Significance
Many bacteria use NO· dioxygenase and NO· reductase to defend themselves against immune-generated NO·. The importance and contribution of these systems under microaerobic conditions, which pathogens are likely to encounter within a host, remain poorly understood. We investigated the NO· response of Escherichia coli throughout the microaerobic regime, and discovered conditions that largely disabled the NO· defenses of E. coli, and environments where the [NO·] oscillated. Components found to comprise the oscillatory circuit are distributed broadly among bacterial species, suggesting that these dynamics could be a characteristic feature of how bacteria respond to NO· in low O2 environments. In support of this hypothesis, analogous oscillations were observed in NO·-stressed cultures of Pseudomonas aeruginosa under low O2 conditions.
Keywords: nitrosative stress, E. coli, microaerobic, Pseudomonas aeruginosa, kinetic modeling
Abstract
The virulence of many pathogens depends upon their ability to cope with immune-generated nitric oxide (NO·). In Escherichia coli, the major NO· detoxification systems are Hmp, an NO· dioxygenase (NOD), and NorV, an NO· reductase (NOR). It is well established that Hmp is the dominant system under aerobic conditions, whereas NorV dominates anaerobic conditions; however, the quantitative contributions of these systems under the physiologically relevant microaerobic regime remain ill defined. Here, we investigated NO· detoxification in environments ranging from 0 to 50 μM O2, and discovered a regime in which E. coli NO· defenses were severely compromised, as well as conditions that exhibited oscillations in the concentration of NO·. Using an integrated computational and experimental approach, E. coli NO· detoxification was found to be extremely impaired at low O2 due to a combination of its inhibitory effects on NorV, Hmp, and translational activities, whereas oscillations were found to result from a kinetic competition for O2 between Hmp and respiratory cytochromes. Because at least 777 different bacterial species contain the genetic requirements of this stress response oscillator, we hypothesize that such oscillatory behavior could be a widespread phenomenon. In support of this hypothesis, Pseudomonas aeruginosa, whose respiratory and NO· response networks differ considerably from those of E. coli, was found to exhibit analogous oscillations in low O2 environments. This work provides insight into how bacterial NO· defenses function under the low O2 conditions that are likely to be encountered within host environments.
Nitric oxide (NO·) plays a critical role in mammalian innate immunity as a potent antimicrobial (1–3), where its broad reactivity contributes to a diverse repertoire of cytotoxic effects including respiratory inhibition, thiol nitrosation, iron–sulfur cluster ([Fe-S]) destruction, DNA deamination, and tyrosine nitration/nitrosylation (4, 5). To cope with this stress, many pathogens harbor defenses to detoxify NO· and its reaction products, and repair NO·-mediated damage to biomolecules (6, 7). Disruption of these defenses has been shown to attenuate virulence in many pathogenic species, such as Neisseria meningitides, Pseudomonas aeruginosa, Yersinia pestis, Mycobacterium tuberculosis, Vibrio cholerae, uropathogenic and enterohemorrhagic Escherichia coli (UPEC and EHEC, respectively), Staphylococcus aureus, and Salmonella enterica serovar Typhimurium (4, 6), which underscores the importance of NO· to immune function and highlights microbial NO· defense networks as a promising source of targets for the development of next-generation antiinfectives (8). This potential has inspired many investigations of bacterial NO· stress, from which major defense systems, such as NO· dioxygenase (NOD) (9, 10) and NO· reductase (NOR) (11, 12), have been identified.
NODs are widely distributed among different bacterial species (13) and provide protection from NO· under oxygenated conditions by catalyzing the reaction between NO· and O2 to form nitrate (9, 10). Although some studies have demonstrated that NODs can detoxify NO· under anaerobic conditions using an NOR function, the rate is orders of magnitude slower than the O2-dependent reaction and the physiological relevance is a matter of debate (14–17). In addition to an NOD, many bacteria also possess a separate NOR, which functions optimally under anaerobic conditions and complements the O2-dependent function of NOD (11, 12, 18, 19). The contributions of these systems to NO· detoxification have been well studied under aerobic and anaerobic conditions (9–12, 15, 16, 20–23), but how they coordinate to remove NO· under the intermediate microaerobic regime is less clear. The fact that O2 tensions in vivo span the entire anaerobic–aerobic spectrum, with ∼120–200 μM dissolved O2 in the lungs, ∼50–130 μM in vasculature, ∼5–50 μM in tissue (muscle, liver, kidney, lymphoid tissues), and ∼0–100 μM in the gastrointestinal tract (24–29), underscores the importance of understanding bacterial NO· detoxification at all O2 levels.
In E. coli, several studies have examined NO· stress under microaerobiosis (12, 30, 31). Gardner et al. (12) measured growth of wild-type (WT), Δhmp, ΔnorV, and ΔhmpΔnorV E. coli in an environment with ∼5 μM O2 and found that ΔhmpΔnorV was the slowest to recover, which suggested that both defense enzymes participate in NO· stress tolerance under low O2 conditions. Two separate studies found that the duration of respiratory inhibition and growth arrest by NO· was inversely proportional to [O2] in E. coli cultures (measured down to ∼25 μM O2), although the cause was uncertain, and was suspected to be the result of reduced NO· autoxidation or decreased abundance of substrate (O2) to drive the Hmp NO· dioxygenation reaction (30, 31). Kinetic characterizations of purified Hmp demonstrated that the rate of NO· dioxygenation was reduced at lower [O2], which was attributed to its relatively low affinity for O2 (measured Km,O2 values ranged from ∼25 to as high as 100 μM O2) (16, 32, 33). Although these studies have provided initial insight into the influence of [O2] on bacterial NO· defenses, they generally focused on the participation of only one network component (namely Hmp), and they did not quantify the distribution of NO· among the available consumption pathways at different [O2]. As such, there remains considerable uncertainty regarding the quantitative contribution of NOD and NOR to NO· detoxification within the microaerobic regime.
Here, we sought to investigate NO· stress in E. coli under low [O2] environments, focusing on the quantitative trade-off between its NOD (Hmp) and NOR (NorV) throughout this regime. To do this, we used a kinetic model to quantitatively characterize E. coli NO· detoxification in environments with [O2] between 0 and 50 μM. Simulations predicted compromised NO· defenses (increased time required to clear NO· from the culture) within this range relative to 50 and 0 μM O2, and this was experimentally confirmed, although the magnitude of the effect was underestimated by the initial predictions. Furthermore, [NO·] oscillations, which were not predicted a priori, were observed at 5 and 10 μM O2. Using an integrated computational and experimental approach, NO· defenses were found to be considerably impaired by decreased substrate (O2) abundance for Hmp, strong O2-mediated suppression of NorV activity, and a general reduction in translation rate at reduced [O2]. Computational dissection of the [NO·] oscillations predicted that they were driven by an imbalanced competition for O2 between Hmp and respiratory cytochromes, the inhibitory effect of NO· on cytochrome quinol oxidase activity, and the NO· detoxification function of Hmp. This hypothesized mechanism was confirmed experimentally with biochemical measurements (quantification of [NO·] and [O2]) from WT and genetic mutants, Δhmp and ΔCYT (mutant devoid of cytochrome bo, bd-I, and bd-II terminal quinol oxidases). Given the modest genetic requirements of this stress response oscillator, 777 different bacterial species cataloged in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (34, 35) have the potential to produce oscillations when stressed with NO· in low O2 environments. To assess the generality of this native oscillator, we chose one of those bacterial species, P. aeruginosa, and found that it exhibited analogous oscillatory behavior when treated with NO· under low O2 conditions. Collectively, this study reveals the existence of metabolic oscillations in bacterial stress response networks under physiologically relevant O2 conditions, quantifies the trade-off between NOD and NOR in this regime, and provides a compelling example of the utility of quantitative modeling in mechanistic investigations of complex phenomena.
Results
NO· Detoxification in E. coli Cultures in Low [O2] Environments.
To begin exploring the dynamics of E. coli NO· detoxification at low [O2], WT E. coli were grown in an O2-control glove box and treated with 50 μM dipropylenetriamine (DPTA) (Z)-1-[N-(3-aminopropyl)-N-(3-ammoniopropyl)amino]diazen-1-ium-1,2-diolate (NONOate), which dissociates with a half-life of ∼2.5 h (at 37 °C and pH 7.4) to release 2 mol of NO· per mol of parent compound. The glove box environment was operated to maintain an atmosphere of 5% or 0% (vol/vol) O2, which corresponded to 50 and 0 μM dissolved [O2], respectively (Materials and Methods). Under 50 μM O2 conditions, the addition of DPTA NONOate produced a peak [NO·] of 0.57 ± 0.03 μM (mean ± SE) and NO· was cleared from the culture by 0.118 ± 0.004 h postdose (tclear,NO·, defined as the time after treatment at which [NO·] dropped below 0.2 μM NO·) (Fig. 1A). Under anaerobic conditions (0 μM O2), the addition of DPTA NONOate yielded a higher [NO·] peak, 1.49 ± 0.04 μM, and a marginally longer tclear,NO· of 0.136 ± 0.009 h (Fig. 1B). Using these data, we used a kinetic model of NO· stress (36) to delineate the O2 concentrations between 0 and 50 μM that were most interesting for experimental investigation. The model was constructed and used in previous studies (36–38), where it was shown to be a valuable tool for quantitative, mechanistic interrogation of the E. coli NO· defense network. A complete list of biochemical species, kinetic parameters, and reactions in the model are presented in Datasets S1 and S2, and SI Appendix, Table S1. Due to the different experimental conditions used here, the model was trained on experimental [NO·] measurements from 50 and 0 μM O2 environments, and a Markov chain Monte Carlo (MCMC) procedure (39, 40) was used to generate an ensemble of plausible models [defined as those exhibiting an evidence ratio (ER) of less than 10] (SI Appendix, Supporting Methods) that accounted for parametric uncertainty and allowed the robustness of forward predictions to be assessed (Dataset S3). Simulated [NO·] curves from the ensemble at 50 and 0 μM [O2] varied little and exhibited excellent agreement with experimental measurements of [NO·] in WT (Fig. 1 A and B), Δhmp, and ΔnorV cultures (SI Appendix, Fig. S1), which gave confidence in the predictive accuracy of the model for these conditions.
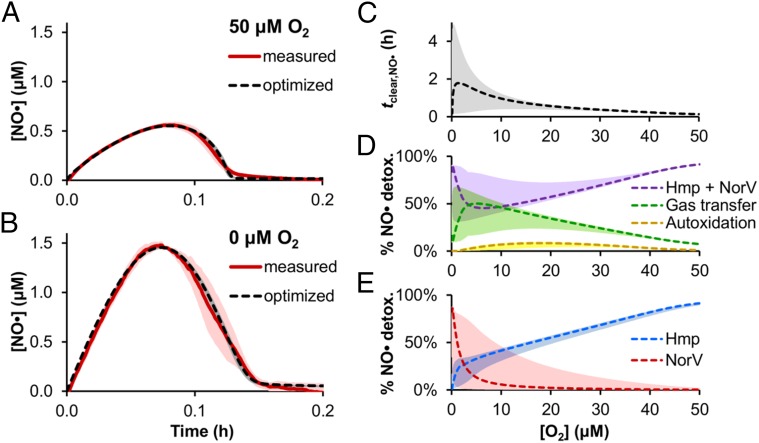
Fig. 1.
Model training and prediction of NO· clearance throughout the microaerobic regime. Exponential-phase cultures of wild-type (WT) E. coli at an OD600 of 0.05 were treated with 50 μM DPTA NONOate at time t = 0 under conditions of (A) 50 μM O2 or (B) 0 μM O2, and the [NO·] was measured (solid red line; mean of three independent experiments, with light red shading representing the SEM). The model was trained on the measured [NO·] curves at 50 and 0 μM O2, where simulations using the resulting optimal parameter set are plotted (dashed black lines). (C) NO· treatment simulations were run for [O2] environments ranging from 0 to 50 μM O2, from which the time of clearance of NO· (tclear,NO·) was calculated (quantified as the time required for [NO·] to drop below 0.2 μM NO·). (D) Predicted contribution of different NO·-consuming pathways (transfer to the gas phase, autoxidation, and the combination of Hmp- and NorV-mediated detoxification) to the total cumulative detoxification of NO· up to time tclear,NO·. (E) Individual contributions of Hmp and NorV to cumulative NO· detoxification. Dashed lines are simulations using the best-fit (minimum SSR, or ER = 1) parameter set, where shading of similar color represents prediction uncertainty (range of viable parameter sets with ER < 10). The O2 concentration corresponds to the concentration of dissolved O2 in cell-free media that was in equilibrium with the atmosphere of the hypoxic chamber.
Simulations of NO· detoxification throughout the microaerobic regime exhibited uncertainty in the 0–20 μM O2 range, where NO· was either cleared similarly across the entire regime with mild inhibition between 10 and 20 μM O2 (∼3-fold increase in tclear,NO· relative to clearance at 0 or 50 μM O2) or NO· detoxification was severely impaired (∼40- to 50-fold increase in tclear,NO· relative to clearance at 0 or 50 μM O2, respectively) with a maximum depression in clearance occurring at ∼0.3 μM O2 (Fig. 1C). The optimal model predicted an effect between these two extremes (∼15-fold increase in tclear,NO· relative to clearance at 0 or 50 μM O2), where the maximum depression in NO· detoxification existed at ∼1.5 μM O2. Using the ensemble of models to dissect NO· consumption into its individual pathways up to the time of NO· clearance, we found that the predicted contribution of autoxidation to NO· clearance remained modest (<10%) throughout the entire range of [O2] explored, whereas Hmp and NorV were the dominant NO· consumption pathways (Fig. 1D). Loss of NO· to the gas phase comprised the majority of the remaining flux, where its maximum contribution was predicted to occur under conditions where Hmp and NorV were least efficient (∼1–15 μM O2). Inspection of individual Hmp and NorV participation revealed that Hmp was predicted to dominate NO· consumption down to ∼0.2–13 μM O2, beyond which (and into anaerobiosis) NorV functioned as the primary NO· sink (Fig. 1E). Given these in silico predictions, and considering the 1 μM [O2] resolution of the glove box O2 controller, we reasoned that additional experiments performed at 5, 10, and 20 μM O2 would resolve uncertainty and determine the extent to which microaerobic environments impair NO· detoxification by E. coli.
NO· detoxification was measured in cultures of WT, Δhmp, and ΔnorV after the addition of 50 μM DPTA NONOate in environments of 5, 10, and 20 μM dissolved O2 (SI Appendix, Fig. S2). Results showed that E. coli was extremely impaired in its ability to remove NO· under microaerobic conditions, exceeding the poorest NO· clearance dynamics predicted by the ensemble. The time needed to clear NO· from the culture was measured to be ∼20-fold (compared with the predicted maximum of ∼15-fold) longer at 5 μM O2 (2.87 ± 0.08 h) than at 50 or 0 μM O2 (0.118 ± 0.004 and 0.136 ± 0.009 h, respectively) (Fig. 2 and SI Appendix, Fig. S3). Another unpredicted behavior was the emergence of oscillations in concentration of NO· at 5 and 10 μM O2. Oscillations began shortly after the initial clearance of NO·, and were damped, eventually stabilizing at ∼0 μM [NO·] by ∼1 h after the initial clearance of NO· (Fig. 2 B and C).
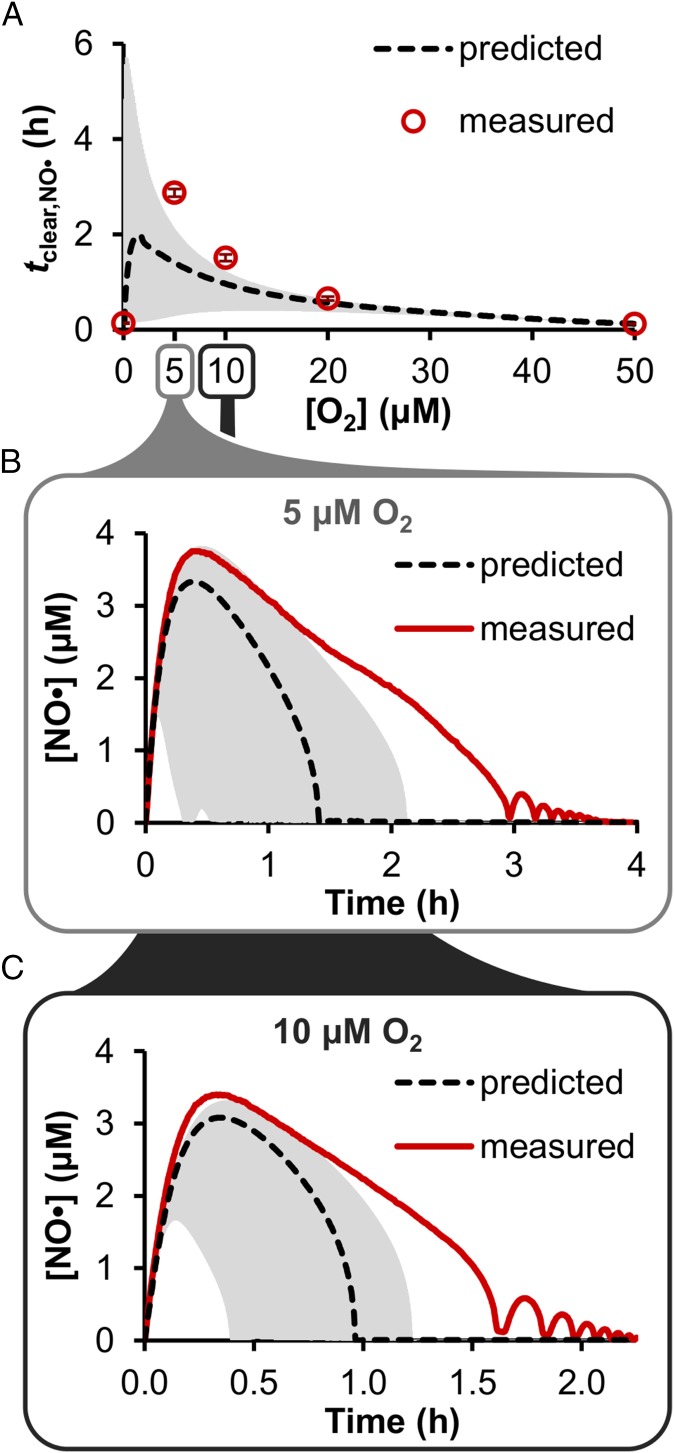
Fig. 2.
Comparison of predicted and measured tclear,NO· and [NO•] at different [O2]. (A) Cultures of WT E. coli (OD600 = 0.05) were treated with 50 μM DPTA NONOate at 50, 20, 10, 5, and 0 μM [O2], and the tclear,NO· (corresponding to initial NO· clearance, in the case of oscillations) was determined (open circles), and compared with the corresponding model-predicted tclear,NO· (dashed black line). The measured data points are the mean of at least three independent experiments, with error bars representing the SEM, and the predicted tclear,NO· was obtained using the best-fit parameter set (from the initial optimization on [NO·] measured at 0 and 50 μM O2), where gray shading represents prediction uncertainty (range of viable parameter sets with ER < 10). The [NO·] curves measured at (B) 5 μM O2 and (C) 10 μM O2 (solid red line) are shown with the corresponding predicted [NO·] for those O2 concentrations (dashed black line). The measured [NO·] curves were obtained from a single experiment that is representative of at least three independent experiments (the results of which are presented in SI Appendix, Fig. S13 A and C); the mean [NO·] was not presented because small deviations in phase and/or period among replicates, when averaged, obscured the oscillatory dynamics. The predicted [NO·] curves were obtained using the best-fit parameter set, with gray shading representing prediction uncertainty (range of viable parameter sets with ER < 10).
We sought to understand the mechanisms underlying the unpredicted severity of low O2 conditions on NO· detoxification and the [NO·] oscillations that were observed. To begin, we considered the possibility that the impairment in NO· defenses at 5 and 10 μM arose from cell death. Both Hmp and NorV require reducing equivalents to detoxify NO·, and if cells were dying it would manifest as extended NO· clearance times. To assess this possibility, samples were removed immediately before the addition of DPTA NONOate and at various times after treatment, and plated on Luria–Bertani (LB) agar to quantify colony-forming units (CFUs). The number of CFUs did not decrease following NO· treatment (SI Appendix, Fig. S4), eliminating cell death as a potential contributor to the poor ability of E. coli to detoxify NO· at low O2 concentrations. In the absence of a contribution from cell death, we assessed whether model simulations and experimentally measured [NO·] dynamics could be reconciled.
Reconciliation of Measured NO· Clearance Times with Model Simulations.
Initially, the 17 model parameters optimized on the WT [NO·] curves at 0 and 50 μM were reoptimized to fit the [NO·] curves from all measured [O2] conditions (0, 5, 10, 20, and 50 μM O2) (Dataset S3). Although the simulated [NO·] profiles at the lower [O2] conditions were greatly improved with the reoptimized parameter set, the model was unable to capture the [NO·] dynamics at all of the conditions simultaneously, and specifically, the fit of the [NO·] curve at 50 μM O2 was compromised (SI Appendix, Fig. S5A). To quantitatively assess the quality of fit at each of the conditions in a manner that was independent of absolute [NO·] or the number of data points describing each curve, the areas under the curve (AUCs) of each measured and simulated [NO·] profile were calculated, AUCmeas and AUCmodel, respectively, and the relative error (AUCRE) was evaluated: AUCRE = |AUCmeas − AUCmodel|/AUCmeas. The fit at 50 μM O2 was the poorest, with an AUCRE just over 2.0, or 200% relative error.
The inability of the model to capture the measured [NO·] dynamics at the five different O2 conditions suggested that constrained parameters, which were those not released during optimization, could be incorrect. We considered this a possibility because parameter values obtained from the literature were not measured under conditions identical to those used in this study. Alternatively, the disagreement between simulations and measurements could implicate a problem with the model structure itself (e.g., missing reactions, incorrect rate equations). To assess whether the existing model structure was sufficient to capture the [NO·] dynamics under the different [O2] conditions, all cellular parameters were allowed to vary within one order of magnitude, and the optimization was repeated. Despite the increased parametric flexibility, the model was still unable to adequately capture all [NO·] curves, where only a small improvement in performance at 50 μM O2 was achieved (AUCRE decreased from 2.0 to 0.67), but at the expense of agreement at 0 μM O2 (AUCRE increased from 0.10 to 0.24) (SI Appendix, Fig. S5B). Furthermore, an assessment of the Akaike information criterion (AIC) (Materials and Methods) suggested that the release of the additional parameters for optimization was not justified by the improvement to model performance, as the AIC was greater when all cellular parameters were relaxed (SI Appendix, Table S2).
The inability of the model to capture the combined set of [NO·] dynamics despite the release of all cellular parameters suggested that a structural change to the model was necessary to accurately describe the observed behavior. Given the noticeably slower growth observed at 0, 5, and 10 μM O2 relative to growth at 20 and 50 μM O2 (SI Appendix, Table S3), and the strong association between growth rate and rate of protein synthesis (41), we reasoned that the rate of translation was likely a function of [O2]. We therefore incorporated an [O2] dependency in the rate equation for translation, where increased [O2] yielded a faster rate of protein production, and vice versa. An optimization was conducted of the original 17 parameters along with the two additional parameters that were introduced with the new [O2]-dependent translation rate equation (kact,O2 and KO2; see Materials and Methods) (Dataset S3). The optimal parameter set with the new model structure was able to capture the [NO·] dynamics at all measured [O2] conditions, with a low AUCER even for the 50 μM O2 [NO·] curve (SI Appendix, Fig. S5C). In addition, the AIC was lower for the new parameter set relative to the previous best fit, despite the penalty of including two additional optimized parameters (SI Appendix, Table S2).
With the improved model performance, we sought to provide direct experimental support for an [O2]-dependent translation rate. Using the model, an experimentally tractable in silico experiment was conducted whereby Hmp was replaced with GFP to allow real-time monitoring of protein expression from the Phmp promoter. NO· treatment of 50 μM DPTA NONOate was simulated for conditions of 50, 10, and 5 μM O2, yielding predicted [GFP] profiles. The original model predicted virtually identical concentrations of GFP among the three O2 environments, whereas the new model structure predicted an increasing rate of GFP production with higher [O2] (SI Appendix, Fig. S6 A and B). When the corresponding experiment was performed (Materials and Methods), the measured [GFP] profiles (SI Appendix, Fig. S6C) were in excellent agreement with those predicted by the model possessing an [O2]-dependent translation rate, providing strong support for the inclusion of this term. To assess whether the effect was specific to NO·-stressed conditions and/or the Phmp promoter, the experiment was repeated in the absence of NO· treatment, and with gfp under the control of an isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible PT5 promoter. The same trend of increasing GFP production with increasing [O2] (while maintaining a constant 1 mM IPTG concentration) was observed (SI Appendix, Fig. S7), providing evidence that the effect was general, and not specific to the NO· response.
Dissection of Oscillations in the NO· Detoxification Network at Low [O2].
In addition to improving the model’s ability to capture the [NO·] dynamics measured at each of the different O2 concentrations, implementation of an [O2]-dependent translation rate resulted in the emergence of oscillations in the simulated [NO·] curve at 5 μM O2 (SI Appendix, Fig. S5C). Although these simulation results were encouraging, the oscillations were not quantitatively accurate at 5 μM O2 (incorrect amplitude, phase, and period), and they were absent from simulations at 10 μM O2. We therefore sought to identify the network components responsible for oscillations at 5 μM O2, and to determine whether any of the parameters governing those processes were preventing oscillations to occur in simulations at 10 μM O2. Using a reaction deletion analysis (Materials and Methods), we identified the minimal set of reactions necessary to sustain [NO·] oscillations at 5 μM O2 (SI Appendix, Table S4). In addition to NO· delivery and exchange of NO· and O2 with the gas phase, reactions contributing to the oscillations were those associated with Hmp and the respiratory cytochromes (cytochromes bo, bd-I, and bd-II, abbreviated collectively here as CYT). Thus, the mechanism underlying the oscillatory [NO·] dynamics was predicted to involve interactions between the O2-consuming activity of Hmp and CYT, the inhibitory effect of NO· on CYT, and NO· consumption by Hmp, as illustrated in Fig. 3A.
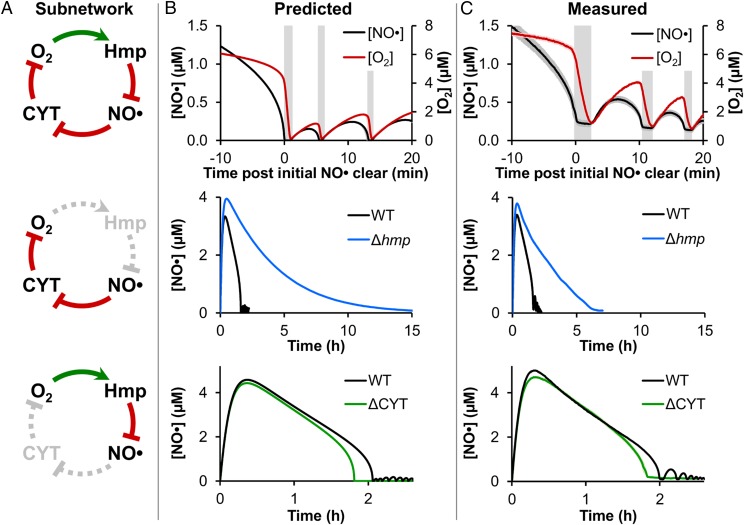
Fig. 3.
Experimental validation of the predicted oscillatory mechanism. (A, Top) Graphical representation of the oscillatory network, where O2 drives Hmp-mediated dioxygenation of NO·, and NO· inhibits CYT reduction of O2. Depiction of how genetic mutants (Middle) Δhmp and (Bottom) ΔCYT impacted the oscillatory network. (B) Predictions and (C) corresponding measurements of (Top) [NO·] and [O2] following treatment of WT E. coli culture with 50 μM DPTA NONOate under a 10 μM O2 environment, showing the region near initial [NO·] clearance (∼1.5 h after DPTA NONOate treatment) when the oscillations begin. Vertical gray bars are to aid visualization of the phase offset between [NO·] and [O2] oscillations, where NO· is depleted first, and then [O2] drops rapidly. (Middle) [NO·] following treatment of WT and Δhmp cultures with 50 μM DPTA NONOate at 10 μM O2, demonstrating the loss of oscillations for Δhmp. (Bottom) [NO·] following treatment of WT and ΔCYT cultures with 50 μM DPTA NONOate at 10 μM O2 (WT culture pH was adjusted to match conditions of ΔCYT; Materials and Methods), demonstrating the elimination of oscillations for ΔCYT. Experimental measurements are the mean of at least three independent experiments, with shading representing the SEM, except for WT [NO·] in the Middle and Bottom, which are representative of at least three independent experiments (the results of which are presented in SI Appendix, Fig. S13 A and D, respectively). Simulations were run using the best-fit parameter set (from optimization “stage 4”), with shading representing prediction uncertainty (range of viable parameter sets with ER < 10).
The minimal network suggested that the value of one or more parameters governing CYT and/or Hmp reactions was likely responsible for the absence of oscillations at 10 μM O2. Given that many of the parameters governing Hmp activity were among those being optimized, whereas the parameters dictating the inhibitory binding of NO· to the cytochromes were obtained from literature and thus fixed, the four parameters associated with the latter process (kCYTbo,NO·-on, kCYTbd,NO·-on, Km,CYTbo,O2, and Km,CYTbd,O2) were relaxed and allowed to vary along with the previous set of 19 parameters in an attempt to capture the [NO·] oscillations at both 5 and 10 μM O2 conditions (Dataset S3). The results of the optimization revealed that the simulated [NO·] curves were now oscillating at 10 μM O2, and the AIC had decreased (SI Appendix, Table S2), indicating that the improvement in sum of the squared residuals (SSR) outweighed the penalty of including additional parameters in the optimization (SI Appendix, Fig. S5D).
Experimental Confirmation of the Predicted Oscillatory Mechanism.
At this stage, the optimized model captured the dynamics of [NO·] under five different [O2] environments (0, 5, 10, 20, and 50 μM O2), exhibited oscillations in [NO·] at 5 and 10 μM O2, and suggested a mechanism involving Hmp and CYT underlying the oscillatory behavior. To experimentally assess the predicted mechanism, we analyzed the system dynamics. Simulations suggested that oscillations should be present in the [O2] curve at a similar time as, but out of phase with, the [NO·] oscillations ([NO·] depletes, then [O2] decreases; Fig. 3B). In addition, we observed in silico that the oscillations could be eliminated through genetic deletion of either hmp or the cytochrome quinol oxidases (Fig. 3B). Each of these three predictions was experimentally tractable, and therefore evaluated.
The NO· treatment assay was repeated at 10 μM O2 conditions, and [O2] was monitored along with [NO·]. In agreement with the model-predicted dynamics, oscillations were observed in [O2] that were appropriately out of phase with those of [NO·] (Fig. 3C). To assess the prediction that deletion of hmp would eliminate [NO·] oscillations following clearance, [NO·] was monitored in a Δhmp culture at 10 μM O2. As predicted, oscillations were not present in the Δhmp [NO·], although clearance was faster than expected (Fig. 3C). The quantitative disagreement between simulations and experiments at longer times (beyond ∼4 h posttreatment) is likely the result of model parameters having been trained on the first ∼2.5 h of treatment at 10 μM O2, which may render extrapolations to long times, such as 15 h, less accurate. Removal of the CYT arm of the proposed oscillatory mechanism was achieved by deleting all E. coli cytochrome quinol oxidases (ΔcyoAΔappBΔcydB; ΔCYT) (Materials and Methods). Measurements of [NO·] in a ΔCYT culture were in excellent agreement with simulations, including an elimination of oscillations (Fig. 3C). Experimental confirmation of model-predicted [O2] dynamics, as well as the impact of Δhmp and ΔCYT deletions, collectively support the proposed mechanism responsible for the [NO·] oscillations.
After confirming the predicted oscillating subnetwork, we investigated differences observed between model and experiment in the period, amplitude, and dampening of the [NO·] oscillations (Fig. 3 B and C). The rapid changes in physiology during the oscillations are likely to be complex and difficult to predict given the parsimonious reaction rate equations used here for transcription and translation. One such effect could be periodic bursts in protein synthesis (e.g., Hmp, NorV) during the brief periods of respiratory relief, which could quickly shift the proteome and influence the rate of NO· detoxification. To determine whether translation during the oscillatory period was contributing to the [NO·] dynamics, NO· treatment assays were performed with the addition of chloramphenicol (CAM) to halt translation before the start of oscillations (Materials and Methods). Oscillations were again observed; however, the amplitudes were increased and the damping was largely reduced in CAM-treated cultures compared with untreated controls (SI Appendix, Fig. S8). To assess the ability of the model to capture the dynamics of the [NO·] oscillations without the effect of complex translational dynamics during the oscillatory period, the 23 model parameters previously optimized to capture the dynamics at all [O2] concentrations were released to fit the measured CAM-treated WT [NO·] curve at 10 μM O2 (Dataset S3). CAM treatment was simulated by setting the translation rate to zero at the time of NO· clearance (1.5 h postdose). For comparison, the same optimization was performed on the WT [NO·] curve at 10 μM O2 without CAM addition (SI Appendix, Fig. S8A). The model was able to capture the measured [NO·] dynamics for CAM-treated cultures far more accurately than those that retained translational activity during the oscillatory period, including much improved agreement in the period, amplitude, and rate of damping (SI Appendix, Fig. S8).
Oscillations in the NO· Response Network of Pseudomonas aeruginosa.
Given that the metabolic stress response oscillator discovered here genetically requires only the co-occurrence of NOD and CYT activities, we performed a bioinformatics analysis of the KEGG database (34, 35). We found that 777 different bacterial species harbor both NOD and CYT functions, and thereby have the capacity to exhibit oscillatory behavior analogous to that of E. coli. To provide experimental evidence that the oscillator can be found in diverse species, we chose to analyze the [NO·] dynamics of P. aeruginosa cultures under low O2 conditions. P. aeruginosa possesses an NOD [flavohemoprotein (Fhp)] and O2-consuming cytochromes, but the network differs considerably from that of E. coli because the Fhp sequence and structure are largely distinct from that of Hmp (<50% amino acid similarity) (42), P. aeruginosa is denitrifying (43, 44), and encodes five aerobic terminal oxidases (Cyo, CIO, Cbb3-1, Cbb3-2, and Aa3) that differ in function (Cbb3-1, Cbb3-2, and Aa3 are cytochrome c oxidases), kinetics, and regulation from the three in E. coli (45).
Initially, we investigated NO· detoxification dynamics of P. aeruginosa at 10 μM O2. Assay conditions were identical to those for E. coli, except cells were grown in basal salts media (BSM) with 15 mM succinate, a minimal media commonly used for P. aeruginosa (46, 47), rather than MOPS [3-(N-morpholino)propanesulfonic acid] media with 10 mM glucose, which was used for experiments with E. coli. Treatment with 50 μM DPTA NONOate yielded an [NO·] curve with a substantially higher peak (9.3 ± 0.5 μM) than was observed for E. coli (3.5 ± 0.1 μM) under the same O2 environment, which can be attributed to the lower pH in BSM compared with MOPS media (7.0 and 7.4, respectively) (SI Appendix, Fig. S9). Although no oscillations were observed for 10 μM O2 conditions, the faster rate of O2 consumption exhibited by P. aeruginosa relative to E. coli (SI Appendix, Fig. S10) suggested that increasing the O2 concentration may yield oscillatory behavior. The P. aeruginosa NO· treatment assay was therefore repeated at 50 μM O2, and NO· oscillations were observed (Fig. 4). Analogous to experiments with E. coli, the [O2] exhibited oscillations that were out of phase with those of [NO·] (Fig. 4). These data, which were collected from a bacterial species that is considerably distinct from E. coli, in media that supports a polar-opposite functioning of central metabolism than that used with E. coli (gluconeogenic compared with glycolytic), provides evidence that the stress response metabolic oscillations described here are broadly distributed.
Fig. 4.
[NO·] and [O2] oscillations in the P. aeruginosa NO· response network. P. aeruginosa in BSM plus 15 mM succinate media at an OD600 of 0.05 was treated with 50 μM DPTA NONOate in an environment of 50 μM O2, and the resulting concentrations of NO· and O2 were measured. The Inset shows a zoomed-in region of oscillatory behavior occurring shortly after the initial NO· clearance. Shaded gray regions are shown to visualize the relief of respiratory inhibition and the phase offset between [NO·] and [O2] oscillations, where NO· is depleted and then [O2] drops rapidly. The data are representative of three independent experiments, the results of which are presented in SI Appendix, Fig. S13 E and F.
Experimental Confirmation of Predicted [NO·] Dynamics Within the 0–5 μM O2 Regime.
Upon demonstration that model simulations were in quantitative agreement with measured [NO·] dynamics in environments with 0, 5, 10, 20, and 50 μM O2, and the oscillatory behavior could be captured, we sought to evaluate the predictive accuracy of the model. Using the ensemble of models that captured the data with ER < 10, NO· treatment simulations were conducted under [O2] ranging from 0 to 50 μM to update the original predictions (Fig. 1C) of tclear,NO·. The simulations exhibited tclear,NO· values that were in excellent agreement with measurements, with a drastic reduction in prediction uncertainty (Fig. 5A). The revised predictions estimated a maximum tclear,NO· occurring within the 0–5 μM O2 regime, peaking at ∼1 μM O2 with a ∼40-fold increase over the tclear,NO· at 0 or 50 μM O2. In addition, the model predicted that [NO·] oscillations would be present for environments with [O2] as low as 3 μM, but not conditions where O2 ≤ 2.5 μM. To experimentally assess predictions within this O2 regime of maximum NO· defense impairment, an NO· treatment assay was conducted at 2.5 μM O2 (±0.5 μM; instrument precision), in the center of the 0–5 μM regime. The measured [NO·] curve was in excellent quantitative agreement with predictions, with an NO· clearance time of 4.0 ± 0.2 h (predicted 3.9–4.6 h for 2.0–3.0 μM O2), including the loss of [NO·] oscillations at this O2 level (Fig. 5B).
Fig. 5.
Revised predictions of tclear,NO· and NO· consumption distribution as a function of [O2]. Using the updated model structure (with an [O2]-dependent translation rate) with parameters trained on WT [NO·] curves measured at 0, 5, 10, 20, and 50 μM O2 (optimization “stage 4”), 50 μM DPTA NONOate treatment was simulated for WT E. coli under [O2] ranging from 0 to 50 μM. (A) The resulting tclear,NO· was predicted (dashed black line) and compared with experimental measurements (open circles; mean of at least three independent experiments with error bars representing the SEM). (B) Comparison of predicted and measured [NO·] at 2.5 ± 0.5 μM O2. The dashed black line represents the simulation at 2.5 μM O2 using the best-fit parameter set, where the dark gray shading (too narrow to see) represents prediction uncertainty (range of viable parameter sets with ER < 10) at 2.5 μM O2, and light gray shading represents the range of simulation results for [O2] conditions spanning 2–3 μM O2, to account for the ±0.5 μM O2 experimental uncertainty in the hypoxic chamber O2 meter. The experimental data (solid red line) are mean of three independent experiments, with light red shading representing the SEM. (C) Predicted distribution of cumulative NO· detoxification (up to time tclear,NO·) for transfer to the gas phase, autoxidation, and combined Hmp- and NorV-mediated detoxification. (D) Individual contributions of Hmp and NorV to cumulative NO· detoxification. Predictions (dashed lines) were obtained using the best-fit parameter set, with shading (which is very small) representing prediction uncertainty (range of viable parameter sets with ER < 10).
Analysis of NO· Flux Distributions Under Microaerobic Conditions.
After confirming the predictive accuracy of the revised model, we explored the underlying distribution of NO· consumption as a function of [O2]. The cumulative distribution of NO· among each of the major pathways (up to tclear,NO·) was calculated using the ensemble of models with ER < 10 for [O2] spanning 0–50 μM (Fig. 5 C and D). The contribution of autoxidation remained relatively minor, accounting for a maximum of 8.7% of the total NO· consumption at ∼20 μM O2 (Fig. 5C). Escape of NO· to the gas phase peaked at 2.8 μM O2 to account for ∼60% of the NO· consumed, and roughly aligned with a minimum in the combined participation of Hmp and NorV, which accounted for ∼36% of NO· detoxification at that O2 concentration. NorV-mediated NO· detoxification fell sharply with increasing [O2] and was surpassed by the Hmp contribution at [O2] > 1.0 μM (Fig. 5D). Above 13 μM O2, Hmp-mediated NO· clearance exceeded that of gas phase transfer and became the dominant NO· consumption pathway, accounting for over 50% of NO· consumption for [O2] > 17 μM.
Discussion
The variability of in vivo O2 tension and its impact on bacterial pathogenesis is illustrated by pathogens whose virulence factors are regulated by O2 [e.g., Shigella flexneri increases expression of its type III secretion system with decreasing [O2] (48)], as well as studies demonstrating attenuated virulence for pathogens lacking machinery optimized for a specific O2 regime [e.g., enterohemorrhagic E. coli lacking aerobic, microaerobic, or anaerobic respiratory pathways exhibited impaired colonization of mouse intestines relative to the WT (49)] (50). As such, understanding how the major bacterial NO· detoxification systems function and interact within the microaerobic regime is critical.
Although NOD and NOR dynamics have been well characterized under aerobic and anaerobic conditions (9–12, 15, 16, 20–23), their behavior is less clear for [O2] lying between these extremes. Previous studies have provided some information on how [O2] impacts bacterial NO· stress (12, 16, 30, 31, 51–53). For example, an increase in the duration of NO·-mediated respiratory inhibition with decreasing [O2] was observed in E. coli (30, 31) and Mycobacterium bovis (51), but the cause was either left unexplored, or assumed to be the result of less available reactant (O2) to drive autoxidation or NOD-mediated NO· detoxification. More recently, [NO·] measurements in cultures of Campylobacter jejuni (52) demonstrated reduced NO· clearance at low aeration rates relative to more O2-rich conditions, and the effect was attributed to lower transcriptional induction of cgb, which encodes for a single-domain hemoglobin known to protect C. jejuni from NO· stress in the presence of O2 (54). Beyond these initial findings, previous studies did not quantify the distribution of NO· among its reaction pathways, leaving the contributions of NOD, NOR, autoxidation, gas phase transport, and other potentially important network components uncertain. To obtain a more quantitative understanding of bacterial NO· defenses under microaerobic conditions, we combined a computational approach with experimentation conducted in environments with precisely controlled O2 tensions.
Here, we demonstrated that E. coli NO· defenses are greatly impaired at low [O2], exhibiting a ∼30-fold increase in NO· clearance time compared with anaerobic and aerobic conditions for the same bolus of DPTA NONOate. A model-facilitated analysis of the distribution of NO· consumption within this regime suggested that a steep drop in NorV contribution with increasing [O2], combined with impaired translational and Hmp activities at low [O2], was producing a suboptimal overlap of these two detoxification systems, resulting in up to a ∼60% loss in their combined NO· detoxification activity (relative to their activity at 0 and 50 μM O2). In addition, we discovered that, at low [O2] conditions, the concentrations of NO· and O2 oscillated. In a previous study by Mukhopadhyay et al. (55), oscillations in norV mRNA transcript levels with a period of ∼0.75–1.5 h were observed in aerobic E. coli cultures treated with acidified nitrite (an NO·-releasing mixture). Other than finding that the oscillations were absent in anaerobic environments, the mRNA oscillations were not further investigated by the authors. The oscillations discovered here were metabolic (NO· and O2), and using a model-guided approach, we dissected these oscillations and found that NOD and CYT were the essential genetic determinants for oscillatory behavior. The relatively modest requirements for oscillations in this stress response network extend its significance beyond E. coli, as any bacterial species possessing an NOD and O2-consuming cytochrome quinol oxidases could potentially exhibit this behavior. Indeed, a bioinformatics analysis of the KEGG database (34, 35) identified 777 bacterial species (spanning 327 genera and 18 phyla) that possessed both NOD and O2-consuming cytochrome quinol oxidase activity (Dataset S4), and we selected one of these species, P. aeruginosa, and found that it exhibited analogous oscillations to those found in E. coli.
The present study provides quantitative insight into E. coli NO· detoxification under physiologically relevant O2 tensions that pathogens are likely to encounter within the host. Better understanding of bacterial NO· response networks and their vulnerabilities under physiologically relevant conditions will provide knowledge that could reveal strategies to disrupt these defenses, and thereby facilitate the development of a new class of antiinfectives that could help alleviate the rising threat of antibiotic resistance.
Materials and Methods
Bacterial Strains.
E. coli K-12 MG1655 and P. aeruginosa PAO1 were used in this study. The Δhmp::kanR and ΔnorV::kanR E. coli mutants were constructed via P1 transduction of the mutation from the corresponding strain in the Keio Collection (56). To construct the O2 respiration-null mutant ΔCYT (ΔcyoAΔappBΔcydB::kanR), the ΔcyoA::kanR mutation was first P1 transduced into the E. coli K-12 MG1655 background. The kanR resistance cassette, flanked by FLP recognition target (FRT) sites, was cured from the mutant by expressing FLP from the pCP20 plasmid (57), after which the ΔappB::kanR mutation was P1 transduced into the cured strain to yield the ΔcyoAΔappB::kanR double mutant. This process was repeated once more with the ΔcydB::kanR mutation to generate the ΔCYT triple mutant. Consumption of O2 in ΔCYT cultures was measured and confirmed to be negligible (SI Appendix, Fig. S11). All mutations were confirmed with colony PCR, using two sets of primers for verification (SI Appendix, Table S5).
Plasmids.
The Phmp-gfp and IPTG-inducible PT5-gfp plasmids were constructed in a previous study (36). Briefly, the Phmp-gfp plasmid possesses a SC101 origin of replication (low copy) and a kanR resistance marker obtained from pUA66 (58), and the Phmp promoter (250 nt immediately upstream of the hmp start codon) followed by the gfp coding sequence. The PT5-gfp plasmid also possesses the SC101 origin of replication and kanR marker, as well as lacIq and IPTG-inducible PT5 promoter transferred from pQE-80L (Qiagen), with gfp expressed from PT5.
Chemicals and Growth Media.
Growth media used in this study was LB Broth (BD Difco), MOPS minimal media (Teknova) with 10 mM glucose as the sole carbon source for E. coli (abbreviated here as “MOPS media”), and BSM minimal media with 15 mM succinate as the sole carbon source for P. aeruginosa. Delivery of NO· was achieved using the chemical donor DPTA NONOate (Cayman Chemical), which releases two NO· molecules per parent compound. Antibiotics used were kanamycin (30 μg/mL) and CAM (100 μg/mL) (Fisher Scientific).
NO· Treatment Assays.
A frozen −80 °C stock of E. coli was used to inoculate 1 mL of LB media in a 14-mL glass test tube, and was grown for 4 h at 37 °C and 250 rpm. A test tube containing 1 mL of fresh MOPS media was inoculated with 10 μL of the LB pregrowth culture, and was transferred to an incubated shaker within a hypoxic chamber (where all subsequent steps took place) to grow overnight (16 h) at 37 °C and 200 rpm. The overnight culture was used to inoculate 20 mL of fresh MOPS media (equilibrated with the chamber atmosphere overnight) in a 250-mL baffled shake flask to an initial OD600 of 0.01, and grown at 37 °C and 200 rpm. When the flask culture reached an OD600 of 0.2 (±0.02), an aliquot was quickly transferred to a bioreactor containing 10 mL of prewarmed (37 °C) MOPS media, from which an appropriate volume was discarded immediately before culture addition, such that the final volume and OD600 of the bioreactor culture was 10 mL and 0.05, respectively. Immediately after inoculation, the bioreactor culture was treated with 50 μM DPTA NONOate, and the concentration of NO· and/or O2 was monitored continuously (≥1 reads per s) for the duration of the assay.
The assay procedure was similar for P. aeruginosa, with a few notable differences. PAO1 cells from a −80 °C were used to inoculate 1 mL of LB media, which was grown overnight (16 h) in a hypoxic chamber at 37 °C and 200 rpm. In a 250-mL baffled shake flask, 20 mL of BSM plus 15 mM succinate was inoculated with 200 μL of the overnight LB culture, and was grown to midexponential phase (OD600 = 0.2) before harvesting cells for DPTA NONOate treatment (which was also conducted in BSM plus succinate media).
NO· and O2 Concentration Measurements.
[NO·] in the culture was measured using an ISO-NOP NO· sensor (World Precision Instruments) suspended ∼5 mm below the surface of the stirred bioreactor culture. The NO· probe was calibrated daily according to the manufacturer’s instructions before measurements. Briefly, the NO· probe tip was suspended in a stirred 10-mL solution of 0.1 M H2SO4 with 0.1 M KI in the bioreactor, and increasing volumes of an aqueous nitrite standard (Fisher) were added to spontaneously generate NO· in 1:1 molar ratio. The resulting signal was used to generate a calibration curve facilitating conversion between probe signal (in picoamperes) and [NO·] (in micromolar concentration).
Dissolved [O2] in the culture was measured using the fiber-optic FireStingO2 robust miniprobe from PyroScience, also suspended ∼5 mm below the surface of the stirred culture in the bioreactor. A temperature probe (PyroScience) was suspended in the water bath to automatically account for any temperature fluctuations. The O2 sensor was calibrated by allowing the signal to stabilize in cell-free media while the chamber was open to the atmosphere, to obtain the air-saturated reading. The air-saturated [O2] for cell-free media (MOPS with 10 mM glucose or BSM with 15 mM succinate) was measured to be identical to that of deionized water at the same conditions (37 °C), and was thus set to 210 μM O2 (59). The factory-set value was used for the zero-point (0% O2) reading, as it was confirmed to be accurate when the chamber environment was anaerobic.
Cell Viability Measurements.
CFUs were quantified before and during NO· stress at 5 and 10 μM O2 conditions to assess whether a significant fraction of the cells were dying. A 10-μL sample was removed from the bioreactor immediately before DPTA NONOate treatment, and every half hour or every hour after treatment for 10 or 5 μM O2 conditions, respectively, for a total of four samples. Each 10-μL aliquot was serially diluted in 1× PBS, delivered in 10-μL spots onto LB plus agar plates, and incubated overnight (16 h) aerobically at 37 °C. Individual colonies were counted the following day to determine the number of CFUs per milliliter at each time point.
Translation Rate Quantification.
The rate of translation was quantified by production of GFP via fluorescence measurements. Plasmids possessing gfp under control of the Phmp promoter or IPTG-inducible PT5 promoter were constructed in a previous study (36), and transformed into Δhmp E. coli. The Phmp-gfp transformants were grown identically as described for the NO· treatment assay, dosed with 50 μM DPTA NONOate at 50, 10, and 5 μM O2, and 200-μL samples were removed every 30 min for 2 h to quantify GFP fluorescence (485-nm excitation, 515-nm emission). The procedure was identical for the PT5-gfp transformants, except the culture was induced with 1 mM IPTG instead of treating with DPTA NONOate. Because cells were growing in the absence of DPTA NONOate treatment, samples were diluted with fresh MOPS media just before fluorescence measurements to maintain a constant OD600 of 0.05.
ΔCYT NO· Treatment Assay.
An in silico deletion of all cytochromes in the model was accomplished by fixing their concentrations to zero, and NO· treatment was simulated. The resulting [NO·] curve revealed a slightly faster NO· clearance in the cytochrome-deficient mutant compared with the WT, as well as an elimination of the [NO·] oscillations (SI Appendix, Fig. S12). To confirm the prediction experimentally, essential subunits of cytochrome bo, bd-I, and bd-II terminal oxidases (cyoA, cydB, and appB) were deleted to generate an O2 respiration-null mutant (ΔCYT). A ΔCYT culture was treated with 50 μM DPTA NONOate, and the resulting [NO·] curve revealed an elimination of the oscillations, as predicted by the model. However, the peak NO· concentration and clearance time were greater than that of the WT culture. A measurement of the culture pH at the time of dose revealed that the ΔCYT cells had acidified the media from a pH of 7.4 (exhibited by cell-free and WT cultures in the presence of O2) to a pH of ∼7.1, which increases the dissociation rate (NO· release rate) of DPTA NONOate. The experiment was thus repeated with WT E. coli, but the culture pH was adjusted to 7.1 with 2 M HCl immediately before DPTA NONOate treatment to replicate the media conditions of the ΔCYT cells. The resulting pH-adjusted WT [NO·] curve exhibited a peak height and clearance time more similar to that of ΔCYT culture, confirming reduced pH as the source of the influence. To reproduce this effect in the model, the DPTA NONOate release rate was increased until the simulated WT NO· clearance time matched that of the pH-adjusted WT measurements (∼2 h). Using this more accurate DPTA NONOate release rate, NO· treatment was simulated for the ΔCYT mutant for comparison with the pH-adjusted WT conditions.
Hypoxic Chamber.
All assays in this study were conducted within a vinyl hypoxic chamber with an anaerobic upgrade that was purchased from Coy Laboratory Products. The chamber environment for microaerobic experiments was maintained at the specified O2 concentration to within ±0.05% O2 (approximately ±0.5 μM dissolved O2 in the culture media), with ∼2,000 ppm (0.2%) CO2, and the balance N2. For anaerobic experiments, a palladium catalyst (Coy Laboratory Products) was placed in the chamber to eliminate trace O2 by reacting with H2 gas, which was maintained at a level of ∼2.0%, with 0.2% CO2 and the balance N2. The environmental O2 concentrations reported in the text refer to the concentration of dissolved O2 in cell-free media that was in equilibrium with the atmosphere of the hypoxic chamber (concentration after >24 h of exposure to atmosphere).
Model Construction and Simulation.
The E. coli NO· model was constructed (37) and further developed (36, 38) in previous studies. Briefly, the model is comprised of a system of ordinary differential equations (ODEs) describing the change in concentration of biochemical species (C) as a function of intensive reaction rates (rI) and reaction stoichiometries (Ŝ):
| [1] |
Reaction rates are a function of species concentrations and kinetic parameters, which were either derived from existing literature, or estimated from experimental measurements in this study. A complete list of all species, reactions, and kinetic parameters in the model is provided in Datasets S1 and S2, and SI Appendix, Table S1. Furthermore, an SBML version of this model was deposited in BioModels (60) and assigned the identifier MODEL1601140000. To facilitate experimental parameterization and validation of the model, the system was partitioned into intracellular and extracellular (growth media) compartments, assuming rapid diffusion of O2 and NO· across the cell membrane (61, 62) (SI Appendix, Supporting Methods). Simulations were run by numerically integrating the system of ODEs in MATLAB R2014b (The MathWorks) using the ode15s function.
Model simulations and analyses were conducted on a Dell OptiPlex 780 desktop workstation with an Intel Core 2 Quad CPU processor at 2.66 GHz. Longer computations, such as parameter optimizations and MCMC parametric analyses, were run on the Princeton Adroit compute cluster, an eight-node Beowulf cluster with twenty 2.50-GHz Intel Ivy Bridge processors and 64-GB RAM per node.
Incorporation of [O2] Term in Translation Rate.
The rate of protein production in the model was a function of the concentration of the associated mRNA transcripts:
| [2] |
where “Protein” is either Hmp, NorV, or NrfA, and “mRNA” is the encoding mRNA (mRNAhmp, mRNAnorV, mRNAnrfA, respectively), and the form of the equation assumes that ribosomes are not limiting. An [O2] dependency was introduced into the rate equation such that increased [O2] resulted in an increased translation rate, the effect saturated at high [O2] (dictated by the added KO2 term), and the rate of translation was nonzero when [O2] was zero:
| [3] |
Quantification of Relative Model Quality.
The error of a simulated [NO·] curve relative to the corresponding experiment ([NO·]sim and [NO·]meas, respectively) was quantified with the SSR between the two datasets:
| [4] |
The SSR was normalized (SSR′) by the experimental variance in [NO·] at each time point:
| [5] |
However, for analyses and optimizations involving oscillatory [NO·] dynamics, the SSR was not normalized, as small shifts in the phase of oscillations greatly increased the variance among experimental replicates within the oscillatory region of [NO·] curves.
The AIC (63) was used to quantitatively assess the relative quality of fit among models (differing in parameter values and/or reaction network structure) (40), which is a standard method that accounts for model–data agreement and parametric complexity to minimize underfitting or overfitting of the data (64). For a given model i, the AIC was calculated (64) using the following equation:
| [6] |
where n is the number of experimental data points, and k is the number of parameters fit to experimental data plus one, to account for the estimation of SSR (64). To account for the relatively small sample sizes that were at times encountered in this study [i.e., approaching less than ∼40 data points per varied parameter (64)], the corrected AIC (AICc) (65) was used (64):
| [7] |
References to “AIC” in the present study refer to this corrected form (AICc). The Akaike weight (or weight of evidence, w) of each model in a set of M models was calculated from the AICc values (64):
| [8] |
where Δi = AICc,i – min(AICc). Using the Akaike weight of each model, their corresponding ERs were determined (66):
| [9] |
where wbest is the weight of the best-fit (lowest AIC) model. The ER, which represents the likelihood of each model relative to the best-fit model (66), was used to quantitatively compare models during the MCMC parametric exploration process, where models with an ER ≥ 10 were considered nonviable (SI Appendix, Supporting Methods) (40).
Parameter Optimization.
Optimization of model parameters was accomplished using the MATLAB lsqcurvefit function, where specified parameters were allowed to vary within set bounds to minimize the SSR between simulated and experimentally measured data. Given the often high dimensionality and nonconvex nature of the optimizations conducted in the present study, each minimization process was repeated 2,000 times with random initial parameter values to improve coverage of the parameter space. In some cases, the viable parameter space was further explored by subsequently using an out-of-equilibrium adaptive Metropolis MCMC method (39) to identify additional parameter sets with similar or improved quality of fit compared with those identified from the initial optimization (SI Appendix, Supporting Methods). The relative quality of fit of the different parameter sets was evaluated using AIC, where optimized parameter sets were ranked according to their ER. The best parameter set (associated with the minimal SSR, and an ER = 1), as well as those exhibiting an ER less than 10 (i.e., at least 10% as likely as the optimal set), were retained as the viable parameter sets for that particular optimization.
Bioinformatic Analysis of KEGG Database.
To estimate the number and diversity of bacterial species possessing NOD and O2-consuming cytochrome oxidoreductase activity, the KEGG database (34, 35) was searched for all unique bacterial species known to possess a NOD (EC 1.14.12.17) and either cytochrome bo or bd quinol oxidase (EC 1.10.3.-). The search yielded 485 species, spanning 167 genera and nine phyla (Dataset S4). Because truncated hemoglobins (trHbNs) have been found to exhibit NOD activity [e.g., the GlbN (51) and GlbO (67) proteins of M. tuberculosis and Mycobacterium leprae, respectively], the search was expanded to include trHbNs, resulting in a total of 777 bacterial species likely to possess both NOD and O2-consuming cytochrome quinol oxidase activity (Dataset S4).
Reaction Deletion (Model Reduction) Analysis.
Random reactions were deleted one-by-one from the model, verifying after each deletion that the quality of fit was not compromised (quantified as maintaining an ER < 10), until no additional reactions could be removed. To avoid influence of the path taken (order in which reactions were deleted) on the final set of reactions obtained, the process was repeated 100 times, and the model with the least number of reactions was selected. Two different models were identified as the smallest (45 reactions, or 23% of the original 196 reactions), differing in only one reaction (one model possessed the NADH-mediated reduction of Hmp, whereas the other contained the NADPH-mediated variant) (SI Appendix, Table S4). The remaining reactions were then deleted individually, manually inspecting the resulting [NO·] curve to determine which reactions eliminated oscillations upon removal. Reactions found to eliminate oscillations upon removal were NO· release from DPTA NONOate, exchange of NO· and O2 with the gas phase, and reactions associated with Hmp, NorV, and CYT. However, simultaneous removal of all remaining NorV-associated reactions had no effect on the [NO·] oscillations, further distilling the set of cellular reactions to include only Hmp and CYT (SI Appendix, Table S4).
Supplementary Material
Acknowledgments
We thank Profs. Vassily Hatzimanikatis, Stanislav Y. Shvartsman, and Ned S. Wingreen for helpful discussions, and acknowledge the National BioResource Project (NIG) for distribution of the Keio collection. We also thank Kristin Adolfsen and Katherine Volzing for assistance with the hypoxic chamber. Some portions of the computational work reported in this study were performed using the Terascale Infrastructure for Groundbreaking Research in Science and Engineering high-performance computer center at Princeton University, which is jointly supported by the Princeton Institute for Computational Science and Engineering and the Princeton University Office of Information Technology’s Research Computing department. This work was supported by National Science Foundation Grants CBET-1453325 and DGE-1148900, and by Princeton University (Forese Family Fund for Innovation, start-up funds).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.S. is a guest editor invited by the Editorial Board.
Data deposition: A Systems Biology Markup Language (SBML) version of the model described in this work was deposited in the BioModels Database (https://www.ebi.ac.uk/biomodels/) with the identifier: MODEL1601140000.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521354113/-/DCSupplemental.
References
- 1.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci USA. 2000;97(16):8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogdan C. Nitric oxide synthase in innate and adaptive immunity: An update. Trends Immunol. 2015;36(3):161–178. doi: 10.1016/j.it.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Fang FC. Antimicrobial reactive oxygen and nitrogen species: Concepts and controversies. Nat Rev Microbiol. 2004;2(10):820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 4.Bowman LAH, McLean S, Poole RK, Fukuto JM. The diversity of microbial responses to nitric oxide and agents of nitrosative stress close cousins but not identical twins. Adv Microb Physiol. 2011;59:135–219. doi: 10.1016/B978-0-12-387661-4.00006-9. [DOI] [PubMed] [Google Scholar]
- 5.Toledo JC, Jr, Augusto O. Connecting the chemical and biological properties of nitric oxide. Chem Res Toxicol. 2012;25(5):975–989. doi: 10.1021/tx300042g. [DOI] [PubMed] [Google Scholar]
- 6.Stern AM, Zhu J. An introduction to nitric oxide sensing and response in bacteria. Adv Appl Microbiol. 2014;87:187–220. doi: 10.1016/B978-0-12-800261-2.00005-0. [DOI] [PubMed] [Google Scholar]
- 7.Poole RK. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem Soc Trans. 2005;33(Pt 1):176–180. doi: 10.1042/BST0330176. [DOI] [PubMed] [Google Scholar]
- 8.Robinson JL, Adolfsen KJ, Brynildsen MP. Deciphering nitric oxide stress in bacteria with quantitative modeling. Curr Opin Microbiol. 2014;19:16–24. doi: 10.1016/j.mib.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner PR, Gardner AM, Martin LA, Salzman AL. Nitric oxide dioxygenase: An enzymic function for flavohemoglobin. Proc Natl Acad Sci USA. 1998;95(18):10378–10383. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hausladen A, Gow AJ, Stamler JS. Nitrosative stress: Metabolic pathway involving the flavohemoglobin. Proc Natl Acad Sci USA. 1998;95(24):14100–14105. doi: 10.1073/pnas.95.24.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomes CM, et al. A novel type of nitric-oxide reductase. Escherichia coli flavorubredoxin. J Biol Chem. 2002;277(28):25273–25276. doi: 10.1074/jbc.M203886200. [DOI] [PubMed] [Google Scholar]
- 12.Gardner AM, Helmick RA, Gardner PR. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J Biol Chem. 2002;277(10):8172–8177. doi: 10.1074/jbc.M110471200. [DOI] [PubMed] [Google Scholar]
- 13.Gardner PR. Hemoglobin: A nitric-oxide dioxygenase. Scientifica (Cairo) 2012;2012:683729. doi: 10.6064/2012/683729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner PR. Nitric oxide dioxygenase function and mechanism of flavohemoglobin, hemoglobin, myoglobin and their associated reductases. J Inorg Biochem. 2005;99(1):247–266. doi: 10.1016/j.jinorgbio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Gardner AM, Gardner PR. Flavohemoglobin detoxifies nitric oxide in aerobic, but not anaerobic, Escherichia coli. Evidence for a novel inducible anaerobic nitric oxide-scavenging activity. J Biol Chem. 2002;277(10):8166–8171. doi: 10.1074/jbc.M110470200. [DOI] [PubMed] [Google Scholar]
- 16.Hausladen A, Gow A, Stamler JS. Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc Natl Acad Sci USA. 2001;98(18):10108–10112. doi: 10.1073/pnas.181199698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SO, Orii Y, Lloyd D, Hughes MN, Poole RK. Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): Reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 1999;445(2-3):389–394. doi: 10.1016/s0014-5793(99)00157-x. [DOI] [PubMed] [Google Scholar]
- 18.Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61(4):533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heiss B, Frunzke K, Zumft WG. Formation of the N-N bond from nitric oxide by a membrane-bound cytochrome bc complex of nitrate-respiring (denitrifying) Pseudomonas stutzeri. J Bacteriol. 1989;171(6):3288–3297. doi: 10.1128/jb.171.6.3288-3297.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vine CE, Cole JA. Nitrosative stress in Escherichia coli: Reduction of nitric oxide. Biochem Soc Trans. 2011;39(1):213–215. doi: 10.1042/BST0390213. [DOI] [PubMed] [Google Scholar]
- 21.Crawford MJ, Goldberg DE. Role for the Salmonella flavohemoglobin in protection from nitric oxide. J Biol Chem. 1998;273(20):12543–12547. doi: 10.1074/jbc.273.20.12543. [DOI] [PubMed] [Google Scholar]
- 22.Field SJ, Thorndycroft FH, Matorin AD, Richardson DJ, Watmough NJ. The respiratory nitric oxide reductase (NorBC) from Paracoccus denitrificans. Methods Enzymol. 2008;437:79–101. doi: 10.1016/S0076-6879(07)37005-5. [DOI] [PubMed] [Google Scholar]
- 23.Vicente JB, et al. Kinetic characterization of the Escherichia coli nitric oxide reductase flavorubredoxin. Methods Enzymol. 2008;437:47–62. doi: 10.1016/S0076-6879(07)37003-1. [DOI] [PubMed] [Google Scholar]
- 24.He G, et al. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc Natl Acad Sci USA. 1999;96(8):4586–4591. doi: 10.1073/pnas.96.8.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 26.Park MK, Myers RA, Marzella L. Oxygen tensions and infections: Modulation of microbial growth, activity of antimicrobial agents, and immunologic responses. Clin Infect Dis. 1992;14(3):720–740. doi: 10.1093/clinids/14.3.720. [DOI] [PubMed] [Google Scholar]
- 27.Sitkovsky M, Lukashev D. Regulation of immune cells by local-tissue oxygen tension: HIF1 alpha and adenosine receptors. Nat Rev Immunol. 2005;5(9):712–721. doi: 10.1038/nri1685. [DOI] [PubMed] [Google Scholar]
- 28.Atkuri KR, Herzenberg LA, Niemi AK, Cowan T, Herzenberg LA. Importance of culturing primary lymphocytes at physiological oxygen levels. Proc Natl Acad Sci USA. 2007;104(11):4547–4552. doi: 10.1073/pnas.0611732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caldwell CC, et al. Differential effects of physiologically relevant hypoxic conditions on T lymphocyte development and effector functions. J Immunol. 2001;167(11):6140–6149. doi: 10.4049/jimmunol.167.11.6140. [DOI] [PubMed] [Google Scholar]
- 30.Yu H, et al. Oxygen-dependent regulation of the respiration and growth of Escherichia coli by nitric oxide. FEBS Lett. 1997;409(2):161–165. doi: 10.1016/s0014-5793(97)00494-8. [DOI] [PubMed] [Google Scholar]
- 31.Stevanin TM, et al. Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo′ or bd, from nitric oxide. J Biol Chem. 2000;275(46):35868–35875. doi: 10.1074/jbc.M002471200. [DOI] [PubMed] [Google Scholar]
- 32.Gardner AM, Martin LA, Gardner PR, Dou Y, Olson JS. Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 tyrosine hydroxyl is essential for dioxygen binding and catalysis. J Biol Chem. 2000;275(17):12581–12589. doi: 10.1074/jbc.275.17.12581. [DOI] [PubMed] [Google Scholar]
- 33.Mills CE, Sedelnikova S, Søballe B, Hughes MN, Poole RK. Escherichia coli flavohaemoglobin (Hmp) with equistoichiometric FAD and haem contents has a low affinity for dioxygen in the absence or presence of nitric oxide. Biochem J. 2001;353(Pt 2):207–213. doi: 10.1042/0264-6021:3530207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M, et al. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014;42(Database issue):D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson JL, Brynildsen MP. An ensemble-guided approach identifies ClpP as a major regulator of transcript levels in nitric oxide-stressed Escherichia coli. Metab Eng. 2015;31:22–34. doi: 10.1016/j.ymben.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Robinson JL, Brynildsen MP. A kinetic platform to determine the fate of nitric oxide in Escherichia coli. PLoS Comput Biol. 2013;9(5):e1003049. doi: 10.1371/journal.pcbi.1003049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson JL, Miller RV, Brynildsen MP. Model-driven identification of dosing regimens that maximize the antimicrobial activity of nitric oxide. Metab Eng Commun. 2014;1:12–18. doi: 10.1016/j.meteno.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zamora-Sillero E, Hafner M, Ibig A, Stelling J, Wagner A. Efficient characterization of high-dimensional parameter spaces for systems biology. BMC Syst Biol. 2011;5:142. doi: 10.1186/1752-0509-5-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adolfsen KJ, Brynildsen MP. A kinetic platform to determine the fate of hydrogen peroxide in Escherichia coli. PLoS Comput Biol. 2015;11(11):e1004562. doi: 10.1371/journal.pcbi.1004562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neidhardt FC, Magasanik B. Studies on the role of ribonucleic acid in the growth of bacteria. Biochim Biophys Acta. 1960;42(1):99–116. doi: 10.1016/0006-3002(60)90757-5. [DOI] [PubMed] [Google Scholar]
- 42.Frey AD, Kallio PT. Bacterial hemoglobins and flavohemoglobins: Versatile proteins and their impact on microbiology and biotechnology. FEMS Microbiol Rev. 2003;27(4):525–545. doi: 10.1016/S0168-6445(03)00056-1. [DOI] [PubMed] [Google Scholar]
- 43.Kalkowski I, Conrad R. Metabolism of nitric oxide in denitrifying Pseudomonas aeruginosa and nitrate-respiring Bacillus cereus. FEMS Microbiol Lett. 1991;66(1):107–111. doi: 10.1016/0378-1097(91)90429-e. [DOI] [PubMed] [Google Scholar]
- 44.Williams DR, Rowe JJ, Romero P, Eagon RG. Denitrifying Pseudomonas aeruginosa: Some parameters of growth and active transport. Appl Environ Microbiol. 1978;36(2):257–263. doi: 10.1128/aem.36.2.257-263.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawakami T, Kuroki M, Ishii M, Igarashi Y, Arai H. Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ Microbiol. 2010;12(6):1399–1412. doi: 10.1111/j.1462-2920.2009.02109.x. [DOI] [PubMed] [Google Scholar]
- 46.Eagon RG, Phibbs PV., Jr Kinetics of transport of glucose, fructose, and mannitol by Pseudomonas aeruginosa. Can J Biochem. 1971;49(9):1031–1041. doi: 10.1139/o71-151. [DOI] [PubMed] [Google Scholar]
- 47.Schook LB, Berk RS. Nutritional studies with Pseudomonas aeruginosa grown on inorganic sulfur sources. J Bacteriol. 1978;133(3):1378–1382. doi: 10.1128/jb.133.3.1378-1382.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marteyn B, et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature. 2010;465(7296):355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones SA, et al. Respiration of Escherichia coli in the mouse intestine. Infect Immun. 2007;75(10):4891–4899. doi: 10.1128/IAI.00484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marteyn B, Scorza FB, Sansonetti PJ, Tang C. Breathing life into pathogens: The influence of oxygen on bacterial virulence and host responses in the gastrointestinal tract. Cell Microbiol. 2011;13(2):171–176. doi: 10.1111/j.1462-5822.2010.01549.x. [DOI] [PubMed] [Google Scholar]
- 51.Ouellet H, et al. Truncated hemoglobin HbN protects Mycobacterium bovis from nitric oxide. Proc Natl Acad Sci USA. 2002;99(9):5902–5907. doi: 10.1073/pnas.092017799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monk CE, Pearson BM, Mulholland F, Smith HK, Poole RK. Oxygen- and NssR-dependent globin expression and enhanced iron acquisition in the response of campylobacter to nitrosative stress. J Biol Chem. 2008;283(42):28413–28425. doi: 10.1074/jbc.M801016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coppola D, et al. Antarctic bacterial haemoglobin and its role in the protection against nitrogen reactive species. Biochim Biophys Acta. 2013;1834(9):1923–1931. doi: 10.1016/j.bbapap.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 54.Elvers KT, Wu G, Gilberthorpe NJ, Poole RK, Park SF. Role of an inducible single-domain hemoglobin in mediating resistance to nitric oxide and nitrosative stress in Campylobacter jejuni and Campylobacter coli. J Bacteriol. 2004;186(16):5332–5341. doi: 10.1128/JB.186.16.5332-5341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukhopadhyay P, Zheng M, Bedzyk LA, LaRossa RA, Storz G. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc Natl Acad Sci USA. 2004;101(3):745–750. doi: 10.1073/pnas.0307741100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaslaver A, et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods. 2006;3(8):623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 59.Montgomery HAC, Thom NS, Cockburn A. Determination of dissolved oxygen by the winkler method and the solubility of oxygen in pure water and sea water. J Appl Chem. 1964;14(7):280–296. [Google Scholar]
- 60.Chelliah V, et al. BioModels: Ten-year anniversary. Nucleic Acids Res. 2015;43(Database issue) D1:D542–D548. doi: 10.1093/nar/gku1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelm M. Nitric oxide metabolism and breakdown. Biochim Biophys Acta. 1999;1411(2-3):273–289. doi: 10.1016/s0005-2728(99)00020-1. [DOI] [PubMed] [Google Scholar]
- 62.Denicola A, Souza JM, Radi R, Lissi E. Nitric oxide diffusion in membranes determined by fluorescence quenching. Arch Biochem Biophys. 1996;328(1):208–212. doi: 10.1006/abbi.1996.0162. [DOI] [PubMed] [Google Scholar]
- 63.Akaike H. Information theory and extension of the maximum likelihood principle. In: Petrov BN, Csaki F, editors. Second International Symposium on Information Theory. Akadémiai Kiadó; Budapest: 1973. pp. 267–281. [Google Scholar]
- 64.Turkheimer FE, Hinz R, Cunningham VJ. On the undecidability among kinetic models: From model selection to model averaging. J Cereb Blood Flow Metab. 2003;23(4):490–498. doi: 10.1097/01.WCB.0000050065.57184.BB. [DOI] [PubMed] [Google Scholar]
- 65.Hurvich CM, Tsai CL. Regression and time-series model selection in small samples. Biometrika. 1989;76(2):297–307. [Google Scholar]
- 66.Burnham KP, Anderson DR. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. 2nd Ed Springer; New York: 2002. [Google Scholar]
- 67.Fabozzi G, Ascenzi P, Renzi SD, Visca P. Truncated hemoglobin GlbO from Mycobacterium leprae alleviates nitric oxide toxicity. Microb Pathog. 2006;40(5):211–220. doi: 10.1016/j.micpath.2006.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.