Significance
Cytokinesis occurs along a cell’s short axis in many organisms, including bacteria, archaea, and eukaryotes. In many protozoa it occurs along the cell’s longitudinal axis. The mechanism underlying this mode of cytokinesis is unknown. We delineate the signaling cascade that regulates cytokinesis along the longitudinal axis of Trypanosoma brucei, which is totally different from that in its human host. Additionally, we discover an alternative cytokinesis pathway that drives trypanosome cell division along the same division plane as the typical cytokinesis, but in an opposite direction. This alternative cytokinesis is activated only when the typical cytokinesis pathway is defective, suggesting that trypanosomes have evolved a backup cytokinesis mechanism to prevent the failure of cell division, thereby ensuring survival of this organism.
Keywords: cytokinesis, Polo-like kinase, Aurora B kinase, backup cytokinesis, Trypanosoma brucei
Abstract
Cytokinesis in Trypanosoma brucei, an early branching protozoan, occurs along its longitudinal axis uni-directionally from the anterior tip of the new flagellum attachment zone filament toward the cell’s posterior end. However, the underlying mechanisms remain elusive. Here we report that cytokinesis in T. brucei is regulated by a concerted action of Polo-like kinase, Aurora B kinase, and a trypanosome-specific protein CIF1. Phosphorylation of CIF1 by Polo-like kinase targets it to the anterior tip of the new flagellum attachment zone filament, where it subsequently recruits Aurora B kinase to initiate cytokinesis. Consistent with its role, CIF1 depletion inhibits cytokinesis initiation from the anterior end of the cell, but, surprisingly, triggers cytokinesis initiation from the posterior end of the cell, suggesting the activation of an alternative cytokinesis from the opposite cell end. Our results reveal the mechanistic roles of CIF1 and Polo-like kinase in cytokinesis initiation and elucidate the mechanism underlying the recruitment of Aurora B kinase to the cytokinesis initiation site at late anaphase. These findings also delineate a signaling cascade controlling cytokinesis initiation from the anterior end of the cell and uncover a backup cytokinesis that is initiated from the posterior end of the cell when the typical anterior-to-posterior cytokinesis is compromised.
In eukaryotes, regulation of cytokinesis, the final step of cell division, involves a complex interplay of numerous proteins at the cytokinesis initiation site and the cleavage furrow. The mechanisms underlying cytokinesis in fungi and metazoa have been well understood, and the core regulatory pathways appear to be evolutionarily conserved (1). Along the cell division plane, which is defined by the position of the central spindle or the nucleus, animals and fungi assemble an actomyosin contractile ring, the cytokinesis apparatus that appeared about 1 billion years ago in the common ancestor of fungi, amoebas, and animals (2). In metazoa, the signaling pathway driving the transition from mitosis to cytokinesis involves two evolutionarily conserved protein kinases, the Polo-like kinase and the Aurora B kinase. Both kinases are concentrated on the central spindle and the midbody during late cell cycle stages and cooperate to recruit the centralspindlin complex to the central spindle and the midbody (3). Subsequently, the centralspindlin complex recruits Ect2, a guanine nucleotide exchange factor, to the midbody, which then recruits and activates the small GTPase RhoA at the midbody. Activation of RhoA further promotes the formation of the actomyosin contractile ring to drive cytokinesis (3).
Unlike many eukaryotic organisms that divide along the cell’s short axis, the early branching protozoan Trypanosoma brucei undergoes cytokinesis along its longitudinal axis (4). The cell division plane in T. brucei is positioned by the newly assembled flagellum and its associated cytoskeletal structure termed the flagellum attachment zone (FAZ) filament (5, 6). Thus, cytokinesis is initiated from the anterior tip of the new FAZ filament, and cleavage furrow ingression occurs uni-directionally along the longitudinal axis toward the posterior end of the cell (4, 7) without forming an actomyosin contractile ring at the cleavage furrow (8).
As in fungi and metazoa, the Polo-like kinase (TbPLK in T. brucei) and the Aurora B kinase (TbAUK1 in T. brucei) are also required for cytokinesis in T. brucei (9, 10). TbPLK is concentrated in the flagellar basal body and the bilobe at late G1 phase, but from early S phase it is concentrated at the new FAZ tip and remains there until early anaphase (11). At the new FAZ tip, TbPLK is believed to promote cytokinesis initiation, but the underlying mechanism is unclear. TbAUK1 forms an unusual chromosomal passenger complex (CPC) with TbCPC1 and TbCPC2, and the complex displays a dynamic localization during the cell cycle. The complex is located in kinetochores from S phase to metaphase and on the central spindle during anaphase, but finally is degraded at the central spindle after late anaphase. However, starting from late anaphase, newly synthesized CPC proteins emerge at the new FAZ tip and then transfer to the cleavage furrow during cytokinesis (12). Localization of TbAUK1 to the new FAZ tip at late anaphase is crucial for cytokinesis initiation (13), but how it is recruited remains mysterious. The sequential recruitment of TbPLK and TbAUK1 to the new FAZ tip led us to hypothesize that an unknown factor is targeted by TbPLK to the new FAZ tip, which subsequently recruits TbAUK1 for cytokinesis initiation.
Here we report the identification of this factor, named CIF1, that links the TbPLK- and TbAUK1-signaling pathways. We also report the delineation of a cytokinesis regulatory pathway in T. brucei, which involves a concerted action of TbPLK, CIF1, and TbAUK1 at the anterior tip of the new FAZ filament. CIF1 is targeted, by TbPLK-mediated phosphorylation, to the anterior tip of the new FAZ filament, where it subsequently recruits TbAUK1 to drive cytokinesis initiation. We also discover a backup cytokinesis pathway that promotes cell division initiation from the posterior end of the cell. T. brucei thus has evolved two distinct cytokinesis pathways that drive cell division along the same division plane but in opposite directions.
Results
CIF1 Colocalizes with TbPLK at Early Cell Cycle Stages and with TbAUK1 at Late Cell Cycle Stages.
To identify the factor(s) that interacts with both TbPLK and TbAUK1, we carried out yeast two-hybrid library screening using the kinase-dead TbPLK (TbPLK-K70R) and TbAUK1 as baits. A hypothetical protein (Tb927.11.15800), which was previously identified as a partner of TbPLK-K70R by us (14), was also identified as a partner of TbAUK1 (Fig. S1 A and B). We named it CIF1 for Cytokinesis Initiation Factor 1 due to its essential role in cytokinesis initiation (see below). CIF1 contains two coiled-coil motifs between residues 125–205 and 218–245 and two putative zinc-finger motifs between residues 680–709 and 758–779 and is conserved in kinetoplastid parasites.
Fig. S1.
Identification of CIF1 by yeast two-hybrid screening with TbPLK-K70R and TbAUK1 as baits and the subcellular localization of CIF1. TbPLK-K70R, the kinase-dead mutant, instead of TbPLK, was used for yeast two-hybrid library screening because wild-type TbPLK does not interact with its substrates in yeast two-hybrid. (A) Confirmation of the interaction between CIF1 and TbPLK-K70R by directional yeast two-hybrid. Growth of yeast on the plates lacking tryptophan (Trp), leucine (Leu), and histidine (His) indicates interaction between the two tested proteins. (B) Confirmation of the interaction between CIF1 and TbAUK1 by directional yeast two-hybrid. Note that CIF1 does not interact with TbCPC1 and TbCPC2. (C) CIF1 is localized to the new FAZ tip from S phase to telophase and the cleavage furrow during cytokinesis. CIF1 was endogenously tagged with a triple HA epitope at its C terminus. Cells were immunostained with FITC-conjugated anti-HA mAb to detect CIF1-3HA and anti-CC2D pAb to label the FAZ filament. Images were all acquired with the same exposure time and were not further adjusted. N, nucleus; K, kinetoplast DNA. (Scale bars: 5 μm.)
We investigated the spatiotemporal distribution of CIF1 and the colocalization of CIF1 with TbPLK and TbAUK1 during the cell cycle. During G1 phase, CIF1 was not detectable, but at early S phase, a small amount of CIF1 was detected at the newly assembled FAZ filament, and its level appeared to increase significantly at the tip of the elongating new FAZ subsequently (Fig. S1C). CIF1 remained at the new FAZ tip until telophase and finally was concentrated at the cleavage furrow during cytokinesis (Fig. S1C). At G1 phase, TbPLK was concentrated at the basal body and the bilobe structure (Fig. 1A), but at early S phase, TbPLK was detected at the newly assembled FAZ filament, where it colocalized with CIF1 (Fig. 1A). TbPLK disappeared from the new FAZ tip at late anaphase, whereas CIF1 remained (Fig. 1A). Intriguingly, when TbPLK disappeared at late anaphase, a small amount of TbAUK1 was detected at the new FAZ tip, where it colocalized with CIF1 (Fig. 1B). Both TbAUK1 and CIF1 finally localized to the cleavage furrow during cytokinesis (Fig. 1B). This temporal relationship of localization patterns suggests that TbPLK may target CIF1 to the new FAZ tip, where CIF1 subsequently recruits TbAUK1 to the new FAZ tip.
Fig. 1.
CIF1 colocalizes with TbPLK during early cell cycle stages and with TbAUK1 during late cell cycle stages. (A) CIF1 colocalizes with TbPLK at the new FAZ tip from early S phase to early anaphase. CIF1-3HA was detected with FITC-conjugated anti-HA mAb and TbPLK was detected with anti-TbPLK pAb. N, nucleus; K, kinetoplast DNA. (B) CIF1 colocalizes with TbAUK1 at the new FAZ tip from late anaphase to telophase and at the cleavage furrow during cytokinesis. CIF1-PTP was labeled with anti-Protein A pAb, and TbAUK1-3HA was stained with FITC-conjugated anti-HA mAb. (Scale bars: 5 μm.)
TbPLK Phosphorylation of CIF1 Is Required for Localizing CIF1 to the New FAZ Tip.
CIF1 is a highly phosphorylated protein, containing 29 in vivo phospho-serine and phospho-threonine sites (15). To test whether CIF1 is an in vivo substrate of TbPLK, we treated the cells with GW843682X, a small-molecule Polo-like kinase inhibitor (16, 17). CIF1 was detected as a doublet on the Western blot, and treatment with GW843682X reduced the level of the upper band of CIF1, which was confirmed to be the phosphorylated form of CIF1, suggesting de-phosphorylation of CIF1 by TbPLK inhibition (Fig. 2A). Similarly, depletion of TbPLK by RNAi (Fig. S2 A–C) also caused CIF1 de-phosphorylation (Fig. 2B). This inhibition of TbPLK activity and depletion of TbPLK protein both disrupted CIF1 localization to the new FAZ tip (Fig. 2 C–F), suggesting that phosphorylation of CIF1 by TbPLK is required to target CIF1 to the new FAZ tip.
Fig. 2.
Phosphorylation of CIF1 by TbPLK targets CIF1 to the new FAZ tip. (A and B). Inhibition of TbPLK activity by GW843682X (A) and depletion of TbPLK (B) caused de-phosphorylation of CIF1. CIF1-3HA was detected with anti-HA antibody. TbPSA6, a subunit of the 26S proteasome, served as the loading control. p-CIF1-3HA, phosphorylated CIF1-3HA. The asterisk indicates a nonspecific band. (C) Inhibition of TbPLK activity with GW843682X abolished CIF1 localization to the new FAZ tip. (D). Quantification of CIF1 localization in control and TbPLK-inhibited binucleate cells. (E) TbPLK depletion disrupted CIF1 localization. (F) Quantification of CIF1 localization in control and TbPLK-depleted binucleate cells. In C and E, CIF1-3HA was detected with anti-HA, and the FAZ filament was labeled with anti-CC2D. N, nucleus; K, kinetoplast DNA. (Scale bars: 5 μm.) In D and F, 300 binucleate cells were counted for each cell line, and results are presented as mean percentage ± SD (n = 3). ***P < 0.001.
Fig. S2.
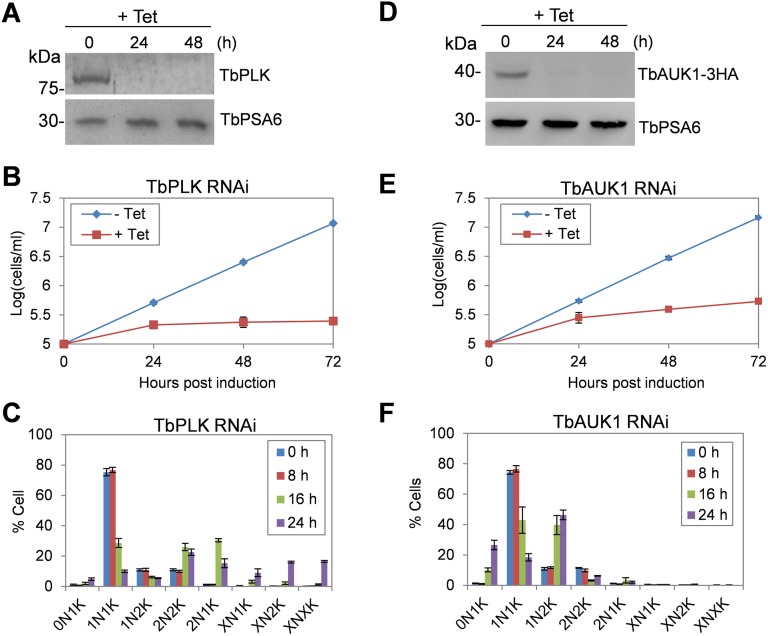
Confirmation of TbPLK and TbAUK1 RNAi cell lines used in the current work. (A) Western blotting to monitor the depletion of TbPLK after RNAi induction with tetracycline (Tet). TbPLK was detected with anti-TbPLK pAb, and TbPSA6 served as the loading control. (B) TbPLK depletion arrested cell growth. Shown is the growth curve of cells incubated without (−Tet) and with (+Tet) tetracycline for 72 h. (C) TbPLK depletion caused defective kinetoplast segregation and cytokinesis. N, nucleus; K, kinetoplast DNA. A total of 300 cells were counted, and error bars indicate SD calculated from three independent experiments. (D) Western blotting to monitor the depletion of TbAUK1 after RNAi induction with Tet. TbAUK1 was endogenously tagged with a triple HA epitope in cells harboring the TbAUK1 RNAi construct. TbAUK1-3HA was detected with anti-HA mAb, and TbPSA6 served as the loading control. (E) TbAUK1 depletion arrested cell growth. Shown is the growth curve of cells incubated without (−Tet) and with (+Tet) tetracycline for 72 h. (F) TbAUK1 depletion caused defective mitosis and cytokinesis. N, nucleus; K, kinetoplast DNA. A total of 300 cells were counted, and error bars indicate SD calculated from three independent experiments.
CIF1 Maintains TbPLK at the New FAZ Tip.
We next investigated the effect of CIF1 depletion on TbPLK localization to the new FAZ tip. Depletion of CIF1 by RNAi (Fig. S3) impaired the localization of TbPLK to the new FAZ tip, leading to the accumulation of TbPLK in the basal body and the bilobe structure in ∼40% of the binucleate cells (Fig. S4 A and B). At the bilobe, TbPLK phosphorylates TbCentrin2 at Ser-54 during late G1 and early S phase, but after TbPLK localizes to the new FAZ tip, TbCentrin2 is de-phosphorylated (18). To test whether TbCentrin2 was phosphorylated in CIF1-deficient binucleate cells, cells were immunostained with the PS54 antibody, which detects the phosphorylated Ser-54 in TbCentrin2 (18), and the results showed that TbCentrin2 was indeed phosphorylated (Fig. S4 C and D). This result provided another line of evidence to confirm that TbPLK was accumulated in the bilobe in a certain population of binucleate cells upon CIF1 depletion. Taken together, these results suggest that CIF1, once targeted to the new FAZ tip by TbPLK phosphorylation, in turn maintains TbPLK at the new FAZ tip. The two proteins thus are interdependent for their localization to the new FAZ tip.
Fig. S3.
Depletion of CIF1 by RNAi caused a cytokinesis defect. (A) Western blotting to monitor the depletion of CIF1-3HA after RNAi induction by tetracycline (Tet). TbPSA6 served as the loading control. (B) CIF1 depletion caused growth defects, but not growth arrest. (C) Depletion of CIF1 caused accumulation of binucleate (2N) and multinucleate (XN, X > 2) cells, suggesting a cytokinesis defect. Anucleate cells include 0N1K and 0N2K cells. N, nucleus; K, kinetoplast DNA. A total of 300 cells were counted, and error bars indicate SD calculated from three independent experiments. (D) Depletion of CIF1 caused cytokinesis furrow ingression from the posterior end of the cell. Cells were immunostained with anti-CC2D antibody to label the FAZ and then counterstained with DAPI to stain nuclear DNA and kinetoplast DNA (small blue dots). (E) Depletion of CIF1 did not affect new FAZ elongation. A total of 200 2N2K cells were analyzed, and error bars indicate SD calculated from three independent experiments. (F) Depletion of CIF1 produced anucleate daughter cells (0N1K and 0N2K), normal daughter cells (1N1K), and multinucleate daughter cells (XN, X > 2). Cells were immunostained with anti-CC2D antibody to label the FAZ and then counterstained with DAPI to stain nuclear DNA and kinetoplast DNA. The posterior end of the cells was aligned to the left. (Scale bars: 5 µm.)
Fig. S4.
CIF1 exerts a feedback control on TbPLK by maintaining TbPLK at the new FAZ tip. (A) Localization of TbPLK in cells containing two nuclei and two kinetoplast DNA (2N2K), which include anaphase and telophase cells. Cells were coimmunostained with anti-TbPLK and 20H5, which labels centrins in the flagellar basal body (BB), the bilobe structure, and the flagellum. N, nucleus; K, kinetoplast DNA. (Scale bar: 5 µm.) (B) Quantification of cells with different TbPLK localization patterns. A total of 200 cells were counted for each cell type, and error bars represent SD calculated from three independent experiments. (C) Effect of CIF1 depletion on TbPLK-mediated TbCentrin2 phosphorylation in 2N2K cells. Noninduced control and CIF1 RNAi-induced cells were immunostained with PS54 for Ser-54-phosphorylated TbCentrin2 and 20H5 for the bilobe structure. The white arrow indicates the cleavage furrow at the posterior end of the cell. (Scale bar: 5 μm.) (D) Quantification of the 2N2K cells with different PS54 localization patterns. A total of 200 cells were counted for each time point, and error bars represent SD calculated from three independent experiments.
CIF1 Is Required for Localizing TbAUK1 to the New FAZ Tip.
Given that CIF1 and TbAUK1 colocalize at the new FAZ tip at late anaphase, we investigated whether CIF1 is required for localization of TbAUK1 to the new FAZ tip. In CIF1 RNAi cells, TbAUK1 localization to the new FAZ tip was disrupted (Fig. 3 A and B), despite the fact that the TbAUK1 protein level was not changed (Fig. 3C), indicating that TbAUK1 was dispersed in the cytosol. These results suggest that CIF1 recruits TbAUK1 to the new FAZ tip at late anaphase. It should be noted that the lack of TbAUK1 in the central spindle in CIF1-deficient binucleate cells was not a direct effect of CIF1 depletion. The fact that CIF1 RNAi arrested cells before cytokinesis initiation (see below) suggests that the CIF1-deficient binucleate cells have already exited mitosis and hence have disassembled the mitotic spindle. In terms of the cell cycle stage, these binucleate cells were equivalent to the telophase cells in wild-type cells, in which TbAUK1 is not detected in the central spindle (Fig. 1B).
Fig. 3.
CIF1 recruits TbAUK1 to the new FAZ tip. (A) CIF1 depletion abolished TbAUK1 localization to the new FAZ tip. TbAUK1-3HA was labeled with FITC-conjugated anti-HA mAb, and the FAZ filament was detected by anti-CC2D pAb. The white arrow in the DIC/DAPI panel indicates the cleavage furrow at the posterior end of the CIF1 RNAi cell. N, nucleus; K, kinetoplast DNA. (Scale bar: 5 μm.) (B) Quantification of TbAUK1 localization in control and CIF1-depleted cells. A total of 300 binucleate cells were counted for each cell line, and results were presented as mean percentage ±SD (n = 3). ***P < 0.001. (C) TbAUK1 protein level was not changed in CIF1 RNAi cells. TbAUK1-3HA was detected with anti-HA antibody. TbPSA6 served as the loading control. (D) Both inhibition of TbAUK1 activity with Hesperadin and depletion of TbAUK1 by RNAi caused de-phosphorylation of CIF1. p-CIF1-3HA, phosphorylated CIF1-3HA. The asterisk indicates a nonspecific band. (E) Inhibition of TbAUK1 activity or depletion of TbAUK1 did not abolish CIF1 localization to the new FAZ tip. CIF1-3HA was detected by anti-HA, and the FAZ filament was labeled by anti-CC2D. (Scale bar: 5 µm.)
We next investigated whether CIF1 is a substrate of TbAUK1 Both inhibition of TbAUK1 activity with Hesperadin, a small-molecule Aurora B kinase inhibitor (19, 20), and depletion of TbAUK1 by RNAi (Fig. S2 D–F) caused CIF1 de-phosphorylation (Fig. 3D), indicating that CIF1 is an in vivo substrate of TbAUK1. However, inhibition of TbAUK1 activity with Hesperadin or depletion of TbAUK1 by RNAi did not affect CIF1 localization to the new FAZ tip (Fig. 3E), suggesting that TbAUK1-mediated phosphorylation of CIF1 is not required to maintain CIF1 at the new FAZ tip from late anaphase to telophase.
TbPLK Is Required for Targeting TbAUK1 to the New FAZ Tip.
If a signaling cascade from TbPLK through CIF1 to TbAUK1 does exist, then inhibition of TbPLK activity or depletion of TbPLK should also impair TbAUK1 localization to the new FAZ tip. To test this possibility, cells were treated with GW843682X, and the localization of TbAUK1 in TbPLK-inhibited cells was examined. This inhibition of TbPLK activity disrupted TbAUK1 localization to the new FAZ tip (Fig. S5 A and B), but did not affect the TbAUK1 protein level (Fig. S5C), suggesting that TbAUK1 was dispersed in the cytosol.
Fig. S5.
TbPLK is required for targeting TbAUK1 to the new FAZ tip. (A) Inhibition of TbPLK activity with GW843682X disrupted TbAUK1 localization to the new FAZ tip. TbAUK1-3HA was labeled with FITC-conjugated anti-HA mAb and CIF1-PTP was detected with anti-Protein A pAb. N, nucleus; K, kinetoplast DNA. (B) Quantification of TbAUK1 localization in control and TbPLK-inhibited cells. (C) TbAUK1 protein level was not changed by TbPLK inhibition. TbAUK1-3HA was detected with anti-HA antibody. TbPSA6 served as the loading control. (D) Depletion of TbPLK impaired TbAUK1 localization to the new FAZ tip. (E) Quantification of TbAUK1 localization in control and TbPLK RNAi cells. (F) TbAUK1 protein level was not changed in TbPLK RNAi cells. In B and E, a total of 300 binucleate cells were counted for each cell line, and results were presented as mean percentage ± SD (n = 3). ***P < 0.001. (Scale bars: 5 μm.)
To further confirm the requirement of TbPLK for TbAUK1 localization to the new FAZ tip, we examined the localization of TbAUK1 in TbPLK RNAi cells, and the results showed that depletion of TbPLK by RNAi also impaired TbAUK1 localization to the new FAZ tip (Fig. S5 D and E). Similar to TbPLK inhibition (Fig. S5C), depletion of TbPLK did not alter the TbAUK1 protein level (Fig. S5F). These results indicate that TbPLK is required for localizing TbAUK1 to the new FAZ tip and also suggest that cytokinesis initiation in T. brucei requires a concerted action of TbPLK, CIF1, and TbAUK1 at the new FAZ tip.
CIF1 Depletion Inhibits Cytokinesis Initiation from the Cell Anterior, but Triggers Cytokinesis Initiation from the Cell Posterior.
CIF1 depletion caused an accumulation of binucleate cells initially and multinucleate (four or more nuclei) cells subsequently (Fig. S3C), suggesting a cytokinesis defect. We therefore investigated the effect of CIF1 depletion on cleavage furrow ingression. In wild-type cells, a division fold is formed along the longitudinal axis of the cell between the two flagella/FAZs before cytokinesis initiation (Fig. 4A). Subsequently, cleavage furrow ingression occurs from the anterior tip of the new FAZ filament and proceeds along the division fold toward the posterior end of the cell (Fig. 4 B–E, Fig. S6A, and Movie S1). At late cytokinesis stage, the two daughter cells are connected at the posterior region by a cytoplasmic bridge (Fig. 4E), which is finally cleaved.
Fig. 4.
CIF1 depletion inhibits cytokinesis initiation from the cell anterior, but leads to cytokinesis from the cell posterior. (A) Formation of a division fold along the longitudinal axis in a control cell. (B–E). Cleavage furrow ingression occurs from cell anterior to cell posterior in control cells. (E) Two daughter cells connected by a cytoplasmic bridge at the posterior region of the cells. (F) Formation of a division fold along the longitudinal axis in a CIF1 RNAi cell. (G and H) Cleavage furrow ingression occurs from cell posterior to cell anterior in CIF1 RNAi cells. (H) Two daughter cells connected by a cytoplasmic bridge at the anterior region of the cells. (Scale bars: 5 μm.) (I) Quantification of cells without and with visible cleavage furrow(s) in control and CIF1 RNAi cells. Binucleate and multinucleate cells (200 cells for each cell type and each time point) were counted. Data were presented as mean percentage ±SD (n = 3). ***P < 0.001. (J) Model for cytokinesis initiation regulated by TbPLK, CIF1, and TbAUK1 and the existence of a backup cytokinesis in T. brucei. A, anterior; P, posterior.
Fig. S6.
Depletion of CIF1 triggered cytokinesis initiation from the posterior end of the cell. (A) Cytokinesis in control cells is initiated from the anterior end of the new FAZ and proceeds to the posterior end of the cell. (B) Cytokinesis in CIF1 RNAi-induced cells is initiated from the posterior end of the cell and proceeds to the anterior end of the cell. White arrows indicate the direction of cleavage furrow ingression. The open arrowhead shows the cleavage of the cytoplasmic bridge that connects the two daughter cells at the anterior ends. The posterior of the cell is up. K, kinetoplast DNA (the small blue dot); N, nucleus. (Scale bars: 5 µm.)
In CIF1 RNAi cells, a similar division fold was also formed along the longitudinal axis of the cell between the two flagella/FAZs (Fig. 4F), but furrow ingression from the anterior tip of the new FAZ filament was inhibited (Fig. 4 G–I and Fig. S7), suggesting that CIF1 is required for cytokinesis initiation from the anterior tip of the new FAZ filament. Strikingly, CIF1-deficient cells started to initiate cytokinesis from the posterior end of the cell toward the anterior end of the cell, albeit at a rate slower than the typical anterior-to-posterior cytokinesis (Fig. 4 G and H, Figs. S6B and S7, and Movie S2). A posterior cleavage furrow was found in ∼15 and ∼37% of the binucleate cells after CIF1 RNAi for 24 and 48 h, respectively, and in ∼61% of the multinucleate cells after CIF1 RNAi for 48 h (Fig. 4I). At late cytokinesis stage, the two daughter cells were also connected by a cytoplasmic bridge (Fig. 4H and Fig. S7 J and K), similar to that in the wild-type cell (Fig. 4E), but at the anterior end. These results suggest that inhibition of the typical anterior-to-posterior cytokinesis activated an alternative, backup cytokinesis from the opposite cell end. This backup cytokinesis appeared to use the same division plane as the typical cytokinesis (Fig. 4 A and F) and was initiated in binucleate cells, but due to its slower rate, these cells became multinucleated (four or more nuclei) before cell division was completed (Fig. 4H and Figs. S6B and S7). Consequently, this alternative cytokinesis appeared to generate two binucleate daughter cells with two flagella (Fig. 4H and Fig. S6B), instead of two uni-nucleate daughter cells as in the wild-type cells (Fig. 4E and Fig. S6A). Moreover, many of the multinucleate cells started to assemble additional cleavage furrows (Figs. S3D and S7 K and L) at the posterior end of the cell and, consequently, were likely to generate multiple (more than two) daughter cells (Figs. S3D and S7K). In some of the multinucleate cells, mitosis and the alternative cytokinesis appeared to be poorly coordinated, leading to the generation of anucleate daughter cells that contained only the kinetoplast DNA, the cell’s unique mitochondrial DNA complex (Figs. S3F and S6B).
Fig. S7.
Scanning electron microscopic analysis of CIF1 RNAi cells showing the initiation of cytokinesis from the posterior end of the cell. (A–I) CIF1 RNAi cells undergoing cytokinesis from the cell posterior at early-to-intermediate stages. (A–E) View of cells from the side of flagella, (F–I) View of cells from the opposite side of the flagella. (J) A CIF1 RNAi cell at the late stage of cytokinesis from the cell posterior, forming a cytoplasmic bridge at the anterior end of the daughter cells. (K) A CIF1 RNAi cell undergoing multiple posterior-to-anterior cytokinesis with one daughter cell connected to the other daughter cells by a cytoplasmic bridge. (L) A CIF1 RNAi cell undergoing posterior-to-anterior cytokinesis started to initiate an additional round of posterior-to-anterior cytokinesis, generating additional posterior ends. CIF1 RNAi was induced for 48 h. A, anterior; P, posterior. (Scale bars: 5 µm.)
Discussion
Trypanosomes initiate cytokinesis from the anterior tip of the new FAZ filament, and it is believed that the regulators playing direct roles in cytokinesis initiation should localize to the new FAZ tip before cytokinesis initiation (21). CIF1 is the fifth protein, after TbAUK1 (12), TbCPC1 (12), TbCPC2 (12), and TbPLK (22), that concentrates at the new FAZ tip, and our RNAi studies demonstrated that CIF1 is essential for cytokinesis initiation from the anterior tip of the new FAZ filament (Fig. 4 and Fig. S3). CIF1 was also identified as a substrate of TbPLK in a recent study and was named TOEFAZ1 for Tip Of Extending FAZ 1 (23). However, given its additional localization to the cleavage furrow during cytokinesis (Fig. 1B and Fig. S1C), this nomenclature does not sufficiently reflect its localization and function. Although McAllaster et al. also showed that RNAi of this protein caused a cytokinesis defect by microscopic inspection, the underlying mechanism was not investigated (23). Our work, however, elucidated the mechanistic role of CIF1 in cytokinesis initiation. It recruits TbAUK1, a crucial regulator of cytokinesis initiation (13), to the new FAZ tip at late anaphase (Fig. 3 A and B) for the latter to initiate cytokinesis from the anterior tip of the new FAZ filament. Moreover, our results also uncovered the mechanism underlying the recruitment of TbAUK1 to the new FAZ tip at late anaphase, which has been a mystery since we first discovered that TbAUK1 emerges at the new FAZ tip at late anaphase (12). It should be noted that CIF1 also localizes to the cleavage furrow during cytokinesis (Fig. 1B and Fig. S1C). Therefore, CIF1 may play additional roles in furrow ingression and cell abscission, but these potential functions of CIF1 cannot be revealed in the RNAi studies because depletion of CIF1 arrested cells before cytokinesis initiation.
It was previously postulated that TbPLK regulates cytokinesis initiation based on the inhibited growth of the RNAi cells (9, 24), but the underlying mechanism has never been addressed. Given that TbPLK disappears from the new FAZ tip before cytokinesis initiation, this raises the question of whether TbPLK plays a direct role in cytokinesis initiation. Here we demonstrated that TbPLK-mediated phosphorylation of CIF1 is required for localizing CIF1 to the new FAZ tip (Fig. 2), which uncovered, for the first time, the mechanistic role of TbPLK in cytokinesis initiation. TbPLK targets CIF1 to the new FAZ tip at early cell cycle stages for the latter to recruit TbAUK1 at later cell cycle stages to initiate cytokinesis from the anterior tip of the new FAZ filament.
We have delineated a signaling cascade that regulates cytokinesis along the longitudinal axis of a trypanosome cell. During early S phase, TbPLK phosphorylates CIF1, thereby targeting CIF1 to the new FAZ tip. In turn, CIF1 maintains TbPLK at the new FAZ tip before it recruits TbAUK1 to the new FAZ tip, where TbAUK1, likely together with CIF1, drives cytokinesis from the anterior tip of the new FAZ filament toward the posterior end of the cell (Fig. 4J). This signaling pathway is totally different from that in metazoa, in which Polo-like kinase and Aurora B kinase cooperate to recruit the centralspindlin complex to the central spindle and the midbody for assembling the actomyosin contractile ring (3) at the cell division plane that is defined by the position of the spindle (25). This mechanism of cytokinesis through the actomyosin contractile ring appeared after T. brucei diverged from the last common eukaryotic ancestor (26). Therefore, T. brucei may have adopted the ancestral cytokinesis pathway or have evolved a trypanosome-specific cytokinesis pathway by adopting some crucial regulators, such as Polo-like kinase and Aurora B kinase, from the ancestral cytokinesis pathway and by incorporating trypanosome-specific factor(s), such as CIF1, in accordance with its unusual cell division plane that is defined by the flagellum and the FAZ filament.
Our findings indicate the existence of two distinct cytokinesis pathways that drive trypanosome cell division along the same division plane but in opposite directions, which is unprecedented. The typical, Polo-like kinase- and Aurora B kinase-dependent cytokinesis occurs from the anterior end of the cell toward the posterior end of the cell under normal growth conditions. The alternative cytokinesis, which is Polo-like kinase- and Aurora B kinase-independent, occurs from the posterior end of the cell toward the anterior end of the cell and is activated only when the typical anterior-to-posterior cytokinesis is inhibited (Fig. 4J). Initiation of cytokinesis from the posterior end of the cell in CIF1 RNAi cells is unlikely to be attributed to the misdirection of the typical cytokinesis machinery, as TbAUK1, the downstream factor of CIF1 and the key regulator of the typical anterior-to-posterior cytokinesis, is not targeted to the posterior end of the cell but is dispersed in the cytosol (Fig. 3 A–C). This alternative cytokinesis appears to be less efficient and slower than the typical cytokinesis (compare Movies S1 and S2) and sometimes generates anucleate daughter cells (Figs. S3C and S6B), but it also generates normal daughter cells (Figs. S6B and S7) and enables trypanosomes to proliferate, albeit at a rate slower than that of the wild-type cells (Fig. S3B). Cytokinesis is one of the most critical steps in the trypanosome cell cycle, and any defect in cytokinesis can lead to cell death. Trypanosomes undergo a complex life cycle by alternating between the insect vector and the mammalian host and must combat with diverse environmental challenges for survival. It is highly likely that under certain adverse conditions in certain life cycle stages, the typical anterior-to-posterior cytokinesis could be defective, and thus a backup plan would be necessary to maintain a critical cell population. Our findings suggest that trypanosomes have evolved a backup cytokinesis mechanism, which is invoked to ensure survival of this organism when the typical cytokinesis is defective.
The existence of a typical cytokinesis and a backup cytokinesis in T. brucei is analogous to the two pathways regulating nonhomologous end joining (NHEJ), an important enzymatic process for repairing DNA double-strand breaks (DSBs) in eukaryotes. In the absence of the canonical NHEJ (c-NHEJ), cells can repair a large portion of DSBs using an alternative pathway that employs a distinct set of factors (27). This alternative NHEJ, termed alt-NHEJ or b-NHEJ (for backup NHEJ), operates with an order of magnitude slower kinetics, frequently joins incorrect ends, and is suppressed by the c-NHEJ (28, 29). Additional examples of the existence of a backup pathway were also found in other cellular processes, such as cell death (30), endoplasmic reticulum-associated degradation (31), and mRNA decay (32). It is believed that evolution ensures the existence of a backup mechanism to prevent the failure of cellular processes that are critically important to life (33). The alternative pathway is cryptic under normal growth conditions, but will be activated once the canonical pathway fails to operate.
Materials and Methods
Trypanosome Cell Lines and RNAi.
The procyclic 427 cell line and 29–13 cell line (34) were cultured according to our published procedures (14). To knock down CIF1, a fragment of CIF1 gene was cloned into the pZJM vector (35) and transfected into the 29–13 cell line. Selection of transfectants, cloning, and RNAi induction were carried out as described previously (14). TbPLK RNAi cell line and TbAUK1 RNAi cell line have been reported previously (12, 17).
Inhibition of TbPLK and TbAUK1 Kinase Activity by Small-Molecule Inhibitors.
The small-molecule PLK inhibitor GW843682X (16) was previously demonstrated to inhibit TbPLK activity in vitro, with an in vitro IC50 of ∼1.3 µM, and in vivo, inhibiting the insect-form trypanosomes with an IC50 of ∼2 µM (17). The small-molecule Aurora B kinase inhibitor Hesperadin (20) was previously demonstrated to inhibit TbAUK1 activity in vitro, with an in vitro IC50 of 40 nM, and in vivo, inhibiting the insect-form trypanosomes with an IC50 of 0.55 µM (19).
Trypanosome cells harboring the pC-CIF1-3HA-PAC construct were treated with 1 µM Hesperadin for 24 h, and cells were collected for Western blotting with anti-HA antibody to monitor the de-phosphorylation of 3HA-tagged CIF1. Cells harboring the pC-CIF1-3HA-PAC construct or both the pC-CIF1-PTP-PAC and the pC-TbAUK1-3HA-NEO constructs were treated with 5 µM GW843682X for 16 h and were then collected for immunostaining with FITC-conjugated anti-HA antibody and anti-Protein A antibody. Cells harboring the pC-CIF1-3HA-PAC construct were treated with 1 µm Hesperadin for 16 h and were then collected for immunofluorescence microscopy with FITC-conjugated anti-HA antibody.
Immunofluorescence Microscopy.
Cells were fixed with cold methanol and incubated with the primary antibody for 1 h at room temperature. The following primary antibodies were used: FITC-conjugated anti-HA antibody, anti-Protein A antibody, anti-CC2D antibody (6), 20H5 (36), and anti-TbPLK antibody (14). Cells were then incubated with FITC-conjugated anti-mouse IgG or Cy3-conjugated anti-rabbit IgG for 1 h at room temperature, mounted with DAPI-containing VectaShield mounting medium (Vector Labs), and imaged using an inverted fluorescence microscope (Olympus IX71) equipped with a cooled CCD camera (model Orca-ER, Hamamatsu) and a PlanApo N 60 × 1.42-NA lens. Images were acquired using Slidebook 5 software.
Scanning Electron Microscopy.
Cells were settled onto coverslips and fixed with 2.5% (vol/vol) glutaraldehyde in PBS for 30 min at room temperature. After washing the cells on the coverslips three times with PBS, cells were dehydrated in a graded series of alcohol [30, 50, 70, 90, and 100% (vol/vol)] for 10 min each. After critical point drying, samples were coated with a 8-nm metal film (Pt:Pd 80:20, Ted Pella Inc.) using a sputter-coater (Cressington Sputter Coater 208 HR, Ted Pella Inc.) and imaged using Nova NanoSEM 230 (FEI). The scanning work distance was at 5 mm, and the accelerating high voltage was at 8 kV.
Time-Lapse Video Microscopy.
Time-lapse video microscopy was carried out as described previously (13). Cells were plated on a 0.6% agarose gel in SDM-79 medium and visualized under an inverted automatic microscope (Olympus IX81) equipped with a cooled CCD camera. Images were taken every 2–3 min, and time-lapse movies were generated using ImageJ.
Statistical Analysis.
Statistical analysis was performed using the t test provided in the Microsoft Excel software. For immunofluorescence microscopy experiments, images were randomly taken and all of the cells in each image were counted.
SI Materials and Methods
Yeast Two-Hybrid Library Screening and Directional Yeast Two-Hybrid Assays.
Construction of the Gal4 activation domain (AD) fusion library for yeast two-hybrid screening and screening of the library with TbPLK-K70R as the bait have been described previously (14). For library screening with TbAUK1 as the bait, the DNA fragment corresponding to the coding region of TbAUK1 was cloned into the pGBKT7 vector for expression of Gal4-binding domain (BD) fusion proteins (bait). Screening of the library and identification of positive clones were essentially the same as described previously (14).
For directional yeast two-hybrid assay, the full-length coding sequence of CIF1 was cloned into the pGADT7 vector for expression of Gal4 AD-fused CIF1 (prey). The full-length coding sequence of TbPLK, TbCPC1, and TbCPC2 were each cloned into the pGBKT7 vector to express Gal4 BD-fused proteins (bait). The prey plasmid was transformed into stain AH109, and the bait plasmids were transformed into strain Y187. The yeast strains carrying both the bait and the prey plasmids were obtained by mating the two haploids at 30 °C overnight, plating the diploid on SD-Leu-Trp plates, and incubating at 30 °C for 2–3 d. Each combination strain was spotted in three 10-fold serial dilutions onto SD-Leu-Trp and SD-Leu-Trp-His plates, and the growth of yeast on the SD-Leu-Trp-His plate indicates the interaction between the bait and the prey proteins.
RNA Interference.
To knock down CIF1 by RNAi, a 558-bp DNA fragment (nucleotides 207–764) corresponding to the N-terminal coding region of CIF1 was PCR-amplified from the genomic DNA and cloned into the pZJM vector (35). The resulting plasmid was linearized with Not I and then transfected into the 29–13 cell line by electroporation according to our published procedures (14). The successful transfectants were selected with 2.5 µg/mL phleomycin in addition to 15 µg/mL G418 and 50 µg/mL hygromycin and then cloned by limiting dilution in a 96-well plate containing SDM-79 medium supplemented with 20% FBS and all three antibiotics. RNAi was induced with 1.0 µg/mL tetracycline. The TbPLK RNAi cell line and the TbAUK1 RNAi cell line have been reported previously (12, 17). Knockdown of TbPLK was confirmed by Western blotting with anti-TbPLK antibody. To confirm the knockdown of TbAUK1, TbAUK1 was endogenously tagged with a triple HA epitope in cells containing the TbAUK1 RNAi construct (pZJM-TbAUK1), and Western blotting was performed with anti-HA antibody. The effect of TbPLK and TbAUK1 knockdown on cell growth was confirmed (see Fig. S2 for details).
In Situ Epitope Tagging of Proteins.
For C-terminal epitope tagging of CIF1 at one of its endogenous loci, a 1,359-bp fragment corresponding to the C-terminal coding region of CIF1, was PCR-amplified from the genomic DNA and cloned into pC-3HA-PAC and pC-PTP-PAC vectors. The resulting plasmids were linearized with XcmI and electroporated into the wild-type 427 cell line and the cell lines harboring pZJM-CIF1, pZJM-TbPLK, or pZJM-TbAUK1. Transfectants were selected with 1 µg/mL puromycin and cloned by limiting dilution in a 96-well plate.
For colocalization of CIF1-PTP and TbAUK1-3HA, the cell line harboring the pC-CIF1-PTP-PAC vector was transfected with pC-TbAUK1-3HA-NEO, which was linearized by NruI digestion. Transfectants were selected with 40 µg/mL G418 in addition to 1 µg/mL puromycin and then cloned by limiting dilution as described above.
For endogenous epitope tagging of TbAUK1, TbCPC1, and TbCPC2 in the CIF1 RNAi cell line, DNA fragments (906 bp of TbAUK1, 669 bp of TbCPC1, and 610 bp of TbCPC2) corresponding to the C-terminal coding region of the three genes were each PCR-amplified from the genomic DNA, cloned into the pC-3HA-PAC vector, linearized with appropriate restriction enzymes (SphI for TbAUK1, XhoI for TbCPC1, and Eco47III for TbCPC2), and transfected into the cell line harboring the pZJM-CIF1 construct. Transfectants were selected under 1 µg/mL puromycin in addition to 15 µg/mL G418, 50 µg/mL hygromycin, and 2.5 µg/mL phleomycin, and clonal cell lines were obtained by limiting dilution as described above.
Supplementary Material
Acknowledgments
We thank Drs. Arthur Günzl, Christopher de Graffenried, and Cynthia He for providing the epitope tagging vectors, the PS54 antibody, and the anti-CC2D antibody, respectively. This work was supported by NIH R01 Grant AI101437 (to Z.L.); National Natural Science Foundation of China Grant 31472058; and Natural Science Foundation of Guangdong Province Grant 2015A030311042 (to Z.-R.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601596113/-/DCSupplemental.
References
- 1.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307(5716):1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD, Wu JQ. Understanding cytokinesis: Lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11(2):149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmena M. Cytokinesis: The final stop for the chromosomal passengers. Biochem Soc Trans. 2008;36(Pt 3):367–370. doi: 10.1042/BST0360367. [DOI] [PubMed] [Google Scholar]
- 4.Vaughan S, Gull K. The structural mechanics of cell division in Trypanosoma brucei. Biochem Soc Trans. 2008;36(Pt 3):421–424. doi: 10.1042/BST0360421. [DOI] [PubMed] [Google Scholar]
- 5.Kohl L, Robinson D, Bastin P. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 2003;22(20):5336–5346. doi: 10.1093/emboj/cdg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Q, Liu B, Sun Y, He CY. A coiled-coil- and C2-domain-containing protein is required for FAZ assembly and cell morphology in Trypanosoma brucei. J Cell Sci. 2011;124(Pt 22):3848–3858. doi: 10.1242/jcs.087676. [DOI] [PubMed] [Google Scholar]
- 7.Wheeler RJ, Scheumann N, Wickstead B, Gull K, Vaughan S. Cytokinesis in Trypanosoma brucei differs between bloodstream and tsetse trypomastigote forms: Implications for microtubule-based morphogenesis and mutant analysis. Mol Microbiol. 2013;90(6):1339–1355. doi: 10.1111/mmi.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.García-Salcedo JA, et al. A differential role for actin during the life cycle of Trypanosoma brucei. EMBO J. 2004;23(4):780–789. doi: 10.1038/sj.emboj.7600094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar P, Wang CC. Dissociation of cytokinesis initiation from mitotic control in a eukaryote. Eukaryot Cell. 2006;5(1):92–102. doi: 10.1128/EC.5.1.92-102.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tu X, Kumar P, Li Z, Wang CC. An aurora kinase homologue is involved in regulating both mitosis and cytokinesis in Trypanosoma brucei. J Biol Chem. 2006;281(14):9677–9687. doi: 10.1074/jbc.M511504200. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda KN, de Graffenried CL. Polo-like kinase is necessary for flagellum inheritance in Trypanosoma brucei. J Cell Sci. 2012;125(Pt 13):3173–3184. doi: 10.1242/jcs.101162. [DOI] [PubMed] [Google Scholar]
- 12.Li Z, et al. Identification of a novel chromosomal passenger complex and its unique localization during cytokinesis in Trypanosoma brucei. PLoS One. 2008;3(6):e2354. doi: 10.1371/journal.pone.0002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Umeyama T, Wang CC. The Aurora kinase in Trypanosoma brucei plays distinctive roles in metaphase-anaphase transition and cytokinetic initiation. PLoS Pathog. 2009;5(9):e1000575. doi: 10.1371/journal.ppat.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu H, Zhou Q, Li Z. A novel basal body protein that is a Polo-like kinase substrate is required for basal body segregation and flagellum adhesion in Trypanosoma brucei. J Biol Chem. 2015;290(41):25012–25022. doi: 10.1074/jbc.M115.674796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urbaniak MD, Martin DM, Ferguson MA. Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J Proteome Res. 2013;12(5):2233–2244. doi: 10.1021/pr400086y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lansing TJ, et al. In vitro biological activity of a novel small-molecule inhibitor of polo-like kinase 1. Mol Cancer Ther. 2007;6(2):450–459. doi: 10.1158/1535-7163.MCT-06-0543. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Umeyama T, Li Z, Wang CC. Polo-like kinase guides cytokinesis in Trypanosoma brucei through an indirect means. Eukaryot Cell. 2010;9(5):705–716. doi: 10.1128/EC.00330-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Graffenried CL, Anrather D, Von Raussendorf F, Warren G. Polo-like kinase phosphorylation of bilobe-resident TbCentrin2 facilitates flagellar inheritance in Trypanosoma brucei. Mol Biol Cell. 2013;24(12):1947–1963. doi: 10.1091/mbc.E12-12-0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jetton N, et al. The cell cycle as a therapeutic target against Trypanosoma brucei: Hesperadin inhibits Aurora kinase-1 and blocks mitotic progression in bloodstream forms. Mol Microbiol. 2009;72(2):442–458. doi: 10.1111/j.1365-2958.2009.06657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauf S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161(2):281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z. Regulation of the cell division cycle in Trypanosoma brucei. Eukaryot Cell. 2012;11(10):1180–1190. doi: 10.1128/EC.00145-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Graffenried CL, Ho HH, Warren G. Polo-like kinase is required for Golgi and bilobe biogenesis in Trypanosoma brucei. J Cell Biol. 2008;181(3):431–438. doi: 10.1083/jcb.200708082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McAllaster MR, et al. Proteomic identification of novel cytoskeletal proteins associated with TbPLK, an essential regulator of cell morphogenesis in Trypanosoma brucei. Mol Biol Cell. 2015;26(17):3013–3029. doi: 10.1091/mbc.E15-04-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammarton TC, Kramer S, Tetley L, Boshart M, Mottram JC. Trypanosoma brucei Polo-like kinase is essential for basal body duplication, kDNA segregation and cytokinesis. Mol Microbiol. 2007;65(5):1229–1248. doi: 10.1111/j.1365-2958.2007.05866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliferenko S, Chew TG, Balasubramanian MK. Positioning cytokinesis. Genes Dev. 2009;23(6):660–674. doi: 10.1101/gad.1772009. [DOI] [PubMed] [Google Scholar]
- 26.Baldauf SL. The deep roots of eukaryotes. Science. 2003;300(5626):1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 27.Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131(2):223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, et al. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 2003;31(18):5377–5388. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrault R, Wang H, Wang M, Rosidi B, Iliakis G. Backup pathways of NHEJ are suppressed by DNA-PK. J Cell Biochem. 2004;92(4):781–794. doi: 10.1002/jcb.20104. [DOI] [PubMed] [Google Scholar]
- 30.Vandenabeele P, Vanden Berghe T, Festjens N. Caspase inhibitors promote alternative cell death pathways. Sci STKE. 2006;2006(358):pe44. doi: 10.1126/stke.3582006pe44. [DOI] [PubMed] [Google Scholar]
- 31.Ushioda R, Hoseki J, Nagata K. Glycosylation-independent ERAD pathway serves as a backup system under ER stress. Mol Biol Cell. 2013;24(20):3155–3163. doi: 10.1091/mbc.E13-03-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamashita A, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12(12):1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 33.Höglund P. DNA damage and tumor surveillance: One trigger for two pathways. Sci STKE. 2006;2006(317):pe2. doi: 10.1126/stke.3172006pe2. [DOI] [PubMed] [Google Scholar]
- 34.Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99(1):89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem. 2000;275(51):40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- 36.He CY, Pypaert M, Warren G. Golgi duplication in Trypanosoma brucei requires Centrin2. Science. 2005;310(5751):1196–1198. doi: 10.1126/science.1119969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.