Significance
The ensemble interactions between the human mismatch repair (MMR) components during mismatch-dependent DNA excision repair remain poorly characterized. We have detailed these interactions in real time to reveal new dynamic collaborations between the four evolutionarily conserved components that ultimately result in a complete MMR event. Our observations demonstrate the stochastic nature of an essential genome maintenance system that ultimately results in robust repair events.
Keywords: Lynch syndrome/HNPCC, single molecule, MSH2–MSH6, MLH1–PMS2, EXOI
Abstract
Mismatch repair (MMR) is activated by evolutionarily conserved MutS homologs (MSH) and MutL homologs (MLH/PMS). MSH recognizes mismatched nucleotides and form extremely stable sliding clamps that may be bound by MLH/PMS to ultimately authorize strand-specific excision starting at a distant 3′- or 5′-DNA scission. The mechanical processes associated with a complete MMR reaction remain enigmatic. The purified human (Homo sapien or Hs) 5′-MMR excision reaction requires the HsMSH2–HsMSH6 heterodimer, the 5′ → 3′ exonuclease HsEXOI, and the single-stranded binding heterotrimer HsRPA. The HsMLH1–HsPMS2 heterodimer substantially influences 5′-MMR excision in cell extracts but is not required in the purified system. Using real-time single-molecule imaging, we show that HsRPA or Escherichia coli EcSSB restricts HsEXOI excision activity on nicked or gapped DNA. HsMSH2–HsMSH6 activates HsEXOI by overcoming HsRPA/EcSSB inhibition and exploits multiple dynamic sliding clamps to increase tract length. Conversely, HsMLH1–HsPMS2 regulates tract length by controlling the number of excision complexes, providing a link to 5′ MMR.
Mismatch repair (MMR) is a highly conserved strand-specific excision-resynthesis process that corrects nucleotide misincorporation errors during replication and nucleotide mismatches arising from recombination between heteroallelic parents or physical damage to the DNA (for review see ref. 1). Mutation of core MMR components results in elevated mutation rates and susceptibility to a variety of cancers (2).
MMR has been reconstituted with purified Escherichia coli, Saccharomyces cerevisae, and human proteins (3–6). The core MutS homologs (MSH) and MutL homologs (MLH/PMS) components direct a strand-specific excision reaction, whereas resynthesis appears to be uniquely performed by the replicative polymerase complex (1). In all organisms the excision process is initiated at a single-strand DNA scission (ssDNA/S) that may be located either 3′ or 5′ and hundreds to thousands of base pairs distant from the mismatch (4, 7). An ssDNA/S positioned on the newly replicated strand ensures accurate correction of replication misincorporation errors (1).
Excision directionality in γ-proteobacteria (E. coli) is linked to the choice of 3′ or 5′ exonucleases that specifically degrade ssDNA generated by the EcUvrD helicase in concert with EcMutS and EcMutL (1). The lack of a helicase distinguishes yeast and human MMR from γ-proteobacteria. Moreover, the eukaryotic 3′- and 5′-excision reactions require different core MMR components and likely occur by different mechanisms (1). For example, the 3′-MMR excision requires the replicative processivity factor PCNA to activate a cryptic MLH/PMS endonuclease activity (8), whereas 5′ MMR uses the only known MMR exonuclease EXOI (3, 5, 6). Unlike the E. coli ssDNA exonucleases, EXOI will initiate 5′ excision from a ssDNA/S in the absence of a helicase (9). Whereas the purified 5′-MMR reaction does not require MLH/PMS or PCNA, complementation studies with cellular extracts displayed a substantial requirement for MLH/PMS (10, 11).
A number of models have been proposed to account for the transmission of mismatch recognition to the ssDNA/S (12) as well as the roles of MMR components in the ensuing excision process (1, 13). However, the ensemble functions of the MMR components during excision in all organisms remain largely unknown. We have applied several single-molecule imaging techniques to visualize the complete human 5′ MMR strand excision process in real time. Our results suggest that dynamic and stochastic processes ultimately control 5′ excision, which may at least partially explain the different factor requirements in crude and purified reactions.
Results
Exonuclease Activity by a Single HsEXOI.
Single molecule flow stretching (smFS) was used to introduce a regulated laminar flow drag force (FD) onto a superparamagnetic (SPM) bead tethered to a flow-cell surface with DNA (SI Appendix, Fig. S1) (14, 15). At an applied FD of 2.5 pN (0.0125 mL/min) we found that a 21.8-kb double-stranded DNA (dsDNA) was almost fully extended across the flow-cell surface (LdsDNA = 7,204 nm), whereas an equivalent denatured 21.8-kilo-nucleotide (knt) single-stranded DNA (ssDNA) remained coiled (LssDNA = 417 nm; SI Appendix, Fig. S1 A and B). This sizeable force–extension difference is consistent with previous work and was used to monitor the production of ssDNA from dsDNA during MMR excision (16).
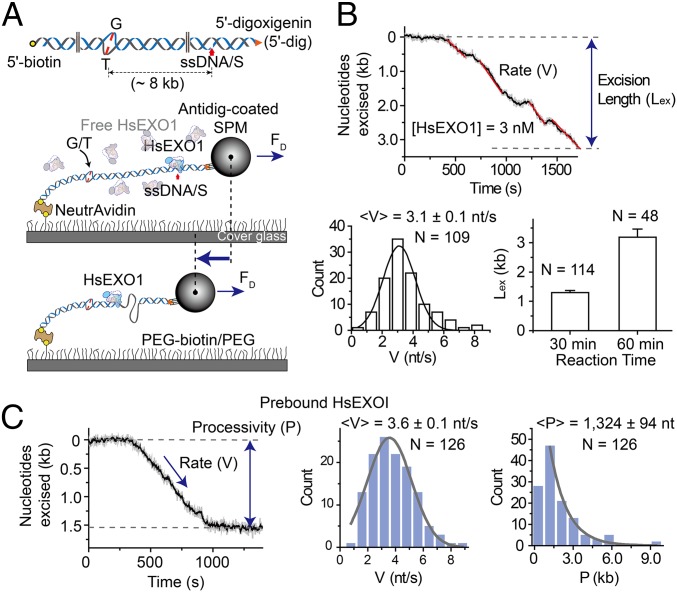
A 15.3-kb DNA substrate was constructed that containing a G/T mismatch and an ssDNA/S located 8 kb on the 5′ side of the mismatch (Fig. 1A; SI Appendix, Table S1). The 5′ ends of this linear DNA substrate were effectively blocked, one with 5′ biotin bound to the surface and the other with by 5′ dig-SPM bead, ensuring that 5′ excision was nearly always initiated at the internal ssDNA/S. A real-time decrease in the dsDNA extension was observed following the addition of Homo sapien (Hs)EXOI (Fig. 1B) (14, 15). The rate appeared relatively constant and corresponds to steady-state 5′-exonuclease activity (3.1 ± 0.1 nt/s; Fig. 1B) that ultimately resulted in an average excision of 1,301 ± 71 nt after 30 min and 3,188 ± 277 nt after 60-min incubations (Fig. 1B; for conversion of nanometers to base pairs, see SI Appendix, Fig. S1). Whereas an examination of steady-state excision scans appeared to reveal pauses (Fig. 1B), both the time and spatial resolution of the smFS system were insufficient to accurately establish any connections to HsEXOI turnover kinetics or excision intermediates.
Fig. 1.
The rate and processivity of HsEXOI exonuclease activity. (A, Top) Illustration of the 15.3-kb DNA substrate containing a G/T mismatch. A single-stranded DNA scission (ssDNA/S) that is located 8 kb 5′ of the mismatch was introduced using Nb.BbvC1 (New England Biolabs). (Middle and Bottom) Schematic illustration of the smFS system to visualize real-time DNA excision by the HsEXOI exonuclease. (B, Top) Representative time trajectory of the SPM bead tethered to the 15.3-kb mismatched DNA during a steady-state HsEXOI excision reaction. The rate (V, red) and excision length (Lex) are shown. (Middle Left) Histogram of binned excision rates that were fit to a Gaussian curve to derive the average rate (<V>). (Middle Right) The average excision length following a 30- and 60-min reaction. (C, Left) Representative time trajectory of the SPM bead tethered to the 15.3-kb mismatched DNA during a single HsEXOI excision event (see text). The rate (V) and processivity (P) are shown. (Middle) Histogram of binned excision rates that were fit to a Gaussian curve to derive the average rate (<V>). (Right) Histogram of binned excision processivity that were fit to a single exponential decay to derive the average processivity (<P>). All error bars and N indicate SE and the number of molecules, respectively.
Previous studies have demonstrated that in presence of Ca2+ cations, HsEXOI remains stably bound to a single-strand scission without catalyzing DNA excision (9, 17). This property was used to examine the excision kinetics of a single HsEXOI exonuclease event by prebinding the protein in the presence of Ca2+ (5 mM) and then initiating the excision reaction by introducing Mg2+ (5 mM) into the flow cell (Fig. 1C). Whereas the catalytic excision rate from single HsEXOI events was not significantly different from the steady-state rate (3.6 ± 0.1 nt/s; Fig. 1C), the average processivity (excision tract length) was 1,324 ± 94 nt (Fig. 1C).
HsRPA Inhibits HsEXOI by Binding to a Nascent Excision Gap.
The eukaryotic ssDNA binding heterotrimeric protein RPA is required for MMR (18). To examine the role of HsRPA during 5′-MMR excision, we first needed to establish whether the smFS system could resolve dsDNA from HsRPA-bound ssDNA (ssDNAHsRPA). We found that saturating HsRPA (>10 nM) extended the 21.8-knt ssDNA to 60% of the length of the corresponding dsDNA (4,200 nm; SI Appendix, Figs. S1 and S2), which was insufficient to clearly resolve exonuclease activity in real time.
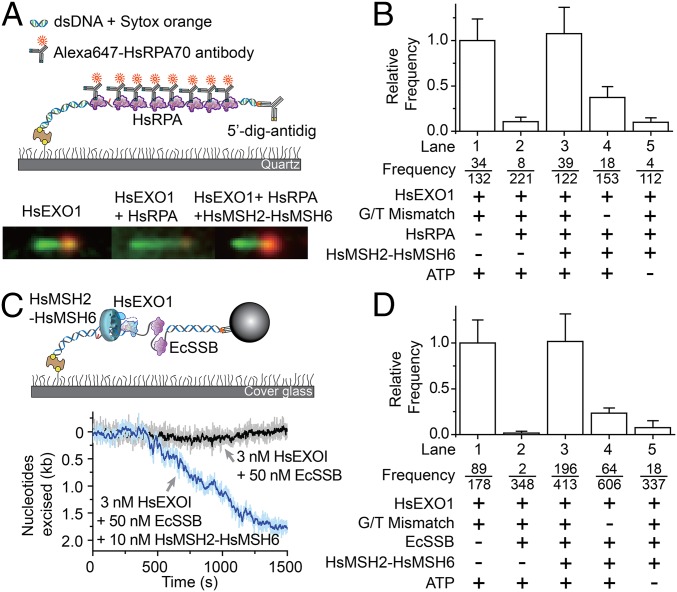
To overcome this technical issue, a static single-molecule total internal reflection fluorescence (smTIRF) analysis was developed with the identical DNA substrate used in the smFS system (Fig. 2A, Top). In this system, the production of ssDNA was visualized by localizing bound HsRPA with Alexa647-conjugated anti-HsRPA70 antibody (Alexa647-HsRPA) and the duplex region was imaged using the dsDNA specific Sytox Orange fluorescent dye. Although not real time, the formation of ssDNA by HsEXOI (alone) was easily observed as a shortening of the Sytox Orange-stained material (green) associated with intense foci that denote coiled ssDNA excision tracts bound by Alexa647-HsRPA added after the excision reaction was terminated (orange; Fig. 2A, Bottom Left). When HsRPA was added together with HsEXOI, we observed small low-intensity Alexa647 foci on numerous DNA molecules (Fig. 2A, Bottom Middle and SI Appendix, Fig. S3, lane 5). We interpret these foci to represent brief ssDNA excision events that by nature are bound by fewer Alexa647-HsRPA. To quantify the frequency of sustained excision events that resulted in long ssDNA tracts with intense Alexa647-HsRPA foci, we first determined the average Alexa647-HsRPA staining intensity of the brief ssDNA excision events produced when HsEXOI and HsRPA were included together (SI Appendix, Fig. S3, lane 5). The number of events that equaled or exceeded one SD above this average intensity (SI Appendix, Fig. S3, red dotted line) relative to the total number of Sytox Orange-stained DNA molecules was then determined for each excision reaction condition (Fig. 2B, see “frequency”). The frequency of sustained ssDNA excision events under these reaction conditions was then normalized to the frequency of sustained ssDNA excision events when HsEXOI was added alone (frequency in lane 1) to obtain the relative frequency (Fig. 2B and SI Appendix, SI Methods). The results clearly indicate that HsRPA dramatically inhibits sustained excision by HsEXOI (Fig. 2B). These observations are similar to bulk studies of human 5′-MMR excision (5), but appear to contrast recombination end-processing studies with the S. cerevisae homologs (19).
Fig. 2.
Reconstitution of 5′-mismatch repair excision on single DNA molecules. (A, Top) Schematic illustration of the static single molecule total internal reflection fluorescence (smTIRF) system used to visualize the 5′-MMR excision process (Methods and SI Appendix, SI Methods). (Bottom Left) Representative molecule following incubation with HsEXOI alone. (Bottom Middle) Representative molecule following incubation with HsEXOI in the presence of HsRPA. (Bottom Right) Representative molecule following incubation with HsEXOI, HsRPA, and HsMSH2–HsMSH6 in the presence of ATP. (B) The relative frequency (±SE) of HsEXOI excision events in the presence of various MMR components determined by smTIRF (3 nM HsEXOI, 10 nM HsMSH2–HsMSH6, 10 nM HsRPA). Frequency is the ratio of the number of corrected excision events (see text and SI Appendix, SI Methods) to the total number of Sytox Orange-stained dsDNA molecules. (C, Top) Schematic Illustration of the real-time smFS system to examine the complete 5′-MMR reaction. (Bottom) Representative time trajectories of SPM beads tethered to the 15.3-kb mismatched DNA. (D) The relative frequency (±SE) of HsEXOI excision events in the presence of various MMR components determined by smFS (Methods). Frequency is the ratio of the number of excision events to the total number of bead-tethered DNA molecules.
There are at least two mechanisms that HsRPA might inhibit HsEXOI excision activity. HsRPA may interfere with HsEXOI binding to the strand scission or HsRPA might bind to a short HsEXOI excision gap ultimately inhibiting the exonuclease. To test these hypotheses, we used single-molecule Förster resonance energy transfer (smFRET) (20, 21). A 73-bp DNA containing a G/T mismatch and a ssDNA/S was attached to a flow-cell surface via a 5′-biotin–NeutrAvidin linkage (SI Appendix, Fig. S4A). The ssDNA/S was located between a Cy3 donor and Cy5 acceptor pair (SI Appendix, Fig. S4A). HsEXOI 5′ → 3′ exonuclease activity beginning at the ssDNA/S will form an ssDNA gap that spontaneously coils placing the Cy3 in proximity for FRET with Cy5 (SI Appendix, Fig. S4 A and B). We observed no difference in the relative frequency of HsEXOI excision-induced FRET in the absence of HsRPA or following preincubation with HsRPA (SI Appendix, Fig. S4C). These results suggest that HsRPA does not inhibit HsEXOI entry or exonuclease activity initiated at a 5′-strand scission.
We examined HsEXOI activity on a substrate DNA containing a mismatch with a recessed 5′ end and a 3′-ssDNA tail (30 nt) that would be a model for a nascent excision gap (SI Appendix, Fig. S4D). A Cy3 donor was located within the dsDNA 15 bp from the recessed 5′ end and a Cy5 acceptor was located on the 3′-ssDNA tail 5 nt from the ssDNA/dsDNA junction. To prevent release of the internal Cy3 by HsEXOI exonuclease activity, two sequential phosphorothioate linkages were introduced into the phosphate backbone (SI Appendix, Fig. S4D). In the absence of HsRPA, we observed ∼17% of the DNA molecules transitioned to a steady high FRET state (Elow = 0.3 → Ehigh = 0.5; SI Appendix, Fig. S4 D and E), indicating HsEXOI-mediated excision of the 15 nt from the recessed 5′ end to the phosphorothioate linkages. In contrast, when HsRPA was preassembled onto the ssDNA tail, we observed only 0.5% of the DNA molecules transition to high FRET (relative frequency = 0.03; SI Appendix, Fig. S4 E and F). These results suggest that HsEXOI is substantially inhibited when HsRPA binds to the ssDNA gap adjacent to the 5′ end that would serve as an entry point for exonuclease activity. Taken together with the small low intensity Alexa647-HsRPA foci in the smTIRF studies, these results suggest that HsEXOI may begin excision from any 5′-strand scission, but is halted when HsRPA binds to the resulting nascent excision gap.
Reconstitution of the Human MMR 5′-Excision Reaction on Single DNA Molecules.
The addition of HsMSH2–HsMSH6 with HsRPA and HsEXOI resulted in significant shortening of the dsDNA (Fig. 2A, Bottom Right) and nearly equivalent frequency of excision events as HsEXOI alone (Fig. 2A, Bottom Right; Fig. 2B, lane 3; and SI Appendix, Fig. S3, lane 4). HsMSH2–HsMSH6 stimulation of HsEXOI excision in the presence of HsRPA was dependent on a mismatch (Fig. 2B, lane 4 and SI Appendix, Fig. S3, lane 6) and ATP (Fig. 2B, lane 5 and SI Appendix, Fig. S3, lane 7). The observation that HsMSH2–HsMSH6, HsEXOI, and HsRPA catalyze a mismatch-dependent 5′-excision reaction is similar to previous studies (3, 5, 6) and suggests that we have fully reconstituted 5′ MMR on single DNA molecules.
Previous studies have suggested that EcSSB may substitute for HsRPA in the human 5′-MMR excision reaction (5). We could not directly compare EcSSB and HsRPA in the smTIRF system because a specific EcSSB antibody does not exist. However, we determined that the 21.8-knt ssDNA was extended 2,013 nm in the presence of saturating EcSSB (50 nM), resulting in a conversion factor of 4.2 nt/nm for the dsDNA → ssDNAEcSSB transition that was well within the resolution of smFS (SI Appendix, Fig. S5). The significant differences in extension when ssDNA is bound by EcSSB compared with HsRPA likely reflects distinct binding mechanisms; whereas the EcSSB tetramer fully wraps ∼65 nt of ssDNA in a “baseball seam” structure (22), HsRPA merely bends ∼30 nt of ssDNA (23). Including EcSSB with HsEXOI completely impeded the contraction of mismatched DNA in the smFS system (Fig. 2 C and D, compare lanes 1 and 2), confirming that EcSSB inhibits HsEXOI exonuclease activity similar to HsRPA (Fig. 2 B and D) (5). The addition of HsMSH2–HsMSH6 overcomes the EcSSB inhibition of HsEXOI, resulting in the real-time observation of 5′ MMR (Fig. 2 C and D, lane 3). The reaction was dependent on a mismatch and ATP (Fig. 2D, lanes 4 and 5) underlining the strong functional similarity between EcSSB and HsRPA in 5′ MMR (compare Fig. 2B and 2D).
HsEXOI Excision Is Dynamic and Controlled by Multiple HsMSH2–HsMSH6 Sliding Clamps.
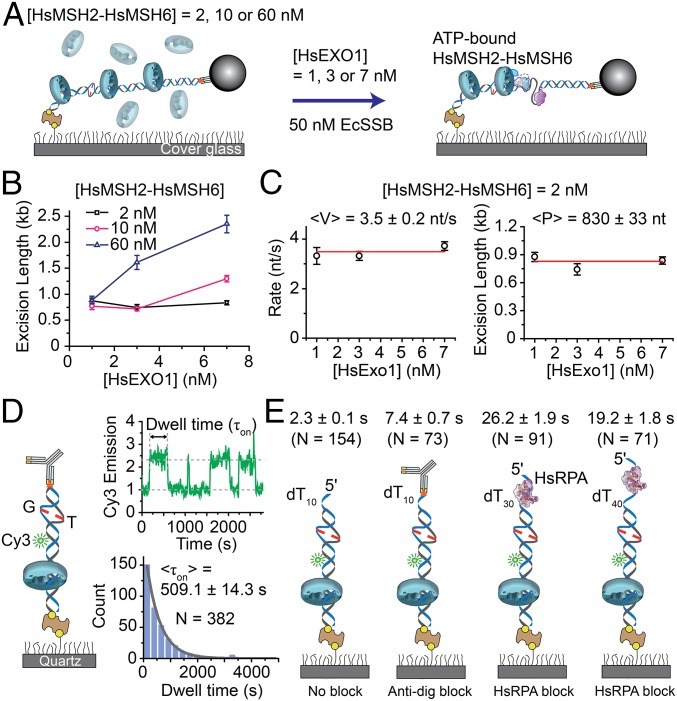
To examine the MMR mechanics, we first determined the concentration-dependent number of HsMSH2–HsMSH6 sliding clamps bound to a single mismatched DNA using mEos3.2–HsMSH2–HsMSH6 that displayed wild-type activity (SI Appendix, Table S2 and Fig. S6). The results suggested that at 2 nM, nearly 90% of the DNA molecules contained a single HsMSH2–HsMSH6 (SI Appendix, Fig. S6). In contrast, at 60 nM, nearly 70% of the DNA molecules contained two to four HsMSH2–HsMSH6 (SI Appendix, Fig. S6). For comparison, the cellular concentration of HsMSH2–HsMSH6 has been estimated to be ∼250 nM (24). All of the HsMSH2–HsMSH6 proteins appeared to diffuse randomly on the mismatched DNA consistent with ATP-bound sliding clamps as described previously (20, 25–27).
We examined the interplay between HsMSH2–HsMSH6 and HsEXOI during 5′-MMR excision (Fig. 3A) and found that the excision length did not vary over a range of HsEXOI concentrations in the presence of 2 nM preassembled HsMSH2–HsMSH6 (Fig. 3B). However, in the presence of 10 nM or 60 nM preassembled HsMSH2–HsMSH6 the excision length significantly increased with increasing HsEXOI (Fig. 3B). These results suggest that increasing the number of HsMSH2–HsMSH6 sliding clamps is correlated with increased total excision length. The rate of 5′-MMR excision at a concentration of HsMSH2–HsMSH6 (2 nM) where most events would be due to a single sliding clamp appeared nearly identical to mismatch-independent HsEXOI excision (3.5 ± 0.2 nt/s; Fig. 3C). This result suggests that HsMSH2–HsMSH6 does not influence the fundamental HsEXOI catalytic process. Interestingly, the excision processivity of the HsMSH2–HsMSH6/HsEXOI complex appeared nearly twofold shorter than HsEXOI alone (830 ± 33 nt; Fig. 3C).
Fig. 3.
Multiple HsMSH2–HsMSH6 sliding clamps enhance HsEXOI excision. (A) Schematic illustration of the single molecule analysis of HsMSH2–HsMSH6 preassembled on mismatched DNA, followed by infusion of HsEXOI in the presence EcSSB. The HsMSH2–HsMSH6 concentration determines the number of sliding clamps loaded on the mismatched DNA (SI Appendix, Fig. S6). (B) The dependence of excision length (±SE) on HsEXOI concentration in the presence of different HsMSH2–HsMSH6 concentrations. The excision lengths were obtained from the analysis of ∼32–160 molecules. (C) The dependence of 5′-MMR excision rate (<V> ± SE) and processivity (<P > ± SE) on HsEXOI concentration in the presence of HsMSH2–HsMSH6 (2 nM) where 88% of the single mismatched DNA molecules contain a single sliding clamp. n = 32, 34, and 126 for 1 nM, 3 nM, and 7 nM HsEXOI. (D, Left) Schematic illustration of the smPIFE 40-bp mismatched DNA substrate (SI Appendix, Table S2). (Right Top) Representative time trace of HsMSH2–HsMSH6 lifetime (τon) analysis. (Right Bottom) Binned histogram fit with a single exponential decay to determine the lifetime (<τon> ± SE) of HsMSH2–HsMSH6 on tightly blocked-end mismatch DNA. (E) Average smPIFE lifetime (<τon>) for HsMSH2–HsMSH6 on various illustrated 40-bp mismatched DNA substrates (SI Appendix, Table S2). The DNA substrates used are (left to right): oligo-dT10 tail; oligo-dT10 tail containing 5′ dig-antidig blocked end; oligo-dT30 tail bound with HsRPA (20 nM); and oligo-dT40 tail bound with HsRPA (20 nM). Dwell times are shown above each illustration (±SE).
One possible explanation for the reduced excision processivity is a decreased dynamic lifetime of the ATP-bound HsMSH2–HsMSH6 sliding clamps. We used single molecule protein-induced fluorescence enhancement (smPIFE) to examine the lifetime of HsMSH2–HsMSH6 sliding clamps on mismatched DNA (Fig. 3D) (28). A 40-bp Cy3-labeled DNA containing a G/T mismatch was attached at the proximal end to the flow-cell surface via a 5′-biotin–NeutrAvidin linkage, whereas the distal end was blocked with a 5′ dig-antidig that traps freely diffusing ATP-bound MSH sliding clamps (Fig. 3D and SI Appendix, Table S3) (20). Time-averaged diffusion of trapped HsMSH2–HsMSH6 sliding clamps on this relatively short DNA produced a PIFE signal that was used to determine dwell time (τon). We found that the lifetime of HsMSH2–HsMSH6 on tightly blocked-end mismatched DNA (τon = 509.1 ± 14.3 s; Fig. 3D) was at least twofold longer than the time required for an average excision tract (830 nt ÷ 3.5 nt/s = 237 s; Fig. 3 C and D). In contrast, HsMSH2–HsMSH6 sliding clamps rapidly dissociated from either unblocked mismatched DNA with blunt end or unblocked mismatched DNA containing an oligo-dT10 ssDNA tail (τon = 2.3 ± 0.1 s; Fig. 3E). The dissociation kinetics was only modestly reduced when the oligo-dT10 ssDNA tail was blocked with dig-antidig (τon = 7.4 ± 0.7 s; Fig. 3E). These observations appear similar to Thermus aquaticus TaMutS (20) and suggest that HsMSH2–HsMSH6 rapidly dissociates from mismatched DNA when it encounters a ssDNA/dsDNA junction.
We speculated that HsMSH2–HsMSH6 sliding clamps might be destabilized in the absence of a fully protected ssDNA gap. HsRPA displays a high affinity 30 nt ssDNA binding mode (29), although structural analysis suggests that only 25 nt are actually bound (23). We found that mismatched DNA containing an oligo-dT30 ssDNA tail prebound by HsRPA significantly increased the lifetime of HsMSH2–HsMSH6 (τon = 26.2 ± 1.9 s) compared with unblocked dT10 or dig-antidig blocked dT10 (Fig. 3E). Increasing the ssDNA tail with oligo-dT40 that is substantially larger than the HsRPA footprint further decreased the lifetime of HsMSH2–HsMSH6 (τon = 19.2 ± 1.8 s; Fig. 3E). Taken as a whole, these observations are consistent with the conclusion that HsMSH2–HsMSH6 stabilizes HsEXOI on mismatched DNA allowing it to catalyze 5′ excision. However, any encounter with an exposed ssDNA gap reduces the lifetime of the HsMSH2–HsMSH6 sliding clamp.
HsMLH1–HsPMS2 Regulates HsMSH2–HsMSH6/HsEXOI Excision.
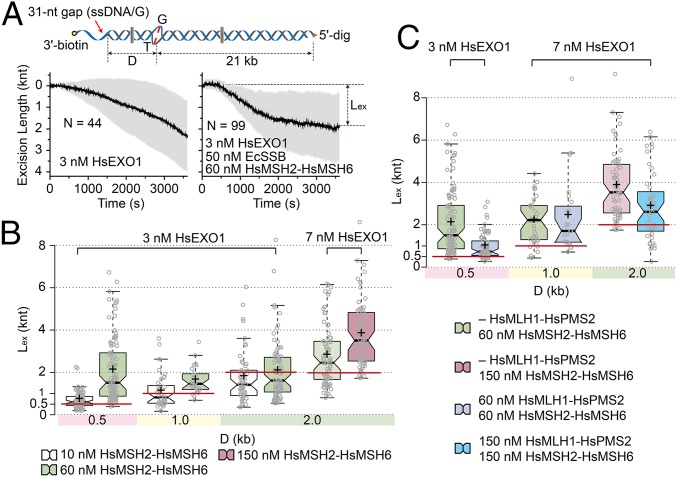
To examine the relationship between the excision tract length and the mismatch location, we constructed a series of single molecule substrates containing a 31-nt ssDNA gap (ssDNA/G) at variable distances (D) from the mismatch (Fig. 4A, Top). Excision by HsEXOI (3 nM) alone appeared relatively continuous over 60 min regardless of a mismatch or its position relative to the ssDNA/G (Fig. 4A, Bottom Left; D = 0.5 kb). The addition of EcSSB dramatically reduced the frequency of excision events nearly 70-fold relative to HsEXOI alone (relative frequency = 1.5%). HsMSH2–HsMSH6 overcomes EcSSB HsEXOI inhibition and the SPM bead approaches an asymptotic maximum excision length (Lex; Fig. 4A, Bottom Right, D = 0.5 kb).
Fig. 4.
The 5′-MMR excision is enhanced by HsMSH2–HsMSH6 and down-regulated by HsMLH1–HsPMS2. (A, Top) Schematic illustration of 21 kb mismatched DNA substrate containing a DNA extension that places a ssDNA 5′ gap at a distance (D) of 0.5, 1.0, and 2.0 kb from the mismatch. (Bottom Left) Time trajectories of the average (black) and SD (gray) during 5′ excision by HsEXOI (3 nM) alone on the 21 kb mismatched DNA with D = 0.5 kb. (Bottom Right) Time trajectories of the average (black) and SD (gray) during 5′ excision on the 21 kb mismatched DNA with D = 0.5 kb. (B) Box plots of the excision length (Lex; knt) vs. D (kb) in the presence of 50 nM EcSSB (from left n = 78, 96, 38, 28, 49, 81, 74, and 64). Concentrations of HsEXOI and box plot colors indicating HsMSH2–HsMSH6 concentration are shown above. (C) Box plots of the excision length (Lex; knt) vs. D (kb) in the presence of 50 nM EcSSB (from left n = 96, 67, 34, 23, 64, and 43). Concentrations of HsEXOI are indicated above plots. Box plot colors indicating HsMSH2–HsMSH6 and HsMLH1–HsPMS2 concentration are shown below. The cross mark and the constriction line indicates the mean and the median, respectively. Solid extensions above and below the median indicate upper and lower quartiles, respectively. Red lines indicate the distance between the mismatch and ssDNA/G.
Using a box plot, we displayed the Lex median (constriction between quartiles), the mean (+), 1.5 times the interquartile range from the first and third quartiles (filled colors) and all Lex observations as whiskers. Importantly, the Lex was dependent on HsMSH2–HsMSH6 concentration but not the distance between the ssDNA/G and the mismatch (Fig. 4B). For example, slightly more than 50% with 10 nM HsMSH2–HsMSH6, and nearly all of excision tracts with 60 nM HsMSH2–HsMSH6, terminated past the mismatch on the DNA substrate containing D = 0.5 kb (Fig. 4B). A similar trend was observed when the ssDNA/G was 1.0 or 2.0 kb from the mismatch, except that in most cases, the excision tract terminated before the mismatch (Fig. 4B, D = 1.0 and 2.0 kb). Additional HsEXOI (7 nM) resulted in more than 50% of the excision tracts terminating past the mismatch with 60 nM HsMSH2–HsMSH6, which could be improved to 90% with 150 nM HsMSH2–HsMSH6. Under nearly all conditions, the excision length could exceed 2–3 times the distance past the mismatch as the distance between the 5′ ssDNA/G and the mismatch.
Introducing HsMLH1–HsPMS2 into the HsMSH2–HsMSH6/HsEXOI 5′-MMR excision reaction reduced the median length of the excision tracts under all conditions examined (Fig. 4C). In most cases, the excision tract terminated just past the mismatch (Fig. 4C). As a control, we found that HsMLH1–HsPMS2 did not activate HsEXOI in the absence of HsMSH2–HsMSH6 nor does a threefold excess of HsMLH1–HsPMS2 decrease the HsEXOI excision tract length activated by a single preassembled HsMSH2–HsMSH6 sliding clamp (SI Appendix, Fig. S7 and see Fig. 3C and SI Appendix, Fig. S6B). Another mechanism that is consistent with previous studies suggests that MLH/PMS might alter the association of MSH with the mismatched DNA (26, 30). We found that the number of mEos3.2–HsMSH2–HsMSH6 sliding clamps was significantly reduced in the presence of HsMLH1–HsPMS2 (SI Appendix, Fig. S8). Combined with studies of the yeast homologs that suggest that the stability of the ScMsh2–ScMsh6/ScMlh1–ScPms1 complex is not significantly different from ScMsh2–ScMsh6 sliding clamps alone (26), these observations are consistent with the hypothesis that HsMLH1–HsPMS2 controls runaway HsMSH2–HsMSH6/HsEXOI excision by stochastically regulating the number of HsMSH2–HsMSH6 sliding clamps associated with the mismatched DNA.
Discussion
We have examined the human 5′-MMR excision process in real time on single DNA molecules containing a G/T mismatch and a single-strand scission or gap. To our knowledge, this is the first reconstitution of a multicomponent DNA repair reaction on single DNA molecules. Several imaging techniques were applied to visualize the mechanics. Our observations expand previous bulk studies with yeast and human MMR proteins (3, 5, 6) by resolving the complex biophysical assemblies between HsRPA, HsEXOI, HsMSH2–HsMSH6, and HsMLH1–HsPMS2.
HsEXOI appeared active on any DNA substrate containing a 5′ single-stranded DNA scission or gap (5, 9, 17). HsRPA dramatically inhibited HsEXOI exonuclease activity (Fig. 2 A and B) (5). However, inhibition only occurred after a ssDNA gap was formed that could be bound by HsRPA (Fig. 2A, Bottom Middle and SI Appendix, Fig. S4). These results suggest a previously unknown very early role for HsRPA in regulating unfettered HsEXOI 5′ excision. In the presence of a mismatch and ATP, HsMSH2–HsMSH6 overcomes the HsRPA inhibition of HsEXOI (3, 5, 6). A surprising result was that EcSSB could fully substitute for HsRPA in all HsEXOI-mediated excision reactions (Figs. 2 C and D, 3, and 4). This observation strongly suggests that there are no explicit interactions between HsMSH2–HsMSH6 and HsEXOI with either HsRPA or EcSSB, and that the singular role of HsRPA during 5′ MMR is as an ssDNA binding protein. These studies appear to clearly explain why no MMR-deficient RPA mutations were identified in yeast mutator genetic screens (31).
The rate of excision by HsEXOI alone was virtually identical to the rate of excision by the HsMSH2–HsMSH6/HsEXOI complex (compare Fig. 1C and Fig. 3C). An attractive hypothesis is that ATP-bound MSH sliding clamps (20, 25, 26) provide a freely diffusible platform that stabilizes EXOI on the 5′ end of an ssDNA scission or gap via well-known protein–protein interaction domains (32). We envision a model where MSH sliding clamps function similarly to PCNA sliding clamps that tether the polymerase machinery to the replication fork (33). By counting the number of HsMSH2–HsMSH6 sliding clamps loaded onto a mismatched DNA molecule, we determined that a single HsMSH2–HsMSH6/HsEXOI complex processively degrades 830 nt during a single 5′-MMR excision event (Fig. 3C). This implies that the lifetime of the HsMSH2–HsMSH6/HsEXOI complex is ∼2 min shorter than HsEXOI alone. It is likely that this reduced lifetime is linked to transient exposure of the ssDNA gap during 5′ excision, which the PIFE studies showed decreased the lifetime of HsMSH2–HsMSH6 sliding clamps 10- to 100-fold depending on the extent of HsRPA occlusion (Fig. 3 D and E). We also note that a significantly decreased HsMSH2–HsMSH6 lifetime on a mismatched DNA containing an exposed ssDNA gap is an alternate explanation for the dramatically decreased MMR efficiency in the absence of HsRPA observed by Genschel et al. (5).
Whereas there is strong biochemical evidence that MSH proteins form multiple extremely stable sliding clamps on mismatch DNA in the presence of ATP (20, 25, 27), one could argue that these are anomalous structures that develop in the absence of additional MMR components. However, we found a near direct correlation between the HsEXOI-dependent 5′-MMR excision tract length (Figs. 3B and 4B) and numbers of HsMSH2–HsMSH6 sliding clamps observed on the mismatched DNA (SI Appendix, Fig. S6). Moreover, at any fixed concentration of HsEXOI, the average excision tract length appeared to be uniquely dependent on HsMSH2–HsMSH6 concentration and was largely independent of the ssDNA/G location relative to the mismatch (Fig. 4B). Together, these observations suggest a concentration-dependent loading of multiple HsMSH2–HsMSH6 sliding clamps that dynamically activate HsEXOI 5′ excision, and argue against different configurations, complexes, or functions of HsMSH2–HsMSH6 during its interaction with HsEXOI.
Previous studies have demonstrated that HsMLH1–HsPMS2 is not required for the 5′-MMR excision reaction (3, 5), but when added, appeared to alter excision termination past the mismatch (6). We found that the HsMSH2–HsMSH6/HsEXOI excision endpoints were shorter in the presence of equivalent HsMLH1–HsPMS2 physiological stoichiometries (Fig. 4C). These observations are consistent with the conclusion that HsMLH1–HsPMS2 inhibits runaway excision by the HsMSH2–HsMSH6/HsEXOI complex. Interestingly, when the mismatch and the ssDNA/G were separated by 2.0 kb, the addition of HsMLH1–HsPMS2 resulted in a significant fraction (40%) of excision tracts that did not reach the mismatch (Fig. 4B, D = 2.0 kb). This was despite relatively high concentrations of HsMSH2–HsMSH6 (150 nM) that should have loaded sufficient sliding clamps capable of activating HsEXOI to easily cover that distance. These results suggest that HsMLH1–HsPMS2 is unlikely to detect the mismatch as part of its excision-regulation role. We also found that the excision tract length of a single HsMSH2–HsMSH6/HsEXOI event was not reduced by HsMLH1–HsPMS2 (SI Appendix, Fig. S7). Instead, we showed that HsMLH1–HsPMS2 appears to reduce the number of dynamic excision events by stochastically modulating the number of HsMSH2–HsMSH6 sliding clamps capable of activating HsEXOI 5′ excision (SI Appendix, Fig. S8). Our data cannot rule out the possibilities that HsMLH1–HsPMS2 might regulate HsMSH2–HsMSH6 loading at the mismatch or titrate an excision component such as HsEXOI independent of the mismatch (32).
Taken together, these studies strongly support a dynamic model for MMR (1, 30) and effectively eliminate static or single-complex models (13, 34). A surprising observation was that even in the absence of a mismatch, HsEXOI efficiently converts a 5′-strand scission into a gap bound by HsRPA before its exonuclease activity is fully inhibited (Fig. 2A, Bottom Middle and SI Appendix, Fig. S4). There is compelling evidence that a combination of RNaseH, the flap endonuclease FEN1, and Polδ polymerase strand-displacement activity ultimately remove the RNA primer from the 5′ end of an Okazaki fragment (35). EXOI has been proposed to play at least a minor role in Okazaki fragment maturation because the rad27Δ(ScFen1) exoIΔ double mutant is lethal in yeast (36). An appealing model would envision HsRPA associated with a transient gap exposed during the removal of the RNA primer that functionally inhibits any potential HsEXOI excision (SI Appendix, Fig. S9A).
The molecular switch/sliding clamp model remains the most consistent mechanism for MMR excision (SI Appendix, Fig. S9) (1, 30). In the presence of a mismatch, multiple ATP-bound HsMSH2–HsMSH6 sliding clamps may be loaded onto the DNA that may then interact with HsMLH1–HsPMS2 (SI Appendix, Fig. S9B). The HsMSH2–HsMSH6 sliding clamp that is closest to the 5′ end initiates 5′ → 3′ excision by stabilizing HsEXOI (SI Appendix, Fig. S9B). A single HsMSH2–HsMSH6/HsEXOI complex generates an ∼800-nt excision tract, which is successively bound by HsRPA before the complex spontaneously dissociates (SI Appendix, Fig. S9C). The next closest HsMSH2–HsMSH6 sliding clamp then stabilizes another HsEXOI at the newly located 5′ end, reinitiating excision (SI Appendix, Fig. S9C). This animated process is proposed to be iterative until the mismatch is released and no additional HsMSH2–HsMSH6 sliding clamps may be loaded onto the DNA (SI Appendix, Fig. S9D). The regulatory role of HsMLH1–HsPMS2 in 5′-MMR excision is likely to be significantly different from its catalytic role on 3′-MMR excision where HsPCNA may activate its intrinsic endonuclease (8). However, the control of HsEXOI by HsMSH2–HsMSH6 and HsMLH1–HsPMS2 appears to provide a dynamic yin and yang for efficient 5′-MMR excision.
Methods
See SI Appendix, SI Methods for comprehensive methods.
Preparation of Proteins and DNA.
HsMSH2–HsMSH6 and HsRPA were purified as previously described (37, 38). HsEXOI and HsMLH1–HsPMS2 were purified with modifications to published methods (SI Appendix, SI Methods) (9, 11).
Single-Molecule Flow Stretching, Total Internal Reflection Fluorescence, and Protein-Induced Fluorescence Enhancement Microscopy.
A 15.3-kb DNA substrate containing a G/T mismatch was constructed as previously described (25). The construction of DNA substrates containing an ssDNA/G at various distances from the G/T mismatched is described in the SI Appendix, SI Methods.
For smFS microscopy, a custom flow chamber was constructed with mismatched DNA attached to the surface and the SPM bead as previously described (39). The SPM bead was imaged with a 10× objective (N.A. = 0.40, Olympus) containing ∼300-tethered DNA molecules in a field of view, recorded with a high-resolution CCD (RETIGA 2000R, Qimaging) using MetaVue (Molecular Devices) imaging software at 1-s time resolution, the bead position determined with high accuracy (∼20 nm) by DiaTrack 3.03 (40), and the data were analyzed using OriginPro8 (OriginLab) and Matlab 2013b (Mathworks).
The smTIRF microscopy analysis was performed as previously described (20, 25). Alexa647-labeled anti-HsRPA70 antibody (Abcam) was infused into the flow chamber following HsRPA (10 nM) incubation in reactions performed in the absence of HsRPA or directly after a 60-min reaction that contained HsRPA. Alexa647 emission was imaged after 10 min and then colocalized with dsDNA following Sytox Orange (Thermo Fisher Scientific) staining.
For smPIFE microscopy, 40 bp of G/T mismatched DNA molecules with various ssDNA tails (0, 10, 30, and 40 oligo-dT) were prepared by annealing paired DNAs (SI Appendix, Table S2 and SI Methods). All of the DNA substrates were labeled with Cy3 (Monofunctional NHS-ester, GE Healthcare), and attached to the flow cell surface via 5′-biotin–NeutrAvidin as previously described (20). DNAs with an oiligo-dT0 or oligo-dT10 tail contained a 5′ dig. Anti-dig antibody (50 nM; Roche) or HsRPA (20 nM) was introduced into the flow chamber followed by HsMSH2–HsMSH6. Emission of Cy3 was imaged by smTIRF as previously described (20, 25). The time resolution was adjusted ranging from 0.2 s to 2 s according to the lifetime of HsMSH2–HsMSH6 using a time-lapse method (SI Appendix, SI Methods). PIFE data were analyzed using IDL 6.4 (EXELIS VIS) and Matlab 2013b (Mathworks).
Supplementary Material
Acknowledgments
We thank Junghyo Jo for helpful discussion and Randal Soukup and Brooke Britton for help in the preparation of mEos3.2–HsMSH2–HsMSH6. This work was supported by NIH Grant CA67007 (to R.F.) and National Research Foundation of Korea Grant 2011-0013901 (to J.-B.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523748113/-/DCSupplemental.
References
- 1.Fishel R. Mismatch repair. J Biol Chem. 2015;290(44):26395–26403. doi: 10.1074/jbc.R115.660142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martín-López JV, Fishel R. The mechanism of mismatch repair and the functional analysis of mismatch repair defects in Lynch syndrome. Fam Cancer. 2013;12(2):159–168. doi: 10.1007/s10689-013-9635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowen N, et al. Reconstitution of long and short patch mismatch repair reactions using Saccharomyces cerevisiae proteins. Proc Natl Acad Sci USA. 2013;110(46):18472–18477. doi: 10.1073/pnas.1318971110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction. J Biol Chem. 2005;280(48):39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12(5):1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, et al. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122(5):693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 7.Grilley M, Griffith J, Modrich P. Bidirectional excision in methyl-directed mismatch repair. J Biol Chem. 1993;268(16):11830–11837. [PubMed] [Google Scholar]
- 8.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126(2):297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 9.Lee BI, Wilson DM., 3rd The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem. 1999;274(53):37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- 10.Geng H, et al. In vitro studies of DNA mismatch repair proteins. Anal Biochem. 2011;413(2):179–184. doi: 10.1016/j.ab.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li G-M, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc Natl Acad Sci USA. 1995;92(6):1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolodner RD, Mendillo ML, Putnam CD. Coupling distant sites in DNA during DNA mismatch repair. Proc Natl Acad Sci USA. 2007;104(32):12953–12954. doi: 10.1073/pnas.0705698104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 14.Lee JB, et al. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439(7076):621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 15.van Oijen AM, et al. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301(5637):1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 16.Bustamante C, Bryant Z, Smith SB. Ten years of tension: Single-molecule DNA mechanics. Nature. 2003;421(6921):423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 17.Orans J, et al. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145(2):212–223. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramilo C, et al. Partial reconstitution of human DNA mismatch repair in vitro: Characterization of the role of human replication protein A. Mol Cell Biol. 2002;22(7):2037–2046. doi: 10.1128/MCB.22.7.2037-2046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cannavo E, Cejka P, Kowalczykowski SC. Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc Natl Acad Sci USA. 2013;110(18):E1661–E1668. doi: 10.1073/pnas.1305166110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong C, et al. MutS switches between two fundamentally distinct clamps during mismatch repair. Nat Struct Mol Biol. 2011;18(3):379–385. doi: 10.1038/nsmb.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JB, et al. Single-molecule views of MutS on mismatched DNA. DNA Repair (Amst) 2014;20:82–93. doi: 10.1016/j.dnarep.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antony E, Weiland EA, Korolev S, Lohman TM. Plasmodium falciparum SSB tetramer wraps single-stranded DNA with similar topology but opposite polarity to E. coli SSB. J Mol Biol. 2012;420(4–5):269–283. doi: 10.1016/j.jmb.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan J, Pavletich NP. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012;26(20):2337–2347. doi: 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem. 1998;273(31):19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 25.Cho WK, et al. ATP alters the diffusion mechanics of MutS on mismatched DNA. Structure. 2012;20(7):1264–1274. doi: 10.1016/j.str.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorman J, et al. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc Natl Acad Sci USA. 2012;109(45):E3074–E3083. doi: 10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gradia S, et al. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3(2):255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 28.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci USA. 2011;108(18):7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34(15):4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell. 2003;12(1):233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 31.Chen C, Umezu K, Kolodner RD. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol Cell. 1998;2(1):9–22. doi: 10.1016/s1097-2765(00)80109-4. [DOI] [PubMed] [Google Scholar]
- 32.Schmutte C, Sadoff MM, Shim KS, Acharya S, Fishel R. The interaction of DNA mismatch repair proteins with human exonuclease I. J Biol Chem. 2001;276(35):33011–33018. doi: 10.1074/jbc.M102670200. [DOI] [PubMed] [Google Scholar]
- 33.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129(4):665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Qiu R, et al. MutL traps MutS at a DNA mismatch. Proc Natl Acad Sci USA. 2015;112(35):10914–10919. doi: 10.1073/pnas.1505655112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng L, Shen B. Okazaki fragment maturation: Nucleases take centre stage. J Mol Cell Biol. 2011;3(1):23–30. doi: 10.1093/jmcb/mjq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tishkoff DX, et al. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94(14):7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amunugama R, et al. RAD51 protein ATP cap regulates nucleoprotein filament stability. J Biol Chem. 2012;287(12):8724–8736. doi: 10.1074/jbc.M111.239426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinen CD, et al. Human MSH2 (hMSH2) protein controls ATP processing by hMSH2-hMSH6. J Biol Chem. 2011;286(46):40287–40295. doi: 10.1074/jbc.M111.297523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park J, et al. Single-molecule analysis reveals the kinetics and physiological relevance of MutL-ssDNA binding. PLoS One. 2010;5(11):e15496. doi: 10.1371/journal.pone.0015496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vallotton P, Olivier S. Tri-track: Free software for large-scale particle tracking. Microsc Microanal. 2013;19(2):451–460. doi: 10.1017/S1431927612014328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.