Significance
Current tissue manufacturing methods fail to recapitulate the geometry, complexity, and longevity of human tissues. We report a multimaterial 3D bioprinting method that enables the creation of thick human tissues (>1 cm) replete with an engineered extracellular matrix, embedded vasculature, and multiple cell types. These 3D vascularized tissues can be actively perfused with growth factors for long durations (>6 wk) to promote differentiation of human mesenchymal stem cells toward an osteogenic lineage in situ. The ability to construct and perfuse 3D tissues that integrate parenchyma, stroma, and endothelium is a foundational step toward creating human tissues for ex vivo and in vivo applications.
Keywords: bioprinting, stem cells, vasculature, tissues, biomaterials
Abstract
The advancement of tissue and, ultimately, organ engineering requires the ability to pattern human tissues composed of cells, extracellular matrix, and vasculature with controlled microenvironments that can be sustained over prolonged time periods. To date, bioprinting methods have yielded thin tissues that only survive for short durations. To improve their physiological relevance, we report a method for bioprinting 3D cell-laden, vascularized tissues that exceed 1 cm in thickness and can be perfused on chip for long time periods (>6 wk). Specifically, we integrate parenchyma, stroma, and endothelium into a single thick tissue by coprinting multiple inks composed of human mesenchymal stem cells (hMSCs) and human neonatal dermal fibroblasts (hNDFs) within a customized extracellular matrix alongside embedded vasculature, which is subsequently lined with human umbilical vein endothelial cells (HUVECs). These thick vascularized tissues are actively perfused with growth factors to differentiate hMSCs toward an osteogenic lineage in situ. This longitudinal study of emergent biological phenomena in complex microenvironments represents a foundational step in human tissue generation.
The ability to manufacture human tissues that replicate the essential spatial (1), mechanochemical (2, 3), and temporal aspects of biological tissues (4) would enable myriad applications, including 3D cell culture (5), drug screening (6, 7), disease modeling (8), and tissue repair and regeneration (9, 10). Three-dimensional bioprinting is an emerging approach for creating complex tissue architectures (10, 11), including those with embedded vasculature (12–15), that may address the unmet needs of tissue manufacturing. Recently, Miller et al. (15) reported an elegant method for creating vascularized tissues, in which a sacrificial carbohydrate glass is printed at elevated temperature (>100 °C), protectively coated, and then removed, before introducing a homogeneous cell-laden matrix. Kolesky et al. (14) developed an alternate approach, in which multiple cell-laden, fugitive (vasculature), and extracellular matrix (ECM) inks are coprinted under ambient conditions. However, in both cases, the inability to directly perfuse these vascularized tissues limited their thickness (1–2 mm) and culture times (<14 d). Here, we report a route for creating thick vascularized tissues (≥1 cm) within 3D perfusion chips that provides unprecedented control over tissue composition, architecture, and microenvironment over several weeks (>6 wk). This longitudinal study of emergent biological phenomena in complex microenvironments represents a foundational step in human tissue generation.
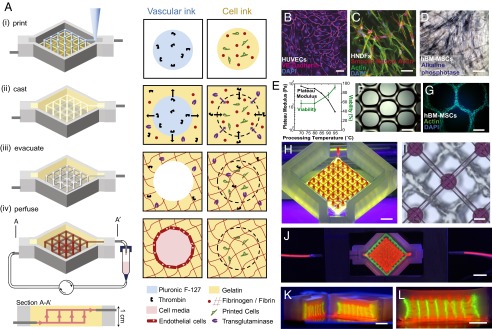
Central to the fabrication of thick vascularized tissues is the design of biological, fugitive, and elastomeric inks for multimaterial 3D bioprinting. To satisfy the concomitant requirements of processability, heterogeneous integration, biocompatibility, and long-term stability, we first developed printable cell-laden inks and castable ECM based on a gelatin and fibrinogen blend (16). Specifically, these materials form a gelatin–fibrin matrix cross-linked by a dual-enzymatic, thrombin and transglutaminase (TG), strategy (Fig. 1 and SI Appendix, Fig. S1). The cell-laden inks must facilitate printing of self-supporting filamentary features under ambient conditions as well as subsequent infilling of the printed tissue architectures by casting without dissolving or distorting the patterned construct (Fig. 1A). The thermally reversible gelation of the gelatin–fibrinogen network enables its use in both printing and casting, where gel and fluid states are required, respectively (SI Appendix, Fig. S2). Thrombin is used to rapidly polymerize fibrinogen (17), whereas TG is a slow-acting Ca2+-dependent enzymatic cross-linker that imparts the mechanical and thermal stability (18) needed for long-term perfusion. Notably, the cell-laden ink does not contain either enzyme to prevent polymerization during printing. However, the castable matrix contains both thrombin and TG, which diffuse into adjacent printed filaments, forming a continuous, interpenetrating polymer network, in which the native fibrillar structure of fibrin is preserved (SI Appendix, Fig. S3). Importantly, our approach allows arbitrarily thick tissues to be fabricated, because the matrix does not require UV curing (19), which has a low penetration depth in tissue (20) and can be readily expanded to other biomaterials, including fibrin and hyaluronic acid (SI Appendix, Fig. S4).
Fig. 1.
Three-dimensional vascularized tissue fabrication. (A) Schematic illustration of the tissue manufacturing process. (i) Fugitive (vascular) ink, which contains pluronic and thrombin, and cell-laden inks, which contain gelatin, fibrinogen, and cells, are printed within a 3D perfusion chip. (ii) ECM material, which contains gelatin, fibrinogen, cells, thrombin, and TG, is then cast over the printed inks. After casting, thrombin induces fibrinogen cleavage and rapid polymerization into fibrin in both the cast matrix, and through diffusion, in the printed cell ink. Similarly, TG diffuses from the molten casting matrix and slowly cross-links the gelatin and fibrin. (iii) Upon cooling, the fugitive ink liquefies and is evacuated, leaving behind a pervasive vascular network, which is (iv) endothelialized and perfused via an external pump. (B) HUVECs growing on top of the matrix in 2D, (C) HNDFs growing inside the matrix in 3D, and (D) hMSCs growing on top of the matrix in 2D. (Scale bar: 50 µm.) (E and F) Images of printed hMSC-laden ink prepared using gelatin preprocessed at 95 °C before ink formation (E) as printed and (F) after 3 d in the 3D printed filament where actin (green) and nuclei (blue) are stained. (G) Gelatin preprocessing temperature affects the plateau modulus and cell viability after printing. Higher temperatures lead to lower modulus and higher HNDF viability postprinting. (H) Photographs of interpenetrated sacrificial (red) and cell inks (green) as printed on chip. (Scale bar: 2 mm.) (I) Top-down bright-field image of sacrificial and cell inks. (Scale bar: 50 µm.). (J–L) Photograph of a printed tissue construct housed within a perfusion chamber (J) and corresponding cross-sections (K and L). (Scale bars: 5 mm.)
The gelatin–fibrin matrix supports multiple cell types of interest to both 2D and 3D culture conditions, including human umbilical vein endothelial cells (HUVECs), human neonatal dermal fibroblasts (HNDFs), and human bone marrow-derived mesenchymal stem cells (hMSCs) (Fig. 1 B–D and SI Appendix, Fig. S5). We find that endothelial cells express vascular endothelial-cadherin (VE-Cad) (Fig. 1B), and HNDFs (Fig. 1C) and hMSCs (Fig. 1D) proliferate and spread on this matrix surface and in bulk. Moreover, the printed cell viability can be as high as 95%, depending on how gelatin is processed before ink formulation. At higher processing temperatures, the average molecular weight of gelatin is reduced from 69 kDa at 70 °C to 32 kDa at 95 °C processing, resulting in softer gels with lower viscosity, shear yield stress, and shear elastic modulus. These cell-laden inks can be printed with ease and accommodate cell densities ranging from 0.1 million per mL to 10 million cells per mL (Fig. 1E and SI Appendix, Fig. S6). Upon printing, hMSCs within this soft gelatin–fibrinogen matrix continue to spread, proliferate, and contract into dense, cellular architectures that align along the printing direction (Fig. 1F), likely arising due to cellular confinement (21) and contraction via the Poisson effect (22).
To construct thick, vascularized tissues within 3D perfusion chips, we coprinted cell-laden, fugitive, and silicone inks (Fig. 1 H and I). First, the silicone ink is printed on a glass substrate and cured to create customized perfusion chips (Movie S1 and SI Appendix, Fig. S1). Next, the cell-laden and fugitive inks are printed on chip, and then encapsulated with the castable ECM (Fig. 1 J–L and Movie S2). The fugitive ink, which defines the embedded vascular network, is composed of a triblock copolymer [i.e., polyethylene oxide (PEO)–polypropylene oxide (PPO)–PEO]. This ink can be removed from the fabricated tissue upon cooling to roughly 4 °C, where it undergoes a gel-to-fluid transition (14, 23). This process yields a pervasive network of interconnected channels, which are then lined with HUVECs. The resulting vascularized tissues are perfused via their embedded vasculature on chip over long time periods using an external pump (Movie S3) that generates smooth flow over a wide range of flow rates (24).
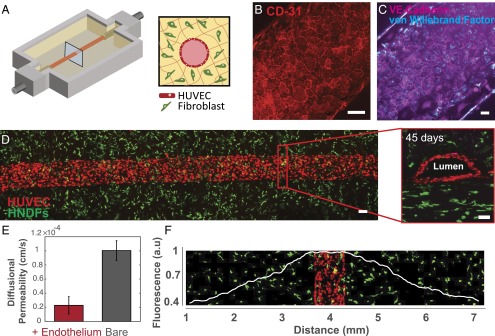
To demonstrate the formation of stable vasculature, we printed a simple tissue construct composed of two parallel channels embedded within a fibroblast cell-laden matrix (Fig. 2). The channels are lined with HUVECs, perfused with 1:1 ratio of endothelial growth media (EGM-2 Bullet kit) and HNDF growth media [DMEM plus 10% (vol/vol) FBS], and subsequently form a confluent monolayer that lines each blood vessel (Fig. 2A). The medium is preincubated for 5 h in the incubator at 37 °C and 5% CO2 and replaced every other day. Importantly, after 6 wk of active perfusion, these endothelial cells maintain endothelial phenotype and remain confluent, characterized by expression of CD31, von Willebrand factor (vWF), and VE-Cad (Fig. 2 B and C). The cross-sectional view of a representative vessel reveals lumen formation (Fig. 2D and Movie S4). Confirming the barrier function of the endothelium, we measured a fivefold reduction in the diffusional permeability compared with unlined (bare) channels (Fig. 2E and SI Appendix, Fig. S7). Stromal HNDFs residing within the surrounding matrix exhibit cell spreading and proliferative phenotypes localized to regions within ∼1 mm of the vasculature (Fig. 2F and SI Appendix, Fig. S8); cells further away from these regions become quiescent likely due to an insufficient nutrient supply. As cell density increases, their viability rapidly decreases at distances beyond 1 mm from the embedded blood vessels (e.g., only 5% of the cells remain viable at 7 mm). Clearly, the perfusable vasculature is critical to support living tissues thicker than 1 mm over long time periods.
Fig. 2.
Three-dimensional vascularized tissues remain stable during long-term perfusion. (A) Schematic depicting a single HUVEC-lined vascular channel supporting a fibroblast cell-laden matrix and housed within a 3D perfusion chip. (B and C) Confocal microscopy image of the vascular network after 42 d, CD-31 (red), vWF (blue), and VE-Cadherin (magenta). (Scale bars: 100 µm.) (D) Long-term perfusion of HUVEC-lined (red) vascular network supporting HNDF-laden (green) matrix shown by top-down (Left) and cross-sectional confocal microscopy at 45 d (Right). (Scale bar: 100 µm.) (E) Quantification of barrier properties imparted by endothelial lining of channels, demonstrated by reduced diffusional permeability of FITC-dextran. (F) GFP-HNDF distribution within the 3D matrix shown by fluorescent intensity as a function of distance from vasculature.
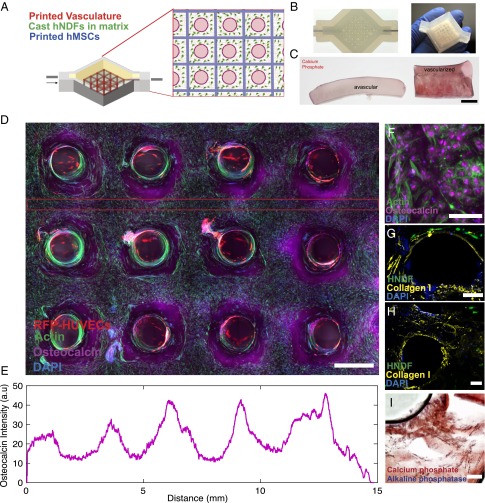
To explore emergent phenomena in complex microenvironments, we created a heterogeneous tissue architecture (>1 cm thick and 10 cm3 in volume) by printing a hMSC-laden ink into a 3D lattice geometry along with intervening in- and out-of-plane (vertical) features composed of fugitive ink, which ultimately transform into a branched vascular network lined with HUVECs. After printing, the remaining interstitial space is infilled with an HNDF-laden ECM (Fig. 3A) to form a connective tissue that both supports and binds to the printed stem cell-laden and vascular features. In this example, fibroblasts serve as model cells that surround the heterogeneously patterned stem cells and vascular network. These model cells could be replaced with either support cells (e.g., immune cells or pericytes) or tissue-specific cells (e.g., hepatocytes, neurons, or islets) in future embodiments. The embedded vascular network is designed with a single inlet and outlet that provides an interface between the printed tissue and the perfusion chip. This network is symmetrically branched to ensure uniform perfusion throughout the tissue, including deep within its core. In addition to providing transport of nutrients, oxygen, and waste materials, the perfused vasculature is used to deliver specific differentiation factors to the tissue in a more uniform manner than bulk delivery methods, in which cells at the core of the tissue are starved of factors (25). This versatile platform (Fig. 3A) is used to precisely control growth and differentiation of the printed hMSCs. Moreover, both the printed cellular architecture and embedded vascular network are visible macroscopically with this thick tissue (Fig. 3B).
Fig. 3.
Osteogenic differentiation of thick vascularized tissue. (A) Schematic depicting the geometry of the printed heterogeneous tissue within the customized perfusion chip, wherein the branched vascular architecture pervades hMSCs that are printed into a 3D lattice architecture, and HNDFs are cast within an ECM that fills the interstitial space. (B) Photographs of a printed tissue construct within and removed from the customized perfusion chip. (C) Comparative cross-sections of avascular tissue (Left) and vascularized tissue (Right) after 30 d of osteogenic media perfusion with alizarin red stain showing location of calcium phosphate. (Scale bar: 5 mm.) (D) Confocal microscopy image through a cross-section of 1-cm-thick vascularized osteogenic tissue construct after 30 d of active perfusion and in situ differentiation. (Scale bar: 1.5 mm.) (E) Osteocalcin intensity across the thick tissue sample inside the red lines shown in C. (F) High-resolution image showing osteocalcin (purple) localized within hMSCs, and they appear to take on symmetric osteoblast-like morphologies. (Scale bar: 100 µm.) After 30 d (G and H), thick tissue constructs are stained for collagen-I (yellow), which appears to be localized near hMSCs. (Scale bars: 200 µm.) (I) Alizarin red is used to stain calcium phosphate deposition, and fast blue is used to stain AP, indicating tissue maturation and differentiation over time. (Scale bar: 200 µm.)
To develop a dense osteogenic tissue, we transvascularly delivered growth media to the tissue during an initial proliferation phase (6 d) followed by an osteogenic differentiation mixture that is perfused for several weeks. Our optimized mixture is composed of BMP-2, ascorbic acid, and glycerophosphate, to promote mineral deposition and alkaline phosphatase (AP) expression (SI Appendix, Fig. S9). To assess tissue maturation, changes in cell function and matrix composition are observed over time. In good agreement with prior studies (21), we find that AP expression in hMSCs occurs within 3 d, whereas mineral deposition does not become noticeable until 14 d, which coincides with visible collagen-1 deposition by hMSCs (SI Appendix, Fig. S9) (21). Fig. 3C shows an avascular tissue produced with comparable hMSC density, in which positive alizarin stains are only observed within a few hundred microns of the tissue surface. By contrast, the thick vascularized tissue stains positive in hMSC regions deep within its core after 30 d of osteogenic differentiation by perfusion. We characterized the mineral deposits, which consist of particulates ∼20–200 nm in size, using SEM/energy-dispersive X-ray spectroscopy (EDS) analysis. Calcium and phosphorous peaks are only observed for vascularized tissues, not the avascular control (SI Appendix, Fig. S9 E and F). The phenotype of hMSCs varies across the printed filamentary features: cells are close-packed, compacted, and exhibit a high degree of mineralization within the filament core, whereas those in the periphery are more elongated and exhibit less mineralization. We observe that subpopulations of HNDFs and hMSCs migrate from their initial patterned geometry toward the vascular channels and wrap circumferentially around each channel (Fig. 3D). After 30 d, the printed hMSCs express osteocalcin within the tissue, and osteocalcin expression is proportional to distance from the nearest vessel (Fig. 3E). Furthermore, we find that collagen deposition is localized within printed filaments and around the circumference of the vasculature (Fig. 3 F–H and SI Appendix, Fig. S9).
In summary, thick, vascularized human tissues with programmable cellular heterogeneity that are capable of long-term (>6-wk) perfusion on chip have been fabricated by multimaterial 3D bioprinting. The ability to recapitulate physiologically relevant, 3D tissue microenvironments enables the exploration of emergent biological phenomena, as demonstrated by observations of in situ development of hMSCs within tissues containing a pervasive, perfusable, endothelialized vascular network. Our 3D tissue manufacturing platform opens new avenues for fabricating and investigating human tissues for both ex vivo and in vivo applications.
Methods
Solution Preparation.
Ink and matrix precursor solutions are prepared before printing the tissue engineered constructs. A 15 wt/vol% gelatin solution (type A; 300 bloom from porcine skin; Sigma) is produced by warming in DPBS (1× Dulbecco’s PBS without calcium and magnesium) to 70 °C (unless otherwise noted) and adding gelatin powder to the solution while vigorously stirring for 12 h at 70 °C (unless otherwise noted), and then the pH is adjusted to 7.5 using 1 M NaOH. The warm gelatin solution is sterile filtered and stored at 4 °C in aliquots for later use (<3 mo). Fibrinogen solution (50 mg⋅mL−1) is produced by dissolving lyophilized bovine blood plasma protein (Millipore) at 37 °C in sterile DPBS without calcium and magnesium. The solution is held at 37 °C for 45 min to allow complete dissolution. The TG solution (60 mg⋅mL−1) is prepared by dissolving lyophilized powder (Moo Glue) in DPBS without calcium and magnesium and gently mixing for 20 s. The solution is then placed at 37 °C for 20 min and sterile filtered before use. A 250 mM CaCl2 stock solution is prepared by dissolving CaCl2 powder in DPBS without calcium and magnesium (Corning). To prepare stock solution of thrombin, lyophilized thrombin (Sigma-Aldrich) is reconstituted at 500 U⋅mL−1 using sterile DPBS and stored at −20 °C. The thrombin aliquots are thawed immediately before use.
Matrix Formulations.
The solutions are mixed together at 37 °C to achieve a final concentration of 10 mg⋅mL−1 fibrinogen, 7.5 wt% gelatin, 2.5 mM CaCl2, and 0.2 wt% TG. For printing, we use 1 wt% TG to account for diffusion and dilution into printed cell filaments. The equilibration time before mixing with thrombin (at a ratio of 500:1) determines optical clarity (SI Appendix, Fig. S3). After mixing, the matrix must be quickly cast, as rapid polymerization ensues. Native fibrin matrix is created by the same procedure without gelatin and TG (SI Appendix, Fig. S4). Alternatively, hyaluronic acid methacrylate can be synthesized and used (26).
Ink Formulations.
A silicone ink, composed of a two-part silicone elastomer (SE 1700; Dow Chemical) with a 10:1 base to catalyst (by weight), is used to create customized perfusion chips. It is homogenized using a mixer (2,000 speed; AE-310; Thinky Corporation) and printed within 2 h of mixing. A fugitive ink, composed of 38 wt% Pluronic F127 (Sigma) and 100 U⋅mL−1 thrombin in deionized, ultrafiltrated water, is used to print the vasculature. A stock solution (40% Pluronic F127) is homogenized using a Thinky mixer and subsequently stored at 4 °C. Before use, 2,000 U⋅mL−1 thrombin solution is added to ink at a ratio of 1:20, homogenized, loaded into a syringe (EFD, Inc.) at 4 °C, and centrifuged to remove any air bubbles. All inks are printed at room temperature.
A cell-laden ink, composed of 7.5 wt/vol% gelatin and 10 mg⋅mL−1 fibrinogen, is prepared for printing. Ink stiffness is tuned by varying the gelatin-processing temperature (70–95 °C) (SI Appendix, Fig. S6). This ink is prepared similarly to the matrix, but without TG and thrombin. Upon printing, cross-linking is achieved by diffusion of these enzymes from the surrounding matrix. To disperse cells in the ink, the fibrinogen–gelatin blend is held at 37 °C, and then cell suspensions are introduced via gentle pipetting. After mixing, the ink is held at 4 °C for 15 min to drive thermal gelation of the gelatin phase. Next, the ink is warmed to room temperature for at least 15 min, where it can be immediately printed for up to 2 h.
Fibrinogen–Fluorophore Conjugation.
To visualize the fibrin network in printed filaments and the cast matrix (SI Appendix, Fig. S3), fibrinogen is conjugated to two fluorophores. Specifically, 1 g of bovine fibrinogen is dissolved in 100 mL of 50 mM borate buffer, pH 8.5 (Thermo Scientific), to form a 10 mg⋅mL−1 solution. N-Hydroxysuccinimide, conjugated with either fluorescein or rhodamine, is added at a 10:1 molar ratio of dye/fibrinogen. After reacting for 2 h at room temperature, the labeled fibrinogen is separated from unconjugated dye by dialysis using 10-kDa MWCO dialysis tubing in a 2-L bath against PBS for 3 d, changing the PBS in the bath twice daily. After dialysis is complete, the fluorescently conjugated fibrinogen is frozen at −80 °C, lyophilized, and stored at −20 °C before use.
Rheological Characterization.
Ink rheology is measured using a controlled stress rheometer (DHR-3; TA Instruments) with a 40-mm diameter, 2° cone and plate geometry. The shear storage (G') and loss (G'') moduli are measured at a frequency of 1 Hz and an oscillatory strain (γ) of 0.01. Temperature sweeps are performed using a Peltier plate over the range from −5 to 40 °C. Samples are equilibrated for 5 min before testing and for 1 min at each subsequent temperature to minimize thermal gradients throughout the sample. Time sweeps are conducted by rapidly placing a premixed solution onto the temperature-controlled Peltier plate held at 37 or 22 °C, unless otherwise noted.
Cell Culture and Maintenance.
hMSCs (Rooster Bio) are cultured in Booster Media (Rooster Bio) and are not used beyond two passages. Green fluorescent protein-expressing HNDFs (GFP-HNDFs) (Angio-Proteomie) are cultured in Dulbecco’s modified Eagle medium containing high glucose and sodium pyruvate (DMEM) (GlutaMAX; Gibco) and supplemented with 10% FBS (Gemini Bio-Products). Primary red fluorescent protein-expressing HUVECs (RFP-HUVECs) (Angio-Proteomie) are cultured in EGM-2 media (complete EGM-2 BulletKit; Lonza). GFP-HNDFs and RFP HUVECs are not used beyond the 15th and 9th passages, respectively.
Three-Dimensional Tissue Fabrication on Perfusable Chips.
All vascularized tissues are created on a custom-designed multimaterial 3D bioprinter equipped with four independently addressable print heads mounted onto a three-axis, motion-controlled gantry with build volume of 725 × 650 × 125 mm (AGB 10000; Aerotech). Each ink is housed in a syringe equipped with a leur-locked nozzle of varying size (i.e., 100-µm to 410-µm diameter) (EFD, Inc.). Inks are deposited by applying air pressure (800 Ultra dispensing system; EFD, Inc.), ranging from 10 to 140 psi, corresponding to print speeds from 1 mm⋅s−1 to 5 cm⋅s−1.
To manufacture the customized perfusion chips, the silicone ink is loaded into a 10-mL syringe, centrifuged to remove air bubbles, and deposited through a tapered 410-µm nozzle. The gasket design is created using custom MATLAB software and the structures are printed onto 50 × 75-mm glass slides. After printing, the chips are cured at 80 °C in an oven for >1 h and stored at room temperature.
To produce thick vascularized tissues, multiple inks are sequentially coprinted within the customized perfusion chips. To form a base layer, a thin film of gelatin–fibrin matrix, containing 0.1 wt% TG, is cast onto the base of the perfusion chip and allowed to dry. Next, the fugitive Pluronic F127 and cell-laden inks are printed onto the surface using 200-µm straight and tapered nozzles, respectively. After printing, stainless metal tubes are fed through the guide channels of the perfusion chip and pushed into physical contact with printed vertical pillars of the fugitive ink positioned at the inlet and outlet of each device (SI Appendix, Fig. S1, and Movie S2). Before encapsulation, TG is added to the molten 37 °C gelatin–fibrin matrix solution and preincubated for 2–20 min depending on the desired matrix transparency (SI Appendix, Fig. S3). To form a cell-laden matrix, the molten 37 °C gelatin–fibrin matrix is first mixed with HNDF-GFP cells and then mixed with thrombin. Next, this matrix is cast around the printed tissue, where it undergoes rapid gelation due to thrombin activity. The 3D tissue chips are stored at 37 °C for 1 h before cooling to 4 °C to liquefy and remove the printed fugitive ink, which is flushed through the device using cold cell media, leaving behind open conduits.
The 3D perfusion chips are loaded onto a machined stainless-steel base, and a thick acrylic lid is placed on top. The lid and base are clamped together by four screws, forming a seal around the silicone 3D printed gasket top. Next, sterile two-stop peristaltic tubing (PharMed BPT) is filled with media and connected to the outlet of a sterile filter that is attached to a 10-mL syringe (EFD Nordson), which serves as a media reservoir. Media that has been equilibrating for >6 h in an incubator at 37 °C, 5% CO2 is added to the media reservoir, and by means of gravity, is allowed to flow through the filter and peristaltic tubing, until all of the air is displaced, before connecting the peristaltic tubing to the inlet of each perfusion chip. Hose pinch-off clamps are added at the inlet and outlet of the perfusion chip to prevent uncontrolled flow when disconnected from the peristaltic pump, which can damage the endothelium or introduce air bubbles to the vasculature. The media reservoir is allowed to equilibrate with atmospheric pressure at all times by means of a sterile filter connecting the incubator environment with the reservoir.
Endothelialization of Vascular Networks.
With the peristaltic tubing removed from the chip outlet, 50–500 µL of HUVEC suspensions (1 × 107 cells per mL) are injected via pipette to fill the vascular network. The silicone tubing is then replaced, and both the outlet and inlet pinch-clamp are sealed. The perfusion chip is incubated at 37 °C to facilitate cell adhesion to the channels under zero-flow conditions. After 30 min, the chip is flipped 180° to facilitate cell adhesion to the other side of the channel, and achieve circumferential seeding of cells in the channel. Finally, the cells are further incubated for between 5 h and overnight at 37 °C before commencing active perfusion.
Active Perfusion.
After endothelial cell seeding, the peristaltic tubing is affixed to a 24-channel peristaltic pump (Ismatec), after which the hose clamps are removed. For single vascular channels, the perfusion rate is set at 13 µL⋅min−1, whereas for thick vascularized tissues, it is set at 27 µL⋅min−1.
Cell Viability Assay.
Cell viability is determined postprinting by printing inks with 2 × 106 cells per mL for each condition. Printed cell-laden filaments (2 × 106 cells per mL for each condition) are deposited onto a glass substrate and then stained using calcein-AM (“live”; 1 µL⋅mL−1; Invitrogen) and ethidium homodimer (“dead”; 4 µL⋅mL−1; Invitrogen) for 20 min before confocal imaging (n = 3 unique samples, imaged n = 10 times). To assess cell viability, live tissue is removed from the perfusion chip, cross-sectioned, and stained using the same staining protocol. Live and dead cell counts are obtained using the 3D objects counter plugin in ImageJ software. The results are averaged and SDs determined for each sample.
Imaging and Analysis.
Photographs and videos of tissue fabrication are acquired using a DSLR camera (Canon EOS, 5D Mark II; Canon). Fluorescent dyes are used to improve visualization of Pluronic F127 (Red, Risk Reactor) and gelatin–fibrin ink (Fluorescein; Sigma-Aldrich). Printed tissue structures are imaged using a Keyence Zoom (VHX-2000; Keyence), an inverted fluorescence (Axiovert 40 CFL; Zeiss), and an upright confocal microscope (LSM710; Zeiss). ImageJ is used to generate composite microscopy images by combining fluorescent channels. Three-dimensional rendering and visualization of confocal stacks are performed in Imaris 7.6.4, Bitplane Scientific Software, and ImageJ software. Cell counting is performed using semiautomated native algorithms in Imaris and ImageJ counting and tracking algorithms.
Immunostaining.
Immunostaining and confocal microscopy are used to assess the 3D vascularized tissues. Printed tissues are first washed with PBS via perfusion for several minutes. Next, 10% buffered formalin is perfused through the 3D tissue for 10–15 min. The tissue is removed from the perfusion chip and bathed in 10% buffered formalin. A 2-h fixation time is required for a 1-cm-thick tissue. The 3D tissues are then washed in PBS for several hours and blocked overnight using 1 wt% BSA in PBS. Primary antibodies to the cell protein or biomarker of interest are incubated with the constructs for 2 d in a solution of 0.5 wt% BSA and 0.125 wt% Triton X-100 (SI Appendix, Table S1). Removal of unbound primary antibodies is accomplished using a wash step against a solution of PBS or 0.5 wt% BSA and 0.125 wt% Triton X-100 in PBS for 1 d. Secondary antibodies are incubated with the constructs for 1 d at the dilutions listed in SI Appendix, Table S1, in a solution of 0.5 wt% BSA and 0.125 wt% Triton X-100 in PBS. Samples are counterstained with NucBlue or ActinGreen for 2 h and then washed for 1 d in PBS before imaging. Confocal microscopy is performed using an upright Zeiss LSM 710 with water-immersion objectives ranging from 10× to 40× using spectral lasers at 405-, 488-, 514-, 561-, and 633-nm wavelengths. Image reconstructions of z stacks are performed in ImageJ using the z-project function with the maximum pixel intensity setting. Three-dimensional image reconstructions are performed using Imaris software.
hMSC Staining.
Fast Blue (Sigma-Aldrich) and alizarin red (SigmaFast; Sigma-Aldrich) are used to visualize AP activity and calcium deposition. One tablet of Fast Blue is dissolved in 10 mL of deionized (DI) water. This solution is stored in the dark and used within 2 h. Cells are washed using 0.05% Tween 20 in DPBS without calcium and magnesium and fixed as described above. The samples are then covered with Fast Blue solution and incubated in the dark for 5–10 min and washed using PBS-Tween buffer. To assess mineralization, 2% alizarin red solution is dissolved in DI water, mixed vigorously, filtered, and used within 24 h. Samples are equilibrated in DI water and incubated with alizarin red solution for a few minutes, then the staining solution is removed, and samples are washed three times in DI water or until background dye is unobservable. Representative slices of both avascular and vascularized, thick tissues are digested using 2 wt% Collagenase I in PBS without Ca2+, Mg2+ at 37 °C for >24 h. The resulting solutions are filtered using a 0.2-µm sterile filter and rinsed with DI water. SEM/EDS is used to carry out elemental analysis on harvested mineral particulates.
FITC-Dextran Permeability Testing.
To assess barrier function of the printed vasculature, diffusional permeability was quantified by perfusing culture media in the vascular channel, while alive, containing 25 µg/mL FITC-conjugated 70-kDa dextran (FITC-Dex; Sigma product 46945) at a rate of 20 µL⋅min−1 for 3 min and 1 µL⋅min−1 thereafter for ∼33 min. The diffusion pattern of FITC-Dex was detected using a wide-field fluorescent microscope (Zeiss Axiovert 40 CFL). Fluorescence images were captured before perfusion and every 3–5 min after for 33 min. Diffusional permeability of FITC-Dex is calculated by quantifying changes of fluorescence intensity over time using the following equation:
Pd is the diffusional permeability coefficient, I1 is the average intensity at an initial time point, I2 is an average intensity after some time (t, ∼30 min), Ib is background intensity (before introducing FITC-Dex), and d is the channel diameter (27). The measurements are performed on embedded channels with and without endothelium (n = 3).
Supplementary Material
Acknowledgments
We thank Donald Ingber, David Mooney, and Christopher Hinojosa for useful discussions; Jessica Herrmann, Humphrey Obuobi, Hayley Price, Nicole Black, Tom Ferrante, and Oktay Uzun for their experimental assistance; and Lori K. Sanders for help with photography and videography. This work was supported by NSF Early-concept Grants for Exploratory Research (EAGER) Award Division of Civil, Mechanical and Manufacturing Innovation (CMMI)-1548261 and by the Wyss Institute for Biologically Inspired Engineering.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521342113/-/DCSupplemental.
References
- 1.Nelson CM, Vanduijn MM, Inman JL, Fletcher DA, Bissell MJ. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314(5797):298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee K, Silva EA, Mooney DJ. Growth factor delivery-based tissue engineering: General approaches and a review of recent developments. J R Soc Interface. 2011;8(55):153–170. doi: 10.1098/rsif.2010.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ingber DE. Mechanobiology and diseases of mechanotransduction. Ann Med. 2003;35(8):564–577. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 4.Abbott RD, Kaplan DL. Strategies for improving the physiological relevance of human engineered tissues. Trends Biotechnol. 2015;33(7):401–407. doi: 10.1016/j.tibtech.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8(10):839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 6.Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21(12):745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol. 2014;32(8):760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 8.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4(4):359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260(5110):920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 10.Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat Biotechnol. 2014;32(8):773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 11.Atala A, Kasper FK, Mikos AG. Engineering complex tissues. Sci Transl Med. 2012;4(160):160rv12. doi: 10.1126/scitranslmed.3004890. [DOI] [PubMed] [Google Scholar]
- 12.Cui X, Boland T. Human microvasculature fabrication using thermal inkjet printing technology. Biomaterials. 2009;30(31):6221–6227. doi: 10.1016/j.biomaterials.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 13.Norotte C, Marga FS, Niklason LE, Forgacs G. Scaffold-free vascular tissue engineering using bioprinting. Biomaterials. 2009;30(30):5910–5917. doi: 10.1016/j.biomaterials.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolesky DB, et al. 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv Mater. 2014;26(19):3124–3130. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 15.Miller JS, et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat Mater. 2012;11(9):768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee KY, Mooney DJ. Hydrogels for tissue engineering. Chem Rev. 2001;101(7):1869–1879. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- 17.Mosesson MW. Fibrinogen structure and fibrin clot assembly. Semin Thromb Hemost. 1998;24(2):169–174. doi: 10.1055/s-2007-995837. [DOI] [PubMed] [Google Scholar]
- 18.Chen R-N, Ho H-O, Sheu M-T. Characterization of collagen matrices crosslinked using microbial transglutaminase. Biomaterials. 2005;26(20):4229–4235. doi: 10.1016/j.biomaterials.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Stoien JD, Wang RJ. Effect of near-ultraviolet and visible light on mammalian cells in culture II. Formation of toxic photoproducts in tissue culture medium by blacklight. Proc Natl Acad Sci USA. 1974;71(10):3961–3965. doi: 10.1073/pnas.71.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayakumar MKG, Idris NM, Zhang Y. Remote activation of biomolecules in deep tissues using near-infrared-to-UV upconversion nanotransducers. Proc Natl Acad Sci USA. 2012;109(22):8483–8488. doi: 10.1073/pnas.1114551109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klumpers DD, Zhao X, Mooney DJ, Smit TH. Cell mediated contraction in 3D cell-matrix constructs leads to spatially regulated osteogenic differentiation. Integr Biol (Camb) 2013;5(9):1174–1183. doi: 10.1039/c3ib40038g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oster GF, Murray JD, Harris AK. Mechanical aspects of mesenchymal morphogenesis. J Embryol Exp Morphol. 1983;78:83–125. [PubMed] [Google Scholar]
- 23.Wu W, DeConinck A, Lewis JA. Omnidirectional printing of 3D microvascular networks. Adv Mater. 2011;23(24):H178–H183. doi: 10.1002/adma.201004625. [DOI] [PubMed] [Google Scholar]
- 24.Giulitti S, Magrofuoco E, Prevedello L, Elvassore N. Optimal periodic perfusion strategy for robust long-term microfluidic cell culture. Lab Chip. 2013;13(22):4430–4441. doi: 10.1039/c3lc50643f. [DOI] [PubMed] [Google Scholar]
- 25.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7(3):211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 26.Burdick JA, Chung C, Jia X, Randolph MA, Langer R. Controlled degradation and mechanical behavior of photopolymerized hyaluronic acid networks. Biomacromolecules. 2005;6(1):386–391. doi: 10.1021/bm049508a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price G, Tien J. In: Methods in Molecular Biology. Khademhosseini A, editor. Humana; Totowa, NJ: 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.