Significance
We report cell-to-cell movement of mitochondria through a graft junction of two tobacco species, Nicotiana tabacum and Nicotiana sylvestris. The flowers of the N. tabacum line we used are male sterile due to a sterility-causing mitochondrial genome, whereas the N. sylvestris flowers are fertile. Grafting created an opportunity for organelle movement during the healing process when cell-to-cell connections at the graft junction were restored. We recognized N. sylvestris mitochondrial DNA transfer by restoration of fertile flower anatomy in plants regenerated from graft junctions. Demonstrating cell-to-cell movement of mitochondria reconstructs the evolutionary process of horizontal mitochondrial DNA transfer and enables modification of mitochondria by DNA acquired from other species.
Keywords: grafting, horizontal gene transfer, mitochondria, plastid, tobacco
Abstract
We report cell-to-cell movement of mitochondria through a graft junction. Mitochondrial movement was discovered in an experiment designed to select for chloroplast transfer from Nicotiana sylvestris into Nicotiana tabacum cells. The alloplasmic N. tabacum line we used carries Nicotiana undulata cytoplasmic genomes, and its flowers are male sterile due to the foreign mitochondrial genome. Thus, rare mitochondrial DNA transfer from N. sylvestris to N. tabacum could be recognized by restoration of fertile flower anatomy. Analyses of the mitochondrial genomes revealed extensive recombination, tentatively linking male sterility to orf293, a mitochondrial gene causing homeotic conversion of anthers into petals. Demonstrating cell-to-cell movement of mitochondria reconstructs the evolutionary process of horizontal mitochondrial DNA transfer and enables modification of the mitochondrial genome by DNA transmitted from a sexually incompatible species. Conversion of anthers into petals is a visual marker that can be useful for mitochondrial transformation.
Horizontal gene transfer (HGT), the acquisition of gene(s) across species mating boundaries, results in a phylogeny of the transferred gene(s) that is incongruent with the phylogeny of the organism. In flowering plants, HGT is relatively rare in the nucleus, but frequently involves mitochondrial DNA (mtDNA); for reviews see refs. 1–3. Pioneering papers described HGT of several mitochondrial genes (4–6) and, in an extreme case, incorporation of six genome equivalents in the 3.9-Mb Amborella trichopoda mtDNA (7). These findings imply that mechanisms exist for DNA delivery between unrelated species. Parasitic plants are frequent participants in HGT, either as donors (8, 9) or recipients (5) of foreign DNA; the DNA exchange between the host and parasite is probably facilitated by the physical connection (for review, see ref. 3). HGT between nonparasites, however, necessitates alternative modes of DNA transfer. Transfer via vectoring agents such as viruses, bacteria, fungi, insects, and pollen; transformational uptake of plant DNA released into the soil; and occasional grafting of unrelated species were proposed (6). The first experimental evidence in support of grafting as a potential mechanism of HGT came from demonstrating exchange of plant DNA in tobacco tissue grafts (10). Movement of entire chloroplast genomes was subsequently demonstrated through tobacco graft junctions, interpreted as evidence for cell-to-cell movement of the organelles (11, 12). However, evidence for cell-to-cell movement of mitochondrial DNA is missing in plants despite the fact that the majority of horizontal gene transfer events involve mitochondrial sequences.
We report here an experimental system for the successful identification of a rare mitochondrial HGT event. Replacing the cytoplasm of Nicotiana tabacum with the cytoplasm of Nicotiana undulata makes the flowers of N. tabacum male sterile due to conversion of anthers to stigmatoid petals (Fig. 1 D–G). Such N. tabacum plants are called alloplasmic substitution lines for carrying an alien cytoplasm and are cytoplasmic male sterile (CMS) because they inherit male sterility only from the maternal parent (13). We reasoned that movement of Nicotiana sylvestris mitochondria into CMS cells should restore anther morphology and pollen production, a change that is easy to detect in plants even if restricted to a few flowers.
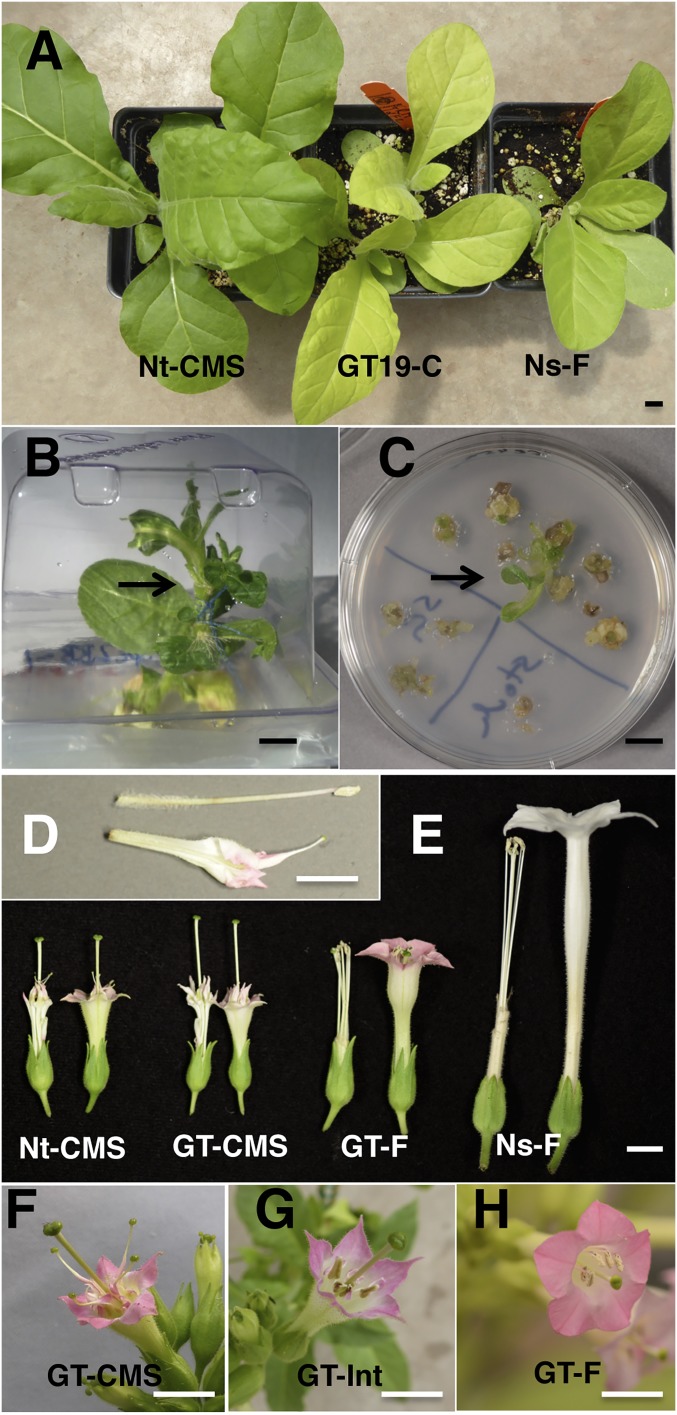
Fig. 1.
Restoration of fertile flower anatomy facilitates identification of mitochondrial graft transmission event. (A) N. tabacum Nt-CMS and fertile N. sylvestris Ns-F graft partners and GT19-C seed progeny. (B) Grafting tobacco in culture. The scion is Nt-CMS, which carries the nuclear gentamycin resistance marker; and the rootstock is Ns-F, which carries the plastid spectinomycin resistance (aadA) and aurea barau genes. Arrow points to graft junction. (C) Selection of gentamycin and spectinomycin double-resistant clones. (Right) Stem slices from the graft region; (Left) from above and below. Arrow points to double-resistant clone. (D) One isolated anther from a wild-type N. tabacum flower (above) and the anther after homeotic conversion of the N. tabacum alloplasmic substitution line (below). (E) Flower morphology of the graft partners and mixed flower anatomy on the GT19-C graft transmission plant. (Right) Flowers are shown with corolla; (Left) with corolla removed. Note homeotic transformation of anthers into stigmatoid petals in Nt-CMS graft partner and the GT-CMS flowers. GT-F and N. sylvestris Ns-F flowers are fertile. The flowers of Nt-CMS graft partner and GT19-C plant (GT-CMS and GT-F) are pink, a nuclear trait; those of the N. sylvestris graft partner are white. A close-up of (F) GT-CMS, (G) GT-intermediate, and (H) GT-F flowers. (Scale bars in Lower Right corners, 10 mm.)
We looked for cell-to-cell movement of mitochondria in stem grafts of two species, N. tabacum and N. sylvestris. We first selected for the nuclear marker from N. tabacum and the chloroplast marker in N. sylvestris and regenerated plants from double-resistant tissue derived from the graft junction. We identified branches with fertile flowers on one of the regenerated plants, indicating presence of fertile mtDNA in the otherwise CMS plant, and analyzed the mtDNA of its fertile and CMS seed progeny. Recombination at alternative sites in the mitochondrial genome facilitated the identification of a candidate mitochondrial gene responsible for homeotic transformation of anthers resulting in CMS.
Results
Plastid Graft Transmission Events.
The CMS N. tabacum graft partner (Nt-CMS) carried a nuclear gentamycin resistance gene (14). The fertile N. sylvestris (Ns-F) graft partner carried two plastid markers: a selectable spectinomycin resistance aminoglycoside-3″-adenylyltransferase (aadA) gene and the visual aurea bialaphos resistance (barau) leaf color gene (15) (Fig. 1A). The two Nicotiana species were grafted (Fig. 1B), the graft junctions were sliced, and resistant shoots from the tissue slices were selected in culture for gentamycin resistance encoded in the nucleus of the CMS N. tabacum graft partner, and spectinomycin resistance encoded in the plastids of the fertile N. sylvestris graft partner. Three clones with resistance to both antibiotics were recovered in the culture of 14 graft junctions (Fig. 1C). The events were designated GT7, GT17, and GT19. The identity of plastids as N. sylvestris in the regenerated plants was confirmed by DNA gel blot analyses of all three lines and sequencing the plastid genome of the GT19-C line (SI Appendix, Fig. S1). The regenerated plants carried only the chromosomes of N. tabacum, the graft partner carrying the nuclear gentamycin resistance gene (SI Appendix, Fig. S2). Therefore, selection for the nuclear marker in N. tabacum and chloroplast marker in N. sylvestris yielded N. tabacum plants with chloroplasts from N. sylvestris, without input of N. sylvestris chromosomes.
From the three double-resistant calli, eight plants were regenerated: GT7-A and GT7-C from callus GT7; GT17-B, GT17-C, and GT17-G from callus GT17; and GT19-A, GT19-B, and GT19-C from callus GT19. All but one had CMS flowers. The GT19-C plant was chimeric, with fertile flowers on two of the four branches, suggesting cotransmission of fertile mitochondria with the N. sylvestris chloroplasts (Fig. 1E). We found three types of flowers on the GT19-C plants: CMS with petaloid-stigmatoid anthers (Fig. 1F), fertile anthers with ample pollen (Fig. 1H), and intermediate with partial conversion of anthers into petals (Fig. 1G). Seeds from fertile flowers were obtained by self-pollination. Seed from sterile flowers was obtained by pollination with wild-type N. tabacum pollen. Fertile or CMS phenotype of the GT19-C seed progeny was stably maintained through three seed generations.
Recombination of Mitochondrial DNA.
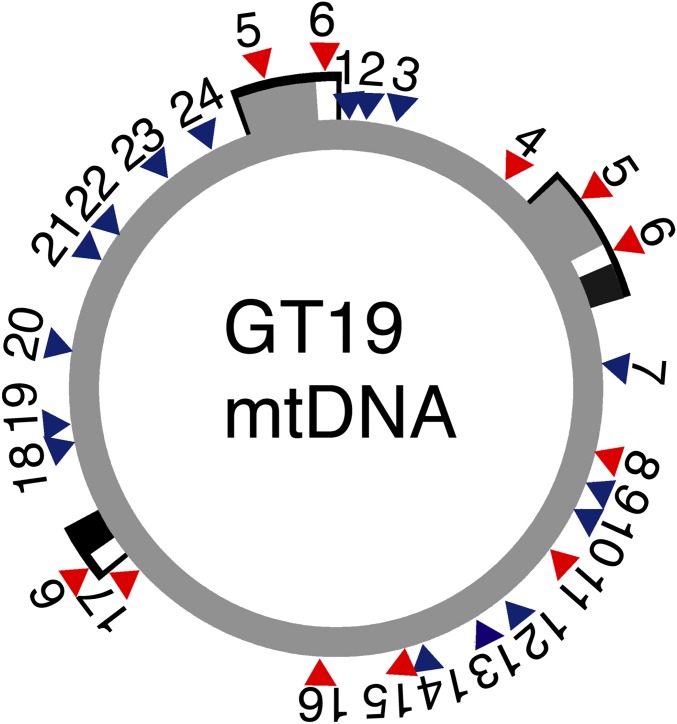
Next, we looked for DNA evidence of mitochondrial movement through the graft junction. We chose N. tabacum plants with undulata cytoplasm as one of the graft partners because the plastid DNA (ptDNA) of N. tabacum and N. undulata differ by 918 ptDNA markers (805 SNPs and 113 short indels) (11), and we expected the mitochondrial DNA to be similarly divergent. Plant mitochondria continuously undergo repeated cycles of fusion and fission (16); therefore, we expected to find recombinant mitochondrial genomes. We tested 24 polymorphic sites in the 430-kb mitochondrial genome (SI Appendix, Table S1). The flower morphology and mtDNA markers of the GT7 (A and C) and GT17 (B, C, and G) plants were N. undulata type. Plants derived from the third event, GT19-A, -B, and -C had chimeric mitochondrial genomes with 8 markers derived from the fertile and 16 from the CMS mitochondria (Fig. 2). However, the mtDNA in the fertile and sterile branches could not be distinguished by the 24 markers.
Fig. 2.
Recombinant GT19 mtDNA is composed of segments of CMS N. undulata and fertile N. sylvestris mtDNA. The origin of 24 markers in the GT19 mtDNA is shown on the N. sylvestris (KT997964) mtDNA map. N. undulata and N. sylvestris markers are in blue and red color, respectively. Note that markers 5 and 6 are located in repeat regions of the mitochondrial genome (48). For polymorphic loci, see SI Appendix, Table S1.
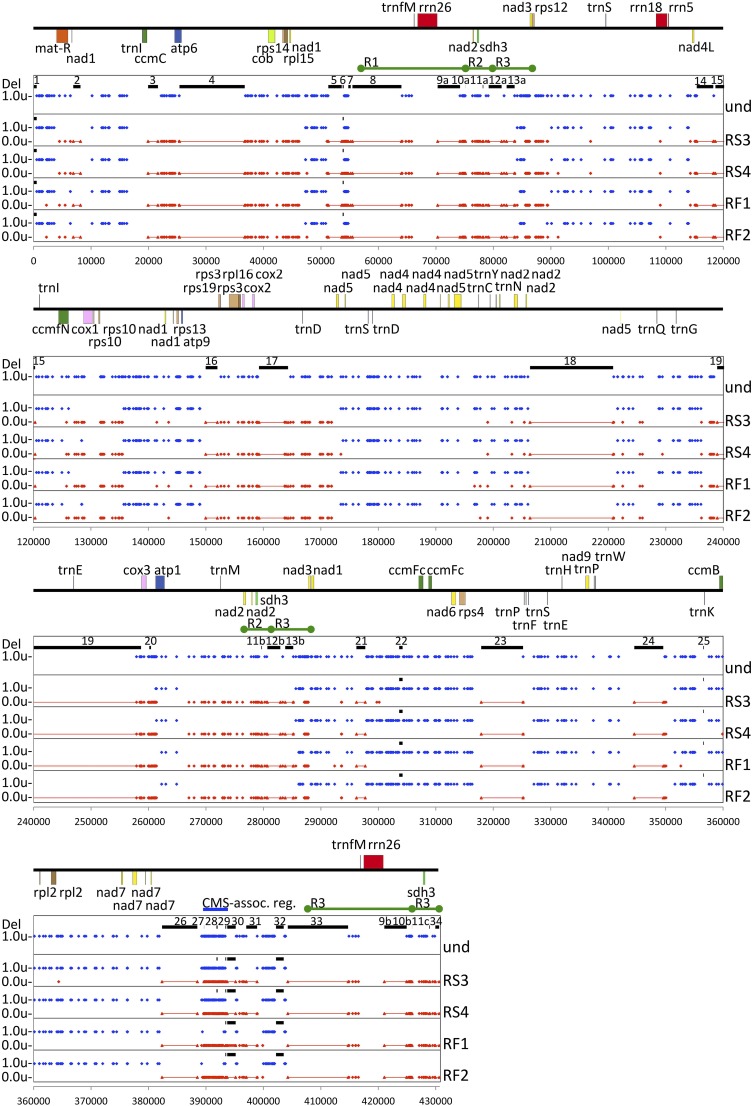
To identify the region of the mitochondrial genome responsible for fertility restoration, we assembled the complete mitochondrial genome of N. sylvestris (GenBank KT997964), and mitochondrial contigs of N. undulata and two fertile and two sterile first seed generation GT19-C progeny (Fig. 3). The 279-kb sequence conserved between the N. sylvestris and N. undulata mitochondrial genomes differ by 977 DNA polymorphic markers (SI Appendix, Table S2) and encode all genes present in the N. sylvestris mtDNA. The only exception is trnE, a gene encoding tRNA-UUC of plastid origin (246,982–247,053, GenBank NC_006581). However, a mitochondrial-derived trnE gene encoding tRNA-UUC is present in the N. undulata mtDNA. Plotted along the N. sylvestris genetic map in Fig. 3 is the origin of mtDNA in the recombinant mitochondrial genomes. The mtDNA in the plants is apparently mosaic, consisting of segments of N. sylvestris and N. undulata mtDNA. The N. sylvestris and N. undulata SNPs are symbolized by red and blue dots, respectively. The crossover sites are not always identical in the recombinant mitochondrial genomes. For example, both the fertile and sterile lines have N. undulata mtDNA at the beginning of the map, but the fertile lines have one N. sylvestris SNP at nucleotide 2,246 (Fig. 3). Based on the switching of red and blue dots in the alignment, the recombinant fertile 1 and fertile 2 (RF1 and RF2) and recombinant sterile 3 and sterile 4 (RS3 and RS4) mitochondrial genomes contain at least 65, 63, 57, and 58 crossover sites (Fig. 3). Five recombination junctions have been PCR amplified and confirmed by Sanger sequencing (SI Appendix, Fig. S4).
Fig. 3.
The mitochondrial genome of the GT19-C seed progeny is a mosaic of the two graft partners. We aligned the parent-specific SNPs in two fertile and two CMS recombinants using the N. sylvestris mtDNA as reference. Red and blue dots identify N. sylvestris- and N. undulata- specific SNPs, respectively. These data representations account for all N. sylvestris genes and exclude sequences unique to N. undulata (und). Deletions in the N. undulata mtDNA are shown as numbered black bars (SI Appendix, Table S5). N. sylvestris sequences absent in N. undulata and present in the recombinant progeny are red lines. The mitochondrial repeats are marked as R1, R2, and R3. The blue bar marks the CMS-associated region between nucleotides 389,706 and 393,005. The gene map was created using the Organellar Genome Draw program (49) based on the N. sylvestris mtDNA annotation (KT997964).
Expression of orf293 Correlates with Homeotic Conversion of Anthers.
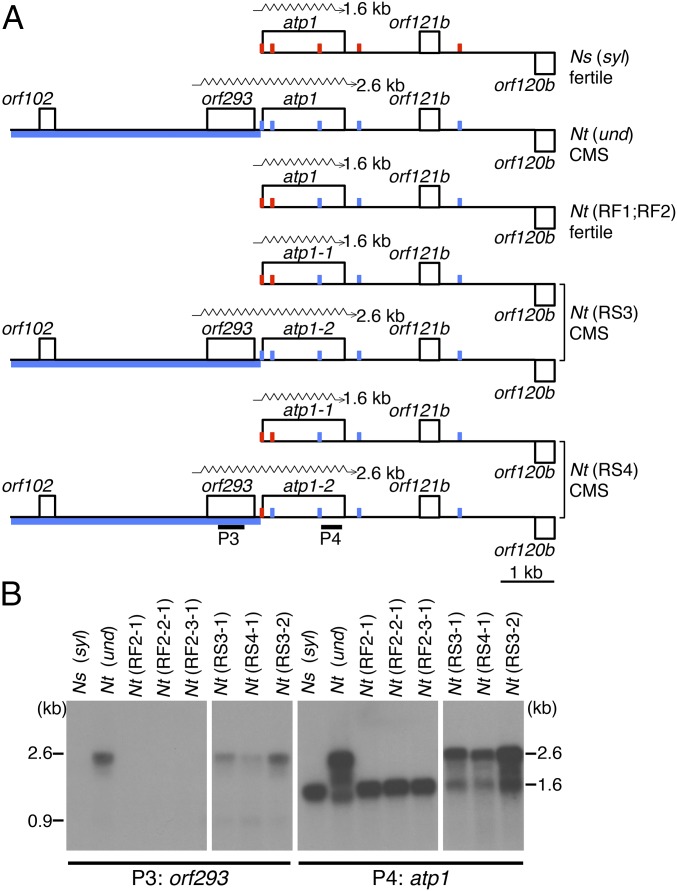
Recombination of mitochondrial genomes at alternative sites facilitated the identification of the region likely to be responsible for CMS that manifests as homeotic conversion of anthers into stigmatoid petals in N. tabacum (Fig. 1D). Sequence alignments revealed that a 3.5-kb region correlates with male sterility: the two recombinants with sterile flowers carried the N. undulata sequence and the N. sylvestris sequence, whereas the fertile recombinants had the cognate region from N. sylvestris alone (CMS-associated region in Fig. 3). Adjacent to the shared region is a 1.2-kb N. undulata-specific sequence encoding orf293 (Fig. 4A). RNA gel blot analyses confirmed that orf293 mRNA accumulates in CMS N. tabacum and sterile recombinants, but is absent in fertile N. tabacum and the fertile recombinants (Fig. 4B). The orf293 transcript is also absent in fertile N. undulata plants, the source of the cytoplasm, although the orf293 gene is present in the mitochondrial genome (SI Appendix, Fig. S3) (GenBank KU180495–KU180498). Absence of sterility-causing mitochondrial transcripts is expected when a nuclear fertility restorer gene is present (17, 18). The predicted protein encoded by orf293 has three transmembrane domains based on the TMHMM transmembrane protein topology program (19). Transmembrane domains are characteristic of sterility-causing mitochondrial genes (17, 18). The flower phenotype of plants carrying orf293 in mitochondria depends on the nuclear background: in N. tabacum the anthers are petaloid, or petaloid-stigmatoid (Fig. 1D); in N. sylvestris, the anthers are converted into stigmatoid structures (11). Based on the flower phenotype, we tentatively named orf293 “homeotic conversion of anthers” (hca) gene.
Fig. 4.
Mitochondrial orf293 expression correlates with homeotic conversion of anthers and CMS. (A) Shown are partial mtDNA maps of N. sylvestris (syl) fertile, N. tabacum (und) CMS, and two fertile (RF1 and RF2) and two sterile (RS3 and RS4) recombinant lines. SNPs derived from the undulata or sylvestris mitochondrial genomes are blue and red dots, respectively. Sixty polymorphisms in the 3.5-kb N. undulata region (Left) are represented by a solid blue line. (B) orf293 mRNA accumulates only in CMS plants. RNA gel blots were hybridized with orf293 (P3) and atp1 (P4) probes. Data are shown for the two graft partners and second and third generation seed progeny of GT19-C marked by adding one more digit for each generation separated by a hyphen, such as Nt(RF2) for the first generation, Nt(RF2-1) for the second, and Nt(RF2-2-1) for the third generation.
Discussion
Fate of Organelles After Cell-to-Cell Movement.
Part of the graft healing process is reconstitution of plasmodesmatal connections (20, 21) (Fig. 5). Plastids and mitochondria meet strikingly different fates subsequent to movement from cell to cell in the graft junction. Thus, far when cell-to-cell movement of plastids was observed, the entire selected plastid genome was recovered, rather than a ptDNA fragment comprising the selected marker. Absence of recombination between the plastid genomes suggested movement of intact organelles rather than naked, fragmented DNA (11, 12). Indeed, chloroplasts rarely fuse (22), precluding recombination that enables new combinations of chloroplast genes.
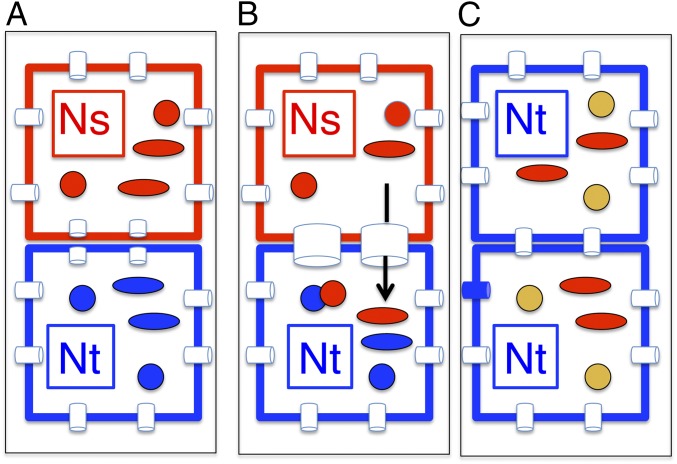
Fig. 5.
Transmission and recombination of mitochondria at graft junctions. (A) Grafting disrupts plasmodesmatal connections. The fertile N. sylvestris nucleus (Ns, red square) partner has fertile mitochondria (red circles) and spectinomycin-resistant chloroplasts (red ovals). The CMS N. tabacum nucleus (Nt, blue square) partner carries a nuclear gentamycin resistance gene, CMS mitochondria (blue circles) and wild-type N. undulata plastids (blue ovals). (B) During primary cell wall formation, organelles (organellar DNA) may enter an adjacent cell at the newly formed plasmodesmatal connections (wide tubes). Graft transmission events are selected by resistance to gentamycin (Nt nucleus, blue square) and spectinomycin-resistant Ns plastids (red ovals). Mitochondria fuse (note touching red and blue circles), whereas chloroplasts do not. (C) The graft transmission plant has the selected N. tabacum (Nt) gentamycin-resistant nucleus (blue square) and spectinomycin-resistant N. sylvestris plastids (red ovals). In fused mitochondria (yellow circles) mtDNA recombination yields chimeric fertile or CMS mitochondrial genomes.
Reconstruction of events that leads to formation of chimeric mitochondrial genomes in cells adjacent to graft junctions is complicated by the propensity of mitochondria for fusion (16, 23, 24). The mtDNA may move as naked DNA, or inside an organelle, but when heterologous mitochondrial genomes are present in a mixed cytoplasm, the mtDNA recombines (25–27). We favor movement of mitochondria as the more likely explanation because the incoming mtDNA is protected inside the organelle, mitochondrial fusion is frequent, and the recombinant genomes carry significant amounts of parental sequences scattered throughout the genome. Because tobacco mitochondria contain 40–160 mtDNA copies in 500–600 organelles per cell (28), recombination may take place among multiple mtDNA copies, yielding alternative recombination products. Alternative recombination in our case yielded fertile and CMS mitochondrial genomes, facilitating the identification of orf293 as a likely CMS (hca) gene. Graft transmission of mitochondria reported here provides a mechanism for horizontal transfer of entire genomes via mitochondrial fusion, which was postulated in evolutionary studies of horizontal gene transfer in mitochondria (7).
Identification of a Tentative CMS Gene in Recombinant Mitochondria.
Orf293 was identified as the tentative CMS gene by linkage to a region derived from N. undulata in the sterile recombinants. Orf293 in N. undulata is adjacent to the ATP synthase F1 subunit 1 (atp1) gene (Fig. 4A). In N. sylvestris and the fertile recombinants the atp1 gene is at a different genomic location. The sterile recombinants carry two atp1 copies: one in the N. sylvestris context (atp1-1) (KU180498) and one linked to orf293 (atp1-2; as in N. undulata) (KU180496 and KU180497) (Fig. 4A). Apparently, at the time of the interspecies mitochondrial fusion, mtDNA carrying both atp1 copies was present. The fertile recombinants formed when orf293 and the linked atp1-2 was lost. Most other parts of the mitochondrial genome are products of homologous recombination with clearly identifiable parental genome segments, where homologous recombination was followed by genome sorting that resulted in preservation of only one of the parental alleles. The exception is the atp1 gene, which has been preserved in both parental genome configurations. The differences between the two atp1 copies are silent mutations that do not involve predicted edited sites (PREP-mt) (29). We therefore believe that CMS is not caused by atp1 coding sequence SNPs in our experimental system.
The predicted ORF293 protein is similar to the predicted protein product of mitochondrial ORF312 present in the alloplasmic CMS eggplant (Solanum melongena, carrying the cytoplasm of S. kurzii) (GenBank AB762696) (30) and to sequences in the pepper mitochondrial genomes, where the sequences are split (GenBank KJ865410; KJ865409). The region of N. undulata mitochondrial genome carrying orf293 is rearranged relative to the N. sylvestris mitochondrial genome. Still, the rearranged region integrated into the chimeric mitochondrial genomes by homologous recombination preserving either the CMS or fertile mtDNA structure. Thus, nonhomologous end joining, the driving force behind mtDNA rearrangement during evolution, is not involved in the formation of mosaic mtDNAs (31, 32). The rearranged mitochondrial genomes were stable for at least three seed generations.
Applications in Biotechnology.
Grafting is a versatile approach that enables recovery of plants with: (i) chloroplasts of the graft partner without the transfer of mitochondria and nuclei (11, 12); (ii) the transfer of both chloroplasts and mitochondria in the absence of any nuclear DNA input (this paper); and (iii) the formation of nuclear hybrids with input of all organelles (33). Cell-to-cell movement of mitochondrial DNA reported here provides an alternative to transformation of mitochondria by naked DNA that has thus far remained elusive (34). An economically important mitochondrial trait that would be a prime target for incorporation by grafting is CMS, simplifying production of hybrid seed (17, 18). CMS transfer by graft transmission is feasible between graft-compatible species, even if they are sexually incompatible. An example would be transfer of CMS-encoding DNA from tobacco to tomato. Tomato is graft compatible with tobacco (35) and currently lacks CMS. The advantage of the graft transmission protocol is that mitochondrial DNA segments, rather than entire mitochondrial genomes, can be transferred in the absence of transferring chromosomal DNA, thereby minimizing nucleus–mitochondrial incompatibility. Furthermore, the orf293 (hca) gene provides a visual marker that can be useful for mitochondrial transformation.
Materials and Methods
Plant Material.
The graft partners were Nt-CMS (Nt-CMS92), a N. tabacum cv. Petit Havana line (15) that carries the cytoplasm of N. undulata and was transformed with Agrobacterium binary vector pPZP221 (14) yielding gentamycin-resistant line Nt-G115 and Ns-F, a fertile Nicotiana sylvestris line, the plastids of which have been transformed with plasmid pCK2 (Ns-pCK2-2) encoding a selectable spectinomycin resistance (aadA) and the visual barau genes (15). Seeds of N. undulata TW145 (PI 306637), TW146 (PI 555575), and TW147 (PI 306637) were obtained from the US Department of Agriculture, Agricultural Research Service National Plant Germplasm System (sun.ars-grin.gov/npgs/). Grafting and selection of graft plastid transmission events was carried out as described (11).
Chromosome Identification by Simple Sequence Repeat Markers.
Total cellular DNA was isolated using the cetyltrimethylammonium bromide (CTAB) method (36). The simple sequence repeat (SSR) markers were adopted from ref. 11, originally described in ref. 37. and are listed in SI Appendix, Table S3. Location of the SSR markers on the N. tabacum chromosomes is described in ref. 38. The PCR program of 94 °C for 5 min; 37 cycles of 94 °C for 45 s, 59 °C for 45 s, 72 °C for 1 min; 72 °C for 10 min was used for all but chromosomes 8, 12, 14, and 16. For chromosomes 8, 12, 14, and 16, the PCR program of 94 °C for 5 min; 37 cycles of 94 °C for 20 s, 54 °C for 20 s, 72 °C for 1 min; 72 °C for 10 min was used. The PCR products were ran on a 2.5% (wt/vol) TAE agarose gel for chromosomes 8, 14, 16–18, and 20 and on a 5% (wt/vol) MetaPhor agarose (Lonza) gel for chromosomes 1–7, 9–13, 15, 19, and 21–24.
Restriction Fragment Analyses of ptDNA.
CTAB purified (36) total cellular DNA was digested with the BamHI restriction enzyme and probed with rrn16, aadA, and bar probes (39). Blots were prepared as described (40).
Next Generation Sequencing of Organellar Genomes.
To determine organelle genome sequences, next generation sequencing (NGS) was performed in the Waksman Genomic Core Facility. Briefly, CTAB purified total cellular DNA (36) was physically sheared with the Covaris system (Covaris) following the manufacturer’s protocol. Sequencing libraries were prepared using the standard TruSeq DNA Library Preparation Kit (Illumina) according to the manufacturer’s protocol. Libraries were size selected at 650 bp with the Egel Agrose Electrophoresis System (Thermo Fisher Scientific) and quantified using the Qubit dsDNA HS (High Sensitivity) Kit (Thermo Fisher Scientific). Finally, libraries were evaluated for fragment size using the Bioanalyzer (Agilent Technologies). Library normalization and sequencing was performed according to the manufacturer’s recommendations with MiSeq v3 (2 × 300 bp) chemistries. Adapters and barcodes were trimmed per the default setting in the Illumina Experiment Manager (v1.8).
GT19-C Plastid Genome Assembly.
The Burrows–Wheeler Alignment Maximal Exact Matches (BWA-MEM) algorithm using default settings (41) was used to map adapter-free quality trimmed reads from four GT19-C offspring, recombinant fertile RF1 and RF2 and recombinant sterile RS3 and RS4, to the N. sylvestris ptDNA (NC_006500). Mapped reads were used to create de novo contigs using the ABySS program, using the paired-end (abyss-pe) option with a k-mer of 90 (42). NC_006500 was used as a guide to map and orient contigs in SeqMan Pro (DNASTAR) to obtain the complete ptDNA sequence. The plastid DNA sequence of the four GT19-C offspring was identical. The mVISTA alignment in SI Appendix, Fig. S1 (43) was prepared using the N. sylvestris ptDNA (NC_007500) as reference with a 300-bp sliding window.
mtDNA Mapping and Contig Assembly.
Adapter-free, quality trimmed reads were mapped to the N. tabacum mtDNA (NC_006581), Hyosciamus niger (KM207685), and two Capsicum annuum cultivars (NC_024624, KJ865409) mtDNAs using the BWA-MEM algorithm, and default settings (41). All mapped reads and their pairs were used to create de novo contigs with the ABySS program v1.9 using the paired-end (abyss-pe) option with a k-mer of 96 (42). N. undulata mitochondrial SNPs were called by the GATK HaplotypeCaller program (44–46) using the BAM file obtained from BWA mapping with default parameters. SNPs and indels called by GATK were filtered, keeping only SNPs with ≥80% SNP ratio and a minimum of 30× coverage in N. undulata. The origin of regions in the seed progeny was assigned by SNPs in the de novo contigs.
RNA Gel Blot Analysis.
Total cellular RNA was isolated from leaves of greenhouse-grown plants using TRIzol (Invitrogen) and dissolved in 20 μL diethylpyrocarbonate (DEPC) water. The isolated RNA was precipitated by adding 2 μL 3M sodium acetate (pH 5.2) and 66 μL 100% ethanol (1 h at −20 °C). RNA was sedimented by centrifugation, washed with 75% ethanol, air dried, and dissolved in 22 μL DEPC water. A total of 3 μg RNA was electrophoresed on 1.5% agarose/formaldehyde gel (6% of 37 wt/vol % formaldehyde) in Mops buffer (47). RNA was transferred to Amersham Hybond-N membranes (GE Healthcare) using capillary transfer. Probes were PCR fragments amplified using total cellular DNA as template, using primers listed in SI Appendix, Table S4. Probing was carried out as described (40).
Supplementary Material
Acknowledgments
C.G. was supported by a Waksman Institute of Microbiology Busch Predoctoral Fellowship and a teaching assistantship from the Division of Life Sciences, Rutgers University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the NCBI GenBank database (accession nos. KT997964 and KU180495–KU180498).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518644113/-/DCSupplemental.
References
- 1.Richardson AO, Palmer JD. Horizontal gene transfer in plants. J Exp Bot. 2007;58(1):1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- 2.Bock R. The give-and-take of DNA: Horizontal gene transfer in plants. Trends Plant Sci. 2010;15(1):11–22. doi: 10.1016/j.tplants.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Davis CC, Xi Z. Horizontal gene transfer in parasitic plants. Curr Opin Plant Biol. 2015;26:14–19. doi: 10.1016/j.pbi.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Won H, Renner SS. Horizontal gene transfer from flowering plants to Gnetum. Proc Natl Acad Sci USA. 2003;100(19):10824–10829. doi: 10.1073/pnas.1833775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis CC, Wurdack KJ. Host-to-parasite gene transfer in flowering plants: Phylogenetic evidence from Malpighiales. Science. 2004;305(5684):676–678. doi: 10.1126/science.1100671. [DOI] [PubMed] [Google Scholar]
- 6.Bergthorsson U, Adams KL, Thomason B, Palmer JD. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424(6945):197–201. doi: 10.1038/nature01743. [DOI] [PubMed] [Google Scholar]
- 7.Rice DW, et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science. 2013;342(6165):1468–1473. doi: 10.1126/science.1246275. [DOI] [PubMed] [Google Scholar]
- 8.Mower JP, Stefanović S, Young GJ, Palmer JD. Plant genetics: Gene transfer from parasitic to host plants. Nature. 2004;432(7014):165–166. doi: 10.1038/432165b. [DOI] [PubMed] [Google Scholar]
- 9.Davis CC, Anderson WR, Wurdack KJ. Gene transfer from a parasitic flowering plant to a fern. Proc Biol Sci. 2005;272(1578):2237–2242. doi: 10.1098/rspb.2005.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stegemann S, Bock R. Exchange of genetic material between cells in plant tissue grafts. Science. 2009;324(5927):649–651. doi: 10.1126/science.1170397. [DOI] [PubMed] [Google Scholar]
- 11.Thyssen G, Svab Z, Maliga P. Cell-to-cell movement of plastids in plants. Proc Natl Acad Sci USA. 2012;109(7):2439–2443. doi: 10.1073/pnas.1114297109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stegemann S, Keuthe M, Greiner S, Bock R. Horizontal transfer of chloroplast genomes between plant species. Proc Natl Acad Sci USA. 2012;109(7):2434–2438. doi: 10.1073/pnas.1114076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlsson J, Leino M, Sohlberg J, Sundström JF, Glimelius K. Mitochondrial regulation of flower development. Mitochondrion. 2008;8(1):74–86. doi: 10.1016/j.mito.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25(6):989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 15.Maliga P, Svab Z. Engineering the plastid genome of Nicotiana sylvestris, a diploid model species for plastid genetics. Methods Mol Biol. 2011;701:37–50. doi: 10.1007/978-1-61737-957-4_2. [DOI] [PubMed] [Google Scholar]
- 16.Logan DC. Mitochondrial fusion, division and positioning in plants. Biochem Soc Trans. 2010;38(3):789–795. doi: 10.1042/BST0380789. [DOI] [PubMed] [Google Scholar]
- 17.Chase CD. Cytoplasmic male sterility: A window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 2007;23(2):81–90. doi: 10.1016/j.tig.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Hanson MR, Bentolila S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell. 2004;16(Suppl):S154–S169. doi: 10.1105/tpc.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 20.Ehlers K, Kollmann R. Primary and secondary plasmodesmata: Structure, origin, and functioning. Protoplasma. 2001;216(1-2):1–30. doi: 10.1007/BF02680127. [DOI] [PubMed] [Google Scholar]
- 21.Melnyk CW, Schuster C, Leyser O, Meyerowitz EM. A developmental framework for graft formation and vascular reconnection in Arabidopsis thaliana. Curr Biol. 2015;25(10):1306–1318. doi: 10.1016/j.cub.2015.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medgyesy P, Fejes E, Maliga P. Interspecific chloroplast recombination in a Nicotiana somatic hybrid. Proc Natl Acad Sci USA. 1985;82(20):6960–6964. doi: 10.1073/pnas.82.20.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheahan MB, McCurdy DW, Rose RJ. Mitochondria as a connected population: Ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J. 2005;44(5):744–755. doi: 10.1111/j.1365-313X.2005.02561.x. [DOI] [PubMed] [Google Scholar]
- 24.Arimura S, Yamamoto J, Aida GP, Nakazono M, Tsutsumi N. Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc Natl Acad Sci USA. 2004;101(20):7805–7808. doi: 10.1073/pnas.0401077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boeshore ML, Lifshitz I, Hanson MR, Izhar S. Novel composition of mitochondrial genomes in Petunia somatic hybrids derived from cytoplasmic male sterile and fertile plants. Mol Gen Genet. 1983;190:459–467. [Google Scholar]
- 26.Belliard G, Vedel F, Pelletier G. Mitochondrial recombination in cytoplasmic hybrids of Nicotiana tabacum by protoplast fusion. Nature. 1979;281:401–403. [Google Scholar]
- 27.Sanchez-Puerta MV, Zubko MK, Palmer JD. Homologous recombination and retention of a single form of most genes shape the highly chimeric mitochondrial genome of a cybrid plant. New Phytol. 2015;206(1):381–396. doi: 10.1111/nph.13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preuten T, et al. Fewer genes than organelles: Extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J. 2010;64(6):948–959. doi: 10.1111/j.1365-313X.2010.04389.x. [DOI] [PubMed] [Google Scholar]
- 29.Mower JP. PREP-Mt: Predictive RNA editor for plant mitochondrial genes. BMC Bioinformatics. 2005;6:96. doi: 10.1186/1471-2105-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshimi M, et al. Variations in the structure and transcription of the mitochondrial atp and cox genes in wild Solanum species that induce male sterility in eggplant (S. melongena) Theor Appl Genet. 2013;126(7):1851–1859. doi: 10.1007/s00122-013-2097-6. [DOI] [PubMed] [Google Scholar]
- 31.Davila JI, et al. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 2011;9:64. doi: 10.1186/1741-7007-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen JO, et al. Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics. 2007;177(2):1173–1192. doi: 10.1534/genetics.107.073312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuentes I, Stegemann S, Golczyk H, Karcher D, Bock R. Horizontal genome transfer as an asexual path to the formation of new species. Nature. 2014;511(7508):232–235. doi: 10.1038/nature13291. [DOI] [PubMed] [Google Scholar]
- 34.Niazi AK, et al. Targeting nucleic acids into mitochondria: Progress and prospects. Mitochondrion. 2013;13(5):548–558. doi: 10.1016/j.mito.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Dawson RF. Accumulation of nicotine in reciprocal grafts of tomato and tobacco. Am J Bot. 1942;29(1):66–71. [Google Scholar]
- 36.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8(19):4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moon HS, Nicholson JS, Lewis RS. Use of transferable Nicotiana tabacum L. microsatellite markers for investigating genetic diversity in the genus Nicotiana. Genome. 2008;51(8):547–559. doi: 10.1139/G08-039. [DOI] [PubMed] [Google Scholar]
- 38.Bindler G, et al. A high density genetic map of tobacco (Nicotiana tabacum L.) obtained from large scale microsatellite marker development. Theor Appl Genet. 2011;123(2):219–230. doi: 10.1007/s00122-011-1578-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kittiwongwattana C, Lutz K, Clark M, Maliga P. Plastid marker gene excision by the phiC31 phage site-specific recombinase. Plant Mol Biol. 2007;64(1-2):137–143. doi: 10.1007/s11103-007-9140-4. [DOI] [PubMed] [Google Scholar]
- 40.Gurdon C, Maliga P. Two distinct plastid genome configurations and unprecedented intraspecies length variation in the accD coding region in Medicago truncatula. DNA Res. 2014;21(4):417–427. doi: 10.1093/dnares/dsu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simpson JT, et al. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009;19(6):1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32(Web Server issue):W273-9. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;11(1110) doi: 10.1002/0471250953.bi1110s43. :1, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green M, Sambrook J. Molecular Cloning: A Laboratory Manual. 4th Ed Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2012. [Google Scholar]
- 48.Sugiyama Y, et al. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: Comparative analysis of mitochondrial genomes in higher plants. Mol Genet Genomics. 2005;272(6):603–615. doi: 10.1007/s00438-004-1075-8. [DOI] [PubMed] [Google Scholar]
- 49.Lohse M, Drechsel O, Kahlau S, Bock R. OrganellarGenomeDRAW: A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41(Web Server issue):W575–581. doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.