Significance
Metabolic engineering is an economically feasible and sustainable alternative for the production of natural products, pharmaceuticals, nutraceuticals, flavors, and fragrances. Of the model systems used to demonstrate and develop this approach, the anticancer agent Taxol stands out for its structural complexity and therapeutic value. A major challenge for the biosynthesis of Taxol and many other natural products is the involvement of cytochrome P450-mediated oxygenation. P450 enzymes are intransigent to functional heterologous expression, especially in Escherichia coli, leading many laboratories to abandon this organism when engineering P450-containing pathways. Here, through a series of optimizations, we demonstrate E. coli as a viable host for P450-mediated oxidative chemistry, advancing Taxol’s biosynthesis through a fivefold increase in oxygenated terpene titers.
Keywords: Taxol, P450, metabolic engineering, natural products, oxygenated taxanes
Abstract
Recent advances in metabolic engineering have demonstrated the potential to exploit biological chemistry for the synthesis of complex molecules. Much of the progress to date has leveraged increasingly precise genetic tools to control the transcription and translation of enzymes for superior biosynthetic pathway performance. However, applying these approaches and principles to the synthesis of more complex natural products will require a new set of tools for enabling various classes of metabolic chemistries (i.e., cyclization, oxygenation, glycosylation, and halogenation) in vivo. Of these diverse chemistries, oxygenation is one of the most challenging and pivotal for the synthesis of complex natural products. Here, using Taxol as a model system, we use nature’s favored oxygenase, the cytochrome P450, to perform high-level oxygenation chemistry in Escherichia coli. An unexpected coupling of P450 expression and the expression of upstream pathway enzymes was discovered and identified as a key obstacle for functional oxidative chemistry. By optimizing P450 expression, reductase partner interactions, and N-terminal modifications, we achieved the highest reported titer of oxygenated taxanes (∼570 ± 45 mg/L) in E. coli. Altogether, this study establishes E. coli as a tractable host for P450 chemistry, highlights the potential magnitude of protein interdependency in the context of synthetic biology and metabolic engineering, and points to a promising future for the microbial synthesis of complex chemical entities.
Metabolic engineering represents a particularly auspicious vehicle for exploiting the power of biological catalysis, offering readily scalable “one-pot” transformations of simple and renewable feedstocks into value-added materials (1). Through the targeted manipulation of genetic material, heterologous enzymatic pathways can be transferred to fermentable microbes, fashioning them into miniature chemical factories (2). A growing number of examples demonstrate the efficacy of this approach and, for a range of complex molecular families, including alkaloids (3), flavonoids (4), and isoprenoids (5), giving hope for a future in which industrial biotechnology improves both our synthetic approach and access to a vast array of even new-to-nature chemical entities (6).
For this future to be realized, however, additional tools and strategies need to be established. Although much attention has been given to developing “parts” that control molecular processes like transcription and translation (7–9), little has been focused on developing different classes of metabolic chemistries (i.e., polymerization, cyclization, oxygenation, and glycosylation) as potential parts. Systematizing the optimization of different metabolic chemistries would bring us closer to an “off the shelf” catalyst approach in metabolic engineering, which would greatly benefit the synthesis of complex molecules such as the blockbuster anticancer agent Taxol. Long captivating chemists and biochemists alike (10), Taxol’s densely functionalized, labyrinthine structure (Fig. 1) provides an excellent model system to probe the capacity of metabolic engineering, and its successful synthesis would likely provide a template for the construction of other complex natural products.
Fig. 1.
Taxol biosynthesis schematic. The schematic depicts an abbreviated overview of the Taxol pathway, beginning with this work's carbon source glycerol. From glycerol, the basic metabolic building blocks of glyceraldehyde-3-phosphate (GAP) and pyruvate (PYR) are made. These building blocks are then carried through the methylerythritol phosphate (MEP) pathway to create the ubiquitous isoprenoid precursors IPP and DMAPP. IPP and DMAPP then are polymerized and cyclized by GGPPS and TxS to form the 20-carbon backbone taxa-4 (5),11 (12)-diene (taxadiene). The blue star indicates the focus of this study, which is the putative next pathway the fifth carbon hydroxylation as performed by the cytochrome P450 CYP725A4 and its reductase partner. Further decoration leads to the advanced intermediate Baccatin III, which combined with group transfer produces the final Taxol compound.
To assemble such biosynthetic routes, however, it is needful to not only elucidate the native enzymatic pathway but to value the underlying chemical logic (11). Nature employs a particular strategy to achieve its vast chemodiversity, beginning with the polymerization and cyclization of basic metabolic building blocks to form a broad range of scaffold molecules (11). After scaffold creation, tailoring enzymes decorate with various functional moieties, effectively multiplying total potential diversity (12). A level of hierarchy exists among functionalizations, with oxygenation claiming a preeminent role (13). Mediated primarily by the cytochrome P450 superfamily of enzymes (14), oxygenation can act not only as an independent decoration, but often serves as the basis for subsequent modification (15). This paradigm of scaffold creation, oxygenation, and further decoration holds especially true for Taxol and the greater isoprenoid family of molecules to which it belongs (16). Built from the five carbon isomers isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) (Fig. 1) (17), a remarkable 97% of isoprenoid scaffolds are oxygenated (18), and the Yew specifically employs up to eight separate P450-mediated oxygenations to create Taxol (19). It is the P450’s unique capacity to selectively oxygenate inactive carbons that provides biosynthesis with its competitive advantage over traditional chemical approaches (20, 21). Accordingly, P450 optimization is seen as a key challenge for successful biosynthesis of Taxol and similar, complex natural products (22).
Taxol’s first P450 was identified nearly two decades ago. After discovering the synthase responsible for the diterpene backbone taxadiene, Croteau and coworkers reported the C-5 hydroxylation and concomitant allylic rearrangement performed by CYP725A4 as Taxol’s first functionalization (Fig. 1) (23). With both terpene synthase and P450 enzymes in hand, the Croteau and Jennewein groups even initiated the construction of heterologous biosynthetic pathways in Escherichia coli (24) and yeast (25), meeting relative success. A significant advance came when a multivariate modular metabolic engineering (MMME) approach was applied to optimize taxadiene production in E. coli (26). By grouping enzymes according to their metabolic branch points (i.e., chemical logic), an expression landscape was quickly explored to identify a balanced pathway construction, resulting in a 15,000-fold improvement in diterpene titer. Upon the addition of CYP725A4 to this strain, however, optimality was lost and titers fell considerably. One hypothesis was that E. coli was simply not an amenable host for P450 chemistry, leading to the design of a yeast coculture in a subsequent study (27). Here, however, we demonstrate E. coli’s suitability through the optimization of the P450 pathway module, identifying a set of conditions that deliver P450-inclusive pathway functionality and achieve a fivefold increase in oxygenated diterpene titer (∼570 mg/L) over previous work. In the process, we observed a problematic coupling between P450 expression and upstream pathway protein expression. The tools and strategies in this work should prove beneficial to other E. coli-based P450 chemistries, including subsequent engineering of the Taxol pathway, which likely includes several other P450s.
Results
Balancing P450 Module Expression.
In previous work (26), the addition of a P450 module into a diterpene-overproducing E. coli strain was found to measurably reduce titers, including the upstream taxadiene molecule. One hypothesized cause was the introduction of a second plasmid to the system. Plasmid-borne, metabolically burdensome pathways are known to be subject to rapid inactivation (28), and the use of a dual-plasmid system carrying an especially burdensome enzyme (P450) may have exacerbated this problem. Therefore, to avoid a dual-plasmid system moving forward, the upstream pathway was chromosomally localized into the E. coli chassis strain. Chromosomal integration was done as two separate modules, designed as operons, and parsed according to metabolic branch points. The first module “MEP” was comprised of the four previously identified rate-limiting enzymes of the IPP-producing methylerythritol phosphate pathway (dxs, idi, ispDF). The second “cyclase” module was comprised of taxadiene synthase (TxS) and geranylgeranyl pyrophosphate synthase (GGPPS). A brief exploration of expression strength was carried out, and single-copy integration under the tightly repressed T7 promoter was identified as optimal for each upstream module (SI Appendix, Fig. S1). Accordingly, this chromosomally integrated T7 MEP T7 TG strain was carried forward as the background for downstream P450 module testing.
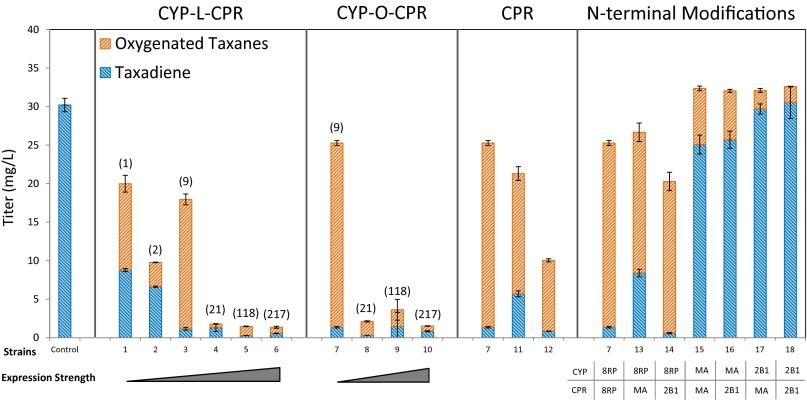
A second hypothesized cause for the poor productivity was that the addition of the P450 module disrupted the pathway’s previously engineered balance. To address this possibility, several strains with a range of P450 module expression strength were created, using our previously described chimeric P450-CPR construction (26). Each construction’s qualitative, relative expression strength was evaluated by using GFP (SI Appendix, Fig. S2), with values assigned by normalizing to a single chromosomal integration under a Trc promoter. Each strain’s relative expression value can be found parenthetical in Fig. 2, strains 1–10. Strain productivity was next evaluated by using small-scale (2-mL) fermentations. Interestingly, this assay shows that each strain with a T7 promoter (Fig. 2, strains 2, 5, and 6) underperforms its copy-matched Trc counterpart (Fig. 2, strains 1, 3, and 4), a weaker, “leaky” promoter unshared by the upstream pathway modules. Second, beyond a relative expression value of nine, the pathway nearly shuts off, including the upstream taxadiene module (Fig. 2, strains 4–6). By simply moving from a five-copy (Fig. 2, strain 3) to 10-copy plasmid system (Fig. 2, strain 4) with the same weak promoter, an order of magnitude decrease in overall terpene titers is observed. Ultimately, a five-copy plasmid under a Trc promoter (Fig. 2, strain 3) is identified as the optimal construction.
Fig. 2.
P450 module optimization screen. The control references the taxadiene producing chromosomally integrated strain that contains no P450 or CPR. The first graph depicts the physically linked chimera strains in order of increasing expression strength: single-copy chromosomal integration with Trc (1) and T7 (2), Trc on a five-copy (3) and 10-copy plasmid (4), and T7 on a five- (5) and 10-copy plasmid (6). Above each strain are given qualitative, relative expression values for the construct collected by GFP experiments and normalized to (1) (SI Appendix, Fig. S2). The second graph depicts the operon construction mimicking the expression strength of strains 3–6 (strains 7–10). The third graph depicts the strains with varying CPR expression, with the P450 held constant at Trc with a five-copy plasmid. Strain 11 corresponds to the CPR expressed from a single chromosomal integration with a T7 promoter. Strain 12 corresponds to the CPR expression with the P450 on the five copy plasmid, but with its own separate T7 promoter. The fourth graph depicts the strains with various N-terminal modifications to the P450 and CPR, ordered in increasing hydrophilicity.
Exploration of Reductase Partner Interactions.
A second major consideration for P450 systems is the inclusion of their reductase partner cytochrome P450 reductase (CPR). Class II microsomal P450s, such as CYP725A4, require a CPR to donate two NADPH-derived electrons during their catalytic cycle (29), with this interaction typically occurring at the endoplasmic reticulum (30). The location of this interaction presents an obstacle because bacteria notably lack such compartmentalization. Accordingly, it has been hypothesized that a physical fusion of these enzymes is necessary to mediate efficient P450/CPR interactions in E. coli (31). Such a construction mimics the highly effective P450 BM3, a natural chimera whose high turnover and native bacterial expression have made it a popular construction to replicate (18, 21). Examples of chimeras include our previous work, wherein CYP725A4 was physically linked to its native Taxus cuspidata CPR (26). Although functional, this approach does present some disadvantages. First, Taxol’s pathway likely includes several P450s, and physically linking each will require significant effort. Moreover, a chimera construction prevents the natural in planta phenomenon of CPR sharing (30), potentially precludes optimal protein–protein interaction through physical constraint, and eliminates the flexibility to modulate P450/CPR ratios. Therefore, motivation existed to examine the efficacy of unlinked systems.
The first approach was to simply create an operon construction of the P450 and CPR. In this format, four plasmids systems with increasing expression strength were tested in small-scale fermentation (Fig. 2, strains 7–10). As seen with the physically linked system, beyond a relative expression of nine (Fig. 2, strains 8–10) the pathway nearly shuts off and little isoprenoid is produced. Additionally, as with the chimera system, the five-copy plasmid Trc system (Fig. 2, strain 7) is found to be an optimal expression level. Interestingly, the overall performance of the operon system not only matched but exceeded the chimera construction, demonstrating that physically linking these enzymes is unnecessary and perhaps suboptimal.
Notably, the operon construction simultaneously modified two parameters with reference to the chimera, both physically unlinking the enzymes and lowering the relative expression of the CPR (32). By lowering CPR expression, it was possible that the reductase partner may have become catalytically limiting as seen in another P450 system (5). To test this hypothesis, two additional constructions were made with varying CPR expression, holding the P450 expression constant with the five-copy plasmid Trc system. Neither the lower expression of a single copy integrated into the chromosome with a T7 promoter nor the higher expression of a separate T7 promoter incorporated into the P450-bearing plasmid shows benefit (Fig. 2, strains 7, 11, and 12). The increased CPR expression of strain 12 had a more dramatic negative impact, possibly due to increased burden, resource competition, or inefficient NADPH utilization. Targeted proteomic analysis, discussed further below, shows the ratio of P450 to CPR in strain 7 to be ∼12. This result follows a trend observed for natural plant systems, wherein multiple P450s have been known to share a single CPR and are found in plant membranes at a ∼15:1 P450 to CPR ratio (30). In addition, it demonstrates another potential shortcoming for the fixed 1:1 ratio of the chimera.
Exploration of N-Terminal Modifications.
Another challenging element for plant P450 utilization in E. coli is their lipophilic N terminus (33). In planta, these hydrophobic tails direct P450 and CPR membrane localization, potentiating the aforementioned electron transfer interactions. However, because E. coli lack such internal membranes, these N termini can become a hindrance. Their lipophilicity decreases solubility, which can lead to increased formation of inclusion bodies and loss of functional protein. To circumvent these effects, N-terminal regions are often truncated and replaced with more hydrophilic peptide sequences. One particularly favored sequence to exchange for a native P450 N terminus has been an eight-residue peptide originally derived from bovine (8RP) (34), which has seen several successful applications (26, 31, 35).
Hypothesizing the benefit of N-terminal modification to predominantly be an effect of greater solubility and expression, we incorporated two increasingly hydrophilic modifications to the P450 and CPR in addition to the 8RP (SI Appendix, Figs. S3 and S4). All three modifications were incorporated into the previously truncated, codon optimized versions of these enzymes (26). The first modification was simply the addition of the two amino acids methionine and alanine (MA), because carrying an alanine at the second amino acid position has consistently shown benefit for P450 expression (36). The second was a hydrophilic leader sequence that had been demonstrated useful for the E. coli expression of an Arabidopsis P450 (2B1) (37).
Interestingly, however, although the MA and 2B1 modifications produced increased P450 enzyme in the soluble protein fraction of a protein gel compared with 8RP (SI Appendix, Figs. S5 and S6), the systems with these modifications show less turnover of taxadiene to its oxygenated downstream products compared with the 8RP modification. This effect is far more pronounced for P450 than for CPR modifications (Fig. 2, strains 7 and 13–18). One hypothesis is that the increased solubility disrupts the P450’s interaction with the lipid membrane and inhibits its ability to accept taxadiene as a substrate. The hydrophobic taxadiene molecule is likely concentrated in the E. coli membrane, and P450s are known to involve the flexible and notably lipophilic F-G loop in substrate acceptance (38). Therefore, if the increasingly hydrophilic modifications were strong enough, they may prevent the P450 from properly interacting with its substrate. This consideration would not present a problem for the 8RP modification, however, because it has been shown to be membrane associated (39, 40). In addition, with respect to the CPR modifications, alternative electrostatic protein–protein interactions (30) may help overcome the increased hydrophilicity of the CPR to allow for continued P450 interaction and electron donation. Lastly, although, the strains containing the more hydrophilic P450s catalyze less oxygenation, they do produce more total terpenes, potentially due to a relief of overall system stress or decrease in membrane crowding.
Targeted omics To Explore the Pathway Balancing in Modular Engineering.
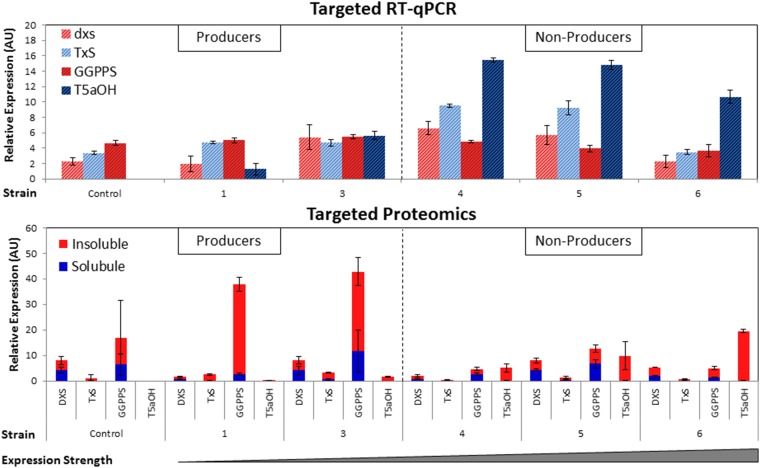
Our previous observation that increased P450 expression disrupted production even of upstream metabolites motivated further examination as to how this interaction was mediated. Accordingly, targeted transcriptomics and proteomics were conducted by using a representative subset of strains to examine the expression of specific pathway genes. For each strain, a time course sampling of 8, 24, 48, and 96 h was completed. From this sampling, 24 h was identified as exponential phase and represented the best differentiation among strains (SI Appendix, Fig. S7).
First, growth and productivity data show that the high expression strength, poor producing chimera strains (4–6) have lower growth during the intermediate time points, but nearly match the better producing strains’ final OD600 values (SI Appendix, Figs. S7 and S8). This finding potentially indicated an inactivating mutation relieving the heterologous pathway’s metabolic burden, but when these plasmids were harvested and sequenced after the fermentation, no mutations were found. Next, targeted quantitative RT-PCR (RT-qPCR) was carried out for several pathway genes. Using at least one representative gene from each pathway module, transcript-level analysis shows that increasing P450 expression seems to have little to no effect on the upstream pathway (Fig. 3A). In fact, two of the poor producing strains (Fig. 3B, strains 4 and 5) actually show higher taxadiene synthase mRNA levels compared with a high producer (Fig. 3B, strain 3). Targeted proteomics reveal a contrasting trend. On the protein level, for the poor producing strains, the upstream “cyclase” pathway decreases in the presence of high P450 expression (Fig. 3B, strains 4–6). In fact, even the further upstream MEP pathway enzymes are down-regulated on the protein level in the poor producing strains (SI Appendix, Fig. S9). It appears that upstream down-regulation is strongest at the earlier time points (SI Appendix, Figs. S9–S12), indicating the need for balanced protein expression from the beginning of the fermentation. This behavior is confirmed in the control, which is a high taxadiene producer without a P450, and shows most of its synthase expression early in the fermentation (SI Appendix, Figs. S9–S12). This observation explains why, although protein levels recover somewhat at later time points in the poor producing strains, their titers remain low, because downstream enzymes would still have little substrate on which to act. Taken together, this data clearly indicates an interdependency of pathway expression and protein level regulation. In addition, the suppressed MEP and cyclase protein levels explain the decrease in even upstream titers. These data shed light on the need to simultaneously balance heterologous enzyme expression for metabolic engineering and synthetic biology applications. Lastly, transcriptomic and proteomic experiments were conducted for the unlinked P450/CPR strains (SI Appendix, Figs. S13–S15), but no obvious trends are observed.
Fig. 3.
Targeted transcriptomic and proteomic data. Upper indicates the targeted transcriptomics results for the 24-h time point. Genes depicted include the first gene in each module’s operon. DOXP synthase (dxs) accounts for the MEP module, TxS for the synthase module, and taxadiene 5-α hydroxylase (T5aOH) for the P450 module. Lower indicates the targeted proteomics results. The insoluble fraction is depicted on top, whereas the soluble fraction is on the bottom. For both transcriptomic and proteomic figures, Left indicates strains with relative productivity, whereas Right indicates strains with poor productivity of terpene molecules.
Bioreactor Scale Fermentation of Oxygenated Taxane-Producing Strain.
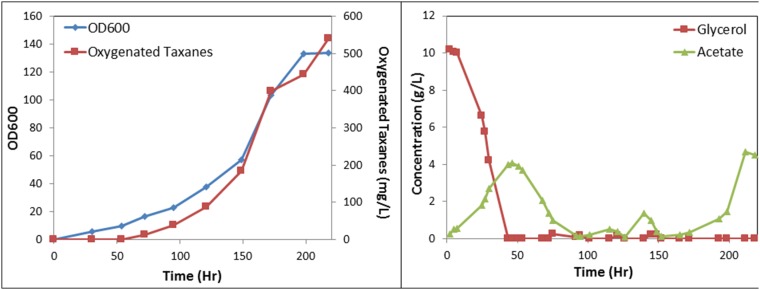
After identifying strain 7 as the optimum construction for oxygenated taxane production by using the microaerobic 2-mL fermentation assays, this strain was carried to scale-up fermentation in a 2-L benchtop fermenter. A brief temperature screen was conducted at 22, 25, and 30 °C, identifying 25 °C to provide faster growth without detriment to P450 catalytic activity (SI Appendix, Fig. S16). Acetate accumulation was minimized by maintaining glycerol concentration near 0 g/L. An initial bolus of 10 g/L was provided for the batch phase. Following batch phase, a modified exponential feeding program was initiated with timed boluses of 0.6 g/L supplemented glycerol and 0.3 g/L yeast extract. Fermentations were grown at 30 °C until an OD600 of 5, at which time the temperature was reduced to 25 °C and the culture was induced. A 20% (vol/vol) dodecane overlay was introduced 4 h after induction. Under these conditions, a titer of 570 ± 45 mg/L was obtained (Fig. 4). Duplicate bioreactor run data can be found in SI Appendix, Fig. S17. This total oxygenated taxane production is significantly higher than our previous work, in which a dual plasmid strain was reported to produce ∼116 mg/L of two oxygenated products. Even more, through our optimization, we observed the accumulation of two additional, previously uncharacterized, oxygenated taxane species (SI Appendix, Figs. S18–S25).
Fig. 4.
Bioreactor performance of strain 7. Left depicts the growth and productivity curves, represented in OD600 and oxygenated taxanes (mg/L) values. Right depicts glycerol and acetate concentrations throughout the duration of the fermentation.
Discussion
Biology’s role in chemical synthesis will increase in coming years. Natural systems possess immense chemical diversity, and harnessing their catalytic power looks to provide long-term environmental and socioeconomic benefits. However, metabolic engineers and synthetic biologists will need to establish new tools and strategies to take full advantage of this potential, especially in terms of developing systematic methods for using different classes of metabolic chemistries. The densely functionalized anticancer agent Taxol provides an excellent model system to approach such development, particularly because of its multiple selective oxygenations.
In nature, selective oxygenation is predominantly mediated by the cytochrome P450 superfamily of enzymes (14). Here, we engineered P450-mediated catalysis in E. coli for the third step in Taxol biosynthesis. By tuning different elements of the CYP725A4 module, E. coli is demonstrated to be a suitable host for plant P450-mediated chemistry. This finding contrasts with a growing expectation that yeast or other eukaryotic hosts are necessary for plant P450 chemistry. Through modular pathway engineering, we identified that the relatively low expression of a five-copy plasmid with a weak promoter was key for P450 and CPR functionality. This result may shed significant light on the perceived failure of previous E. coli P450 systems, because it is common to use higher copy plasmids and stronger promoters. Through altering reductase partner interactions, we found that chimera linkages are not necessary for bacterial expression of plant P450s and may even negatively impact overall function. In addition, we observed that lower CPR to P450 ratios are favorable. Through modulating the N termini of the P450 and CPR, we found that greater hydrophilicity relieved overall system stress, but that membrane association benefited high turnover. This membrane association may either be needed for substrate acceptance or to localize the P450 to the area of highest hydrophobic substrate concentration. Taken together, a strain was identified that rendered high-level oxygenated taxane production. Ultimately, this work advances Taxol metabolic engineering, which has seen stagnation in titer increases over the past half-decade.
In addition to strain construction, diagnostic targeted omics studies revealed that suboptimal strains were regulated on the protein level. In fact, an unprecedented interdependency of heterologous proteins was observed. High expression of a single pathway module (P450) broadly affected upstream pathway enzyme expression, undermining overall system performance. The exact cause of this behavior is unknown, however, because it manifests on the protein and not transcript level, it may be a result of the P450 enzyme monopolizing rare tRNA, chaperones, or dominating ribosome residence time. This understanding would concert with recent studies that found a limit to the “free ribosome pool,” (41–43) especially considering the growth retardation of our poor performing strains. This observation likely has relevance outside of the Taxol pathway and ought to be considered for other synthetic biology and metabolic engineering applications. As the field assembles increasingly complex systems, it will be paramount to effectively decouple protein interdependency, and this study highlights the potential magnitude of these interactions.
Work remains to fully elucidate the Taxol pathway, which increasingly appears more complex than originally thought. Crucial to the success of this and other complex natural product biosynthesis projects will be improved tools to identify, screen, and engineer the enzymes of plant-specialized metabolisms, especially the N-terminally anchored cytochrome P450s. Systematic approaches will greatly expedite progress and will especially benefit from an improved understanding of the key elements for catalytic function of each class of enzyme. This work lays a foundation for faster and more efficient expression of P450 enzymes in vivo in E. coli, with the hope that in the near future we will see wide-scale application of biosynthesis of complex and valuable natural products.
Materials and Methods
For details, see SI Appendix, SI Materials and Methods.
MEP, TG, and P450 Module Construction.
Construction of the MEP and TG module has been described (26). For the P450 module, various configurations for the P450 and CPR were made. First, the physically linked “chimera” with a GSTGS linker (l) and the operon 5′-GGATCCAAGGAGATATACC-3′ (o) structure were constructed. Second, the different N-terminal modifications, included the following: MA (incorporation of two residue peptide MA to the 42-aa N-terminally truncated P450 and to the 74-aa N-terminally truncated CPR), 8RP (incorporation of eight-residue peptide MALLLAVF to the 24-aa N-terminally truncated P450 and to the 74-aa N-terminally truncated CPR), and 2B1 (incorporation of 16-residue peptide MAKKTSSKGKLPPGPS to the 59-aa N-terminally truncated P450 and to the 89-aa N-terminally truncated CPR). A pictorial summary of these modifications can be found in SI Appendix, Figs. S3 and S4. Plasmids p5Trc, p5T7, p10Trc, and p10T7 (26) were used to clone the different configurations of P450 and CRP by two rounds of cloning using NdeI/SalI sites for P450 and BamHI/SalI sites for CPR, respectively. All primers used for this work can be found in SI Appendix, Table S2.
Chromosomal Integration.
Site-directed chromosomal integration was accomplished by homologous recombination mediated by λ-Red recombinase (induced from pKD46). Linear DNA for homologous recombination was generated by amplifying the pathway module with the FRT-flanked antibiotic resistance cassette from the appropriate template [pMB1CmFRT-TrcGFP and pMB1CmFRT-T7GFP for MMME expression level investigation; p10T7-TG-KmFRT for taxadiene expression; pMB1CmFRT-Trc-(8RP)P450-l-(MA)CPR, pMB1CmFRT-T7-(8RP)P450-l-(MA)CPR and pMB1CmFRT-T7-(8RP)CPR for oxygenated taxadiene expression] using primers to build in site-specific homologies toward lacY or malT loci. Positive transformants were cured from the antibiotic resistance cassette by using FLP recombinase (induced from pCP20). Successful chromosomal integration was confirmed by colony PCR and subsequent sequencing. All primers used are listed in SI Appendix, Table S2.
Small-Scale “Hungate” Fermentation.
For small-scale fermentation assays, the oxygenated taxane producing E. coli strains were first streaked on LB agar plates containing corresponding antibiotic and incubated overnight at 37 °C. Individual colonies were then picked and inoculated into 3 mL of LB media with appropriate antibiotic and incubated overnight (250 rpm, 30 °C) to achieve an approximate OD600 of 3. These LB precultures were then used to inoculate 2-mL microaerobic fermentations, completed in Chemglass Hungate tubes. Hungate 2-mL culture tubes and septas were purchased from ChemGlass. Screw caps for Hungate tubes were purchased from Kimble Chase. PrecisionGlide 26G × 3/8 [0.45 mm × 10 mm] needles were purchased from BD. Media components and concentrations can be found in SI Appendix, Tables S4 and S5. Briefly, basal R-media was prepared by mixing the components of SI Appendix, Table S4 with 1.5 mL of Antifoam B and adjusting the pH to 7.6 by using 6 M NaOH. Basal media components were then autoclaved at 121 °C for 30 min. The “poststerilization” media was then prepared as outlined in SI Appendix, Table S5, including the addition of 0.1 mM IPTG and corresponding antibiotic diluted 1000× from stock concentration. Total fermentation volume was comprised of 1.950 mL of complete R-media, 50 μL of LB preculture, and 200 μL of dodecane (10% overlay). Each vial was sealed with a rubber septa and screwcap, vortexed, and a single PrecisionsGlide Needle was inserted into the septa to allow for gaseous exchange. Initial production screening fermentations were run at 22 °C, 250 rpm for 4 d. After the fermentations were completed, OD600 values were measured and the culture was centrifuged to collect the dodecane overlay. This overlay was subsequently diluted into hexane for analytical procedures described below. All experiments were performed in triplicate.
Total RNA Quantification.
RT-PCR was performed by using the QuantiTect Multiplex RT-PCR kit using 50 ng of RNA per well, in accordance to the modified Quantitect Multiplex RT-PCR listed above. All RNA was isolated freshly before analysis to ensure that RNA did not degrade during storage. Threshold data were exported to analyze with the reference gene (hcaT), which was present in each well. The minimum information for publication of quantitative real-time PCR experiments (qPCR MIQE) guidelines were followed for all assay conditions and assay characteristics. MIQE checklist can be found with SI Appendix, SI Materials and Methods.
Targeted profiling of select proteins was performed by using an Agilent 6460 Triple Quadrupole Mass Spectrometer equipped with a jet-stream electrospray ionization source. Dynamic multiple reaction experiments were performed by using positive ion-spray mode. The source-dependent MS parameters such as gas temperature, gas flow, sheath gas temperature, and sheath gas flow were set at 325 °C, 12 L/min, 350 °C, and 11 L/min, respectively. Nebulizer pressure was set at 45 psi. Capillary and nozzle voltages were set to 4,500 V and 1,100 V, respectively. Peptide selection was performed by using Skyline 2.6 software, and collision energy, fragmentation voltage, and parent and product ion were determined and optimized for each specific protein.
Scale-Up Fermentation.
Benchtop scale-up fermentations were carried out in 2-L Sartorius-Stedim B-DCU II process units. A modified exponential time-dependent feeding program was used for fed-batch phase. This feeding program and other process parameters were controlled by using Sartorius S88 MFCS-win software package.
Using strain 7, fermentations were prepared as follows: a cell bank stock is first removed from −80 °C and streaked onto an LB agar plate containing 50 μg/L spectinomycin and incubated overnight at 37 °C. Single colonies are then picked and inoculated into 10 mL of LB broth containing 50 μg/L spectinomycin in a 50-mL Falcon tube and grown at 30 °C, 250 rpm until reaching ∼3 OD600. Next, 3 mL of this LB broth preculture is used to inoculate 100 mL of R-media complete in a 500-mL round bottom shake flask. Media compositions can be found in SI Appendix, Tables S4 and S5. The R-media preculture is then grown to ∼3 OD600, and 50 mL of this R-media preculture is used to inoculate the benchtop bioreactor.
Basal bioreactor R-media components can be found in SI Appendix, Table S6. For bioreactor preparation, these components are first dissolved into 500 mL of deionized (DI) H2O, the volume is filled to 925 mL, and then added into the bioreactor. Next, the bioreactor and basal R-media are autoclaved together. Following autoclaving, “poststerilization” components and 20% (vol/vol) dodecane overlay are added. A table of poststerilization components can found in SI Appendix, Tables S7 and S8. During fermentation, pH is maintained at 7.00 (±0.05) via a controller with 17% phosphoric acid and 3 M NaOH used as acid and base feeds, respectively. Initial conditions for agitation and aeration were 400 rpm and 0.75 liters per minute (lpm), respectively. Dissolved oxygen was maintained at ∼15%. Each bolus fed consisted of 0.6 g of supplemented glycerol and 0.3 g of yeast extract. Supplemented glycerol feed composition can be found in SI Appendix, Table S9. Feed yeast extract concentration is 400 g/L.
GC-MS Analysis.
GC-MS analysis was conducted by using an Agilent Technologies 7890A GC System with a 240 Ion Trap GC/MS, 7693 Autosampler, DB-5 column (Agilent), a splitless single taper glass wool ultrainert inlet liner, and 11-mm Restek premium nonstick center guiding septa. Injections were made in pulsed splitless mode, with an inlet temperature of 250 °C and pressure of 7.652 psi. The GC method proceeds as follows, 50 °C with a hold time of 1 min, followed by a 10 °C/min ramp to 200 °C (15 min), and a 5 °C/min ramp to 270 °C (14 min), for a total run time of 30 min. The postrun temperature is increased to 320 °C for 5 min between each sample. MS is collected between 30–350 m/z and from 17 to 30 min for each sample. Carrier gas is helium with a flowrate of 1 mL/min.
Samples were prepared by taking clear centrifuged overlay and diluting 100–200× into n-Hexanes and vortexing vigorously. Before each set of samples were run, washes of methanol and hexane were carried out twice. Per every six samples, and additional hexane wash was carried out. Serial dilutions of taxadiene were used to quantify approximate titer, adding 50-μL samples of taxadiene standards into GC vial inserts. Analysis was completed by using the Agilent MS Workstation software package. Oxygenated titer was counted by summing the integrated values for the four major oxygenated taxane species (m/z 288), whose retention times correspond 20.838 min, 21.114 min, 21.536 min, and 21.800 min. Figures depicting oxygenated taxane GC-MS can found in the SI Appendix, Figs. S14–S21).
Supplementary Material
Acknowledgments
We thank Prof. Keith E. J. Tyo, Dr. Christine N. S. Santos, and Dr. Ryan Philippe for their careful readings of this manuscript and Kang Zhou for his assistance in protein gel work.
Footnotes
Conflict of interest statement: C.G.L., S.S., G.S., M.D.M., and P.K.A. have financial interests in Manus Biosynthesis, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515826113/-/DCSupplemental.
References
- 1.Woolston BM, Edgar S, Stephanopoulos G. Metabolic engineering: Past and future. Annu Rev Chem Biomol Eng. 2013;4:259–288. doi: 10.1146/annurev-chembioeng-061312-103312. [DOI] [PubMed] [Google Scholar]
- 2.Jullesson D, David F, Pfleger B, Nielsen J. Impact of synthetic biology and metabolic engineering on industrial production of fine chemicals. Biotechnol Adv. 2015;33(7):1395–1402. doi: 10.1016/j.biotechadv.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Thodey K, Galanie S, Smolke CD. A microbial biomanufacturing platform for natural and semisynthetic opioids. Nat Chem Biol. 2014;10(10):837–844. doi: 10.1038/nchembio.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos CNS, Koffas M, Stephanopoulos G. Optimization of a heterologous pathway for the production of flavonoids from glucose. Metab Eng. 2011;13(4):392–400. doi: 10.1016/j.ymben.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Paddon CJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496(7446):528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 6.Paddon CJ, Keasling JD. Semi-synthetic artemisinin: A model for the use of synthetic biology in pharmaceutical development. Nat Rev Microbiol. 2014;12(5):355–367. doi: 10.1038/nrmicro3240. [DOI] [PubMed] [Google Scholar]
- 7.Shiue E, Prather KLJ. Synthetic biology devices as tools for metabolic engineering. Biochem Eng J. 2012;65:82–89. [Google Scholar]
- 8.Jones JA, et al. ePathOptimize: A combinatorial approach for transcriptional balancing of metabolic pathways. Sci Rep. 2015;5:11301. doi: 10.1038/srep11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones JA, Toparlak ÖD, Koffas MA. Metabolic pathway balancing and its role in the production of biofuels and chemicals. Curr Opin Biotechnol. 2015;33:52–59. doi: 10.1016/j.copbio.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Guerra-Bubb J, Croteau R, Williams RM. The early stages of taxol biosynthesis: An interim report on the synthesis and identification of early pathway metabolites. Nat Prod Rep. 2012;29(6):683–696. doi: 10.1039/c2np20021j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anarat-Cappillino G, Sattely ES. The chemical logic of plant natural product biosynthesis. Curr Opin Plant Biol. 2014;19:51–58. doi: 10.1016/j.pbi.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh CT, Fischbach MA. Natural products version 2.0: Connecting genes to molecules. J Am Chem Soc. 2010;132(8):2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizutani M. Impacts of diversification of cytochrome P450 on plant metabolism. Biol Pharm Bull. 2012;35(6):824–832. doi: 10.1248/bpb.35.824. [DOI] [PubMed] [Google Scholar]
- 14.Podust LM, Sherman DH. Diversity of P450 enzymes in the biosynthesis of natural products. Nat Prod Rep. 2012;29(10):1251–1266. doi: 10.1039/c2np20020a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pateraki I, Heskes AM, Hamberger B. Cytochromes P450 for terpene functionalisation and metabolic engineering. Adv Biochem Eng Biotechnol. 2015;148:107–139. doi: 10.1007/10_2014_301. [DOI] [PubMed] [Google Scholar]
- 16.Boutanaev AM, et al. Investigation of terpene diversification across multiple sequenced plant genomes. Proc Natl Acad Sci USA. 2015;112(1):E81–E88. doi: 10.1073/pnas.1419547112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohlmann J, Keeling CI. Terpenoid biomaterials. Plant J. 2008;54(4):656–669. doi: 10.1111/j.1365-313X.2008.03449.x. [DOI] [PubMed] [Google Scholar]
- 18.Renault H, Bassard J-E, Hamberger B, Werck-Reichhart D. Cytochrome P450-mediated metabolic engineering: Current progress and future challenges. Curr Opin Plant Biol. 2014;19:27–34. doi: 10.1016/j.pbi.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Kaspera R, Croteau R. Cytochrome P450 oxygenases of Taxol biosynthesis. Phytochem Rev. 2006;5(2-3):433–444. doi: 10.1007/s11101-006-9006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernhardt R, Urlacher VB. Cytochromes P450 as promising catalysts for biotechnological application: Chances and limitations. Appl Microbiol Biotechnol. 2014;98(14):6185–6203. doi: 10.1007/s00253-014-5767-7. [DOI] [PubMed] [Google Scholar]
- 21.Jung ST, Lauchli R, Arnold FH. Cytochrome P450: Taming a wild type enzyme. Curr Opin Biotechnol. 2011;22(6):809–817. doi: 10.1016/j.copbio.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang MCY, Keasling JD. Production of isoprenoid pharmaceuticals by engineered microbes. Nat Chem Biol. 2006;2(12):674–681. doi: 10.1038/nchembio836. [DOI] [PubMed] [Google Scholar]
- 23.Hefner J, et al. Cytochrome P450-catalyzed hydroxylation of taxa-4(5),11(12)-diene to taxa-4(20),11(12)-dien-5alpha-ol: The first oxygenation step in taxol biosynthesis. Chem Biol. 1996;3(6):479–489. doi: 10.1016/s1074-5521(96)90096-4. [DOI] [PubMed] [Google Scholar]
- 24.Huang Q, Roessner CA, Croteau R, Scott AI. Engineering Escherichia coli for the synthesis of taxadiene, a key intermediate in the biosynthesis of taxol. Bioorg Med Chem. 2001;9(9):2237–2242. doi: 10.1016/s0968-0896(01)00072-4. [DOI] [PubMed] [Google Scholar]
- 25.Engels B, Dahm P, Jennewein S. Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metab Eng. 2008;10(3–4):201–206. doi: 10.1016/j.ymben.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Ajikumar PK, et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330(6000):70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou K, Qiao K, Edgar S, Stephanopoulos G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol. 2015;33(4):377–383. doi: 10.1038/nbt.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyo KEJ, Ajikumar PK, Stephanopoulos G. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat Biotechnol. 2009;27(8):760–765. doi: 10.1038/nbt.1555. [DOI] [PubMed] [Google Scholar]
- 29.Munro AW, Girvan HM, Mason AE, Dunford AJ, McLean KJ. What makes a P450 tick? Trends Biochem Sci. 2013;38(3):140–150. doi: 10.1016/j.tibs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Jensen K, Møller BL. Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry. 2010;71(2-3):132–141. doi: 10.1016/j.phytochem.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 31.Leonard E, Koffas MAG. Engineering of artificial plant cytochrome P450 enzymes for synthesis of isoflavones by Escherichia coli. Appl Environ Microbiol. 2007;73(22):7246–7251. doi: 10.1128/AEM.01411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim HN, Lee Y, Hussein R. Fundamental relationship between operon organization and gene expression. Proc Natl Acad Sci USA. 2011;108(26):10626–10631. doi: 10.1073/pnas.1105692108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitaoka N, Wu Y, Xu M, Peters RJ. Optimization of recombinant expression enables discovery of novel cytochrome P450 activity in rice diterpenoid biosynthesis. Appl Microbiol Biotechnol. 2015;99(18):7549–7558. doi: 10.1007/s00253-015-6496-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barnes HJ, Arlotto MP, Waterman MR. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc Natl Acad Sci USA. 1991;88(13):5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang MCY, Eachus RA, Trieu W, Ro D-K, Keasling JD. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol. 2007;3(5):274–277. doi: 10.1038/nchembio875. [DOI] [PubMed] [Google Scholar]
- 36.Zelasko S, Palaria A, Das A. Optimizations to achieve high-level expression of cytochrome P450 proteins using Escherichia coli expression systems. Protein Expr Purif. 2013;92(1):77–87. doi: 10.1016/j.pep.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 37.Morrone D, Chen X, Coates RM, Peters RJ. Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin biosynthesis. Biochem J. 2010;431(3):337–344. doi: 10.1042/BJ20100597. [DOI] [PubMed] [Google Scholar]
- 38.Cojocaru V, Winn PJ, Wade RC. The ins and outs of cytochrome P450s. Biochim Biophys Acta. 2007;1770(3):390–401. doi: 10.1016/j.bbagen.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Tang Z, Salamanca-Pinzón SG, Wu Z-L, Xiao Y, Guengerich FP. Human cytochrome P450 4F11: Heterologous expression in bacteria, purification, and characterization of catalytic function. Arch Biochem Biophys. 2010;494(1):86–93. doi: 10.1016/j.abb.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shukla A, Huang W, Depaz IM, Gillam EM. Membrane integration of recombinant human P450 forms. Xenobiotica. 2009;39(7):495–507. doi: 10.1080/00498250902934884. [DOI] [PubMed] [Google Scholar]
- 41.Ceroni F, Algar R, Stan G-B, Ellis T. Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat Methods. 2015;12(5):415–418. doi: 10.1038/nmeth.3339. [DOI] [PubMed] [Google Scholar]
- 42.Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of cell growth and gene expression: Origins and consequences. Science. 2010;330(6007):1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 43.Carrera J, Rodrigo G, Singh V, Kirov B, Jaramillo A. Empirical model and in vivo characterization of the bacterial response to synthetic gene expression show that ribosome allocation limits growth rate. Biotechnol J. 2011;6(7):773–783. doi: 10.1002/biot.201100084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.