Abstract
The Fusarium species are a widely spread phytopathogen identified in an extensive variety of hosts. The Fusarium genus is one of the most heterogeneous fungi and is difficult to classify. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis is a useful method in detection of DNA polymorphism in objective sequences. The aim of the present study was to identify the phylogenetic associations and usefulness of the internal transcribed spacer (ITS) region as a genetic marker within the most clinically important strain of the Fusarium species. A total of 50 strains of Fusarium spp. were used in the study, including environmental, clinical and reference isolates. The primers ITS1 and ITS4 were used in the study. Two restriction enzymes, HaeIII and SmaI, were assessed for the digestion of PCR products. A PCR product of ~550-base pairs was generated for each Fusarium species. The digested products with HaeIII and SmaI demonstrated that the bands generated for the medically significant Fusarium species, including F. solani, F. oxysporum, F. verticillidea, F. proliferatum and F. fujikuri, have different restriction enzyme patterns. In conclusion, it appears that the PCR-RFLP method used in the present study produces a sufficient restriction profile for differentiation of the most medically significant Fusarium species.
Keywords: Fusarium, polymerase chain reaction-restriction fragment length polymorphism, internal transcribed spacer region
Introduction
The genus Fusarium members are ubiquitous fungi frequently found in soils and plants (1). Fusarium species have been distinguished as a cause of localized infections (1,2). Due to bone marrow grafts and immunosuppressive treatment, invasive Fusarium infections have increased during the last few decades. The host immunological status and the level of the infection are the most significant aspects for the clinical effect of Fusarium infections (3,4).
The Fusarium species are a widely spread phytopathogen found in an extensive variety of hosts (5). It causes wilts and root rot disease, which produces secondary metabolites such as T2-toxin, zearalenone and trichothecene, causing huge economic problems through losing crops (6,7). The genus Fusarium is seldom able to cause human infections, such as onychomycosis, skin infections or keratitis. The Fusarium genus is one of the most heterogeneous fungi and is difficult to classify. Conversely, identification at the species level is required for biological and epidemiological reasons.
Conventional diagnostic techniques for identification of the Fusarium species in culture or in infected tissues are based on morphological features. This procedure is time consuming, and it can frequently be difficult to discriminate between similar species. Molecular approaches are more sensitive and quicker. Furthermore, they are employed to the specific detection of the Fusarium species. For these reasons, the molecular biological technique has been established in Fusarium systematics and the molecular variation at the DNA level has been investigated in numerous studies (8–10). The use of polymerase chain reaction (PCR) with primers targeted to the internal transcribed spacer (ITS) region of the ribosomal DNA (11,12) for identification of the Fusarium species has been reported. The ITS region sequences have shown to be highly variable in Fusarium genus (13).
PCR-restriction fragment length polymorphism (RFLP) analysis is a useful method in the detection of DNA polymorphism in objective sequences. In the present study, the amplified ITS region of rDNA was digested with 2 restriction endonucleases for genetic variation among Fusarium spp. The aim of the study was to identify the phylogenetic associations and usefulness of the ITS region as a genetic marker within the most clinically important of the Fusarium species.
Materials and methods
Microorganisms
A total of 50 strains of Fusarium spp. were used in the study including environmental, clinical and reference isolates. The following strains were used as a reference: F. solani PFCC 5284, F. solani PFCC 5285, F. oxysporum PFCC 30067, F. oxysporum PFCC 5115, F. verticillidea PFCC 53–131, F. verticillidea PFCC 15–89, F. proliferatum PFCC 48–125, F. proliferatum PFCC 12–86 and F. fujikuri PFCC 5144. Enviromental strains were recovered from soil. Two strains were clinical strains, including F. solani PFCC 5284.
DNA extraction
In total, 100 ml of YEPD medium in Erlenmeyer flasks was inoculated with 1 ml of thick spore suspension. The flasks were incubated at 200 rpm under agitation at 25°C for 72 h to obtain mycelium growth. The mycelia were harvested, washed with 0.5 M EDTA and sterile dH2O and ground into a fine powder with a pestle and mortar using liquid nitrogen.
Approximately 100 mg of the mycelium powder was transferred into a 1.5-ml tube and 400 µl of lysis buffer [100 mM Tris-HCl (pH 8.0), 30 mM EDTA (pH 8.0) and 5% SDS w/v] was added.
Following the incubation of the tubes at 100°C for 20 min, 150 µl of 3 M acetate potassium was added. The suspension was maintained for 10 min at −20°C, and centrifuged at 14,000 × g for 10 min in 4°C. The supernatant was transferred to a 1.5-ml Eppendorf tube (Eppendorf AG, Hamburg, Germany), and subsequently, 250 µl of phenol-chloroform-isoamyl alcohol (25:24:1, v/v) was added and the mixture was briefly vortexed and centrifuged at 14,000 × g for 10 min. Subsequent to transferring the upper aqueous phase to a new 1.5-ml tube, 250 µl chloroform-isoamyl alcohol was added. The tubes were briefly vortexed and centrifuged in 4°C at 14,000 × g for 10 min. The supernatant was transferred to a new tube, an equal volume of ice-cold 2-propanol was added and the mixture was incubated at −20°C for 10 min and subsequently centrifuged at 14,000 × g for 10 min. The upper aqueous phase was removed and the pellet was washed with 300 µl of ethanol 70%. The ethanol was removed and the DNA pellet was air-dried and dissolved in 50 µl of dH2O.
PCR amplification
The primer sets (ITS1, 5′-TCCGTAGGTGAACCTGCGG-3′) and (ITS4, 5′-TCCTCCGCTTATTGATATGC-3′) were used to amplify an ~600-base pair (bp) DNA fragment of the ITS region. PCR reactions were prepared to a final volume of 50 µl, containing reaction buffer, 2.2 mM MgCl2, 200 µM of each dNTP, 2.5 unit of Taq DNA polymerase (CinnaGen, Karaj, Iran), a 30-ng DNA template and 50 pmol of each primer.
An initial denaturation step for 5 min at 95°C was followed by 30 cycles of denaturation at 94°C for 40 sec, annealing at 58°C for 40 sec and extension at 72°C for 40 sec, with a final extension at 72°C for 5 min. The PCR product was run on a 1% agarose gel in Tris-base, acetic acid and EDTA (TAE) buffer, and stained with ethidium bromide. The PCR amplification of the ITS region resulted in ~595-bp of fragment.
RFLP analysis
Two restriction enzymes were analyzed. HaeIII and SmaI had restriction sites in the ITS region of the Fusarium species. The reaction mixture for each enzyme was carried out in a total volume of 20 µl containing 10 units of the enzyme, 2 µl of the related buffer, 5 µl of the PCR product and Ultrapure water to create the 20-µl volume. Digested PCR products were subjected to electrophoresis on a 1.5% agarose gel in TAE buffer, and stained with ethidium bromide.
Results
PCR amplification of the ITS regions
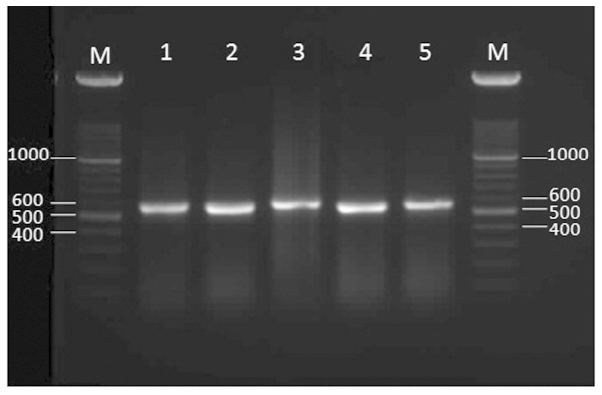
PCR amplification of the ITS region with primers ITS1 and ITS4 resulted in an ~550-bp band (Fig. 1). The fragment was obtained from all 50 Fusarium strains. The ITS-region products were sequenced from several isolates, including the reference strains. A Basic Local Alignment Search Tool search showed that the ITS PCR products from 5 medically significant Fusarium reference strains, which were F. solani, F. oxysporum, F. verticillidea, F. proliferatum and F. fujikuri, exhibited a 99% homology with the associated sequences in the GenBank.
Figure 1.
Agarose gel electrophoresis of the internal transcribed spacer region base pair (bp) products of the Fusarium species. M, 100 bp ladder; lane 1, F. oxysporum PFCC 5115; lane 2, F. verticillidea PFCC 15–89; lane 3, F. proliferatum PFCC 12–86; lane 4, F. fujikuri PFCC 5144; lane 5, F. solani PFCC 5284.
Restriction patterns for the Fusarium strains
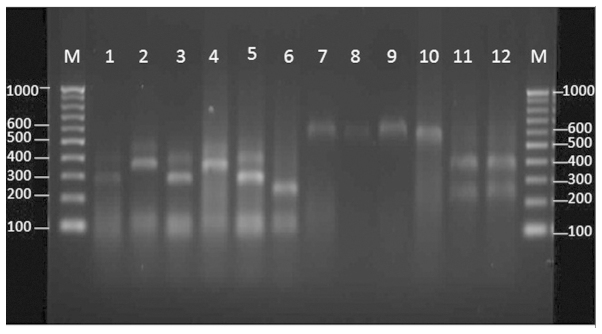
Different restriction patterns were obtained following digestion with the HaeIII and SmaI enzymes for the reference Fusarium strains, including F. solani, F. oxysporum, F. verticillidea, F. proliferatum and F. fujikuri (Table I and Fig. 2).
Table I.
Restriction fragment size of the ITS region of the Fusarium species following digestion with HaeIII and SmaI, according to the sequencing of the strains.
| Fusarium species | ITS size prior to digestion, bp | HaeIII, bp | SmaI, bp |
|---|---|---|---|
| F. oxysporum | 550 | 347, 88, 86 | 550 |
| F. verticillidea | 550 | 347, 89, 85 | 550 |
| F. proliferatum | 570 | 281, 90, 89, 77 | 570 |
| F. fujikuri | 550 | 281, 91, 77, 61 | 324, 210 |
| F. solani | 570 | 241, 123, 91, 65 | 325, 217 |
ITS, internal transcribed spacer; bp, base pairs.
Figure 2.
Agarose gel electrophoresis of the internal transcribed spacer region base pair (bp) products of the Fusarium species following digestion with HaeIII (lanes 1–6) and SmaI (lanes 7–12). M, 100 bp ladder; lane 1, F. oxysporum PFCC 5115; lane 2, F. verticillidea PFCC 15–89; lane 3, F. proliferatum PFCC 12–86; lane 4, F. verticillidea PFCC 53–131; lane 5, F. fujikuri PFCC 5144; lane 6, F. solani PFCC 5284; lane 7, F. oxysporum PFCC 5115; lane 8, F. verticillidea PFCC 15–89; lane 9, F. proliferatum PFCC 12–86; lane 10, F. verticillidea PFCC 53–131; lane 11, F. fujikuri PFCC 5144; lane 12, F. solani PFCC 5284.
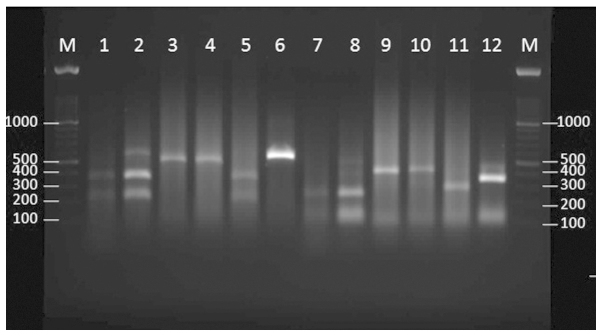
The restriction patterns of 6 environmental Fusarium strains with HaeIII and SmaI are shown in Fig. 3. Strain E42 was recognized as F. oxysporum, based on the sequencing and restriction enzymes pattern.
Figure 3.
Agarose gel electrophoresis of the internal transcribed spacer region base pair (bp) products of the Fusarium species (environmental isolates) following digestion with SmaI (lanes 1–6) and HaeIII (lanes 7–12). M, 100 bp ladder; lane 1, E20; lane 2, E34; lane 3, E35; lane 4, E36; lane 5, E39; lane 6, E42; lane 7, E20; lane 8, E34; lane 9, E35; lane 10, E36; lane 11, E39; lane 12, E42.
Discussion
The ITS region was confirmed in the present study to be particularly suitable for the purpose of providing target genes for molecular identification of the Fusarium species. Variation in the nucleotide composition of the ITS region was successfully employed for recognition among the species (11–13). A variety of targets have been employed for DNA-based identification and dicrimination of pathogenic Fusarium species. DNA diversity in intergenic spacer (14) or ITS regions (13), β-tubulin, calmodulin, elongation factor 1α (15) and mycotoxins biosynthetic genes (16) as target genes for identification of Fusarium species have been tested with the PCR method. Several studies have shown that ITS1 and ITS2 are useful targets for detection of certain Fusarium species (13,17). The study by O'Donnell (18) reported an unexpected level of divergence for ITS sequences within the F. sambucinum species. There are certain advantages for the ITS region being a good target for detection reasons. The ITS region is comparatively conserved within numerous fungal species. It is present as multiple copies in the genome of fungi, and yields adequate taxonomic resolution for the majority of fungi (19). Furthermore, there are a large number of sequences from this locus in the GenBank, which enable the comparison of the obtained sequences. Therefore, nucleotide sequence heterogeneity in this region could be employed for classification of the majority of pathogenic fungi (20).
In the present study, the ITS1 and ITS4 primers were used to amplify the 5.8S rDNA gene. RFLP using HaeIII and SmaI restriction enzymes provided a genus-specific assay for the rapid identification of medically significant Fusarium genus. According to the RFLP result, five species of Fusarium, which were F. solani, F. oxysporum, F. verticillidea, F. proliferatum and F. fujikuri, were divided into five RFLP types when used with the HaeIII and SmaI enzymes.
In conclusion, it appears that the present PCR-RFLP method produces a restriction profile for the differentiation of the most medically significant Fusarium species.
Acknowledgements
The present study was supported by the Health Research Institute (Infectious and Tropical Diseases Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran) (grant no. 92118).
References
- 1.Kuruvilla TS, Dias M. Fusarium Solani: A causative agent of skin and nail infections. Indian J Dermatol. 2012;57:308–309. doi: 10.4103/0019-5154.97680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varon AG, Nouer SA, Barreiros G, Trope BM, Magalhães F, Akiti T, Garnica M, Nucci M. Superficial skin lesions positive for Fusarium are associated with subsequent development of invasive fusariosis. J Infect. 2014;68:85–89. doi: 10.1016/j.jinf.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Nucci M, Anaissie E. Fusarium infections in immunocompromised patients. Clin Microbiol Rev. 2007;20:695–704. doi: 10.1128/CMR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan R, Ng KP, Gan GG, Na SL. Fusarium sp. infection in a patient with Acute Lymphoblastic Leukaemia. Med J Malaysia. 2013;68:479–480. [PubMed] [Google Scholar]
- 5.Ma LJ, Geiser DM, Proctor RH, Rooney AP, O'Donnell K, Trail F, Gardiner DM, Manners JM, Kazan K. Fusarium pathogenomics. Annu Rev Microbiol. 2013;67:399–416. doi: 10.1146/annurev-micro-092412-155650. [DOI] [PubMed] [Google Scholar]
- 6.Tamura M, Mochizuki N, Nagatomi Y, Harayama K, Toriba A, Hayakawa K. A method for simultaneous determination of 20 Fusarium toxins in cereals by high-resolution liquid chromatography-Orbitrap mass spectrometry with a pentafluorophenyl column. Toxins (Basel) 2015;7:1664–1682. doi: 10.3390/toxins7051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barros G, Zanon MS, Palazzini JM, Haidukowski M, Pascale M, Chulze S. Trichothecenes and zearalenone production by Fusarium equiseti and Fusarium semitectum species isolated from Argentinean soybean. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2012;29:1436–1442. doi: 10.1080/19440049.2012.698397. [DOI] [PubMed] [Google Scholar]
- 8.Arif M, Zaidi NW, Haq QM, Singh YP, Taj G, Kar CS, Singh US. Morphological and comparative genomic analyses of pathogenic and non-pathogenic Fusarium solani isolated from Dalbergia sissoo. Mol Biol Rep. 2015;42:1107–1122. doi: 10.1007/s11033-014-3849-3. [DOI] [PubMed] [Google Scholar]
- 9.Datta J, Lal N. Application of molecular markers for genetic discrimination of Fusarium wilt pathogen races affecting chickpea and pigeonpea in major regions of India. Cell Mol Biol (Noisy-le-grand) 2012;58:55–65. [PubMed] [Google Scholar]
- 10.Short DP, O'Donnell K, Geiser DM. Clonality, recombination, and hybridization in the plumbing-inhabiting human pathogen Fusarium keratoplasticum inferred from multilocus sequence typing. BMC Evol Biol. 2014;14:91. doi: 10.1186/1471-2148-14-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubey SC, Priyanka K, Singh V. Phylogenetic relationship between different race representative populations of Fusarium oxysporum f. sp. ciceris in respect of translation elongation factor-1α, β-tubulin, and internal transcribed spacer region genes. Arch Microbiol. 2014;196:445–452. doi: 10.1007/s00203-014-0976-0. [DOI] [PubMed] [Google Scholar]
- 12.Chang SC, Macêdo DP, Souza-Motta CM, Oliveira NT. Use of molecular markers to compare Fusarium verticillioides pathogenic strains isolated from plants and humans. Genet Mol Res. 2013;12:2863–2875. doi: 10.4238/2013.August.12.2. [DOI] [PubMed] [Google Scholar]
- 13.Mirete S, Patiño B, Jurado M, Vázquez C, González-Jaén MT, Puertas M. Structural variation and dynamics of the nuclear ribosomal intergenic spacer region in key members of the Gibberella fujikuroi species complex. Genome. 2013;56:205–213. doi: 10.1139/gen-2013-0008. [DOI] [PubMed] [Google Scholar]
- 14.Konstantinova P, Yli-Mattila T. IGS-RFLP analysis and development of molecular markers for identification of Fusarium poae, Fusarium langsethiae, Fusarium sporotrichioides and Fusarium kyushuense. Int J Food Microbiol. 2004;95:321–331. doi: 10.1016/j.ijfoodmicro.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Van Poucke K, Monbaliu S, Munaut F, Heungens K, De Saeger S, Van Hove F. Genetic diversity and mycotoxin production of Fusarium lactis species complex isolates from sweet pepper. Int J Food Microbiol. 2012;153:28–37. doi: 10.1016/j.ijfoodmicro.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, Bouras N, Yang J, Howard RJ, Strelkov SE. Mycotoxin production by isolates of Fusarium lactis from greenhouse sweet pepper (Capsicum annuum) Int J Food Microbiol. 2011;151:150–156. doi: 10.1016/j.ijfoodmicro.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 17.O'Donnell K, Sutton DA, Fothergill A, McCarthy D, Rinaldi MG, Brandt ME, Zhang N, Geiser DM. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J Clin Microbiol. 2008;46:2477–2490. doi: 10.1128/JCM.02371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Donnell K. Ribosomal DNA internal transcribed spacers are highly divergent in the phytopathogenic ascomycete Fusarium sambucinum (Gibberella pulicaris) Curr Genet. 1992;22:213–220. doi: 10.1007/BF00351728. [DOI] [PubMed] [Google Scholar]
- 19.Balajee SA, Borman AM, Brandt ME, Cano J, Cuenca-Estrella M, Dannaoui E, Guarro J, Haase G, Kibbler CC, Meyer W, et al. Sequence-based identification of Aspergillus, Fusarium, and Mucorales species in the clinical mycology laboratory: Where are we and where should we go from here? J Clin Microbiol. 2009;47:877–884. doi: 10.1128/JCM.01685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwen PC, Hinrichs SH, Rupp ME. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med Mycol. 2002;40:87–109. doi: 10.1080/mmy.40.1.87.109. [DOI] [PubMed] [Google Scholar]