Significance
Sex ratio is an important indicator of reproductive health, and its skewing reflects disturbed embryonic development. This study focused on sex skewing, which has recently identified in human in vitro fertilization (IVF) babies. We reported herein that the skewed sex ratio in mouse IVF offspring was due to the impaired imprinted X chromosome inactivation via suppressing the ring finger protein 12 (Rnf12)/X-inactive specific transcript (Xist) pathway; the sex skewing can be corrected by overexpressing Rnf12 or by supplementation of retinoic acid in embryo culture medium. Hence, our study not only identified a major epigenetic error responsible for sex skewing in IVF offspring, but also implicated a potential strategy for preventing sex skewing and IVF-associated complications by targeting erroneous epigenetic modifications induced by IVF.
Keywords: in vitro fertilization, sex ratio, X chromosome inactivation, Xist, Rnf12
Abstract
Dynamic epigenetic reprogramming occurs during normal embryonic development at the preimplantation stage. Erroneous epigenetic modifications due to environmental perturbations such as manipulation and culture of embryos during in vitro fertilization (IVF) are linked to various short- or long-term consequences. Among these, the skewed sex ratio, an indicator of reproductive hazards, was reported in bovine and porcine embryos and even human IVF newborns. However, since the first case of sex skewing reported in 1991, the underlying mechanisms remain unclear. We reported herein that sex ratio is skewed in mouse IVF offspring, and this was a result of female-biased peri-implantation developmental defects that were originated from impaired imprinted X chromosome inactivation (iXCI) through reduced ring finger protein 12 (Rnf12)/X-inactive specific transcript (Xist) expression. Compensation of impaired iXCI by overexpression of Rnf12 to up-regulate Xist significantly rescued female-biased developmental defects and corrected sex ratio in IVF offspring. Moreover, supplementation of an epigenetic modulator retinoic acid in embryo culture medium up-regulated Rnf12/Xist expression, improved iXCI, and successfully redeemed the skewed sex ratio to nearly 50% in mouse IVF offspring. Thus, our data show that iXCI is one of the major epigenetic barriers for the developmental competence of female embryos during preimplantation stage, and targeting erroneous epigenetic modifications may provide a potential approach for preventing IVF-associated complications.
There have been more than 5 million people born from in vitro fertilization (IVF) since the birth of Louise Brown on July 25, 1978 (1); this makes IVF as one of the most effective and successful assisted reproductive technologies that are routinely used for treating infertility that affects ∼15% couples globally (2). Nowadays, IVF contributes to 1–5% of all newborns in developed countries (2). In addition, IVF is one of the most promising technologies for accelerating the intensive propagation or improving intensity of genetic selection from genetically superior individuals in domestic animals (3). Although the great majority of IVF-conceived offspring are in good health, epidemiologic analyses in humans and laboratory studies in animals show that IVF is associated with various short- or long-term consequences, such as miscarriage, preterm birth, lower birth weight, skewed sex ratio, and higher disease risks in later life (4–8).
Dynamic epigenetic reprogramming occurs during preimplantation development (9). Environmental perturbations such as manipulation and culture of embryos in vitro during IVF are expected to influence the epigenetic programming of the developing embryos during this critical period, often leading to nonrandom epigenetic errors such as aberrant DNA methylation and histone modifications (10, 11). Indeed, aberrant DNA methylation of some genes has been linked to certain complications, including vascular dysfunction and imprinting disorders, in IVF offspring (12, 13).
Skewed sex ratio is an issue of great concern among the IVF-associated consequences. The first case of IVF-associated sex skewing was reported in bovine as early as 1991 (14), and this phenomena has been confirmed repeatedly in bovine and porcine IVF embryos (15–17). Recent epidemiologic data from Oceania and United Kingdom also show that IVF can result in sex skewing in humans with a higher male birth rate (5, 6). Sex ratio is an important indicator of reproductive health that tends to be sensitive or even adaptive to many factors (18, 19), and its skewing has a potential effect on long-term social and behavior consequences (20).
IVF-associated sex skewing reflects potential sex-biased embryonic developmental defects (21). The sex-biased embryonic defects are reminiscent of the disrupted sex-specific epigenetic events. X chromosome inactivation (XCI) is a female-specific epigenetic event that occurs during early development for balancing the X-linked gene dosage between males and females. Previous studies show that disturbed XCI caused by mutation or deletion of related genes results in sex skewing in mice (22–24). Thus, we hypothesized that the IVF process may disturb the XCI status, thereby leading to the skewed sex ratio. We report herein that skewed sex ratio in mouse IVF offspring is a result of female-biased developmental defects due to impaired XCI (iXCI) via reduced ring finger protein 12 (Rnf12)/X-inactive specific transcript (Xist) expression. Accordingly, correcting this epigenetic error by overexpression of Rnf12 to compensate iXCI via Xist up-regulation or by incubation of the IVF embryos with an epigenetic modulator retinoic acid (RA) redeemed the skewed sex ratio. Thus, our data show that iXCI is one of the major epigenetic barriers for the developmental competence of female embryos during preimplantation stage and targeting erroneous epigenetic modifications may provide a potential approach for preventing IVF-associated complications.
Results and Discussion
Sex Skewing in IVF Offspring Is Linked to Female-Biased Developmental Defects During Peri-Implantation Stage.
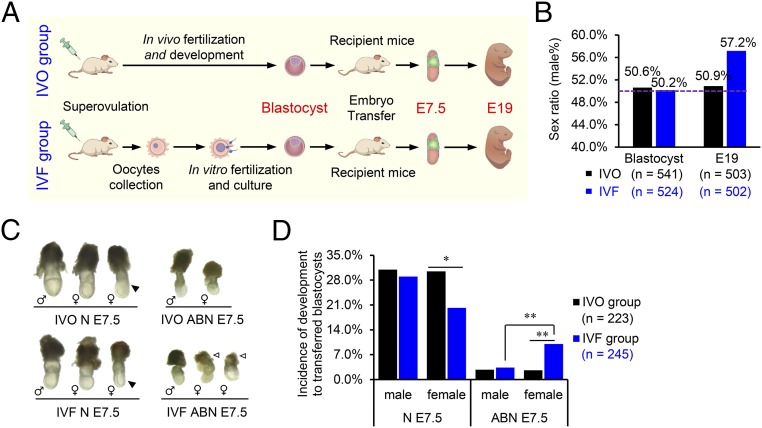
To test the temporal patterns of IVF-associated sex skewing, we generated mouse embryos under standardized IVF conditions (25) and used in vivo fertilized embryos as the control (IVO group; Fig. 1A). We found that IVF resulted in a significantly higher male birth rate (57.17% vs. 50.89%; Fig. 1B and Fig. S1A); further analysis showed that sex skewing did not occur before implantation, because sex ratios of both IVF and IVO blastocysts were near 50% (Fig. 1B and Fig. S1B). After implantation, IVF embryos had a lower survival rate that could be observed as early as embryonic day (E)7.5 (Fig. S2A). At E7.5, IVF female embryos showed significantly greater incidence in morphological abnormalities than their male counterparts (10.01% vs. 2.60%), accompanied by a higher frequency of abnormal extraembryonic tissues (Fig. 1 C and D and Figs. S1C and S2B). These results show that IVF embryos have female-biased developmental defects during the peri-implantation stage, which is responsible for the skewed sex ratio in their offspring.
Fig. 1.
Sex ratio is skewed in IVF embryos due to female-biased abnormal embryonic development. (A) Schematic diagram of the experimental design. Embryos after IVF and development (IVO group, control) or IVF and culture (IVF group) were sampled at the blastocyst stage, E7.5, and at birth (E19), followed by detailed examinations. (B) Sex ratio (proportion of males) of blastocysts and E19 fetuses in IVO and IVF groups. The dotted line indicates the expected natural sex ratio of 50%. (C) Representative images of IVO and IVF E7.5 embryos with normal (N) or abnormal (ABN) morphologies. Solid arrowhead indicates developmentally delayed embryos with normal morphology. A large proportion of IVF female ABN E7.5 embryos displayed severe disorganized extraembryonic tissues (open arrowhead). (D) Incidence of development of male and female embryos with different morphologies at E7.5. The number (n) of embryos examined in each group is indicated. *P < 0.05, **P < 0.01.
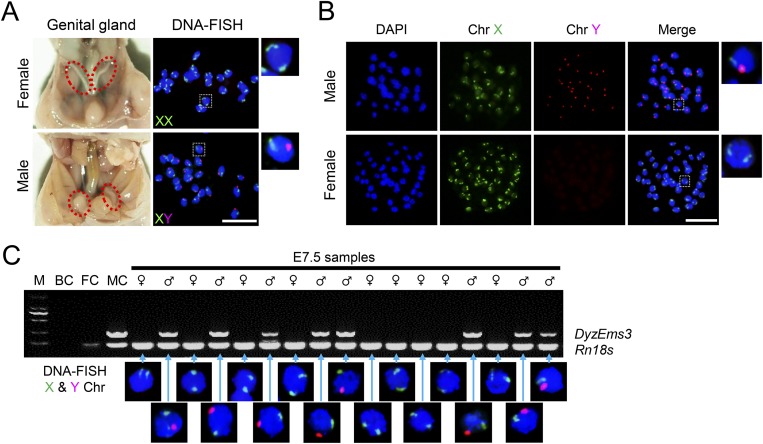
Fig. S1.
Methods for sex determination. (A) At E19, the sex of fetuses was determined based on anatomical observation of the genital glands. To validate the results, 20 randomly selected fetuses were examined using DNA-FISH against the X and Y chromosomes; these results agreed with the anatomical analyses. (B) At blastocyst stage, the sex of blastocysts was determined using DNA-FISH. (Rightmost panels) Higher magnification of boxed regions. (C) At E7.5, epiblasts were isolated from embryos by microdissection, and the sex was determined by PCR. In female samples, only one amplicon corresponding to the Rn18S gene was detected. In male samples, an additional amplicon corresponding to the Y chromosome-specific DyzEms3 segment was detected. Template DNA samples from adult male and female somatic cells were used as the male control (MC) and the female control (FC), respectively. A blank control (BC) without template (water blank) was also examined. To exclude possible cross-contamination between samples, 17 randomly selected embryos were further validated using DNA-FISH. Results are positioned relative to the corresponding PCR sample. M, marker. (Scale bar, 50 μm.)
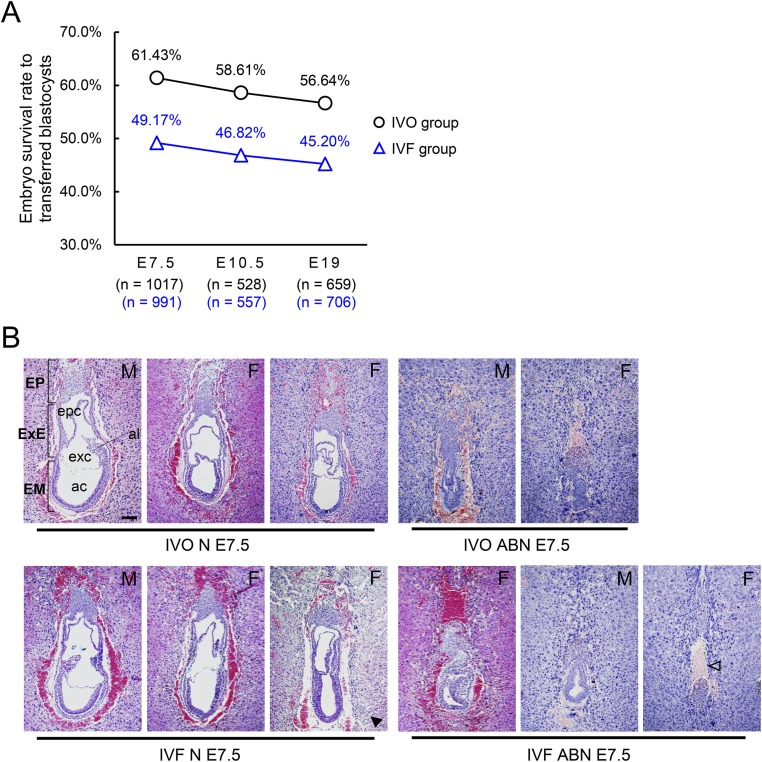
Fig. S2.
IVF embryos have lower survival rates after embryo transfer. (A) Survival rates of IVO and IVF embryos at E7.5, E10.5, and E19. The number (n) of embryos or fetuses in each group was indicated. (B) H&E staining of sagittal sections of IVO and IVF embryos with normal (N) or abnormal (ABN) morphology. ac, amniotic cavity; al, allantois; EM, embryo; EP, ectoplacental cone; epc, ectoplacental cone cavity; exc, exocoelomic cavity; ExE, extraembryonic ectoderm; F, female; M, male. Solid arrowhead indicates developmentally delayed embryos with normal morphology. Open arrowhead indicates a representative IVF female ABN E7.5 embryo that is missing extraembryonic tissues. (Scale bar, 100 μm.)
iXCI Is Impaired in IVF Female Embryos.
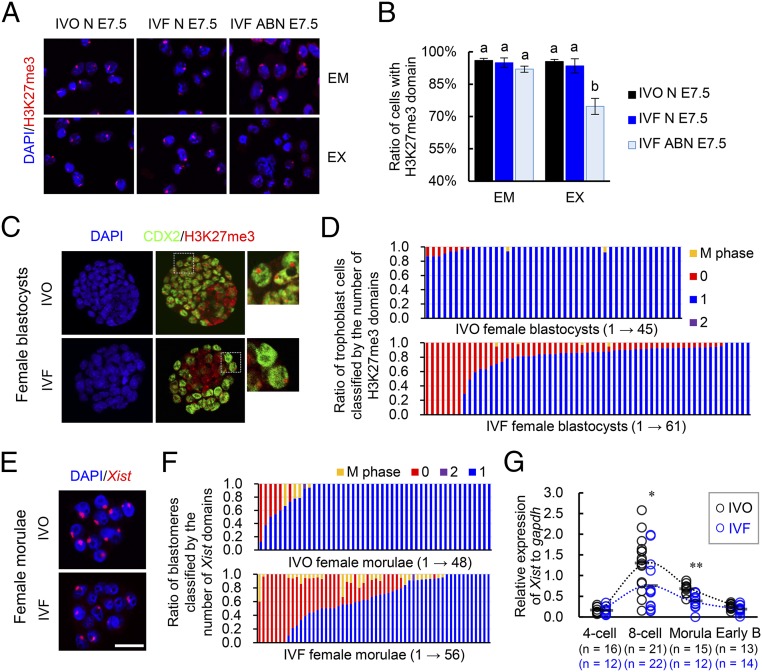
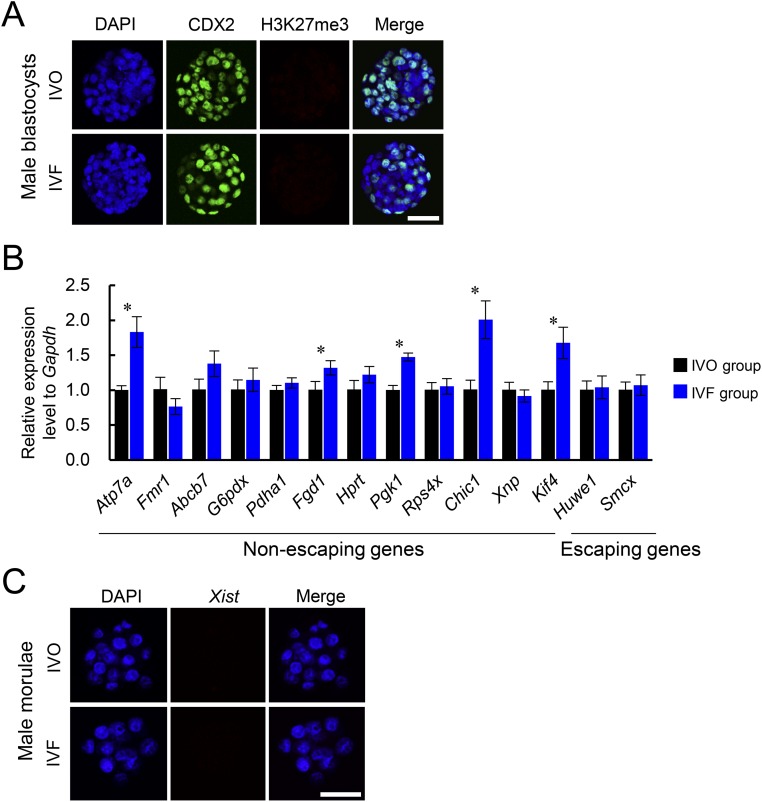
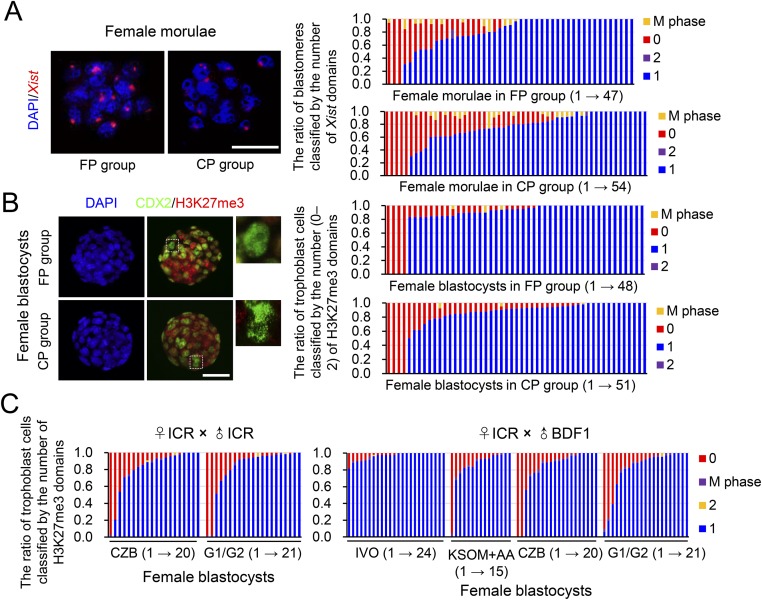
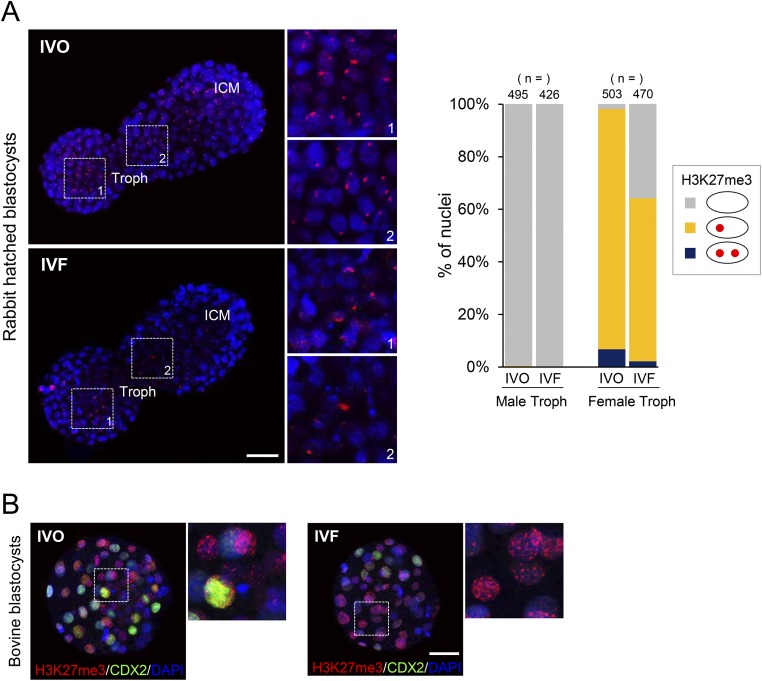
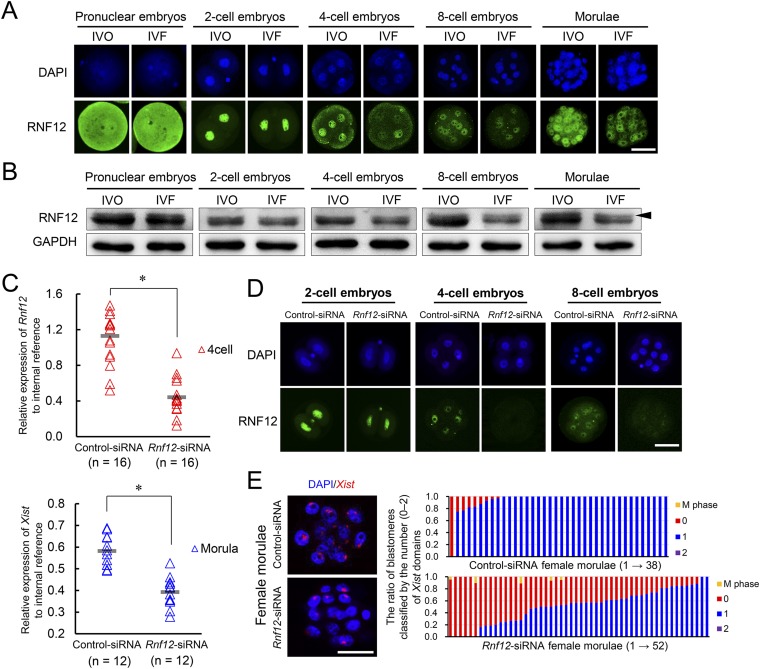
The observed female-biased developmental defects led us to hypothesize that a certain sex-specific epigenetic event was disturbed by IVF. Because the observed female-biased defects are similar to that previously described in mouse embryos with disturbed XCI caused by paternally inherited Xist mutations (22, 23), we postulated that the IVF embryos may have impaired XCI. Mice undergo two waves of XCI: iXCI initiates in early preimplantation embryos and persists in extraembryonic tissues, and random XCI (rXCI) occurs in the embryonic cells shortly after implantation (26–28). By detecting the H3K27me3, a classic marker for establishment of XCI by achieving X chromosome wide heterochromatinization of transcriptional depression (28), we first confirmed that rXCI was not impaired in either morphologically normal or abnormal E7.5 IVF female embryos because H3K27me3 enrichment was normal in nearly all epiblast cells (Fig. 2 A and B). In contrast, iXCI was impaired in abnormal E7.5 IVF female embryos because H3K27me3 domains were significantly decreased in the extraembryonic tissues of these embryos (Fig. 2 A and B). We then examined whether iXCI was affected by IVF during the preimplantation stage when it establishes (27). As expected, nearly all IVO female blastocysts (n = 45) consistently showed a single H3K27me3 domain in each trophoblast cell (Fig. 2 C and D). In contrast, a substantial proportion of the IVF female blastocysts (n = 61) contained trophoblast cells lacking the H3K27me3 domain, with a variable frequency ranging from 4.8% to 100.0% (Fig. 2 C and D). Both IVO and IVF male blastocysts had no H3K27me3 domain (Fig. S3A). These results confirmed that iXCI is impaired in a proportion of preimplantation IVF female embryos, whereas male embryos have no ectopic iXCI. This notion is further supported by the fact that mRNAs of several X-linked nonescaping genes, including ATPase, Cu++ transporting, alpha polypeptide (Atp7a), FYVE, RhoGEF and PH domain-containing 1 (Fgd1), phosphoglycerate kinase 1 (Pgk1), cysteine-rich hydrophobic domain 1 (Chic1), and kinesin family member 4 (Kif4), were up-regulated in IVF female embryos (Fig. S3B). We also found that both the fertilization process (FP) and culture process (CP) of IVF contributed to iXCI impairment (Fig. S4 A and B). In comparison with FP or CP alone, the IVF process caused the most severe loss of H3K27me3 domain in the embryos, suggesting a synergetic effect of CP and FP on iXCI impairment. Furthermore, we also found that the IVF-induced iXCI impairment was not limited to a specific genotype or culture condition (Fig. S4C). In rabbits, H3K27me3 immunostaining revealed impaired XCI in IVF female blastocysts as well (Fig. S5A). In cows, H3K27me3 is less useful for determining XCI status in blastocysts (Fig. S5B); up-regulation of representative X-linked nonescaping glucose-6-phosphate dehydrogenase (G6pd) and Pgk genes (16) suggests impaired XCI in IVF female bovine embryos. In humans, although comparisons cannot be made with naturally conceived embryos due to ethical considerations, the observed incomplete initiation of XCI at the blastocyst stage in IVF embryos may be a reflection of impaired XCI (29, 30). Thus, despite different mechanisms of XCI among species, these data suggest that IVF-induced XCI impairment seems to be common in these species.
Fig. 2.
iXCI is impaired in IVF female embryos. (A) Representative H3K27me3 immunostaining in cells isolated from embryonic (EM) and extraembryonic (EX) tissues of normal IVO (IVO N) and IVF (IVF N), as well as abnormal IVF (IVF ABN) female embryos at E7.5. (B) Percentage of H3K27me3-positive cells. Values with different letters are significantly different (P < 0.05). (C) Representative immunostaining for H3K27me3 (red) in the nuclei (DAPI) of IVO and IVF female blastocysts colabeled with CDX2 (green)-positive trophoblast cells. (Rightmost panels) Higher magnification of boxed regions. (D) The ratio of trophoblast cells classified by the number (0–2) of H3K27me3 domains. (E) Representative localization of Xist expression in the nuclei (DAPI) of IVO and IVF female morulae. (F) The ratio of blastomeres classified by the number (0–2) of Xist domains. (D and F) Each bar represents one female embryo. (G) Expression levels of Xist in female IVO and IVF embryos during preimplantation stage. Circles represent the relative expression value in each embryo. Horizontal gray lines represent the mean values. The number of embryos in each group is indicated. Early B, early blastocyst. *P < 0.05, **P < 0.01. (Scale bar, 50 μm.)
Fig. S3.
iXCI status of IVF male embryos and expression patterns of X-linked genes in IVF female embryos. (A) Representative nuclear H3K27me3 immunostaining in IVO and IVF male blastocysts. Trophoblast cells were positively labeled with CDX2 staining. (B) Relative expression levels of representative X-linked genes to internal reference in pooled IVF and IVO female blastocysts. (C) Representative nuclear Xist noncoding RNA expression in IVO and IVF male morulae. (Scale bar, 50 μm.) *P < 0.05.
Fig. S4.
IVF-induced iXCI impairment is not limited to specific process or culture condition of IVF, and genotype of embryos. The fertilization process (FP) group: pronuclear embryos after IVF were transferred to fallopian tubes of pseudo-pregnant surrogate mice and developed to the morula or blastocyst stage in vivo. The culture process (CP) group: in vivo fertilized pronuclear embryos were recovered from donors and developed to the morula or blastocyst stage under in vitro culture conditions. (A) Representative nuclear Xist noncoding RNA localization in FP and CP female morulae. The ratio of blastomeres classified by the number of Xist domains (0–2) in the nucleus is presented on the right. Each bar represents one female morula. (B) Representative nuclear H3K27me3 immunostaining in FP and CP female blastocysts. (Rightmost panels) Higher magnification of boxed regions. The ratio of trophoblast cells (CDX2-positive) classified by the number of H3K27me3 domains (0–2) in the nucleus is presented on the right. Each bar represents one female blastocyst. (C) Scoring of H3K27me3 immunostaining in the trophoblast cells of blastocysts produced under various conditions. Scoring was based on the ratio of trophoblast cells classified by the number (0–2) of H3K27me3 domains in the nucleus. Each bar represents one female embryo. (Scale bars, 50 μm.)
Fig. S5.
XCI status of IVF rabbit and bovine embryos. (A) Representative nuclear H3K27me3 immunostaining (red) in rabbit IVO and IVF hatched female blastocysts. The boxed regions indicate the trophoblasts. (Rightmost panels) hHigher magnification of boxed regions. ICM, inner cell mass; Troph, trophoblasts. Proportion of cells showing the ratio of trophoblast cells classified by the number (0–2) of H3K27me3 domains in the nucleus. The number (n) of cells in each group was indicated. (B) Nuclear H3K27me3 immunostaining in bovine IVO and IVF female blastocysts. (Rightmost panels) Higher magnification of boxed regions. H3K27me3-positive domains are localized clearly and randomly within the nucleus (DAPI) in both groups, agreeing with the results of a previous study on bovine IVF blastocysts (47). [Scale bars, 100 (A) and 50 μm (B).]
iXCI via Reduced Rnf12/Xist Expression in IVF Female Embryos Is Responsible for the Skewed Sex Ratio at Birth.
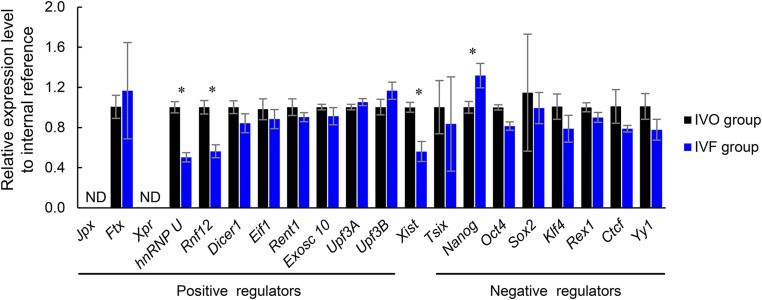
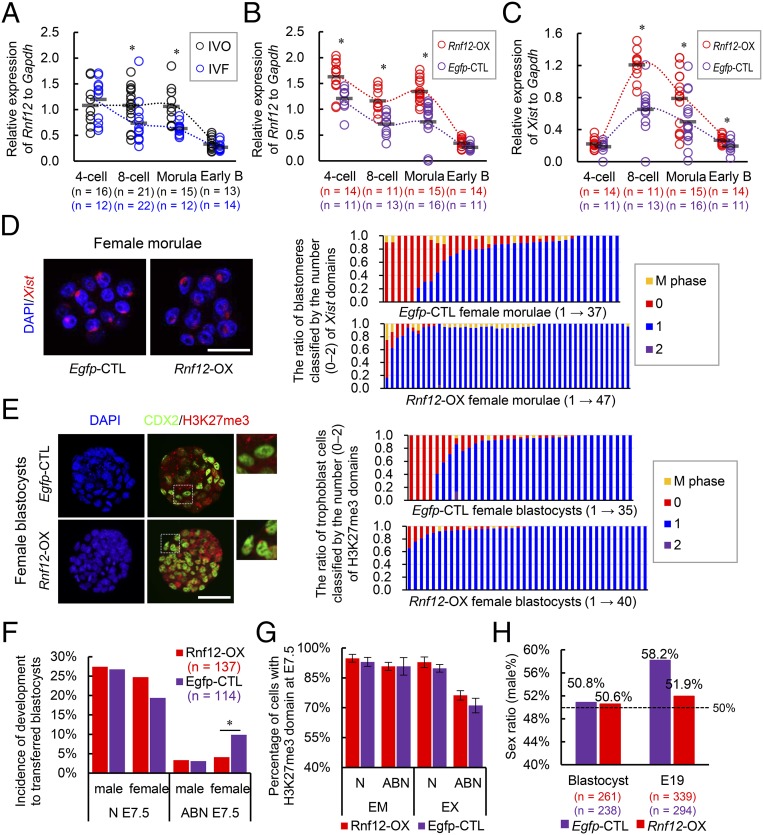
Next, we asked whether the impaired iXCI is responsible for the female-biased developmental defects and the sex skewing in IVF embryos. Because XCI is initiated by Xist RNA coating (31), we first examined Xist mRNA expression during preimplantation development. A substantial proportion of IVF female morulae showed degrees of absence of Xist domains within the nucleus of each blastomere (Fig. 2 E and F). Both IVO and IVF males had no Xist expression (Fig. S3C). Single embryo quantitative RT-PCR (qRT-PCR) analysis also confirmed that Xist expression was significantly inhibited in a proportion of preimplantation IVF female embryos from eight-cell to morula stages (Fig. 2G), suggesting insufficient initiation and establishment of iXCI because Xist expression is essential for initiating iXCI (31). Because Xist overexpression would induce transcriptional repression not only in the X chromosome but also in autosomes (32), we assessed whether expression of other upstream regulators of Xist could be used to compensate for impaired iXCI (Fig. S6). Among them, Rnf12 was down-regulated in preimplantation IVF female embryos (Fig. 3A) in association with decreased Xist mRNA expression (Fig. 2G). We also confirmed that RNF12 protein was decreased in IVF preimplantation embryos by both immunofluorescence microscopy and immunoblotting (Fig. S7 A and B). Moreover, Rnf12 knockdown by siRNA (Fig. S7 C and D) led to Xist down-regulation and loss of Xist domain within the nucleus of female embryos (Fig. S7 C and E), suggesting a crucial role of Rnf12 transcription in the expression of Xist during preimplantation stage. In addition, RNF12 can specifically activate Xist expression in the paternal X chromosome during iXCI and also prevent excessive up-regulation of Xist with a rapid feedback mechanism (24, 33). Together, these data show that Rnf12 overexpression can be used to compensate impaired iXCI by fine-tuning Xist expression in IVF female embryos. As expected, overexpression of Rnf12 (Fig. 3B) resulted in increased Xist expression (Fig. 3C) to the levels comparable to that in IVO embryos (Fig. 2G) during XCI initiation. Rnf12 overexpression also significantly improved iXCI status in IVF female embryos as revealed by Xist RNA-FISH and H3K27me3 immunostaining, without inducing ectopic iXCI in both females and males (Fig. 3 D and E and Fig. S8 A–C).
Fig. S6.
Relative expression levels of upstream regulators of Xist to internal reference in pooled in IVF and IVO female embryos. *P < 0.05. ND, not detected.
Fig. 3.
iXCI impairment in IVF female embryos is responsible for the skewed sex ratio at birth. (A) Rnf12 expression levels in female IVO and IVF embryos during the preimplantation stage. (B) Rnf12 and (C) Xist expression levels in female embryos overexpressing Rnf12 (Rnf12-OX) and control (Egfp-CTL) during the preimplantation stage. (A–C) Circles represent the relative expression level in each embryo. Horizontal gray lines represent the mean values. Early B, early blastocyst. (D) RNA-FISH of Xist expression in female morulae. The ratio of blastomeres classified by the number of Xist domains is presented on the right. (E) H3K27me3 immunostaining in female blastocysts. The ratio of trophoblast cells classified by the number of H3K27me3 domains is presented on the right. (D and E) Each bar represents one female embryo. (F) Incidence of development of Rnf12-OX and Egfp-CTL embryos with normal (N) or abnormal (ABN) morphology at E7.5. (G) Percentage of H3K27me3-positive cells from the embryonic (EM) and extraembryonic (EX) tissues of E7.5 female embryos. (H) Sex ratios in Rnf12-OX and Egfp-CTL groups. The dotted line indicates the expected proportion of 50%. The number of embryos/fetuses in each group is indicated. *P < 0.05. (Scale bar, 50 μm.)
Fig. S7.
Rnf12 transcription is essential for Xist expression in female preimplantation embryos. (A) Representative RNF12 immunostaining in preimplantation IVO and IVF female embryos. (B) Western blot analyses for RNF12 in preimplantation IVO and IVF embryos. Arrowhead (the upper band) indicates the running positions of the phosphorylated form of RNF12 (48). (C) Relative expression levels of Rnf12 (four-cell stage) and Xist (morula stage) in female embryos injected with Rnf12-specific siRNA (Rnf12-siRNA) and control siRNA (control-siRNA). Each triangle represents the expression value in each embryo. Horizontal gray lines represent the mean values. The number of embryos in each group is indicated. (D) Immunostaining of RNF12 in mouse female preimplantation embryos after knockdown of Rnf12. (E) RNA-FISH analyses of Xist noncoding RNA in Rnf12 and control siRNA-injected female morulae. The ratio of blastomeres classified by the number of Xist domains (0–2) in the nucleus is presented on the right. Each bar represents one female morula. (Scale bar, 50 μm.) *P < 0.05.
Fig. S8.
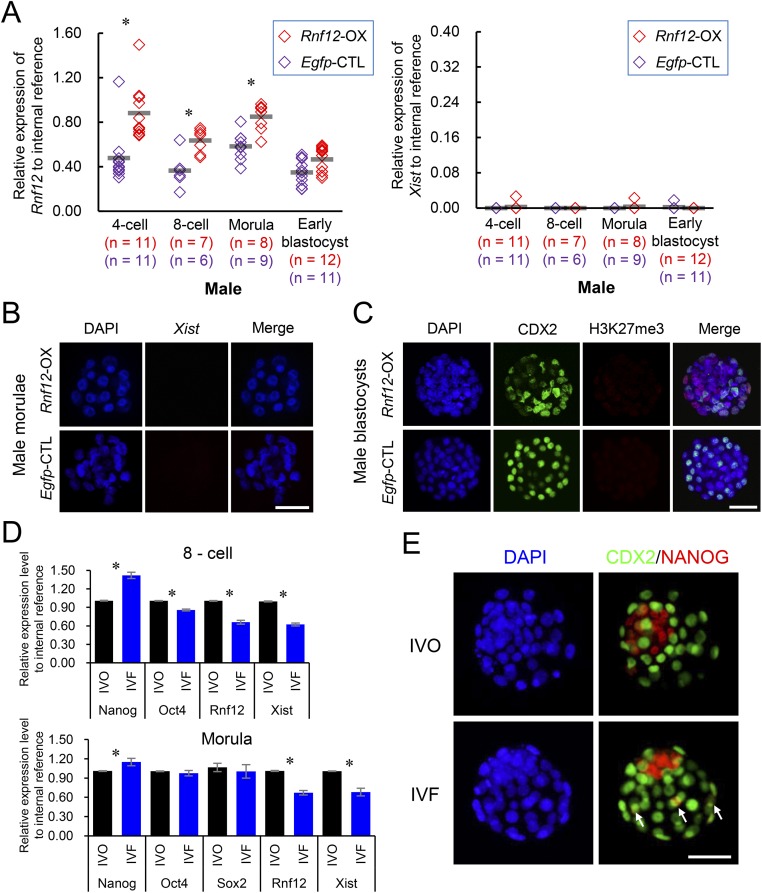
Rnf12 overexpression does not induce an ectopic iXCI in male embryos. (A) Relative expression levels of Rnf12 and Xist in male embryos overexpressing Rnf12 (Rnf12-OX) and Egfp (Egfp-CTL) during the preimplantation stage. Each diamond represents the expression value detected by single embryo qRT-PCR. Horizontal gray lines represent mean values. The number of embryos in each group is indicated. (B) RNA-FISH analyses of Xist noncoding RNA in Rnf12-OX and Egfp-CTL male embryos. (C) H3K27me3 immunostaining in Rnf12-OX and Egfp-CTL male embryos; trophoblast cells were labeled with CDX2. (D) Relative mRNA expression levels of pluripotency factors (Nanog, Oct4, and Sox2), Rnf12, and Xist in pooled IVO and IVF female embryos. Sox2 was not detected at the eight-cell stage. (E) NANOG immunostaining in IVO and IVF female blastocysts. White arrows indicate the mislocalization of NANOG in trophoblast cells. (Scale bar, 50 μm.) *P < 0.05.
We further determined whether improvement of iXCI could rescue the developmental defects in IVF female embryos. Rnf12 overexpression had no evident side effects on blastocyst development as it did not affect the blastocyst formation rate (83.5% vs. 84.2%) and their sex ratio (Fig. 3H). However, the proportion of morphologically abnormal female embryos was significantly decreased at E7.5 after overexpressing Rnf12 (4.09% vs. 9.89%; Fig. 3F), although the iXCI status remained impaired in abnormal embryos (Fig. 3G). These results showed that improved iXCI status due to Rnf12/Xist up-regulation can rescue the developmental defects in IVF female embryos. Furthermore, Rnf12 overexpression resulted in correction of the skewed sex ratio (58.16%) of the nontreated IVF offspring to 51.92% at birth (Fig. 3H), comparable to that of IVO group (50.89%; Fig. 1B). Although the exact causes of reduced Rnf12 expression in IVF female embryos remain to be determined, we found that negative Rnf12 regulators (34), i.e., Nanog homeobox (Nanog) and POU domain, class 5, transcription factor 1 (Oct4), were dysregulated in IVF female embryos (Fig. S8D). We found Nanog expression to be higher in IVF female embryos (Fig. S6); however, immunofluorescence microscopy analysis showed evident mislocalization of NANOG protein in some individual trophoblast cells of IVF female blastocysts (Fig. S8E). These findings show that Nanog expression in IVF embryos is dysregulated in a spatiotemporal manner, which may partially explain the inhibited Rnf12/Xist expression in IVF embryos. Nonetheless, our findings show that iXCI impairment is a major epigenetic error responsible for the female-biased developmental defects and skewed sex ratio at birth in mouse IVF offspring.
Supplementation of RA in Embryo Culture Medium Improved iXCI Status and Corrected Skewed Sex Ratio of IVF Offspring.
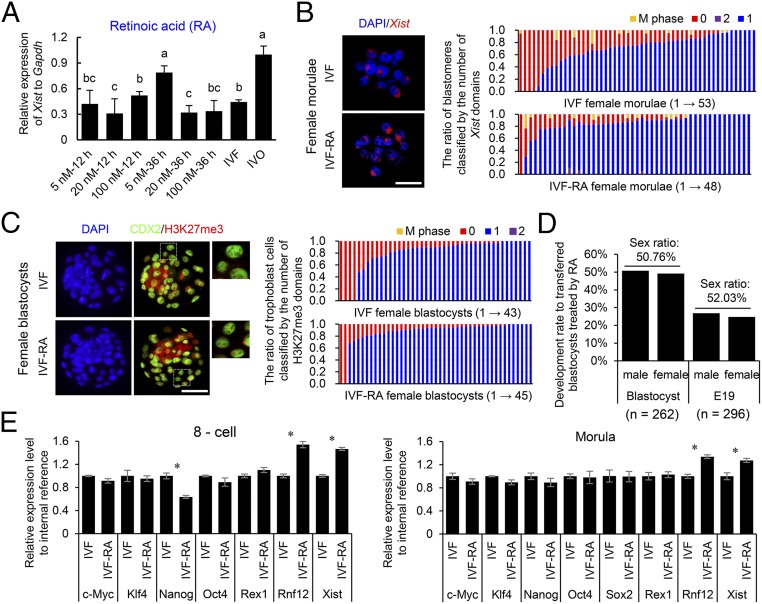
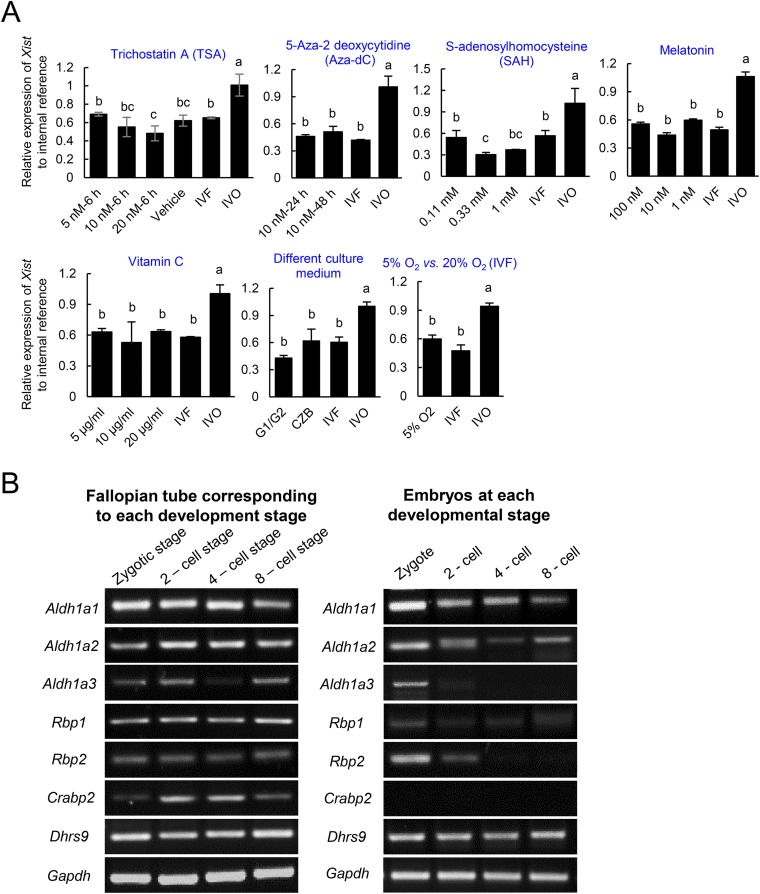
Having determined a causal role of impaired iXCI in the skewed sex ratio at birth, we then asked whether improving iXCI status via supplementation of epigenetic modulators in culture medium could prevent sex skewing in mouse IVF offspring. Because establishment and maintenance of XCI involves Xist coating, histone modifications, and DNA methylation (35), we screened several well-known related modulators as potential candidates (Fig. 4A and Fig. S9A). We found that supplementation of RA in the embryo culture medium significantly up-regulated Rnf12/Xist expression and improved the iXCI status in IVF female embryos (Fig. 4 A–C and E). Moreover, RA supplementation effectively corrected the skewed sex ratio (57.17%) in nontreated mouse IVF offspring to 52.03% at birth (Fig. 4D). In mouse ES cells, RA up-regulates Xist expression by inhibiting the expression of pluripotency factors (36). Similarly, we found that RA suppressed Nanog and Oct4 expression in preimplantation mouse IVF female embryos (Fig. 4E), and these effects were reversed by RA withdrawal from the culture medium (Fig. 4E). RA had no significant adverse effect on the developmental potential as the cell number ratio of trophoblast to inner cell mass in treated IVF embryos did not differ from that of untreated ones (2.54 ± 0.61 vs. 2.50 ± 0.58). Interestingly, we found that the expression of key genes involved in the synthesis and metabolism of RA can be detected in preimplantation embryos and fallopian tube (Fig. S9B). Thus, it is possible that dysregulation of fallopian tube-derived RA and other factors such as cytokines and hormones contribute to the compromised development of IVF embryos, probably via epigenetic involved mechanisms such as XCI.
Fig. 4.
Supplementation of RA in the culture medium improves the iXCI status and corrects the skewed sex ratio in IVF offspring. (A) The relative Xist expression in RA-treated IVF female morulae. Values with different letters are significantly different (P < 0.05). At least 50 embryos were evaluated in each group for each replicate. (B) Representative nuclear Xist noncoding RNA localization in RA-treated female morulae. The ratio of blastomeres classified by the number (0–2) of Xist domains is presented on the right. (C) Representative nuclear H3K27me3 immunostaining in RA-treated female blastocysts. The ratio of trophoblast cells classified by the number (0–2) of H3K27me3 domains is presented on the right. (Rightmost panels) Higher magnification of boxed regions. (B and C) Each bar represents one female embryo. (D) Development rates of RA-treated male and female embryos at the blastocyst stage and E19. The number of embryos or fetuses in each group is indicated. (E) The effect of RA on the expression levels of the pluripotency factors, Rnf12 and Xist, in pooled IVF female embryos at the eight-cell and morula stages. Sox2 was not detected at the eight-cell stage. *P < 0.05. (Scale bar, 50 μm.)
Fig. S9.
(A) Effects of supplementations on the relative Xist expression in IVF female morulae. After IVF, the zygotes were further cultured in culture medium supplemented with different compounds. Values with different letters are significantly different (P < 0.05). At least 50 embryos were evaluated in each group for each replicate. DMSO [0.01% (vol/vol)] was used as the vehicle control (Vehicle). (B) The expression patterns of genes involved in synthesis and metabolism of RA in early preimplantation embryos and fallopian tube corresponding to each development stage.
Conclusions
Sex ratio, as an important indicator of reproduction health, tends to be skewed by many endogenous or exogenous factors, including altered nutrition [e.g., high-fat diet (37, 38) and glucose concentration (39)], environmental conditions [e.g., crowded conditions (40)], and other factors (e.g., IVF processes). However, the underlying mechanisms are likely to be complex and remain largely unknown.
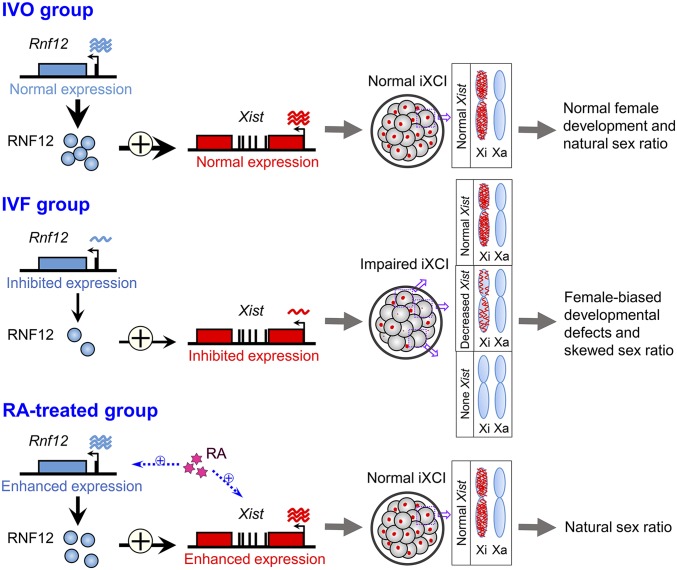
Our current study, focusing on the sex skewing induced by IVF conditions, identified that this skewed sex ratio is due to impaired iXCI via reduced Rnf12/Xist expression during early embryonic development; supplementation of an epigenetic modulator RA in embryo culture medium can correct the skewed sex ratio in mouse IVF offspring (Fig. 5). Our current findings have suggested a potential strategy for preventing sex skewing and IVF-associated complications by targeting erroneous epigenetic modifications induced by IVF. Moreover, considering the crucial role of epigenetic reprogramming in early embryonic development, the changed epigenetic modifications may also involve in sex skewing induced by other factors.
Fig. 5.
A model illustrating the causal role of iXCI in sex skewing in mouse IVF offspring. IVF induces impaired iXCI via reduced Rnf12/Xist expression during early mouse embryonic development, resulting in female-biased developmental defects at the peri-implantation stage and skewed sex ratio at birth. Supplementation of an epigenetic modulator RA can correct the skewed sex ratio by improving the iXCI status through up-regulating the Rnf12/Xist expression in IVF female embryos.
Materials and Methods
Details are in the SI Materials and Methods, including experimental procedures and reagents.
Animals.
ICR female mice aged 7–8 wk, ICR male mice aged 10–12 wk, and (C57BL/6 × DBA/2) F1 (BDF1) male mice aged 10–12 wk were fed ad libitum and housed under controlled lighting conditions (12 light:12 dark). They were maintained under specific pathogen-free conditions. All animal experiments were approved by and performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of China Agricultural University.
Preparation of Mouse Embryos.
All experiments involved in embryo preparation were performed as previously described (25), with minor modifications. ICR female mice were superovulated by an i.p. injection of 5 IU pregnant mare serum gonadotropin and a further i.p. injection 48 h later of 5 IU human chorionic gonadotropin (hCG). In the IVO group, superovulated ICR females were cocaged individually with ICR males after the hCG injection. The following morning, successful mating was confirmed by the presence of a vaginal plug. In some experiments, superovulated ICR females were mated individually with BDF1 males after the hCG injection. In the IVF group, sperm were obtained from the cauda epididymis and capacitated for 1 h in human tubal fluid (HTF; SAGE) medium at 37 °C in 5% CO2. Oocytes were collected from the ampullae at 14 h after hCG treatment. Gametes were then coincubated in HTF medium for 4 h at 37 °C in 5% CO2. After 4 h in the incubator, zygotes were washed and cultured to the blastocyst stage in potassium simplex optimization medium containing amino acids (KSOM+AA; Millipore) under mineral oil at 37 °C in 5% CO2.
Pseudo-pregnant female mice (recipients) were cocaged individually with vasectomized males 3.5 d before embryo transfer. The morning after mating, the recipients were checked for the presence of a vaginal plug. The day of plugging was considered to be day 0.5 of the pseudo-pregnancy. Well-developed blastocysts with similar morphologies were selected for embryo transfer. Twelve blastocysts were transferred to each recipient, with six embryos in each uterine horn. For some experiments, pronuclear stage embryos were transferred into the oviducts of day 0.5 recipients.
Collection of Mouse Embryos.
IVO embryos at the four-cell, eight-cell, and morula stages (54–56, 68–70, and 78–80 h after hCG, respectively) were recovered from donors by flushing the fallopian tubes with M2 medium. Embryos at the blastocyst stage (92–94 h after hCG) were recovered from donors by flushing the uterus with M2 medium (25). IVF embryos at different preimplantation stages were collected based on their developmental progress and morphology. Before the final collection, samples were washed with acidic Tyrode’s solution (Sigma) to eliminate the zona pellucida. Any residual contaminants were depleted via a short series of additional washes in PBS/0.1% polyvinyl alcohol (PVA).
At E7.5, the conceptuses covered with the decidual mass were gently teased away from the uterus. The decidua in which the conceptus embedded was peeled off, and the parietal yolk sac was opened to expose the visceral yolk sac endoderm layer. Embryonic and extraembryonic parts of the E7.5 embryos were separated and washed in PBS before further analyses.
SI Materials and Methods
Animals.
ICR female mice aged 7–8 wk, ICR male mice aged 10–12 wk, and (C57BL/6 × DBA/2) F1 (BDF1) male mice aged 10–12 wk were fed ad libitum and housed under controlled lighting conditions (12 light:12 dark). They were maintained under specific pathogen-free conditions. All animal experiments were approved by and performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of China Agricultural University.
Preparation of Mouse Embryos.
All experiments involved in embryo preparation were performed as previously described (25), with minor modifications. ICR female mice were superovulated by an i.p. injection of 5 IU pregnant mare serum gonadotropin and a further i.p. injection 48 h later of 5 IU hCG. In the IVO group, superovulated ICR females were cocaged individually with ICR males after the hCG injection. The following morning, successful mating was confirmed by the presence of a vaginal plug. In some experiments, superovulated ICR females were mated individually with BDF1 males after the hCG injection. In the IVF group, sperm were obtained from the cauda epididymis and capacitated for 1 h in HTF (SAGE) medium at 37 °C in 5% (vol/vol) CO2. Oocytes were collected from the ampullae at 14 h post-hCG treatment. Gametes were then coincubated in HTF medium for 4 h at 37 °C in 5% (vol/vol) CO2. After 4 h in the incubator, zygotes were washed and cultured to the blastocyst stage in potassium simplex optimization medium containing amino acids (KSOM+AA; Millipore) under mineral oil at 37 °C in 5% (vol/vol) CO2.
Pseudo-pregnant female mice (recipients) were cocaged individually with vasectomized males 3.5 d before embryo transfer. The morning after mating, the recipients were checked for the presence of a vaginal plug. The day of plugging was considered to be day 0.5 of the pseudo-pregnancy. Well-developed blastocysts with similar morphologies were selected for embryo transfer. Twelve blastocysts were transferred to each recipient, with six embryos in each uterine horn. For some experiments, pronuclear stage embryos were transferred into the oviducts of day 0.5 recipients.
Collection of Mouse Embryos.
IVO embryos at the four-cell, eight-cell, and morula stages (54–56, 68–70, and 78–80 h after hCG, respectively) were recovered from donors by flushing the fallopian tubes with M2 medium. Embryos at the blastocyst stage (92–94 h after hCG) were recovered from donors by flushing the uterus with M2 medium (25). IVF embryos at different preimplantation stages were collected based on their developmental progress and morphology. Before the final collection, samples were washed with acidic Tyrode’s solution (Sigma) to eliminate the zona pellucida. Any residual contaminants were depleted via a short series of additional washes in PBS/0.1% PVA.
At E7.5, the conceptuses covered with the decidual mass were gently teased away from the uterus. The decidua in which the conceptus embedded was peeled off, and the parietal yolk sac was opened to expose the visceral yolk sac endoderm layer. Embryonic and extraembryonic parts of the E7.5 embryos were separated and washed in PBS before further analyses.
Preparation of bovine blastocysts.
In the IVO group, blastocysts were obtained after superovulation and artificial insemination (AI) of donor females, as described previously (41). Estrous cycles of donors were synchronized, and a total of 400 mg NIH-FSH-p (Bioniche Animal Health Canada) was given i.m. to the donors, in 50 mg doses, twice a day, for 4 consecutive days, starting on day 9 or 10 of the estrous cycle. A dose of prostaglandin (PG) F2α was injected i.m. into the donors along with the sixth injection of FSH-p. Estrus detection was performed every 4–6 h, and AI was performed twice at 10–12 and 22–24 h after the onset of estrus. On day 7 of development, blastocysts were nonsurgically recovered by uterine flushing.
In the IVF group, as described previously (42), bovine oocytes were collected from ovaries obtained from a slaughterhouse, and matured in Tissue Culture Medium-199 (TCM-199) plus 10% (vol/vol) FBS (HyClone), 1% antibiotic-antimycotic (Gibco BRL), and 10 ng/mL epidermal growth factor (22–24 h). In vitro fertilization was conducted in Bracket and Oliphant's (BO) medium. Briefly, matured oocytes with multiple layers of expanded cumulus cells were washed and then in BO fertilization medium supplemented with 6 mg/mL essential fatty acid-free (FAF)-BSA and 10 mg/mL heparin. Fifteen to 20 cumulus-oocyte complexes (COCs) were placed in 50 µL BO medium, under mineral oil, containing frozen-thawed sperm (1–2 × 106 sperm/mL) for 24 h in 5% (vol/vol) CO2 in air at 38.5 °C. Cumulus cells were removed by pipetting, and presumptive zygotes were cultured in 20-µL drops of Bovine VitroCleave (IVF Vet Solutions) under mineral oil for 5 d. On day 5, embryos were transferred in groups of 5–10 to 20-µL drops of Bovine VitroBlast (IVF Vet Solutions) under mineral oil.
Preparation of rabbit hatched blastocysts.
All rabbits (New Zealand White rabbit) were maintained under the same environmental, nutritional, and general management conditions. In the IVO group, as described previously (29), females were superovulated and mated with normal males. Superovulation was performed by five s.c. injections of NIH-FSH-p (Bioniche Animal Health Canada) for 3 d before mating: two 6-mg injections on day 1 at 12-h intervals, two 12-mg injections on day 2 at 12-h intervals, and one 6-mg injection on day 3, followed 12 h later by an i.v. injection of 30 IU hCG at mating time. Uterus flushing was at 120 h postcoitum (h.p.c.) for the recovery of hatched blastocysts.
In the IVF group, the donors were killed at 14 h after hCG injection, and COCs were recovered from oviducts by flushing with Dulbecco's PBS (DPBS) medium (Gibco BRL). Ejaculated sperm was collected using an artificial vagina. Ten to 15 COCs were placed in 200 µL Tyrode medium (adding albumin [6 g/L] and sodium lactate [0.3% (vol/vol)], sodium pyruvate [22 μg/L], heparin [10 μg/L], hypotaurine [11 μg/L], and adrenalin [1.8 μg/L]), under mineral oil, containing sperm (1–2 × 106 sperm/mL) for 6 h in 5% (vol/vol) CO2 in air at 37 °C. Cumulus cells were removed by pipetting, and presumptive zygotes were washed and cultured in 100 μL TCM-199 plus 10% (vol/vol) FBS (HyClone).
DNA-FISH.
DNA-FISH was performed using XCyting DNA Probes (MetaSystems) based on the manufacturer’s instructions. Briefly, blastocysts were individually transferred to the microscope slide and treated with 0.5 µL 0.01 N HCl/0.1% Tween-20 solution. After air-drying for 20 min, the slides were incubated for 2 min at 37 °C in HCl/pepsin solution (0.01 N HCl/0.01% pepsin in double distilled water) and then fixed in 80% (vol/vol) methanol at −20 °C for 20 min. The slides were then incubated with XCyting mouse probes for chromosomes X (FITC) and Y (Orange) and denatured for 3 min at 75 °C. Hybridization took place overnight at 37 °C, and the slides were washed the next day in wash buffer at 45 °C without agitation. The samples were then counterstained with DAPI (Vector Laboratories). Fluorescent signals were acquired on an upright microscope (BX51; Olympus) using an attached digital microscope camera (DP72; Olympus).
Real-Time Quantitative PCR Analysis.
RNA extraction and reverse transcription were performed according to the manufacturer’s protocol of the Cells-to-cDNA TM II Kit (Ambion). Real-time PCR was performed using SsoFast EvaGreen Supermix (Bio-Rad) in a CFX96 real-time PCR machine (Bio-Rad) using primers in Table S1. The amplification conditions were as follows: initial denaturation for 30 s at 95 °C and 40 cycles of 5 s at 95 °C, annealing, and elongation for 5 s at 60 °C. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was used as internal reference as reported in previous related studies (43, 44). At least five independent experiments were performed.
Table S1.
Primer information
| Gene name | Forward (5′ to 3′) | Reverse (5′ to 3′) |
| Gapdh | TGCCCCCATGTTTGTGATG | TGTGGTCATGAGCCCTTCC |
| Actb | AGGTCATCACTATTGGCAAC | ACTCATCGTACTCCTGCTTG |
| Xist | GGCGGTGCAAACTAAAACTC | CAGTAGGCTTAGAGAACCGC |
| Rnf12 | TTTGTCGCAGGGCAGTCTTA | GTTTGCCCATCACTATTCCAGC |
| Atp7a | GCTTGATTGCTATTGCTGAT | CATCTCCTACCATTGCTACA |
| Abcb7 | CTGTCCTCTTCCATAATACCA | TCCTCGTTCTCCTACTTGT |
| Chic1 | GACCTATCACATTCCTTCTACA | TACAAGCACACTTCCATTCA |
| Xnp | GACATCACTTCTTCATCTTCTG | GTCGTCGTCGTCATCATC |
| Fmr1 | GACGAACTCAGTGATTGGT | CCTCTTCCTCTTCCTCCTT |
| Kif4 | TTGTAGACCTCGCTGGAT | CATAAGAGTGTGGCTATTGC |
| Rps4x | TGACTGGAGGTGCTAACTT | CAATGGTGAGGCGGATTC |
| Smcx | CCGAATCCAGAGGTTGAAT | ATCCGCCGTTCTACATTG |
| Huwe1 | CTACCACCACCACCACTA | TATACCACTGCCTCCTTCA |
| G6pdx | ACCAGACTTCCCAGAGGCTA | AGGCAGCATGAGGTCTCTTC |
| Pgk1 | CAGTTGCTGCTGAACTCAAATC | CCCTTCCCTTCTTCCTCTACAT |
| Fgd1 | GACAGCAAGGATGCCCAGAA | AAGGAGGCGGTCGTTGAATA |
| Pdha1 | GGCATAAACCCTACGGACCA | CTCCTCTTCGTCCTGTTAGC |
| Hprt1 | CAGCGTCGTGATTAGCG | GCCTCCCATCTCCTTCAT |
| c-Myc | TGAGCCCCTAGTGCTGCAT | TCTTGCTCTTCTTCAGAGTCGCT |
| Nanog | AGGATGAAGTGCAAGCGGTG | TGCTGAGCCCTTCTGAATCAG |
| Oct4 | CCCCAATGCCGTGAAGTTG | TCAGCAGCTTGGCAAACTGTT |
| Sox2 | CACAGATGCAACCGATGCA | GGTGCCCTGCTGCGAGTA |
| Klf4 | CCAGCAAGTCAGCTTGTGAA | GGGCATGTTCAAGTTGGATT |
| Rex1 | CTAAAGCAAGACGAGGCAAG | AGAATGGGTTCGGAAAACTC |
| Rbp2 | ATGGAGAGTAATGAGAAC | TCTTGAGTGATGATCTTC |
| Crabp2 | CACGGAGATTAACTTCAA | ATTTCACCAAACTCTTACA |
| Aldh1a1 | GGCTACACATCTCCTATA | CGCTTATATCTATCCTTCTAT |
| Aldh1a2 | TCACATCGGCATAGACAA | TCTTCAGGTTACTTCTTCCA |
| Aldh1a3 | TCTACCACATTCAATAAC | GAAGACTACAGAACTATG |
| Dhrs9 | ACTGGTATGAACTGAGAG | GTATGAGGCTTGTATTGG |
| Rbp1 | AACCTCCATTCCAGTGAA | AATCATCTAGTGTAAGACATTGT |
| DYzEms3 | TAGGATGGTAAGCCCAATGC | TTGGTTGGTTAATTGTTTGGG |
| Rn18s | AGAAACGGCTACCACATCCAA | CCTGTATTGTTATTTTTCGTCACTACCT |
| Rnf12-IVT | TAATACGACTCACTATAGGGGCC | TCACACAACACTTTCTCTGTTCCC |
| ACCATGGAGAACTCAGATTCTAACG | AGAAGATAAGACTGCCCTGCGAC |
For qRT-PCR on single female or male embryos, the embryo was collected and treated with 7 μL ice-cold Cell Lysis II Buffer (plus the Armored RNA Control). Next, 2 μL of the lysis product was digested with proteinase K at 55 °C for 2 h and used for sex determination with TaKaRa Ex Taq Hot Start Version (TaKaRa) according to the manufacturer’s instructions. The amplification conditions were as follows: initial denaturation for 3 min at 95 °C; 40 cycles of 10 s at 95 °C, annealing for 15 s at 52 °C, and elongation for 15 s at 72 °C s; and a 5-min extension at 72 °C. The remaining 5 μL of the lysis product was used to perform reverse transcription according to the manufacturer’s protocol of the Cells-to-cDNA TM II Kit (Ambion).
RNA-FISH.
RNA-FISH was performed using QuantiGene ViewRNA (Affymetrix) based on the manufacturer’s instructions. Fixed embryos were transferred to a microscope slide individually and then fixed in 10% (vol/vol) formaldehyde, boiled in pretreatment solution, and digested with proteinase K. Slides were hybridized for 2 h at 40 °C with custom-designed QuantiGene ViewRNA probes against Xist. The bound probes were amplified using PreAmp and Amp molecules. Label Probe-AP was added, and Fast Red Substrate was used to produce a signal (red dots, Cy3 fluorescence). The embryos were then counterstained with DAPI, and images were acquired using a confocal laser scanning microscope (Digital Eclipse C1; Nikon). Afterward, embryos were removed from the slides and digested individually with proteinase K at 55 °C for 10 h. Sex determination was performed by PCR according to the methods described above.
Immunofluorescence Microscopy.
Embryos were fixed with 4% (wt/vol) paraformaldehyde in PBS (pH 7.4) at 4 °C for 1 h and then permeabilized with PBS/0.5% Triton X-100/0.1% PVA (PBST-PVA) at room temperature for 1 h. After three washes with PBS/0.1% PVA at 37 °C for 10 min, the embryos were blocked in PBS/1% BSA at 4 °C for 6 h. Fixed embryos were then incubated with primary anti-H3K27me3 antibodies (1:1,000; Millipore), anti-RNF12 antibodies (1:400; Abnova), and anti-CDX2 antibodies (1:200; Santa Cruz Biotechnology) overnight at 4 °C. Following three washes with PBST-PVA, the embryos were incubated with secondary antibodies conjugated with Alexa Fluor 488 (anti-mouse; Invitrogen) or Alexa Fluor 594 (anti-rabbit; Invitrogen) for 1 h at room temperature. Finally, the samples were counterstained with DAPI. The fluorescence signals were imaged using a confocal laser scanning microscope (Digital Eclipse C1; Nikon). The sex of each embryo was determined by PCR according to the method described above.
E7.5 IVF embryos were obtained and classified into two subgroups: abnormal (ABN) embryos and normal (N) embryos, according to well-established morphological landmarks (45). As previously described (23), embryonic and extraembryonic parts of E7.5 embryos were separated and washed in PBST and then incubated with trypsin at 37 °C for 15 min. The embryos were disaggregated into single cells by vigorous pipetting and fixed in 4% (wt/vol) paraformaldehyde at 4 °C. A small proportion of the isolated cells were used to determine the sex of the embryos by PCR according to the methods described above. Female embryos were then used for the following procedures. Isolated cells from embryonic and extraembryonic parts were each pooled. These cells were transferred to microscope slides and air dried. Immunofluorescence staining was performed according to the methods described above. Scoring of immunofluorescence experiments was carried out on at least 200 cells in each experiment.
Western Blot.
Preimplantation embryos were collected, and ice-cold 2× Laemmli buffer (Bio-Rad) with 5% (vol/vol) β-mercaptoethanol (β-ME) and protease inhibitors were added. The samples were maintained with constant agitation for 10 min at 4 °C and subsequently heated at 100 °C for 10 min. The proteins were separated on 7.5% (wt/vol) or 12% (wt/vol) acrylamide gels containing 0.1% SDS and then transferred onto hydrophobic PVDF membranes (Amersham). The membranes were blocked with 5% nonfat dried milk in TBS containing 0.05% Tween-20 for 2 h at room temperature and then incubated overnight at 4 °C with diluted anti-RNF12 antibodies (1:1,000; Abnova), followed by three washes with TBS containing 0.05% Tween-20 for 20 min. Peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch) were added for 1.5 h, and protein bands were detected using an ImageQuant LAS 4000 (GE Healthcare). Quantification of the blots was performed using ImageJ. No fewer than 100 embryos were pooled for each time point in each group. Similar results were obtained from three independent experiments.
RNA Injection.
mRNAs for Rnf12 and enhanced green fluorescent protein (Egfp) were synthesized by a T7 mMESSAGE mMACHINE Kit (Ambion) using primers in Table S1 and then dissolved in nuclease-free water to a final concentration of 200 μg/mL Synthetic siRNA duplexes were designed by Stealth Designer (Life Technologies): Stealth RNAi siRNA for Rnf12 (ID: MSS208660) and Stealth RNAi siRNA Negative Control LO GC (12935-200) for the negative control. siRNA duplex mixtures were prepared as 200 μM stock solutions and stored at −80 °C until use. BLOCK-iT Alexa Fluor Red Fluorescent Oligo (Life Technologies) was used as an indicator.
Microinjection of mRNA or siRNA was performed as previously described (44). For mRNA microinjections, to prevent excessive overexpression of Rnf12 that may lead to ectopic iXCI (33), a moderate injection dosage was determined by injecting Rnf12 mRNA with different concentrations. Approximately 10 pL Egfp mRNA solution alone or a mixture [1:1 (vol/vol)] of Rnf12 mRNA solution and Egfp mRNA solution was injected into the cytoplasm (46) of IVF embryos at the pronuclear stage. EGFP-positive embryos were selected at the two-cell stage for further culture and analyses. For siRNA microinjections, ∼5 pL siRNAs was microinjected into the cytoplasm of IVO embryos at the pronuclear stage. After incubation at 37 °C for 20 min, embryos were transferred to the oviducts of pseudopregnant female mice.
Statistical Analysis.
All of the data are presented as the mean ± SD. One-way ANOVA was used to compare differences among groups using IBM SPSS 20 (Statistical Package for the Social Sciences) statistics software. The sex ratio was calculated as the proportion of males (number of males/total number of embryos or fetuses) and compared with the naturally expected ratio (50%) using a corrected χ2 procedure (16, 39).
Acknowledgments
We thank Atsuo Ogura for reading the manuscript and providing helpful comments. This work was supported by National Natural Science Foundation of China Grant 31472092, the Earmarked Fund for the Innovative Teams of Beijing Swine Industrialization Research Program, and National High-Tech R&D Program Grants 2011AA100303 and 2013AA102506 (to J.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523538113/-/DCSupplemental.
References
- 1.Steptoe PC, Edwards RG. Birth after the reimplantation of a human embryo. Lancet. 1978;2(8085):366. doi: 10.1016/s0140-6736(78)92957-4. [DOI] [PubMed] [Google Scholar]
- 2.ESHRE 2014 ART fact sheet. Available at https://www.eshre.eu/Guidelines-and-Legal/ART-fact-sheet.aspx. Accessed April 1, 2015.
- 3.IETS 2014. 2013 Statistics of Embryo Collection and Transfer in Domestic Farm Animals (International Embryo Transfer Society, Champaign, IL)
- 4.Servick K. Unsettled questions trail IVF’s success. Science. 2014;345(6198):744–746. doi: 10.1126/science.345.6198.744. [DOI] [PubMed] [Google Scholar]
- 5.Maalouf WE, Mincheva MN, Campbell BK, Hardy IC. Effects of assisted reproductive technologies on human sex ratio at birth. Fertil Steril. 2014;101(5):1321–1325. doi: 10.1016/j.fertnstert.2014.01.041. [DOI] [PubMed] [Google Scholar]
- 6.Dean JH, Chapman MG, Sullivan EA. The effect on human sex ratio at birth by assisted reproductive technology (ART) procedures: An assessment of babies born following single embryo transfers, Australia and New Zealand, 2002-2006. BJOG. 2010;117(13):1628–1634. doi: 10.1111/j.1471-0528.2010.02731.x. [DOI] [PubMed] [Google Scholar]
- 7.Hart R, Norman RJ. The longer-term health outcomes for children born as a result of IVF treatment: Part I—General health outcomes. Hum Reprod Update. 2013;19(3):232–243. doi: 10.1093/humupd/dms062. [DOI] [PubMed] [Google Scholar]
- 8.Källén B, et al. Cancer risk in children and young adults conceived by in vitro fertilization. Pediatrics. 2010;126(2):270–276. doi: 10.1542/peds.2009-3225. [DOI] [PubMed] [Google Scholar]
- 9.Mann MR, Bartolomei MS. Epigenetic reprogramming in the mammalian embryo: Struggle of the clones. Genome Biol. 2002;3(2):S1003. doi: 10.1186/gb-2002-3-2-reviews1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T, et al. IVF results in de novo DNA methylation and histone methylation at an Igf2-H19 imprinting epigenetic switch. Mol Hum Reprod. 2005;11(9):631–640. doi: 10.1093/molehr/gah230. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Gonzalez R, Ramirez MA, Pericuesta E, Calle A, Gutierrez-Adan A. Histone modifications at the blastocyst Axin1(Fu) locus mark the heritability of in vitro culture-induced epigenetic alterations in mice. Biol Reprod. 2010;83(5):720–727. doi: 10.1095/biolreprod.110.084715. [DOI] [PubMed] [Google Scholar]
- 12.Sutcliffe AG, et al. Assisted reproductive therapies and imprinting disorders--a preliminary British survey. Hum Reprod. 2006;21(4):1009–1011. doi: 10.1093/humrep/dei405. [DOI] [PubMed] [Google Scholar]
- 13.Rexhaj E, et al. Mice generated by in vitro fertilization exhibit vascular dysfunction and shortened life span. J Clin Invest. 2013;123(12):5052–5060. doi: 10.1172/JCI68943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avery B, Madison V, Greve T. Sex and development in bovine in-vitro fertilized embryos. Theriogenology. 1991;35(5):953–963. doi: 10.1016/0093-691x(91)90306-x. [DOI] [PubMed] [Google Scholar]
- 15.Torner E, Bussalleu E, Briz MD, Yeste M, Bonet S. Embryo development and sex ratio of in vitro-produced porcine embryos are affected by the energy substrate and hyaluronic acid added to the culture medium. Reprod Fertil Dev. 2014;26(4):570–577. doi: 10.1071/RD13004. [DOI] [PubMed] [Google Scholar]
- 16.Wrenzycki C, et al. In vitro production and nuclear transfer affect dosage compensation of the X-linked gene transcripts G6PD, PGK, and Xist in preimplantation bovine embryos. Biol Reprod. 2002;66(1):127–134. doi: 10.1095/biolreprod66.1.127. [DOI] [PubMed] [Google Scholar]
- 17.Gutiérrez-Adán A, Granados J, Pintado B, De La Fuente J. Influence of glucose on the sex ratio of bovine IVM/IVF embryos cultured in vitro. Reprod Fertil Dev. 2001;13(5-6):361–365. doi: 10.1071/rd00039. [DOI] [PubMed] [Google Scholar]
- 18.Rosenfeld CS, Roberts RM. Maternal diet and other factors affecting offspring sex ratio: A review. Biol Reprod. 2004;71(4):1063–1070. doi: 10.1095/biolreprod.104.030890. [DOI] [PubMed] [Google Scholar]
- 19.Trivers RL, Willard DE. Natural selection of parental ability to vary the sex ratio of offspring. Science. 1973;179(4068):90–92. doi: 10.1126/science.179.4068.90. [DOI] [PubMed] [Google Scholar]
- 20.Hesketh T, Xing ZW. Abnormal sex ratios in human populations: Causes and consequences. Proc Natl Acad Sci USA. 2006;103(36):13271–13275. doi: 10.1073/pnas.0602203103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orzack SH, et al. The human sex ratio from conception to birth. Proc Natl Acad Sci USA. 2015;112(16):E2102–E2111. doi: 10.1073/pnas.1416546112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoki Y, et al. Incomplete X-inactivation initiated by a hypomorphic Xist allele in the mouse. Development. 2011;138(13):2649–2659. doi: 10.1242/dev.061226. [DOI] [PubMed] [Google Scholar]
- 23.Senner CE, et al. Disruption of a conserved region of Xist exon 1 impairs Xist RNA localisation and X-linked gene silencing during random and imprinted X chromosome inactivation. Development. 2011;138(8):1541–1550. doi: 10.1242/dev.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin J, et al. Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice. Nature. 2010;467(7318):977–981. doi: 10.1038/nature09457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagy A, Gerstenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo—A Laboratory Manual. 3rd Ed Cold Spring Harbor Press; Plainview, NY: 2003. [Google Scholar]
- 26.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 27.Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256(5519):640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 28.Huynh KD, Lee JT. X-chromosome inactivation: A hypothesis linking ontogeny and phylogeny. Nat Rev Genet. 2005;6(5):410–418. doi: 10.1038/nrg1604. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto I, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472(7343):370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- 30.van den Berg IM, et al. X chromosome inactivation is initiated in human preimplantation embryos. Am J Hum Genet. 2009;84(6):771–779. doi: 10.1016/j.ajhg.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379(6561):131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 32.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5(4):695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 33.Jonkers I, et al. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139(5):999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 34.Navarro P, Moffat M, Mullin NP, Chambers I. The X-inactivation trans-activator Rnf12 is negatively regulated by pluripotency factors in embryonic stem cells. Hum Genet. 2011;130(2):255–264. doi: 10.1007/s00439-011-0998-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21(3):359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 36.Ahn JY, Lee JT. Retinoic acid accelerates downregulation of the Xist repressor, Oct4, and increases the likelihood of Xist activation when Tsix is deficient. BMC Dev Biol. 2010;10:90. doi: 10.1186/1471-213X-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenfeld CS, et al. Striking variation in the sex ratio of pups born to mice according to whether maternal diet is high in fat or carbohydrate. Proc Natl Acad Sci USA. 2003;100(8):4628–4632. doi: 10.1073/pnas.0330808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green MP, et al. Nutritional skewing of conceptus sex in sheep: Effects of a maternal diet enriched in rumen-protected polyunsaturated fatty acids (PUFA) Reprod Biol Endocrinol. 2008;6:21. doi: 10.1186/1477-7827-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura K, Spate LD, Green MP, Roberts RM. Effects of D-glucose concentration, D-fructose, and inhibitors of enzymes of the pentose phosphate pathway on the development and sex ratio of bovine blastocysts. Mol Reprod Dev. 2005;72(2):201–207. doi: 10.1002/mrd.20342. [DOI] [PubMed] [Google Scholar]
- 40.Krackow S. Effects of mating dynamics and crowding on sex ratio variance in mice. J Reprod Fertil. 1997;110(1):87–90. doi: 10.1530/jrf.0.1100087. [DOI] [PubMed] [Google Scholar]
- 41.Bertolini M, et al. Growth, development, and gene expression by in vivo- and in vitro-produced day 7 and 16 bovine embryos. Mol Reprod Dev. 2002;63(3):318–328. doi: 10.1002/mrd.90015. [DOI] [PubMed] [Google Scholar]
- 42.Nedambale TL, Dinnyés A, Yang X, Tian XC. Bovine blastocyst development in vitro: Timing, sex, and viability following vitrification. Biol Reprod. 2004;71(5):1671–1676. doi: 10.1095/biolreprod.104.027987. [DOI] [PubMed] [Google Scholar]
- 43.Inoue K, et al. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science. 2010;330(6003):496–499. doi: 10.1126/science.1194174. [DOI] [PubMed] [Google Scholar]
- 44.Matoba S, et al. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci USA. 2011;108(51):20621–20626. doi: 10.1073/pnas.1112664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rugh R. The Mouse: Its Reproduction and Development. Oxford Science Publications; Oxford: 1968. [Google Scholar]
- 46.Ohashi S, Naito K, Liu J, Sheng Y, Yamanouchi K, Tojo H. Expression of exogenous proteins in porcine maturing oocytes after mRNA injection: Kinetic analysis and oocyte selection using EGFP mRNA. J Reprod Dev. 2001;47(6):351–357. [Google Scholar]
- 47.Breton A, LE Bourhis D, Audouard C, Vignon X, Lelièvre JM. Nuclear profiles of H3 histones trimethylated on Lys27 in bovine (Bos taurus) embryos obtained after in vitro fertilization or somatic cell nuclear transfer. J Reprod Dev. 2010;56(4):379–388. doi: 10.1262/jrd.09-182a. [DOI] [PubMed] [Google Scholar]
- 48.Gontan C, et al. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature. 2012;485(7398):386–390. doi: 10.1038/nature11070. [DOI] [PubMed] [Google Scholar]