Significance
In recent years, next-generation sequencing has facilitated the discovery of thousands of nonprotein-coding RNAs (ncRNAs). Some of these ncRNAs have been shown to interact with transcription regulators and may thus form a new class of ncRNAs that function in controlling gene expression. Identifying proteins that interact with this class of ncRNAs will be instrumental in elucidating their poorly understood molecular mode of action. The ncRNA EBV-encoded RNA 2 (EBER2) expressed by the oncogenic Epstein–Barr virus interacts with the cellular transcription factor paired box protein 5 (PAX5) to impact viral gene expression. In this study, we identified additional functionally important protein components of the EBER2–PAX5 complex. Our findings pave the way for investigating whether other transcription-regulating ncRNAs use the same protein components to exert their function.
Keywords: noncoding RNA, Epstein–Barr virus, paraspeckle, RNA binding protein
Abstract
Epstein–Barr virus (EBV) produces a highly abundant noncoding RNA called EBV-encoded RNA 2 (EBER2) that interacts indirectly with the host transcription factor paired box protein 5 (PAX5) to regulate viral latent membrane protein 1/2 (LMP1/2) gene expression as well as EBV lytic replication. To identify intermediary proteins, we isolated EBER2–PAX5-containing complexes and analyzed the protein components by mass spectrometry. The top candidates include three host proteins splicing factor proline and glutamine rich (SFPQ), non-POU domain-containing octamer-binding protein (NONO), and RNA binding motif protein 14 (RBM14), all reported to be components of nuclear bodies called paraspeckles. In vivo RNA–protein crosslinking indicates that SFPQ and RBM14 contact EBER2 directly. Binding studies using recombinant proteins demonstrate that SFPQ and NONO associate with PAX5, potentially bridging its interaction with EBER2. Similar to EBER2 or PAX5 depletion, knockdown of any of the three host RNA-binding proteins results in the up-regulation of viral LMP2A mRNA levels, supporting a physiologically relevant interaction of these newly identified factors with EBER2 and PAX5. Identification of these EBER2-interacting proteins enables the search for cellular noncoding RNAs that regulate host gene expression in a manner similar to EBER2.
Epstein–Barr virus (EBV)-encoded RNA 2 (EBER2) is one of two highly abundant nuclear noncoding RNAs (ncRNAs) expressed during both latent and lytic infection of human B cells by the gamma herpesvirus EBV (1). Previous genome-wide location analysis of EBER2 using capture hybridization analysis of RNA targets (CHART) (2) revealed that EBER2 binds to the so-called terminal repeat (TR) regions of the double-stranded EBV genome (3). After DNA circularization to form the latent episome, these repeats are located in the first intron of the viral transcripts encoding latent membrane protein (LMP) 2A and 2B that are generated by alternative promoter use (4). Until recently, only the RNA chaperone La was known to interact with EBER2 through the stretch of uridylates at its 3′ end (5). Prompted by chromatin colocalization of EBER2 and the host transcription factor paired box protein 5 (PAX5) at the TR regions (6), we showed that PAX5 associates with EBER2 as well (3). However, negative results in electrophoretic mobility shift assays (EMSAs) using recombinantly expressed EBER2 and PAX5 suggested that the interaction between the ncRNA and host transcription factor might be bridged by an intermediate protein(s).
Within EBER2 are two accessible regions available for hybridization with complementary nucleic acids based on ribonuclease H sensitivity (3). One of these sites (Fig. 1A, Top) engages in RNA–RNA interactions with nascent transcripts from the TR regions, thus facilitating the recruitment and accumulation of PAX5 at this locus (3). Loss of either EBER2 or PAX5 binding to the TR regions results in up-regulation of the nearby LMP1, 2A, and 2B genes, with LMP2A showing the most pronounced increase. We have hypothesized that the binding of EBER2–PAX5 organizes viral chromatin at the TR locus such that transcription through this region becomes impaired (7). As the TRs are also critical for viral lytic replication (8, 9), knockdown of either EBER2 or PAX5 leads to diminished viral replication (3).
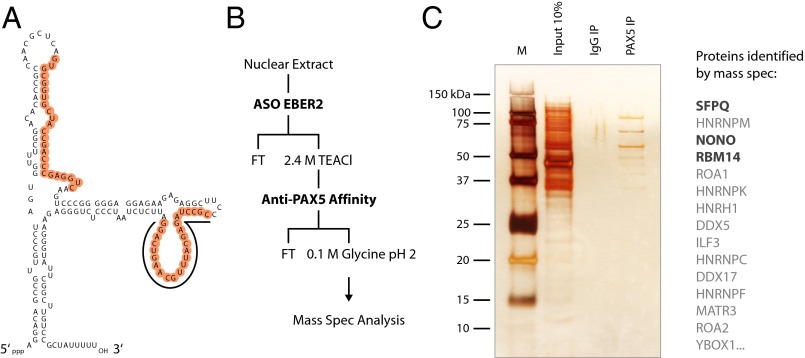
Fig. 1.
Identification of EBER2-interacting proteins. (A) Secondary structure of EBER2 is shown with the two accessible regions marked in orange (3). The underlined region is targeted by the ASO for EBER2 selection. The other marked region engages in RNA–RNA interactions with nascent transcripts from the TR regions. (B) Purification scheme for EBER2–PAX5-containing complexes. Nuclear extract from BJAB-B1 cells was subjected to EBER2–RNP selection using a biotinylated ASO and streptavidin beads (3). The EBER2–RNP was eluted with 2.4 M tetraethylammonium chloride (TEACl) and further purified by immunoprecipitation (IP) using anti-PAX5 antibody. Bound proteins were eluted with 0.1 M glycine pH 2 and subjected to mass spectrometry analysis. (C) Eluted proteins from the second purification step (PAX5 IP) shown in B were silver stained. The input material obtained from the 2.4 M TEACl elution and a control immunoprecipitation with IgG antibody (IgG IP) are also shown. The top 15 proteins identified by mass spectrometry are listed to the Right. Candidate proteins analyzed further are in bold. M, marker.
The RNA recognition motif (RRM) is widely found in ∼1% of protein-coding genes in the human genome (10). This abundant class of proteins has been implicated in a diverse range of cellular processes, such as alternative splicing, RNA export, and regulation of RNA stability (11, 12). RRM-containing proteins also execute functions in which their RNA-binding ability is not intuitively apparent, such as transcription regulation. Several RRM-containing proteins have been reported to carry out multiple functions (13). For example, splicing factor proline and glutamine rich (SFPQ), non-POU domain-containing octamer-binding protein (NONO), and RNA binding motif protein 14 (RBM14) each have well-documented roles in alternative splicing (13, 14) and contribute to transcription regulation (15–17). They are also essential for the formation of paraspeckles, which are subnuclear bodies consisting of more than 30 proteins that form on the long ncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) (18–20). Interestingly, RRM proteins not only play roles in overlapping cellular processes, but can also act in concert with each other, as they exhibit physical interaction (21, 22).
Here, we isolated ribonucleoprotein (RNP) complexes containing both EBER2 and PAX5 with the goal of identifying the protein(s) that bridges their interaction. As in our EBER2 CHART experiments (3), we used an antisense oligonucleotide (ASO) complementary to one of the accessible sites in EBER2 for selection, followed by immunoprecipitation using anti-PAX5 antibodies; the selected RNP complexes were subsequently analyzed by mass spectrometry. Using UV crosslinking experiments and protein binding studies, we generated an interaction map of the identified proteins SFPQ, NONO, and RBM14 with EBER2 and PAX5. Importantly, our results show that these three proteins are functionally intertwined with EBER2 and PAX5, as they converge in the regulation of viral LMP2A gene expression. Our findings facilitate the search for cellular ncRNAs that, similar to EBER2, may be involved in recruiting interacting transcription factors to their target sites on chromatin through RNA–RNA interactions with nascent transcripts.
Results
Purification of an EBER2–PAX5-Containing Complex.
To identify proteins that associate with EBER2, we started with a nuclear extract from BJAB-B1 cells, an EBV-positive lymphoma cell line that expresses EBER2 as a result of EBV infection, and added a biotinylated ASO complementary to one of EBER2’s accessible sites (3) (underlined region in Fig. 1A) that was coupled to streptavidin beads. The selected EBER2-containing RNP complexes were then subjected to a subsequent purification step using anti-PAX5 antibodies, yielding a complex that contains both EBER2 and PAX5 (Fig. 1B). Bound proteins were eluted from the Protein G beads and subjected to mass spectrometry analysis. As expected, PAX5 was among the identified proteins, validating our experimental procedure. Most other proteins present were already well known for their RNA-binding ability (Fig. 1C, Fig. S1A, and Dataset S1). For further analysis, we chose to focus on SFPQ, heterogeneous nuclear RNP M (HNRNPM), NONO, and RBM14 based on their top mass spectrometry scores. We also included HNRNPK and matrin 3 (MATR3), as they had previously been functionally linked to SFPQ, NONO, or RBM14 (18, 21).
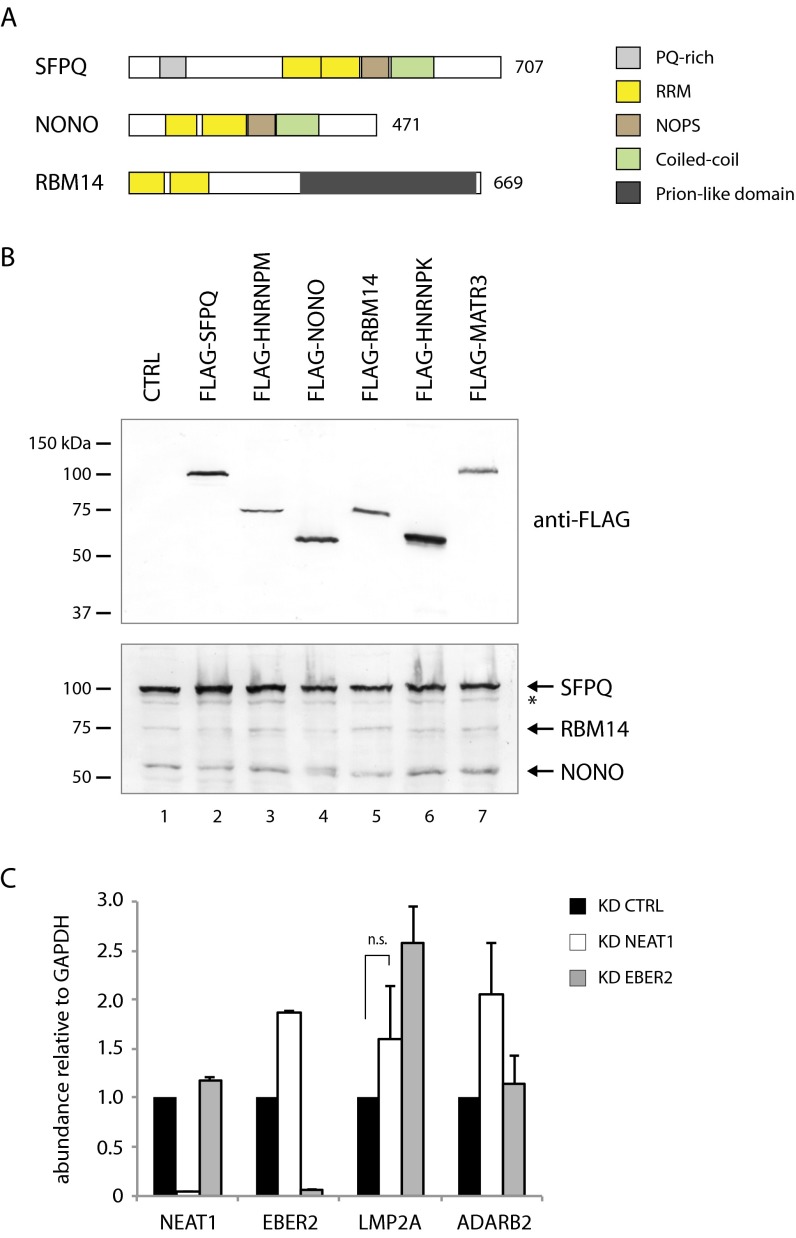
Fig. S1.
(A) Schematics of the SFPQ, NONO, and RBM14 proteins. The RRMs, proline-glutamine-rich (PQ-rich) domain, NOPS (NONA/paraspeckle) domains, predicted coiled-coil domains, and prion-like domain are indicated. SFPQ and NONO are predicted to form a heterodimer via their RRMs, NOPS domain, and coiled-coil region (36). The prion-like domain of RBM14 interacts with NONO (22). Schematics are adapted from refs. 18, 22, and 36. (B, Top) FLAG-tagged proteins expressed in HEK 293T cells were detected by Western blot analysis using anti-FLAG antibody to demonstrate comparable expression after transient transfection. Vectors expressing FLAG-tagged constructs were transfected using Effectene (Qiagen) according to the manufacturer’s instructions, except for MATR3 where twice the recommended transfection reagent was used. CTRL, untransfected HEK 293T cell lysate. (Bottom) The overall protein levels of SFPQ, NONO, and RBM14 under these transfection conditions were examined by Western blot using anti-SFPQ, anti-NONO, and anti-RBM14 antibodies, respectively. Levels of other proteins were not examined due to unavailability of antibodies. Expression of FLAG-tagged SFPQ, NONO, or RBM14 does not markedly increase the overall levels of these proteins. Note that FLAG-tagged NONO migrates slower than the endogenous protein, as indicated by a doublet (lane 4), whereas FLAG–SFPQ and FLAG–RBM14 do not (lanes 2 and 5). A weaker faster migrating band (indicated by *) is also detected by anti-SFPQ antibodies. (C) Knockdown of NEAT1 does not result in significant up-regulation of LMP2A. NEAT1 and EBER2 were knocked down with previously described antisense oligonucleotides (NEAT1 ASO: mC*mC*mC*mU*mC*T*A*G*T*C*T*T*G*G*C*mU*mC*mA*mU*mU; EBER2 ASO: mC*mC*mU*mG*mA*mC*T*T*G*C*A*A*A*T*G*C*T*C*mU*mA*mG*mG*mC*mG) (3, 28) using Lonza Nucleofector Device II, Kit T, and program T-016 (for detailed procedure see Materials and Methods). RNA levels were measured by qRT-PCR analysis. ADARB2 levels were examined as a positive control for efficient NEAT1 depletion as described (28). The following real-time PCR primer pairs were used: hNEAT1 TCGGGTATGCTGTTGTGAAA and TGACGTAACAGAATTAGTTCTTACCA; ADARB2 GCCATCCCACATACCTCCAA and GGGGTGCTACTGCCTCTCCT (28, 37). All data represent the mean of three independent experiments ± SD. n.s., statistically not significant.
In Vivo and in Vitro Interactions with EBER2.
To ask which of these proteins directly interacts with EBER2 in vivo, we expressed in HEK 293T cells FLAG-tagged versions of each of the proteins along with EBER2. EBER2 was expressed at a level comparable to that in the EBV-infected cell line BJAB-B1 (∼2.5 × 105 copies per cell); the FLAG-tagged proteins were expressed at levels comparable to each other (Fig. S1B). After UV irradiation, we immunoprecipitated nuclear extracts using anti-FLAG antibodies under denaturing conditions that disrupt noncovalent interactions (23). Thus, EBER2 will be detected only if it has been covalently UV crosslinked to the tagged protein. We observed EBER2 binding directly to SFPQ, HNRNPM, NONO, and RBM14, but not to HNRNPK or MATR3 (Fig. 2A).
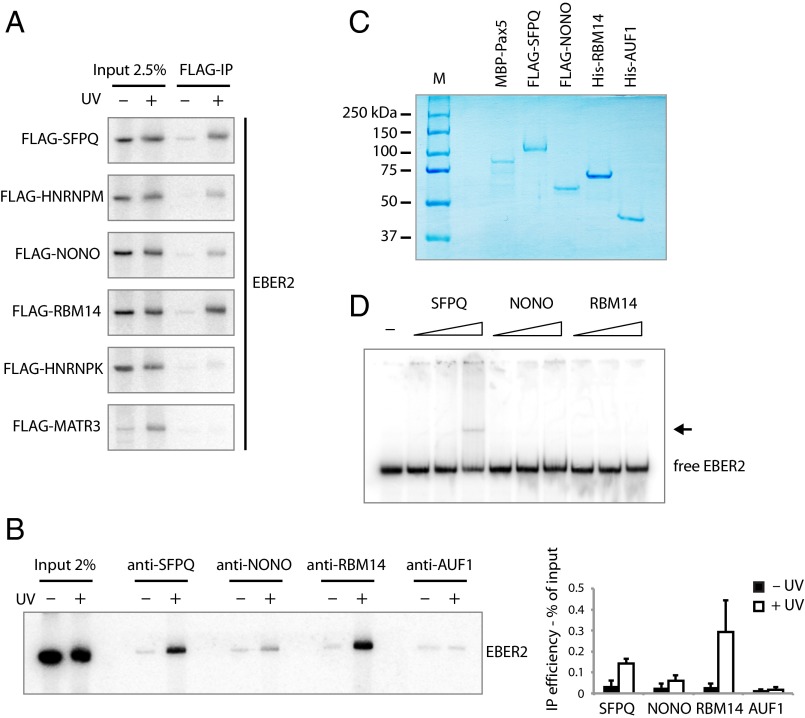
Fig. 2.
UV-crosslinking experiments and EMSAs identify proteins interacting with EBER2. (A) HEK 293T cells were transfected with plasmids expressing EBER2 and FLAG-tagged SFPQ, HNRNPM, NONO, RBM14, HNRNPK, or MATR3. EBER2 was expressed at levels comparable to those in BJAB-B1 cells (∼2.5 × 105 copies per cell); FLAG-tagged proteins were expressed at levels comparable to each other (Fig. S1B). Intact cells were irradiated with UV light where indicated. EBER2 was coimmunoprecipitated using anti-FLAG antibodies and detected by Northern blot analysis, showing direct interactions with SFPQ, HNRNPM, NONO, and RBM14. For the input lanes, RNA was isolated from 2.5% the amount of lysate used in the FLAG-IP. (B) Intact BJAB-B1 cells were irradiated with UV light where indicated and subjected to immunoprecipitation using antibodies against SFPQ, NONO, RBM14, and AUF1 as a negative control. EBER2 was detected by Northern blot analysis. EBER2 interacts directly with SFPQ, RBM14, and weakly with NONO. EBER2 quantification from three independent experiments (mean ± SD) is shown to the Right. (C) Coomassie-stained gel of recombinantly expressed proteins MBP–Pax5, FLAG–SFPQ, FLAG–NONO, His–RBM14, and His–AUF1p40 used for EMSA. SFPQ and NONO were purified from baculovirus-infected Sf9 cells, whereas the other proteins were purified from E. coli. (D) EMSA of EBER2 with SFPQ, NONO, and RBM14. EBER2 (1 nM in vitro transcribed) was incubated with increasing amounts of recombinant protein (5, 25, 125 molar excess) for 30 min on ice in EMSA buffer and resolved on a native polyacrylamide gel. An EBER2 bandshift is observed after incubation with recombinant SFPQ at the highest molar ratio (1:125). Free EBER2 is indicated; the arrow points to the EBER2–SFPQ complex.
We next examined which of these native proteins interact with EBER2 in EBV latently-infected cells. We used specific antibodies to target proteins endogenously expressed in BJAB-B1 cells. We did not include HNRNPM in our analyses, as this protein and specific antibodies against it have not been well characterized, whereas the other three proteins have been widely studied. After UV crosslinking, both SFPQ and RBM14 exhibit clear direct in vivo binding to EBER2 (Fig. 2B). EBER2 is weakly immunoprecipitated by anti-NONO antibodies, whereas no EBER2 enrichment is observed with negative control antibodies against AU-rich element binding factor 1 (AUF1), a well-characterized RNA-binding protein (24).
To confirm their direct interaction with EBER2, recombinantly expressed tagged RNA-binding proteins, as well as PAX5, were purified from baculovirus-infected insect Sf9 cells or from Escherichia coli (Fig. 2C) for use in EMSAs with full-length EBER2. We observed an EBER2 bandshift after incubation with SFPQ at the highest molar ratio (1:125), whereas no bandshift was observed for NONO or RBM14 (Fig. 2D). Because RBM14 was purified from E. coli, it may lack posttranslational modifications necessary for interaction with EBER2 (Discussion). This notion is supported by the previous observation that phosphorylation of the C-terminal region of RBM14 is essential for protein–protein interactions (22).
We then asked whether any of the three proteins interact with PAX5 in coimmunoprecipitation experiments using BJAB-B1 nuclear extracts followed by Western blot analysis (Fig. 3A). We were able to recapitulate previously reported interactions between SFPQ and NONO, as well as between NONO and RBM14 (21, 22). SFPQ and RBM14, and to a lesser extent NONO, exhibited interactions with PAX5, suggesting the presence of a complex containing all four proteins. Coimmunoprecipitation using IgG or anti-AUF1 antibodies provided negative controls. Prior treatment of the nuclear extract with RNase A, which was required to reduce background, argues that the observed protein–protein interactions are independent of RNA. To construct a more detailed interaction map, recombinantly expressed tagged SFPQ, NONO, and RBM14 were subjected to binding assays with PAX5 fused N terminally to maltose binding protein (MBP–Pax5) and immobilized on amylose resin. SFPQ and NONO bound to MBP–Pax5, whereas RBM14 and AUF1, the negative control, did not (Fig. 3B). Taken together, these results argue that SFPQ and RBM14 make direct contacts with EBER2, whereas SFPQ and NONO interact specifically with PAX5 (Fig. 4D).
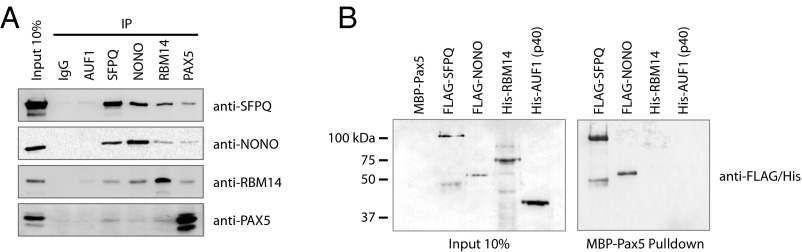
Fig. 3.
Protein–protein interaction studies. (A) Coimmunoprecipitation experiments using EBV-positive BJAB-B1 cell lysate and antibodies to SFPQ, NONO, RBM14, and PAX5 described in refs. 6, 18. Isotype control (IgG) and anti-AUF1 antibodies were included as negative controls. RNase A treatment was carried out before immunoprecipitation (Materials and Methods). PAX5 interaction with SFPQ, NONO, and RBM14 is observed, suggesting the presence of a complex containing all four proteins. (B) Selection by MBP–Pax5 (Pulldown) of recombinant proteins shown in Fig. 2C. Western blot analyses using anti-FLAG and anti-His antibodies to detect FLAG–SFPQ, FLAG–NONO, His–RBM14, or His–AUF1p40 were carried out to examine binding to PAX5. SFPQ and NONO bind PAX5, whereas RBM14 and AUF1 exhibit no interaction. Input samples (Left) and proteins bound to MBP–Pax5 (Right) are shown.
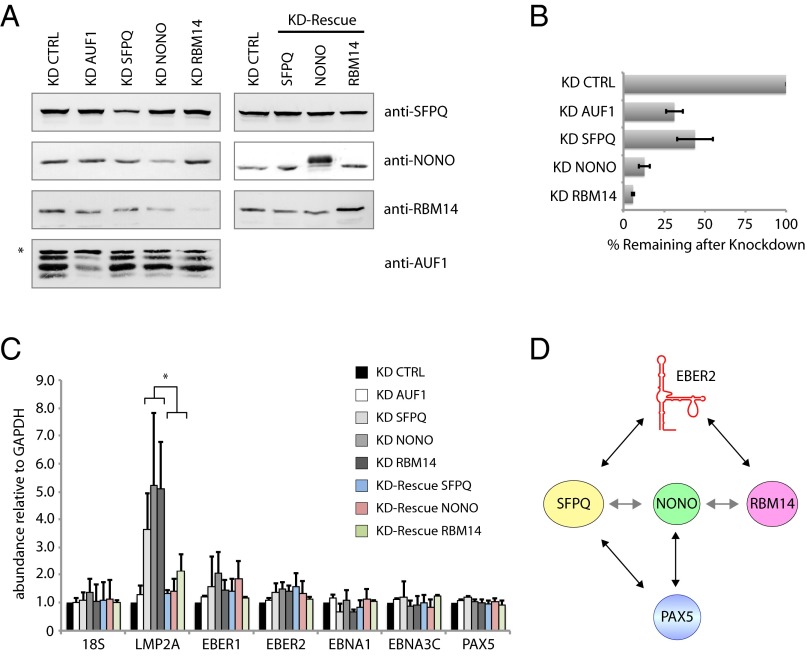
Fig. 4.
Depletion of SFPQ, NONO, and RBM14 results in up-regulation of LMP2A. (A) AUF1, SFPQ, NONO, and RBM14 were knocked down in BJAB-B1 cells with specific shRNAs. Western blots show knockdown (KD) efficiencies (Left). RNAi-resistant constructs containing an N-terminal FLAG-tag were coexpressed with shRNAs to rescue depletion (Right). The FLAG-tagged rescue construct of NONO exhibits slower electrophoretic migration, whereas FLAG-tagged SFPQ and RBM14 are indistinguishable from the endogenous proteins (Fig. S1B). * indicates a nonspecific band recognized by the anti-AUF1 antibody. (B) Quantification of three independent knockdown experiments as in A. Band intensities of Western blots were quantified using the software ImageJ (NIH). To determine the knockdown efficiency of AUF1, the second most rapidly migrating band in A, representing the p40 and p42 isoforms, was quantified. (C) RNA levels of several EBV genes were measured by quantitative RT-PCR analysis and normalized to GAPDH levels. LMP2A expression is up-regulated upon depletion of SFPQ, NONO, and RBM14; up-regulation is reversed upon rescue of RNAi depletion. All data represent the mean of three independent experiments ± SD. *P < 0.04. (D) Proposed interaction map of the EBER2–PAX5 RNP. Shaded arrows indicate previously reported protein–protein interactions (21, 22).
Knockdown of SFPQ, NONO, or RBM14 Results in Up-Regulation of LMP2A Expression.
To assess whether the presence of SFPQ, NONO, and RBM14 in the EBER2–PAX5 RNP is physiologically relevant, we probed LMP2A gene expression, which is down-regulated by the EBER2–PAX5 complex (3). We designed short-hairpin RNA (shRNA) constructs that specifically target SFPQ, NONO, or RBM14 and confirmed their knockdown efficiencies in BJAB-B1 cells by Western blot analyses (Fig. 4A, Left; quantitations in 4B). An shRNA construct targeting AUF1 (23) was included as a negative control. RNA-interference (RNAi)-resistant cDNA constructs were also generated for use in rescue experiments to ascertain that the observed effects are due to specific knockdown of each protein (Fig. 4A, Right). Upon nucleofection of shRNA constructs into BJAB-B1 cells, the transcript levels of several EBV genes (three mRNAs and two ncRNAs) were assessed by qRT-PCR analysis. Of the genes tested, LMP2A was the only markedly up-regulated gene after SFPQ, NONO, or RBM14 depletion (Fig. 4C), similar to the up-regulation observed after EBER2 or PAX5 depletion (3). LMP2A up-regulation was not due to concomitant reduction in PAX5 or EBER2 levels after knockdown of SFPQ, NONO, or RBM14 (Fig. 4C). Knockdown of AUF1 did not affect viral gene transcription. Importantly, the increase in LMP2A levels was reversed when RNAi-resistant constructs were coexpressed with the corresponding shRNAs. We conclude that depletion of SFPQ, NONO, or RBM14 individually results in up-regulation of LMP2A expression, mimicking depletion of EBER2 or PAX5 (3) and substantiating a functional link and concerted action of these physically interacting factors.
Discussion
Our previous indications that EBER2 interacts indirectly with the host transcription factor PAX5 (3) prompted us to search for additional components of the EBER2–PAX5 RNP complex. We began by using mass spectrometry to identify several RNA-binding proteins that copurify with EBER2 and PAX5 from nuclear extracts of latently infected B cells (Fig. 1 and Dataset S1). We focused our study on paraspeckle components SFPQ, NONO, and RBM14 and were able to verify direct interactions with EBER2 and/or PAX5 (Figs. 2B and 3B). A schematic of the intermolecular contacts within the EBER2 RNP is shown in Fig. 4D. We did not recover a majority of paraspeckle proteins by mass spectrometry, in particular the major paraspeckle protein PSP1, arguing against the EBER2–PAX5 complex exerting its function through association with paraspeckles. PAX5 and EBER2, as judged by immunostaining and in situ hybridization, respectively, localize diffusely in the nucleus (25, 26), further indicating a spatial independence from paraspeckles. SFPQ has been shown to escape sequestration in paraspeckles to regulate host gene expression (27, 28). Similarly, SFPQ, NONO, and RBM14 may associate with EBER2 and PAX5 outside of paraspeckles to regulate viral gene expression.
Most importantly, we observed that knockdown of these newly identified EBER2/PAX5-interacting proteins affects expression of the latent viral genome (Fig. 4C), phenocopying the knockdown of EBER2 or PAX5 (3, 6) and confirming a functional link in addition to physical interaction. EBER2 and PAX5 were previously shown to be necessary for efficient replication and packaging of EBV DNA as well (3). It remains unknown whether SFPQ, NONO, and RBM14 are likewise essential for viral lytic replication. Intriguingly, these proteins also play a role in the life cycle of another virus, the HIV (29–31), suggesting a more common involvement of these factors in viral regulation. Detailed studies of these RRM-containing proteins are complicated by the fact that all three proteins function in multiple cellular processes, including transcription regulation, paraspeckle formation, and notably alternative splicing (13, 15–18). Thus, further studies are required to exclude the possibility that the observed up-regulation of LMP2A mRNA upon knockdown of SFPQ, NONO, and RBM14 results from enhanced splicing and generation of mature mRNA, independent of the EBER2–PAX5 complex. Similarly, because these proteins have been implicated in promoter activation, we cannot exclude the possibility that SFPQ, NONO, and RBM14 regulate the transcription of LMP2A directly. Localization of these factors by chromatin immunoprecipitation analyses of the LMP2 gene locus should shed light on the exact mechanism of LMP2A up-regulation.
Our studies revealed a discrepancy between the in vivo direct UV crosslinking of EBER2 to RBM14 and the lack of apparent interactions in EMSAs using purified factors. RBM14 contains a C-terminal prion-like domain that is subject to posttranslational modification and required for protein–protein interaction (Fig. S1A) (22). Unlike SFPQ and NONO, we were unable to purify recombinant RBM14 from Sf9 cells due to its limited solubility in cell lysate, probably because of the behavior of its prion-like domain. We therefore used recombinant RBM14 purified from E. coli, but it is possible that the missing posttranslational modifications are necessary for EBER2 binding. In the same vein, we cannot exclude the possibility that the lack of posttranslational modifications in RBM14 abrogates interactions with PAX5. An alternative explanation for the lack of in vitro interactions between EBER2 and RBM14 could be that an auxiliary protein is required to load EBER2 onto RBM14. Such a protein might also be required for efficient binding of EBER2 to SFPQ; a bandshift was observed only at a 125-fold molar excess of SFPQ (Fig. 2D). We therefore asked whether the combined presence of any of the other factors would enhance the binding of EBER2 in EMSAs, but found that no combination of SFPQ, NONO, and RBM14 resulted in a stronger bandshift.
Protein–protein interactions with PAX5 appear to be RNA independent because coimmunoprecipitation of SFPQ, NONO, and RBM14 was observed following RNase A treatment of nuclear extracts (Fig. 3A). We thus propose a model in which a complex containing these three proteins assembles first with PAX5. Subsequent addition of EBER2, which associates directly with the TR regions of the EBV genome through RNA–RNA interactions with nascent transcripts, would serve to recruit the protein components to these genomic binding sites. It would be interesting to learn whether this protein assembly occurs in normal host B cells, or only after EBV infection. The poorly characterized protein HNRNPM warrants further examination as a putative essential component of the EBER2–PAX5 complex, as it was one of the top EBER2 binding candidates in our mass spectrometry analysis and exhibits robust in vivo UV crosslinking to EBER2 (Fig. 2A and Dataset S1).
SFPQ, NONO, and RBM14 are the protein factors most critical for paraspeckle formation (18). Moreover, these proteins are required for the stability of the paraspeckle RNA component NEAT1, around which paraspeckles form, as their depletion greatly diminishes NEAT1 levels (18). These proteins do not appear to be required for the stabilization of EBER2 (Fig. 4C). Furthermore, the integrity of paraspeckles, as evidenced by depletion of NEAT1, does not affect EBER2–PAX5-mediated gene regulation of LMP2A (Fig. S1C). The function of paraspeckles is not fully understood, but an emerging theme suggests a role in nuclear retention of A-to-I edited RNAs. For example, the CTN–RNA contains a 3′ UTR that is targeted for editing and is responsible for its localization to paraspeckles (32). Furthermore, transcripts containing inverted repeat Alu elements in their 3′ UTR, which are also targets of editing, accumulate in paraspeckles when bound to NONO, but can undergo cytoplasmic export when NONO is replaced by Staufen1 binding to these mRNAs (33). Even though NEAT1, and thus paraspeckles, are mammalian specific (19), similar RNA retention mechanisms appear to be present in other organisms. In the Xenopus oocyte system, A-to-I edited RNAs are retained in the nucleus by a ternary complex consisting of SPPQ, NONO, and MATR3 (21). In light of the strictly nuclear localization of EBER2 (26), it is interesting that several paraspeckle components are present in the EBER2 RNP. The question arises as to whether the nuclear localization of EBER2 can also be attributed to its association with these paraspeckle proteins, exploiting a host mechanism for nuclear RNA retention. Whether the related EBER1 is retained in the nucleus possibly via the same mechanism remains to be addressed.
Materials and Methods
Purification of EBER2–PAX5 Complex.
A biotinylated ASO complementary to EBER2 nucleotides 101–124 (underlined region in Fig. 1A) was coupled to streptavidin beads and incubated with nuclear extract from BJAB-B1 cells. Cells were crosslinked with 0.25% formaldehyde before generation of nuclear extract as described (3). Bound EBER2–RNP complexes were eluted from streptavidin beads with 2.4 M tetraethylammonium chloride at 40 °C for 15 min. The eluate was diluted 10-fold with PBS and incubated with anti-PAX5 antibody (Santa Cruz sc-1974) and Protein G Sepharose for a second purification step. The bound proteins were eluted with 0.1 M glycine pH 2 and subjected to mass spectrometry analysis. A control immunoprecipitation was carried out with normal mouse IgG antibodies (Santa Cruz sc-2025) and Protein G Sepharose.
UV Crosslinking and Immunoprecipitation.
Complementary DNA (cDNA) clones were obtained from Addgene or Open Biosystems (SFPQ: ID 46320; HNRNPM: ID 2900532; NONO: ID 35379; RBM14: ID 2819856; HNRNPK: ID 2964383; MATR3: ID 32880) and coding sequences were cloned into vector pcDNA-FLAG. HEK 293T cells were transfected with EBER2 expression plasmid (pBS-5×EBER2) together with each of the FLAG expression plasmids using Effectene (Qiagen). UV irradiation followed by immunoprecipitation was carried out as described (23). Antibodies used were: mouse anti-FLAG (Sigma M2), mouse anti-SFPQ (Sigma clone 6D7), rabbit anti-NONO (Bethyl A300-587A), rabbit anti-RBM14 (Bethyl A300-845A), and rabbit anti-AUF1 (34).
Coimmunoprecipitation and Western Blot Analysis.
A total of 107 BJAB-B1 cells were resuspended in 500 μL nuclei lysis buffer (10 mM Tris pH 8.0, 0.32 M sucrose, 3 mM CaCl2, 0.1 mM EDTA, 0.1% Nonidet P-40) in the presence of 1× protease inhibitor cocktail (Calbiochem) and 1 mM PMSF and incubated for 10 min on ice followed by a 3-min centrifugation step at 850 × g in a table-top centrifuge to pellet nuclei. A total of 1 mL RIPA buffer was added to the nuclei and incubated for 15 min at 37 °C after addition of 4 μg RNase A (Sigma). Debris was cleared by centrifugation, and 250 μL of lysate was used for each immunoprecipitation reaction with 1 μg of antibody and 20 μL of either Protein A or G Sepharose. The following antibody dilutions were used for Western blot analysis: anti-SFPQ (1:1,000), anti-NONO (1:2,500), anti-RBM14 (1:2,500), anti-PAX5 (1:200), and mouse anti-AUF1 (1:2,000, kind gift of Gideon Dreyfuss, University of Pennsylvania, Philadelphia) (35).
Protein Purification and EMSA.
The coding sequences of SFPQ and NONO were cloned into the pFastBac vector (Invitrogen) including an N-terminal FLAG-tag. Proteins were expressed in baculovirus-infected Sf9 cells using the Bac-to-Bac Expression System (Invitrogen). After initial purification from Sf9 cell lysate with anti-FLAG M2 beads (Sigma), the eluate was further purified over a Superose 6 and Mono Q column.
RBM14 did not express well in Sf9 cells and exhibited low solubility in cleared lysate (Western blot signal in whole cell lysate was much stronger than in cleared lysate). Therefore, RBM14 cDNA was cloned into the pET28a vector to include a C-terminal His-tag. The protein was expressed in E. coli BL21 cells and purified using nickel affinity chromatography followed by subsequent cleanup by gel filtration. MBP–Pax5 was expressed as described (3).
EMSAs were carried out as described (23). In brief, full-length EBER2 was in vitro transcribed with T7 RNA polymerase and 5′ end labeled with γ[32P]ATP and T4 polynucleotide kinase. Purified proteins were incubated on ice for 30 min with 1 nM EBER2 at the indicated molar ratios in 10 μL EMSA buffer (10 mM Tris pH 7.4, 50 mM NaCl, 0.5 mM DTT, 0.1 mM ZnSO4, 1 mM MgCl2, 4% glycerol, 50 ng tRNA). Reactions were resolved on a 6% nondenaturing polyacrylamide gel in 0.5× TBE buffer at 200 V for 2 h at 4 °C. Gels were dried and exposed to a phosphor imaging screen.
Protein–Protein Interaction Experiments.
A total of 0.5 μg of MPB–Pax5 was immobilized on 5 μL of packed amylose resin (NEB) by incubating for 4 h at 4 °C in 250 μL binding buffer (20 mM Hepes pH 7.9; 150 mM NaCl; 0.2 mM EDTA; 0.5 mM DTT) containing 5 μg BSA to block nonspecific binding. A total of 0.5 μg of recombinant FLAG-SFPQ, FLAG-NONO, His-RBM14, or His-AUF1p40 was added and incubated overnight with shaking. Beads were washed five times with 1 mL binding buffer and then resuspended in SDS loading buffer. Proteins were detected by Western blot analysis using anti-FLAG (Sigma, 1:1,000 dilution) and anti-His antibodies (Santa Cruz, 1:200 dilution).
RNA Interference and Quantitative RT-PCR.
shRNA constructs against SFPQ, NONO, and RBM14 were cloned downstream of the murine U6 promoter in pBluescript vector. The following shRNA sequences were used (loop sequence is underlined): atggttcaggaggccagaaatttcaagagaatttctggcctcctgaaccat (SFPQ); gaacagggttactgtatactgaattcaagagattcagtatacagtaaccctgttc (NONO); and gtctgcagcctcctcactagcttattcaagagataagctagtgaggaggctgcagac (RBM14). RNAi-resistant constructs were generated by site-directed mutagenesis of pcDNA-FLAG expression vectors to contain the following silent mutations (underlined): SFPQ atacggcagcggcggacaaaagt and RBM14 aagcgccgctagtagcttggccta. The shRNA against NONO targets the 3′ UTR region; hence coexpression of pcDNA-FLAG-NONO is sufficient to rescue the knockdown.
For each knockdown experiment, 1.25 μg shRNA plasmid was nucleofected into 2.5 × 106 BJAB-B1 cells using the Lonza 4D-Nucleofector System (SF solution; program EN-150). Nucleofection was repeated after 48 h, and cells were harvested after 72 h of knockdown. For knockdown-rescue experiments, 1.25 μg of RNAi-resistant expression construct was conucleofected. For Western blot analysis, cells were lysed in RIPA buffer before SDS-gel electrophoresis. For qRT-PCR analysis, RNA was purified using TRIzol (Invitrogen) and cDNA was generated using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Primers for qPCR analysis have been described (3).
Supplementary Material
Acknowledgments
We thank Drs. Johanna Withers and Mingyi Xie for helpful comments and Angela Miccinello for editorial assistance. This work was supported by Grants CA16038 and CA193300 from the National Cancer Institute. J.A.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601773113/-/DCSupplemental.
References
- 1.Lerner MR, Andrews NC, Miller G, Steitz JA. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1981;78(2):805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon MD, et al. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci USA. 2011;108(51):20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee N, Moss WN, Yario TA, Steitz JA. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell. 2015;160(4):607–618. doi: 10.1016/j.cell.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raab-Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47(6):883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 5.Tycowski KT, et al. Viral noncoding RNAs: More surprises. Genes Dev. 2015;29(6):567–584. doi: 10.1101/gad.259077.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arvey A, et al. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe. 2012;12(2):233–245. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee N, Steitz JA. Noncoding RNA-guided recruitment of transcription factors: A prevalent but undocumented mechanism? BioEssays. 2015;37(9):936–941. doi: 10.1002/bies.201500060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feederle R, Shannon-Lowe C, Baldwin G, Delecluse HJ. Defective infectious particles and rare packaged genomes produced by cells carrying terminal-repeat-negative epstein-barr virus. J Virol. 2005;79(12):7641–7647. doi: 10.1128/JVI.79.12.7641-7647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmermann J, Hammerschmidt W. Structure and role of the terminal repeats of Epstein-Barr virus in processing and packaging of virion DNA. J Virol. 1995;69(5):3147–3155. doi: 10.1128/jvi.69.5.3147-3155.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finn RD, et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006;34(Database issue):D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lunde BM, Moore C, Varani G. RNA-binding proteins: Modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8(6):479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 13.Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO--multi-functional nuclear proteins. FEBS Lett. 2002;531(2):109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 14.Auboeuf D, et al. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol Cell Biol. 2004;24(1):442–453. doi: 10.1128/MCB.24.1.442-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosonina E, et al. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol. 2005;25(15):6734–6746. doi: 10.1128/MCB.25.15.6734-6746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amelio AL, et al. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc Natl Acad Sci USA. 2007;104(51):20314–20319. doi: 10.1073/pnas.0707999105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwasaki T, Chin WW, Ko L. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM) J Biol Chem. 2001;276(36):33375–33383. doi: 10.1074/jbc.M101517200. [DOI] [PubMed] [Google Scholar]
- 18.Naganuma T, et al. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31(20):4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2(7):a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13(1):95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106(4):465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 22.Hennig S, et al. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol. 2015;210(4):529–539. doi: 10.1083/jcb.201504117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee N, Pimienta G, Steitz JA. AUF1/hnRNP D is a novel protein partner of the EBER1 noncoding RNA of Epstein-Barr virus. RNA. 2012;18(11):2073–2082. doi: 10.1261/rna.034900.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White EJ, Brewer G, Wilson GM. Post-transcriptional control of gene expression by AUF1: Mechanisms, physiological targets, and regulation. Biochim Biophys Acta. 2013;1829(6-7):680–688. doi: 10.1016/j.bbagrm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cattoretti G, et al. Stages of germinal center transit are defined by B cell transcription factor coexpression and relative abundance. J Immunol. 2006;177(10):6930–6939. doi: 10.4049/jimmunol.177.10.6930. [DOI] [PubMed] [Google Scholar]
- 26.Fok V, Friend K, Steitz JA. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J Cell Biol. 2006;173(3):319–325. doi: 10.1083/jcb.200601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imamura K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53(3):393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Hirose T, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25(1):169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zolotukhin AS, et al. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol Cell Biol. 2003;23(18):6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Budhiraja S, et al. Mining the human complexome database identifies RBM14 as an XPO1-associated protein involved in HIV-1 Rev function. J Virol. 2015;89(7):3557–3567. doi: 10.1128/JVI.03232-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kula A, Gharu L, Marcello A. HIV-1 pre-mRNA commitment to Rev mediated export through PSF and Matrin 3. Virology. 2013;435(2):329–340. doi: 10.1016/j.virol.2012.10.032. [DOI] [PubMed] [Google Scholar]
- 32.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123(2):249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 33.Elbarbary RA, Li W, Tian B, Maquat LE. STAU1 binding 3′ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev. 2013;27(13):1495–1510. doi: 10.1101/gad.220962.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dempsey LA, Hanakahi LA, Maizels N. A specific isoform of hnRNP D interacts with DNA in the LR1 heterodimer: Canonical RNA binding motifs in a sequence-specific duplex DNA binding protein. J Biol Chem. 1998;273(44):29224–29229. doi: 10.1074/jbc.273.44.29224. [DOI] [PubMed] [Google Scholar]
- 35.Piñol-Roma S, Choi YD, Matunis MJ, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988;2(2):215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- 36.Passon DM, et al. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc Natl Acad Sci USA. 2012;109(13):4846–4850. doi: 10.1073/pnas.1120792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clemson CM, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.