) |
| Sequist et al, 2008 | Direct sequencing | 25–35 | Insensitive | 2.04 | NA | (22) |
| Chen et al, 2009 | Scorpion ARMS | 1 | Expensive | 0 | 48.3 | (11) |
| Maheswaran et al, 2008 | 38 (CTC) | 64 (CTC) | (4) | |||
| Taniguchi et al, 2011 | BEAMing | 0.01 | Complicated, time-consuming | 4.8 (plasma) | 43.5 (plasma) | (19) |
| He et al, 2013 | Direct sequencing | NA | Time-consuming | NA | 6.1 (plasma) | (9) |
| Mutant-enriched PCR | 0.1 | 36.4 (plasma) | ||||
| Arcila et al, 2011 | PCR-sequencing/FA | 12.5 | Post-PCR procedure | NA | 52 | (12) |
| LNA-PCR-sequencing | 0.1 | 68 | ||||
| Rosell et al, 2011 | PNA-TaqMan PCR | NA | Acceptable | 35–38 | NA | (3) |
| Miyazawa et al, 2008 | PNA-LNA PCR | 0.1 | Acceptable | 0 | NA | (13) |
| Oh et al, 2011 | PNA-clamping PCR | 0.01 | Acceptable | 8.2 | NA | (14) |
| Oh et al, 2010 | Molecular beacon-PCR | 2 | Insensitive | NA | NA | (18) |
| Li et al, 2009 | COLD-PCR | 0.8 | Acceptable | NA | NA | (17) |
| Kim et al, 2013 | Pyrosequencing | NA | Insensitive | 0.49 | NA | (23) |
| Inukai et al, 2006 | Direct sequencing | NA | Acceptable | 0.36 | NA | (10) |
| Mutant-enriched PCR | 0.1 | 3.2 | ||||
| Guha et al, 2013 | DISSECT-PNA-LNA PCR | 0.01 | Complicated | NA | NA | (15) |
| Fujita et al, 2012 | PCR-clone hybridisation | NA | Complicated | 79 | NA | (5) |
| Su et al, 2012 | Direct sequencing | 25–35 | Special equipment | 2.7–2.8 | 33.3 | (2) |
| MALDI-TOF-MS | 1.5 | required | 25.2–31.5 | 83.3 | ||
FA, fragment analysis; BEAMing, beads, emulsion, amplification, and magnetics; PCR, polymerase chain reaction; DISSECT, differential strand separation at critical temperature; COLD-PCR, co-amplification at lower denaturation temperature-PCR; NA, not available; CTC, circulating tumour cell; LNA, locked nucleic acid; PNA, peptide nucleic acid; ARMS, amplification refractory mutation system; MALDI-TOF-MS, matrix-assisted laser desorption/ionisation-time of flight mass spectrometry.
The method used to screen such a small fraction of mutant alleles as the template should be highly specific and sensitive. Based on the mimic human genomic DNA panel that consisted of serially diluted plasmids containing T790M mutation, the present study results revealed that the ACB-ARMS PCR assay can detect 0.01% mutant alleles in total genomic DNA, which corresponds to <10 copies. This method is comparable to PNA-clamping PCR (14,15) or the BEAMing assay (20), and is more sensitive than any other documented method (Table III) (2–5,9–15,17–19,22,23). Regarding specificity, the purine/purine and pyrimidine/pyrimidine mismatches are considerably more selective than the purine/pyrimidine mismatches (24). Thus, the relatively weak G/T mismatch of the wild-type template/primer leads to base misincorporation during PCR and false-positive results when the ARMS method is used. Hence, in the present study, another mismatched site (C>T) was designed at the third base of the 3′-ends of the forward primer of the T790M mutation to decrease the non-specific combination of the primer with wild-type template alleles. This method was used to screen 10 different concentrations of human wild genomic DNA (Sigma-Aldrich China Inc.), and no specific amplifications were found, which suggested that this assay is highly specific (data not shown).
In clinical practice, 27 TKI-naïve clinical samples were successfully screened in the present study using this method and 6 mutant samples of T790M (22.2%) were found. These results were confirmed by performing ACB-ARMS PCR in triplicate. The positive examples were further identified by clone sequencing, and all mutant cases harboured T790M mutant colonies in different proportions. These results also strongly suggested the high specificity of this method. Furthermore, the result was consistent with the majority of previous studies (2–4), in which the incidence of de novo T790M mutation ranged from 25.2 to 38% in TKI pre-treated specimens, as detected by relatively sensitive methods. These findings suggested that T790M mutation is more predominant than expected in pre-treated samples, and indicated the high practicability of this protocol.
The present study showed that the incidence of T790M mutations was not associated with any other clinical factors, including gender, age, smoking status, pathological type and EGFR-activating mutations, which was similar to results reported in a study by Oxnard et al (25). However, certain studies have suggested that the T790M mutation is associated with a late disease stage (10) and EGFR 19 Del mutations (3), which was not confirmed by the present study due to the limited sample size. In TKI-naïve specimens, the predicted value of T790M mutation for TKI response duration was contradictory. In the studies of Su et al (2) or Rosell et al (3), patients with the T790M mutation experienced a shorter PFS time in response to EGFR-TKI treatment in comparison to patients without T790M in TKI-naïve samples. However, Fujita et al (5) reported that a high proportion of T790M alleles may define a clinical subset with a relatively favourable prognosis. However, the present study also did not reveal significant differences in the PFS time based on the T790M mutation status in NSCLC patients harbouring EGFR-activating mutations (3.2 vs. 19.5 months in patients with or without the mutation, respectively; P=0.256). Certain important issues regarding the association between T790M mutation and PFS time should to be considered in the present study. First, this retrospective study examined a limited simple size, and a selection bias did exist. Second, a number of additional factors regulate the TKI response duration beyond EGFR T790M mutation. Intratumoural EGFR mutational heterogeneity (26) and other EGFR-related genetic aberrances and downstream pathways (27), such as C-Met amplification, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α mutation and EGFR amplification (1), have been identified as associated with TKI resistance, which confounded the effect of T790M on PFS time. Although the start time for OS was defined as the time of diagnosis, not the time of informed consent or EGFR-TKI treatment, in the present study, the median OS time in the different groups did not significantly differ (10 vs. 20 months, respectively; P=0.689), which was consistent with the study by Su et al (2). However, more recent studies have also revealed contradictory effects of T790M mutation on OS time. Oxnard et al (25) and Hata et al (28) reported that patients who acquired TKI resistance and harboured the T790M mutation experienced a significantly longer post-progression survival time than patients without the T790M mutation (19 vs. 12 months; P=0.036). However, Lee et al (29) reported that the patients with T790M-positive tumours experienced a shorter overall survival time than those with T790M-negative tumours (median, 9.1 vs. 18.7 months; P=0.018). Numerous factors affect the OS, including the patient's performance status score, cancer therapy, disease stage, and importantly, organ metastasis. In basic studies, it has been revealed that tumour cells carrying the T790M mutation grow slowly and present with indolent biological behaviours (30). Therefore, the true prognostic value of T790M mutation should be further investigated. Furthermore, large prospective randomised clinical trials with excellent designs to balance the confounding factors in different groups are warranted to validate the clinical significance of T790M mutation in TKI-naïve specimens.
In summary, the ACB-ARMS PCR assay considerably improves the sensitivity and specificity of T790M mutation detection, and could be a promising method to screen for this mutation in lung cancer samples that contain only a small number of mutant cells. The clinical significance of de novo T790M mutation should be further investigated in future studies.
References
- 1.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su KY, Chen HY, Li KC, Kuo ML, Yang JC, Chan WK, Ho BC, Chang GC, Shih JY, Yu SL, Yang PC. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol. 2012;30:433–440. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]
- 3.Rosell R, Molina MA, Costa C, Simonetti S, Gimenez-Capitan A, Bertran-Alamillo J, Mayo C, Moran T, Mendez P, Cardenal F, et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res. 2011;17:1160–1168. doi: 10.1158/1078-0432.CCR-10-2158. [DOI] [PubMed] [Google Scholar]
- 4.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujita Y, Suda K, Kimura H, Matsumoto K, Arao T, Nagai T, Saijo N, Yatabe Y, Mitsudomi T, Nishio K. Highly sensitive detection of EGFR T790M mutation using colony hybridization predicts favorable prognosis of patients with lung cancer harboring activating EGFR mutation. J Thorac Oncol. 2012;7:1640–1644. doi: 10.1097/JTO.0b013e3182653d7f. [DOI] [PubMed] [Google Scholar]
- 6.Janne PA, Ramalingam SS, Yang JC, et al. Clinical activity of the mutant-selective EGFR inhibitor AZD9291 in patients (pts) with EGFR inhibitor-resistant non-small cell lung cancer (NSCLC) J Clin Oncol. 2014;32(Suppl 15):508s Abstract 8009. doi: 10.3978/j.issn.2218-6751.2014.08.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sequist LV, Soria J, Gadgeel SM, et al. First-in-human evaluation of CO-1686, an irreversible, highly selective tyrosine kinase inhibitor of mutations of EGFR (activating and T790M) J Clin Oncol. 2014;32(Suppl 15):508s Abstract 8010. [Google Scholar]
- 8.Kim D, Lee D, Kang J, et al. Clinical activity and safety of HM61713, an EGFR-mutant selective inhibitor, in advanced non-small cell lung cancer (NSCLC) patients (pts) with EGFR mutations who had received EGFR tyrosine kinase inhibitors (TKIs) J Clin Oncol. 2014;32(Suppl 15):508s Abstract 8011. [Google Scholar]
- 9.He C, Zheng L, Xu Y, Liu M, Li Y, Xu J. Highly sensitive and noninvasive detection of epidermal growth factor receptor T790M mutation in non-small cell lung cancer. Clin Chim Acta. 2013;425:119–124. doi: 10.1016/j.cca.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Inukai M, Toyooka S, Ito S, Asano H, Ichihara S, Soh J, Suehisa H, Ouchida M, Aoe K, Aoe M, et al. Presence of epidermal growth factor receptor gene T790M mutations as a minor clone in non-small cell lung cancer. Cancer Res. 2006;66:7854–7858. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 11.Chen HJ, Mok TS, Chen ZH, Guo AL, Zhang XC, Su J, Wu YL. Clinicopathologic and molecular features of epidermal growth factor receptor T790M mutation and c-MET amplification in tyrosine kinase inhibitor-resistant Chinese non-small cell lung cancer. Pathol Oncol Res. 2009;15:651–658. doi: 10.1007/s12253-009-9167-8. [DOI] [PubMed] [Google Scholar]
- 12.Arcila ME, Oxnard GR, Nafa K, Riely GJ, Solomon SB, Zakowski MF, Kris MG, Pao W, Miller VA, Ladanyi M. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res. 2011;17:1169–1180. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyazawa H, Tanaka T, Nagai Y, Matsuoka M, Huqun Sutani A, Udagawa K, Zhang J, Hirama T, Murayama Y, et al. Peptide nucleic acid-locked nucleic acid polymerase chain reaction clamp-based detection test for gefitinib-refractory T790M epidermal growth factor receptor mutation. Cancer Sci. 2008;99:595–600. doi: 10.1111/j.1349-7006.2007.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh JE, An CH, Yoo NJ, Lee SH. Detection of low-level EGFR T790M mutation in lung cancer tissues. APMIS. 2011;119:403–411. doi: 10.1111/j.1600-0463.2011.02738.x. [DOI] [PubMed] [Google Scholar]
- 15.Guha M, Castellanos-Rizaldos E, Makrigiorgos GM. Dissect method using PNA-LNA clamp improves detection of EGFR T790m mutation. PLoS One. 2013;8:e67782. doi: 10.1371/journal.pone.0067782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuang Y, Rogers A, Yeap BY, Wang L, Makrigiorgos M, Vetrand K, Thiede S, Distel RJ, Jänne PA. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res. 2009;15:2630–2636. doi: 10.1158/1078-0432.CCR-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Wang L, Jänne PA, Makrigiorgos GM. Coamplification at lower denaturation temperature-PCR increases mutation-detection selectivity of TaqMan-based real time PCR. Clin Chem. 2009;55:748–756. doi: 10.1373/clinchem.2008.113381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh YH, Kim Y, Kim YP, Seo SW, Mitsudomi T, Ahn MJ, Park K, Kim HS. Rapid detection of the epidermal growth factor receptor mutation in non-small-cell lung cancer for analysis of acquired resistance using molecular beacons. J Mol Diagn. 2010;12:644–652. doi: 10.2353/jmoldx.2010.090208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, Higashiyama M, Kodama K, Imamura F, Kato K. Quantitative detection of EGFR mutations in circulating tumor DNA derived form lung adenocarcinomas. Clin Cancer Res. 2011;17:7808–7815. doi: 10.1158/1078-0432.CCR-11-1712. [DOI] [PubMed] [Google Scholar]
- 20.Board RE, Thelwell NJ, Ravetto PF, Little S, Ranson M, Dive C, Hughes A, Whitcombe D. Multiplexed assays for detection of mutations in PIK3CA. Clin Chem. 2008;54:757–760. doi: 10.1373/clinchem.2007.098376. [DOI] [PubMed] [Google Scholar]
- 21.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V, Sobin L. International Association for the Study of Lung Cancer International Staging Committee; Participating Institutions: The IASLC Lung Cancer Staging Project: Proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 22.Sequist LV, Martins RG, Spigel D, Grunberg SM, Spira A, Jänne PA, Joshi VA, McCollum D, Evans TL, Muzikansky A, et al. First-line gefitinib in patients with advanced non-small cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 23.Kim HJ, Oh SY, Kim WS, Kim SJ, Yoo GH, Kim WD, Lee KY. Clinical investigation of EGFR mutation detection by pyrosequencing in lung cancer patients. Oncol Lett. 2013;5:271–276. doi: 10.3892/ol.2012.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oxnard GR, Arcila ME, Sima CS, Riely GJ, Chmielecki J, Kris MG, Pao W, Ladanyi M, Miller VA. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: Distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17:1616–1622. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai H, Wang Z, Wang Y, Zhuo M, Zhou Q, Duan J, Yang L, Wu M, An T, Zhao J, Wang J. Detection and clinical significance of intratumoral EGFR mutational heterogeneity in Chinese patients with advanced non-small cell lung cancer. PLoS One. 2013;8:e54170. doi: 10.1371/journal.pone.0054170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HR, Cho BC, Shim HS, Lim SM, Kim SK, Chang J, Kim DJ, Kim JH. Prediction for response duration to epidermal growth factor receptor-tyrosine kinase inhibitors in EGFR mutated never smoker lung adenocarcinoma. Lung Cancer. 2014;83:374–782. doi: 10.1016/j.lungcan.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 28.Hata A, Katakami N, Yoshioka H, Takeshita J, Tanaka K, Nanjo S, Fujita S, Kaji R, Imai Y, Monden K, et al. Rebiopsy of non-small cell lung cancer patients with acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitor: Comparison between T790M mutation-positive and mutation-negative populations. Cancer. 2013;119:4325–4332. doi: 10.1002/cncr.28364. [DOI] [PubMed] [Google Scholar]
- 29.Lee Y, Lee GK, Lee YS, Zhang W, Hwang JA, Nam BH, Kim SH, Kim JH, Yun T, Han JY, et al. Clinical outcome according to the level of preexisting epidermal growth factor receptor T790M mutation in patients with lung cancer harboring sensitive epidermal growth factor receptor mutations. Cancer. 2014;120:2090–2098. doi: 10.1002/cncr.28711. [DOI] [PubMed] [Google Scholar]
- 30.Chmielecki J, Foo J, Oxnard GR, Hutchinson K, Ohashi K, Somwar R, Wang L, Amato KR, Arcila M, Sos ML, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]