Abstract
Limited tools exist that are capable of monitoring nucleic acid conformations, fluctuations, and distributions in free solution environments. Single molecule free solution hydrodynamic separation enables the unique ability to quantitatively analyze nucleic acid biophysics in free solution. Single molecule fluorescent burst data and separation chromatograms can give layered insight into global DNA conformation, binding interactions, and molecular distributions. First, we show that global conformation of individual DNA molecules can be directly visualized by examining single molecule fluorescent burst shapes and that DNA exists in a dynamic equilibrium of fluctuating conformations as it is driven by Poiseuille flow through micron-sized channels. We then show that this dynamic equilibrium of DNA conformations is reflected as shifts in hydrodynamic mobility that can be perturbed using salt and ionic strength to affect packing density. Next, we demonstrate that these shifts in hydrodynamic mobility can be used to investigate hybridization thermodynamics and binding interactions. We distinguish and classify multiple interactions within a single sample, and demonstrate quantification amidst large concentration differences for the detection of rare species. Finally, we demonstrate that these differences can resolve perfect complement, 2bp mismatched, and 3bp mismatched sequences. Such a system can be used to garner diverse information about DNA conformation and structure, and potentially be extended to other molecules and mixed-species interactions, such as between nucleic acids and proteins or synthetic polymers.
INTRODUCTION
Many common methods to analyze nucleic acids, study their conformation, and monitor binding interactions rely on differences in electrophoretic mobility. DNA hybridization and electrophoretic mobility are commonly used in Southern blotting (or RNA hybridization in Northern blotting) to detect specific DNA sequences1,2. However, these techniques are labor and time intensive, expensive, and require large sample volumes. Electrophoretic mobility shift assays (EMSAs) have been used for qualitative conformational analysis of DNA-protein binding and to monitor large scale conformational changes but these assays are only considered to be semi-quantitative and the behavior of molecules in the gel can differ from that in native solution3.
Other methods have been developed that can more directly determine nucleic acid properties. Crystallography has been used to determine precise molecular conformation, but the crystallization process itself can influence the observed conformation, often requires strict solution conditions, is complicated and time consuming, and only provides a population average conformation4,5. Fluorescence Correlation Spectroscopy (FCS) provides an alternative method for detecting molecular concentration, hydrodynamic size, and mass change due to binding. Experimentation is faster and more quantitative than EMSA, but data analysis is complicated, size resolution is limited, and, like crystallography, FCS provides only a population average, making individual discrimination of multiple species difficult 6.
Hydrodynamic separation provides an alternative method to analyze nucleic acids in free solution7. Rather than relying on differences in electrophoretic mobility, hydrodynamic separations occur according to differences in the molecules’ size in solution7-9. Hydrodynamic chromatography performed in columns packed with non-porous beads has been particularly useful for particle and polymer characterization, but open microcapillary tubes have been demonstrated effective for the separation of biomacromolecules including DNA10,11. Initially, open tubular hydrodynamic separations could only be performed on large macromolecules or by labeling small molecules with drag tags, but recent studies have shown that by reducing the diameter of the separation channel to approach the radii of the molecules to be separated (i.e. nominally 1 µm), high resolution sizing can be performed on diverse biomolecules including small oligonucleotides, large DNA molecules, and proteins over a wide dynamic range12-14. The use of hydrodynamic chromatography enables separation by size independent of charge and allows for studies of molecular interactions in native environments without a gel matrix.
Previously, we combined hydrodynamic separation with single-molecule fluorescence spectroscopy to perform highly sensitive and quantitative analysis using <100 molecules of DNA15. Single molecule analysis is performed by analyzing the individual fluorescent bursts generated as separated DNA molecules traverse a confocal laser detection curtain16. Whereas hydrodynamic separation can determine the average size of a molecular ensemble based on peak retention time, single molecule spectroscopy provides the opportunity to look at individual molecules within that ensemble. By examining the size and shape of each single molecule fluorescent burst, it is possible to determine molecular information regarding individual DNA conformations and distributions. The combination of these methods allows layered information regarding ensemble conformations and their underlying single molecule distributions. The high sensitivity and wide dynamic range of single molecule spectroscopy further enables concurrent analysis of rare and abundant species.
In this study, we perform single molecule free solution hydrodynamic separation to probe global DNA conformation changes and binding interactions. Molecular properties are determined by coupled analysis of single molecule fluorescent burst characteristics and hydrodynamic mobilities. First, we analyze single molecule fluorescent bursts to determine DNA conformation and define DNA packing density. We then show that DNA packing density and hydrodynamic mobility are related by investigating the effects of buffer ionic strength and salt concentration on DNA packing. Next, we demonstrate the ability to monitor hybridization thermodynamics and binding interactions under different denaturing conditions. Finally, we demonstrate that hybridization-induced mobility shifts are a function of target ssDNA oligo length and hybridization bond strength. We show that we can identify and quantify multiple binding events within a single sample, enabling accurate calculation of the hybridization efficiency. We also demonstrate the dynamic range to detect rare species despite excess concentrations of background molecules.
EXPERIMENTAL SECTION
CICS Instrumentation and Data Acquisition
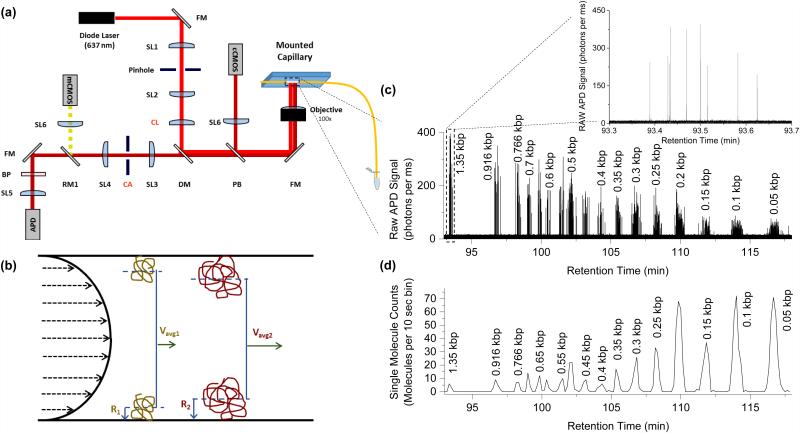
Data was collected on a custom built instrument designed to be made at a reduced cost and smaller footprint as compared to our previous CICS design16. A schematic of the optical components is shown in Figure 1a. This system contains one laser diode (OBIS 640-40LX, Coherent Inc.) which emits at 640 nm. The beam was expanded using two doublet spherical lenses SL1 and SL2 (f=19mm and f=75mm, Thorlabs) and a 25 μm pinhole. A cylindrical lens CL (f=150 mm) was used to expand the beam in one dimension and a full mirror FM was used to direct the optical path to focus on the back focal plane of a 100X oil objective (1.3 NA, Olympus, UPlanFLN). The objective also collected the emitted light. A dichroic mirror DM (Semrock, Di01-R635) separated excitation light from emitted light, which then passed through a confocal slit aperture CA (National Aperture) before being passed through an emission filter BP (Semrock, FF01-676/37) and focused with a doublet lens SL5 onto a silicon avalanche photodiode (APD, Excelitas, SPCM-AQR10). Two CMOS cameras were used for focusing and alignment of the capillary lumen. A pellicle beamsplitter PB split a fraction of the optical path before the DM to focus (SL6) on the first camera (cCMOS, Thorlabs), which was used for course alignment and maintaining focus during experiments. The camera after the CA (mCMOS, Thorlabs) was used to align the capillary lumen within the rectangular aperture and only received signal when the removable mirror (RM1) was in place. The detection window of the capillary was mounted on a glass slide, which was held by vacuum to a 3-axis piezo stage (9063-XYZ-PPP, Newport Corp.). The entire footprint of all optical components is confined to a 2 ft by 2 ft benchtop optical breadboard (Thorlabs). A single power supply and control system was built to power the laser, stage, and APD, and was connected to a laptop computer for control of all components.
Figure 1.
CICS optical detection platform and the hydrodynamic separation principle. (a) Schematic of the CICS optical detection platform with the cylindrical lens (CL) providing the beam expansion over the burned viewing window of the mounted separation capillary. Further description of the optical components is provided in the Methods section. (b) Schematic of the hydrodynamic separation principle: larger molecules are more excluded from the slower moving flow regimes near the wall, resulting in a faster average velocity for larger molecules than smaller molecules. (c) Separation of a 50 bp ladder with single molecules detected for each fragment size. The inset provides a closer look at the 9 detected molecules of the largest fragment size. (d) Single molecule bursts from the raw trace are summed over a larger bin size to form a single molecule chromatogram where each peak is a subpopulation of uniform fragment sizes. Every chromatogram peak in (d) corresponds to a fragment species present in the ladder, but only enough species are labeled to orient the reader.
Photon counts were collected from the APD in 0.1 or 1 ms bins using a DAQ card (National Instruments) and software written in LabView (National Instruments). Labview software was also used to control the laser power (8mW) and the flipper mount that positioned the removable mirror (Thorlabs).
Separation
A schematic illustrating the hydrodynamic separation mechanism is shown in Figure 1b. Laminar flow in a capillary has a parabolic velocity profile due to the no-slip boundary condition at the capillary walls. Molecules in the capillary are free to diffuse across the cross section of the channel and sample many different flow streams. Over a long capillary length, this diffusion results in a flow velocity that is an average of the sampled flow streams. The average velocity of the solute is thus the integrated average of all velocity streams. The center of mass of a larger molecule, however, is excluded from the walls by its effective radius. This prevents larger molecules from sampling the slowest flow regimes near the wall, resulting in a faster average velocity. Thus, larger molecules will move at a faster average velocity than smaller molecules, with the solute moving the slowest. An example of the collected photon data from the separation of a 50bp ladder is shown in Figure 1c and 1d. The relative mobility was calculated using the following equation:
where m is the relative mobility of the DNA fragment, vDNA and tDNA are the average velocity and retention time of the fragment, respectively, and vdye and tdye are the average velocity and retention time of the free Alexa dye, which was assumed to be the same as the average retention time of the solute.
All separations were performed at room temperature in fused silica capillaries purchased from PolyMicro (Molex), with inner diameters ranging from 1.6 µm (as measured by SEM) to 10 µm. Capillaries were cut to length and a short section of the polyimide coating was burned from the capillary to form a detection window. The capillary length L was measured as the distance between the capillary inlet and the center of the detection window.
Pressure was provided by compressed argon gas through an electronic dual valve pressure controller (Alicat Scientific) for differential pressures up to 500 psi. LabView software was designed to control and record the pressure provided by the valves for precise timing and pressure control.
Data Analysis
Thresholding was used to detect single molecule fluorescent bursts above the background and were analyzed in bins of various sizes depending on the flow rate. OriginPro (OriginLab, version 9) was used to fit the chromatograms to a set of Gaussian peaks, from which the peak center could be extracted. This was used as the average retention time of each species and was used to calculate the relative mobility. The width of each fragment peak was calculated as 4 times the standard deviation (σ) of the fitted Gaussian peak and was used to determine the retention time range for each fragment size. The peak width is reported as error bars in plots of relative mobility.
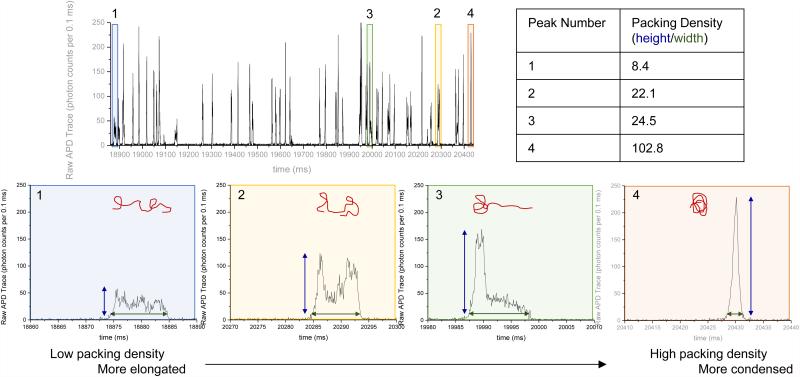
The retention time range for each fragment size was then used to sort the single molecule burst data by fragment size. The packing density for a single burst was calculated from the single molecule fluorescence data by dividing the burst height by the burst width, as illustrated in Figure 2. The mean packing density of each fragment size was calculated by averaging the packing densities of all single molecule bursts of molecules identified as that fragment size.
Figure 2.
Single molecule bursts of 23 kbp species separated in high salt conditions (25 mM NaCl) display a large variety in burst shapes ranging from stretched (1) to condensed (4), as characterized by the packing density (burst height divided by burst width).
Buffer Preparation
EACA (6-Aminocaproic acid) and Bis-tris were purchased from Sigma and dissolved in water to 500 mM stock concentrations. These stock solutions were used to generate the base 20 mM EACA/Bis-tris (EB) buffer at a final concentration of 10 mM EACA and 10 mM Bis-tris. Stock solutions of 5 M NaCl and 1 M MgCl2 were used to create buffers with added salt. 1 M NaOH was used as stock solution for denaturation experiments.
Sample Preparation
Lambda DNA, HindIII digested Lambda DNA, 1 kb DNA Ladder, and Supercoiled DNA Ladder (all from New England Biolabs, Inc.) were used as double stranded DNA samples. Staining was performed at 5 or 10 ng/μL total dsDNA concentration and 1 μM TOTO-3 Iodide (Life Technologies) for at least 1 hour in the dark. Single stranded DNA was purchased from IDT. Labeled oligos were ordered with either Alexa 647 or Cy5 covalently attached. The covalently labeled 25 bp DNA strand was synthesized and hybridized by IDT. Cy5 was covalently attached to one 24 nt long strand, which was hybridized to an unlabeled complementary strand, 26 nt long. Free Alexa was purchased as dry Alexa Fluor 647 NHS Ester (Life Technologies) and diluted in water. For more specific details outlining the sample and separation parameters for the experiments presented in each figure, please refer to the Supplemental Table S4.
Denaturation
Denaturation by heat and low ionic strength: DNA samples were prepared in 20 mM EB buffer and heated to 95 °C for 10 minutes before cooling to 20 °C (room temperature). Separations were performed in 20 mM EB buffer in 1.6 µm inner diameter capillary with a length of 40 cm.
Denaturation by pH: The DNA samples were prepared in 10 mM NaOH (pH 12). This was allowed to sit for at least 5 minutes before diluting the sample ten-fold in water to a final concentration of 1 mM NaOH. The separation was performed in 1 mM NaOH buffers in 1.6 µm inner diameter capillary with a length of 80 cm.
Hybridization
DNA samples were prepared at 100X concentration in 100 mM NaCl, heated to 95 °C for 10 minutes, then cooled to 20 °C (room temperature) for at least 2 minutes, and then diluted 100-fold into 20 mM EB buffer. The final sample buffer contained 20 mM EB + 1 mM NaCl. The separations were performed in 20 mM EB with 1 mM NaCl buffer in a 1.6 µm inner diameter capillary with a length of 40 cm.
RESULTS AND DISCUSSION
Direct Visualization of DNA Conformation via Single Molecule Fluorescence
The experimental platform (a) and separation mechanism (b) are illustrated in Figure 1. Hydrodynamic separations utilize the parabolic flow profile generated by pressure-driven flow in small, micron-sized capillaries to separate molecules by their effective radii. Separation is performed by first injecting a picoliter-sized sample plug at the beginning of the capillary and then driving the sample plug down the capillary. As the plug travels down the length of the capillary, radial diffusion allows molecules within the plug to sample various flow streams throughout the cross-section, resulting in an average velocity, or mobility, that is size and conformation dependent. Molecules with larger effective radii will have faster average velocities and shorter average retention times than smaller ones.
Once the separated molecules reach the detection region, cylindrical illumination confocal spectroscopy (CICS) is used to individually count and analyze the DNA (Figure 1a) 16. Figure 1c and 1d show separation of a 50 bp DNA ladder. The APD yields a raw fluorescence time trace consisting of a train of single molecule fluorescent bursts (Figure 1c) that can be analyzed and counted to form a chromatogram containing a series of DNA peaks resolved by size (Figure 1d). The chromatogram peaks can then be analyzed to determine the peak retention time, which is a reflection of the ensemble average length and conformation of all molecules within that peak. Repeated separations of the same sample demonstrate that the relative mobility of a DNA fragment population is highly repeatable when separated under the same experimental conditions (Supplemental Fig. S1 and Table S1).
In addition, the single molecule fluorescent bursts can be analyzed to give layered information regarding molecular distributions and provide insight into the properties of individual DNA molecules. For example, burst size or height can be used to determine the size of a DNA molecule17 or the DNA mass content within a polymer/DNA complex18. By further analyzing the burst shapes19, we are able to directly visualize DNA conformation while passing through the 1 μm long detection region. Figure 2 shows representative single molecule bursts arising from 23 kb DNA fragments in a separation of λ HindIII digest. The bursts display diverse shapes despite being generated by identical 23 kb DNA fragments. Highly condensed DNA molecules (Figure 2, peak 4) show narrow burst widths and tall burst heights. Conversely, highly stretched or elongated molecules (Figure 2, peak 1) show wide burst widths and short burst heights.
To quantify these conformational differences, we developed a metric called packing density, calculated as the ratio of burst height (photons per 0.1 ms bin) to burst width (ms). Condensed molecules result in high packing densities while elongated molecules result in low packing densities (Figure 2, table). When the single molecule bursts of all DNA corresponding to a single chromatogram peak are analyzed, a wide distribution of packing densities can be seen (Supplemental Figure S2). Furthermore, no obvious correlation is seen between packing density and retention time (i.e. temporal position within the peak). This diversity of burst shapes and lack of correlation between packing density and retention time imply that the DNA is likely in dynamic fluctuation between different conformations rather than fixed in a single conformation. These fluctuations are thermodynamically driven by molecular properties such as length and stiffness and separation conditions such as buffer properties and shear rate. A mean packing density for a single fragment size is found by taking the average packing density of all of the molecules identified as a single fragment size. Analysis of repeated separations demonstrate less than 6% variation in mean packing density for a single fragment size between three separation experiments (Supplemental Fig. S1 and Table S1).
Monitoring Perturbations on DNA Conformation via Mobility Shifts
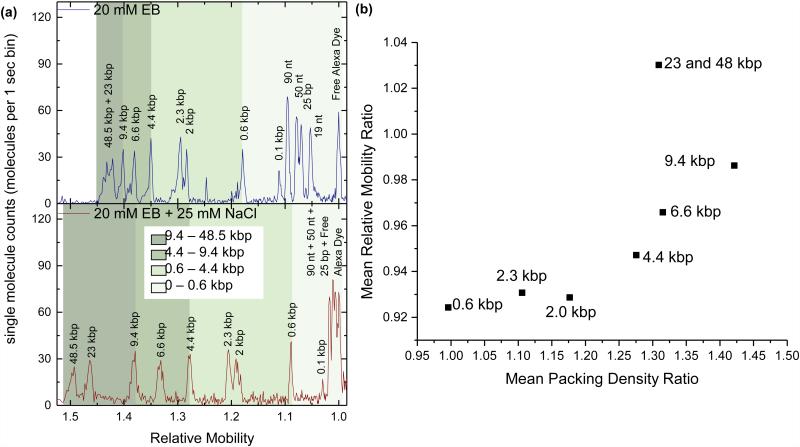
Perturbations in buffer ionic strength affect DNA packing densities, which are then reflected as changes in the relative mobility. In Figure 3a, DNA separations were performed on λ HindIII digested DNA using a 1.6 µm ID capillary and either a low ionic strength 20 mM EB elution buffer (top) or a high ionic strength 20 mM EB elution buffer with 25 mM NaCl added (bottom). The chromatograms are plotted as a function of relative mobility, which was calculated by normalizing retention time to the reference Alexa dye. In the low ionic strength buffer, the short DNA peaks (<0.6 kb) are well resolved while the long DNA peaks (>4.4 kb) are closely spaced. The 48 kb and 23 kb peaks cannot be individually resolved and appear as a single broad peak. However, when ionic strength is increased by the addition of 25 mM NaCl, the long DNA peaks (>4.4 kb) all become well resolved while the short DNA peaks (<0.1 kb) become closely spaced and unresolvable. Increasing the ionic strength creates perturbations in DNA conformation that affect large DNA quite differently than small DNA.
Figure 3.
A sample comprised of 13 DNA fragments ranging in size from 19 nt to 48 kbp is separated in the same 1.6 μm diameter capillary under both low ionic strength (20 mM EB buffer) and high salt conditions (20 mM EB + 25 mM NaCl). (a) Single molecule chromatograms of both separation conditions reveal that adding salt to the separation buffer induces relative mobility changes for small and large fragments. (b) Comparison of the effect of the added salt on ratiometric changes in relative mobility and packing density of the dsDNA fragments suggests a positive correlation between the two metrics.
Single molecule burst analysis further supports the conclusions drawn from the relative mobility shifts. The single molecule chromatogram was used to identify the fluorescent bursts belonging to each fragment size in order to calculate the average packing density of each DNA fragment. In Figure 3b, the change in DNA packing density due to 25 mM NaCl salt perturbation is plotted against the change in relative mobility. A strong correlation is seen between changes in packing density and relative mobility, indicating that the two measures are indeed related. The greater the change in packing density (i.e. the tighter the DNA condenses), the faster the DNA travels in high salt relative to low salt. This result was verified by comparing the separations of supercoiled and linear DNA fragments, where the ratiometric plot of supercoiled to linear DNA also displays a positive correlation between mean relative mobility and mean packing density (Fig. S4c). Interestingly, we note that if the DNA undergoes no change in packing density, it travels slower in high salt than low salt. This suggests that conformational changes alone do not account for this shift in relative mobility, and factors such as spatial distribution and flow profile may be affected as well.
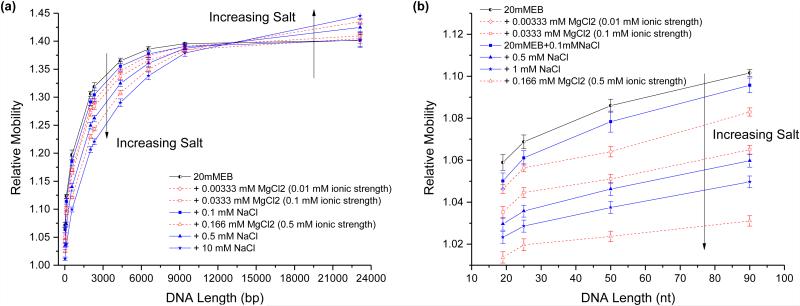
We further perturbed DNA conformation by testing the effects of a range of monovalent (sodium) and divalent (magnesium) cation concentrations on relative mobility and packing density. Separations were performed on double stranded (Figure 4a) and single stranded DNA (Figure 4b) using the low ionic strength 20 mM EB buffer20 with various amounts of added NaCl and MgCl2. Increasing salt concentration generally resulted in decreasing relative mobilities for most ssDNA and dsDNA species. These cations interact with the DNA’s negatively charged backbone to decrease electrostatic repulsion between nucleotides, reducing the effective radius of the DNA fragments and decreasing relative mobility. This is also supported by the higher average packing density displayed by the double stranded DNA fragments in high ionic strength buffer compared to low ionic strength buffer (Supplemental Figure S3b and d). Previous studies have further shown that the persistence length of both single stranded21 and double stranded DNA22 decreases with higher salt concentration. Moreover, even at the same ionic strength, magnesium cations had a greater effect on relative mobility than sodium cations. Magnesium has a higher affinity for DNA than sodium and results in a greater reduction in persistence length than an ionic strength equivalent concentration of sodium21-23.
Figure 4.
The effects of both sodium chloride (blue) and magnesium chloride (red) on the packing density of double stranded and single stranded DNA is probed by comparing their relative mobilities in the same 1.6 μm diameter capillary. (a) HindIII digested λ dsDNA is separated in a buffer with added ionic strengths of 0.01 mM (Diamond), 0.1 mM (square), 0.5 mM (triangle), and 10 (star). (b) ssDNA oligos (19 nt, 24 nt, 50 nt, and 90 nt) are separated in added ionic strengths of 0.01 mM (Diamond), 0.1 mM (square), 0.5 mM (triangle), and 1 mM (star). Error bars represent the width of each fragment’s elution peak, which was calculated as 4 times the standard deviation of the fitted Gaussian peak.
In Figure 4, all the DNA fragments exhibited a decrease in relative mobility with increased salt except for the large 23 kbp dsDNA fragment which behaved in the opposite manner. The average packing densities (Supplemental Figure S3b) also display opposing trends for large DNA fragments (23 and 48 kbp) compared to the rest of the smaller fragments. We attribute this behavior to the size-dependent effects of shear and weak confinement on the relatively compliant DNA molecules. Molecules with larger effective radii experience a larger velocity gradient, and thus shear gradient, across the molecule (refer to Fig. 1b). As a result, the larger molecules begin to elongate along the capillary length24. This molecular deformation also results in a larger-than-anticipated depletion region near the capillary walls25-29. The crossover size at which large DNA and small DNA begin behaving differently appears to be the same (< 15 kbp) in both the packing density and relative mobility plots (Supplemental Figure S3), suggesting that both measures are sensitive to perturbations in DNA conformation. Interestingly, the topology-induced crossover size between supercoiled and linear DNA appears to be much smaller (< 5 kbp) than the salt-induced crossover size (Supplemental Fig. S4), which could be due to differences in polymer properties between linear and supercoiled DNA, such as flexibility and elasticity, that affect the molecules’ responses to shear.
The results shown in Figures 2 and 3 were performed in a 1.6 µm ID capillary, but similar trends were also observed in 5 µm and 10 µm ID capillaries (Supplemental Figure S5). One notable observation is that the crossover size, where increasing salt concentration leads to increasing relative mobility, increases with increasing capillary radius. Thus, it is not the absolute DNA size and compliance that results in increased mobility for large DNAs, but its size relative to the capillary diameter and compliance that is important.
Monitoring DNA Hybridization and Denaturation
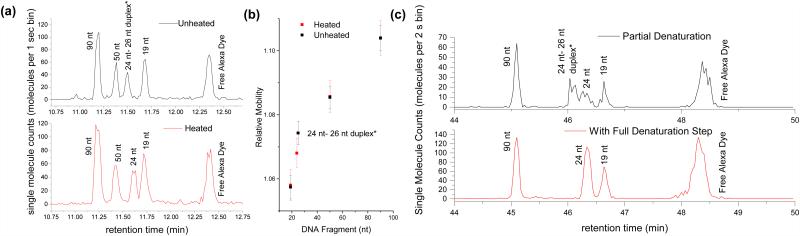
Hybridization and denaturation events also result in mass and structural changes that induce hydrodynamic mobility shifts. As seen in Figure 5a, heat was used to denature a DNA duplex into its single stranded components: a 24 nt fluorophore-labeled oligo and a 26 nt unlabeled oligo that cannot be seen. The resulting 24 nt peak has a longer retention time than the original duplex peak due to the decreased rigidity of ssDNA and its smaller effective radius. By calculating the relative mobilities (Figure 5b), it can be seen that only the double stranded species exhibits a mobility shift due to the heating step; the mobilities of the single stranded species (90, 50, and 19 nt) remain unchanged. Such perturbations can also be made using pH. In Figure 5c, separations were performed using 1 mM NaOH and 10 mM NaOH to denature the DNA. Using 1 mM NaOH (pH ~11), both single stranded and double stranded species were seen, suggesting that the sample was only partially denatured. When 10 mM NaOH (pH ~12) was used instead, only the single-stranded 24 nt peak was seen, indicating full denaturation. No mobility shifts were seen in the single stranded 90 nt and 19 nt DNA species.
Figure 5.
Denaturation of a double stranded DNA duplex into single strands results in a relative mobility shift. (a) A sample consisting of three single stranded species (19 nt, 50 nt, and 90 nt) and one double stranded species (24 nt – 26 nt duplex*) is separated in a 1.6 μm capillary at room temperature (top) and after heating the sample to 95 C for 5-10 min (bottom). (b) The average relative mobilities from the chromatograms in (a) is plotted against species size to illustrate that only the duplex mobility is affected by heating. (c) High pH environments (top – 1 mM NaOH; bottom 10 mM NaOH) also exhibit denaturation-induced mobility shifts for the duplex species only. Error bars represent the width of each fragment’s elution peak, which was calculated as 4 times the standard deviation of the fitted Gaussian peak.
Measuring Hybridization Efficiency and Binding Strength
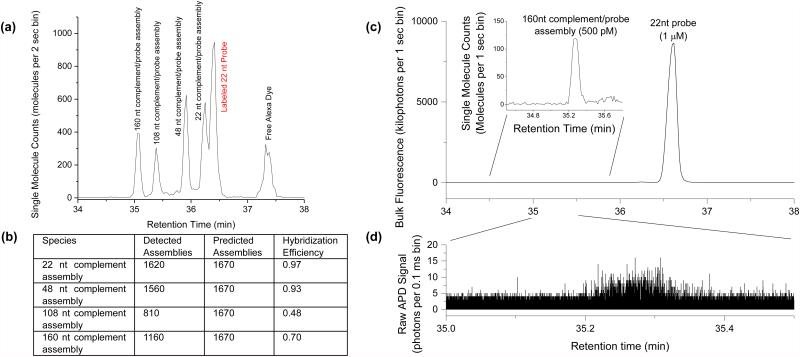
Making use of mobility shifts to separate species and single molecule counting for quantification, we then examined hybridization efficiency under competitive binding conditions. In Figure 6a, separations were performed on a sample containing a 22 nt Cy5 labeled probe and four unlabeled oligos ranging in length from 22 nt to 160 nt. Each oligo contained an identical 22 nt complementary region at the 5’ end. All five of the expected species could be clearly resolved in the separation chromatogram, including the 22 bp duplex species and the 22 nt single stranded probe. Binding of an unlabeled complement to the 22 nt probe resulted in a mobility shift proportional to the length of the complementary strand. As the chromatogram is generated by direct single molecule counting, the exact number of molecules contained in each of these peaks can be easily and accurately quantified (Figure 6b). Although the complementary regions on each oligo were identical in sequence and expected to have identical enthalpic binding contributions, the number of detected assemblies for each complement was not equal and generally decreased with length. This decrease in hybridization efficiency with length suggests an underlying entropic folding effect despite the oligos being designed to have minimal stable secondary structures.
Figure 6.
Hybridization of a fluorescently tagged single stranded probe to a complementary oligo is accompanied by a relative mobility shift. (a) Equal ratios of four complementary targets (lengths 22 nt, 48 nt, 108 nt, and 160 nt) are combined with a 22 nt ssDNA probe at five-times higher concentration and separated. (b) The hybridization efficiency of the competing hybridization reactions in (a) is calculated by comparing the quantity of a detected assemblies (determined by single molecule counting) with the number predicted (determined from the input ratios and the total number of fluorescent molecules present). (c) Separation and detection of the 160 nt complement/probe assembly (inset) in 2000-fold excess concentration of labeled probe. (d) The raw APD trace of the single fluorophore bursts that correspond to the hybridized 160 nt complement/probe assembly.
In the previous experiment, the probe and complementary oligos were input at similar concentrations. However, in many assays and separations, molecules are often present at widely varying levels. In Figure 6c and 6d, separation was performed on a mixture containing 1 µM 22 nt Cy5 labeled probe and 500 pM 160 nt unlabeled complement oligo. Despite this large mismatch in concentration, we could easily count 670 molecules of the hybridized assembly while simultaneously detecting a 2000-fold excess of unhybridized probes. By leveraging single molecule sensitivity and bulk fluorescence, we are able to quantify DNA across wide dynamic ranges, unmatched by other highly sensitive binding characterization methods such Fluorescence Correlation Spectroscopy (FCS)30.
Detecting Hybridization Mismatches
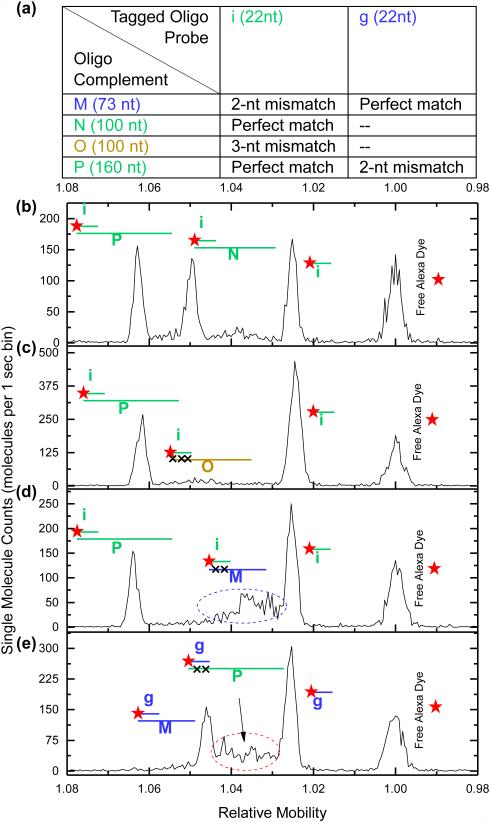
Finally, we investigated the effect of hybridization mismatches on relative mobility shifts. Separations were performed on sample mixtures containing 22 nt Cy5-labeled DNA probes, mimicking the sequence of either microRNA let-7g (g) or let-7i (i), and various perfect match, 2 bp mismatch, and 3 bp mismatch oligos (Figure 7a). As shown in Figure 7b, when probe i is hybridized to a mixture containing 160 nt and 100 nt perfect match oligos (P and N, respectively), three sharp peaks are seen in the chromatogram (Figure 7b). If the experiment is repeated by swapping oligo N (perfect match) for oligo O, which is the same length but possesses a 3 nt mismatch, binding stability is greatly reduced and the 100 nt peak entirely vanishes (Figure 7c). When oligo M, which is 73 nt oligo and contains a 2 nt mismatch, is swapped in, a broad smear, indicative of partial or weak hybridization, is seen (Figure 7d). In all cases, sharp peaks corresponding to the 160 nt perfect match complement and 22 nt free probe i are unaffected (Figure 7b-d). The same trend can be seen in Figure 7e when probe g is mixed with a 73 nt perfect match and a 160 nt oligo containing 2 bp mismatch (oligos M and P, respectively). Whereas the perfect match pair results in stable binding over the duration of a separation and appears as a sharp peak, the 2 bp mismatch pair results in weak hybridization that dynamically fluctuates over the duration of a separation and appears as a broad smear. 3 bp mismatch pairs result in binding that is too weak to be detected.
Figure 7.
Mimic sequences from the Let-7 microRNA family are used to investigate the sensitivity of hybridization-induced mobility shifted peaks to sequence complementarity and bond stability. (a) The length and complementarity of the oligo combinations used in the separations shown in (b)-(e) are listed: i and g are fluorescently tagged mimics of Let-7i and Let-7g respectively; M, N, O, and P are unlabeled oligos of varying complementarity. The species present in the chromatograms shown in (b)-(e) are labeled pictorially with probe i and its complementary oligos in green, probe g and its complements in blue, and oligo O, which does not have a fully complementary probe, in gold. Red stars designate the fluorescently labeled species: probes i and g and the free alexa dye. (b) Separation chromatogram of probe i combined with perfectly complementary oligos P (160 nt) and N (100 nt) shows distinct mobility shifted peaks for each complementary oligo. (c) No 100 nt peak is observed in the separation chromatogram when oligo P is replaced with oligo O (3 bp mismatch). (d) Separation chromatogram of probe i combined with oligos P (perfect complement) and M (73 nt oligo with 2 bp mismatch). (e) Separation chromatogram of probe g with the same M and P oligos used in (d), making M a perfect complement and P a 2 bp mismatch.
CONCLUSION
We have demonstrated a platform capable of assessing changes in global DNA conformation and DNA interactions by coupling hydrodynamic separation with single molecule analysis. We relate the global conformation of stained dsDNA fragments in solution to both the shape of single molecule fluorescent bursts as well as the relative hydrodynamic mobility of the fragment. We validated the effects of cation interactions on global DNA conformation, and propose utilizing this technique to assess conformational changes due to other factors, such as protein binding or histone modifications. With the resolution to separate single and double stranded species, we also demonstrated the ability to classify and quantify hybridization interactions. Not only could we quantify the competitive hybridization efficiency to different length targets, but we could also distinguish the strong bond of a perfectly complementary sequence from weak binding due to sequence mismatches, a cause of cross-talk and background in hybridization assays. Assay development requires careful and time consuming testing and optimization, to some extent because theoretical hybridization efficiencies can differ from experimental results. This platform is capable of quantifying hybridization under experimental conditions, distinguishing false-positives from true-positives, and has the wide dynamic range to detect large concentration ranges, suggesting its utility in both assay optimization as well as end detection for multiplexed analysis. The high sensitivityand quantitative nature of single molecule detection would also allow for detection and analysis of weak interactions and rare hybrid species that are difficult to study with current methods. Finally, since the separation and detection method are electric field free, intermolecular interactions beyond DNA-DNA hybridization could be studied using this platform, including other nucleic acids, proteins, or even synthetic polymer-nucleic acid interactions, for which examination at a single molecule level in free solution would be beneficial.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank funding sources from National Institutes of Health (R01CA155305, R21CA173390) and National Science Foundation (1033744). S.M. Friedrich is also grateful for the support from the National Science Foundation Graduate Research Fellowship Program under Grant No. (1232825).
Footnotes
Associated Content
Supporting Information
Experimental details and additional experimental data and results. This information is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- (1).Southern EM. J. Mol. Biol. 1975;98:503. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- (2).Alwine JC, Kemp DJ, Stark GR. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5350. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Hellman LM, Fried MG. Nat. Protoc. 2007;2:1849. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Xu Y, Colletier JP, Jiang H, Silman I, Sussman JL, Weik M. Protein Sci. 2008;17:601. doi: 10.1110/ps.083453808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).DiGabriele AD, Sanderson MR, Steitz TA. Proc. Natl. Acad. Sci. U. S. A. 1989;86:1816. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Meseth U, Wohland T, Rigler R, Vogel H. Biophys. J. 1999;76:1619. doi: 10.1016/S0006-3495(99)77321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Tijssen R, Bos J, Van Kreveld ME. Anal. Chem. 1986;58:3036. [Google Scholar]
- (8).Iki N, Kim Y, Yeung ES. Anal. Chem. 1996;68:4321. [Google Scholar]
- (9).Wang X, Wang S, Veerappan V, Byun CK, Nguyen H, Gendhar B, Allen RD, Liu S. Anal. Chem. 2008;80:5583. doi: 10.1021/ac800549k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Striegel AM. Anal. Bioanal. Chem. 2012;402:77. doi: 10.1007/s00216-011-5334-3. [DOI] [PubMed] [Google Scholar]
- (11).Striegel AM, Brewer AK. Annu. Rev. Anal. Chem. 2012;5:15. doi: 10.1146/annurev-anchem-062011-143107. [DOI] [PubMed] [Google Scholar]
- (12).Tijssen R, Bleumer JPA, Vankreveld ME. J. Chromatogr. 1983;260:297. [Google Scholar]
- (13).Wang X, Cheng C, Wang S, Zhao M, Dasgupta PK, Liu S. Anal. Chem. 2009;81:7428. doi: 10.1021/ac901265t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wang X, Veerappan V, Cheng C, Jiang X, Allen RD, Dasgupta PK, Liu S. J. Am. Chem. Soc. 2009;132:40. doi: 10.1021/ja909233n. [DOI] [PubMed] [Google Scholar]
- (15).Liu KJ, Rane TD, Zhang Y, Wang TH. J. Am. Chem. Soc. 2011;133:6898. doi: 10.1021/ja200279y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Liu KJ, Wang TH. Biophys. J. 2008;95:2964. doi: 10.1529/biophysj.108.132472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Liu KJ, Brock MV, Shin LM, Wang TH. J. Am. Chem. Soc. 2010;132:5793. doi: 10.1021/ja100342q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Beh CW, Pan D, Lee J, Jiang X, Liu KJ, Mao HQ, Wang TH. Nano Lett. 2014;14:4729. doi: 10.1021/nl5018404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Reccius CH, Stavis SM, Mannion JT, Walker LP, Craighead HG. Biophys. J. 2008;95:273. doi: 10.1529/biophysj.107.121020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Bier M, Ostrem J, Marquez RB. Electrophoresis. 1993;14:1011. doi: 10.1002/elps.11501401161. [DOI] [PubMed] [Google Scholar]
- (21).Chen H, Meisburger SP, Pabit SA, Sutton JL, Webb WW, Pollack L. Proc. Natl. Acad. Sci. U. S. A. 2012;109:799. doi: 10.1073/pnas.1119057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6185. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Korolev N, Lyubartsev AP, Rupprecht A, Nordenskiold L. Biophys. J. 1999;77:2736. doi: 10.1016/s0006-3495(99)77107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Graham MD. Annu. Rev. Fluid Mech. 2011;43:273. [Google Scholar]
- (25).Shafer RH, Laiken N, Zimm BH. Biophys. Chem. 1974;2:180. doi: 10.1016/0301-4622(74)80040-2. [DOI] [PubMed] [Google Scholar]
- (26).Shafer RH. Biophys. Chem. 1974;2:185. doi: 10.1016/0301-4622(74)80041-4. [DOI] [PubMed] [Google Scholar]
- (27).Jendrejack R, Dimalanta E, Schwartz D, Graham M, de Pablo J. Phys. Rev. Lett. 2003;91 doi: 10.1103/PhysRevLett.91.038102. [DOI] [PubMed] [Google Scholar]
- (28).Jendrejack RM, Schwartz DC, de Pablo JJ, Graham MD. J. Chem. Phys. 2004;120:6315. doi: 10.1063/1.1637331. [DOI] [PubMed] [Google Scholar]
- (29).Raghunath C, Roland GW, Gerhard G. J. Phys.: Condens. Matter. 2011;23:184117. [Google Scholar]
- (30).Rigler R, Elson E. Fluorescence Correlation Spectroscopy - Theory and Applications. Springer; Berlin ; New York: 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.