Significance
Mitochondria are major cellular energy suppliers and have to cope with changes in nutrient supply and energy demand that naturally occur throughout the day. We obtained the first, to our knowledge, comprehensive mitochondrial proteome around the clock and identified extensive oscillations in mitochondrial protein abundance that predominantly peak during the early light phase. Remarkably, several rate-limiting mitochondrial enzymes that process different nutrients accumulate in a diurnal manner and are dependent on the clock proteins PER1/2. Concurrently, we uncovered daily oscillations in mitochondrial respiration that are substrate-specific and peak during different times of the day. We propose that the circadian clock PERIOD proteins regulate the diurnal utilization of different nutrients by the mitochondria and thus, optimize mitochondrial function to daily changes in energy supply/demand.
Keywords: circadian rhythm, proteomics, metabolism, mitochondria, PERIOD proteins
Abstract
Mitochondria are major suppliers of cellular energy through nutrients oxidation. Little is known about the mechanisms that enable mitochondria to cope with changes in nutrient supply and energy demand that naturally occur throughout the day. To address this question, we applied MS-based quantitative proteomics on isolated mitochondria from mice killed throughout the day and identified extensive oscillations in the mitochondrial proteome. Remarkably, the majority of cycling mitochondrial proteins peaked during the early light phase. We found that rate-limiting mitochondrial enzymes that process lipids and carbohydrates accumulate in a diurnal manner and are dependent on the clock proteins PER1/2. In this conjuncture, we uncovered daily oscillations in mitochondrial respiration that peak during different times of the day in response to different nutrients. Notably, the diurnal regulation of mitochondrial respiration was blunted in mice lacking PER1/2 or on a high-fat diet. We propose that PERIOD proteins optimize mitochondrial metabolism to daily changes in energy supply/demand and thereby, serve as a rheostat for mitochondrial nutrient utilization.
Mitochondria serve as major suppliers of cellular energy through nutrient oxidation. One of the major challenges that mitochondria face is the adaptation to changes in nutrient supply and energy demand. An inability of mitochondria to deal with altered nutrient environment is associated with metabolic diseases, such as diabetes and obesity (1, 2).
Mitochondria oxidize carbohydrates and lipids to generate ATP by a process known as oxidative phosphorylation. Pyruvate and fatty acids are transported from the cytoplasm into the mitochondrial matrix, where they are catabolized into acetyl CoA. Pyruvate is converted to acetyl CoA through the action of the pyruvate dehydrogenase complex (PDC), whereas fatty acids are oxidized through a cycle of reactions that trim two carbons at a time, generating one molecule of acetyl CoA in each cycle [i.e., fatty acid oxidation (FAO)]. The acetyl groups are then fed into the Krebs cycle for additional degradation, and the process culminates with the transfer of acetyl-derived high-energy electrons along the respiratory chain.
Mounting evidence suggests that circadian clocks orchestrate our daily physiology and metabolism (3–6). The mammalian circadian timing system consists of a central pacemaker in the brain that is entrained by daily light–dark cycles and synchronizes subsidiary oscillators in virtually all cells of the body, in part by driving rhythmic feeding behavior. The core clock molecular circuitry relies on interlocked transcription–translation feedback loops that generate daily oscillations of gene expression in cultured cells and living animals (7). Many transcriptomes (8–12) and more recently, several proteomics (13–15) and metabolomics studies (16–21) highlighted the pervasive circadian control of metabolism.
Rest–activity and feeding–fasting cycles that naturally occur throughout the day impose pronounced changes in nutrient supply and energy demand that need to be addressed by mitochondrial function. However, very little is known at the molecular level how mitochondria handle these fluctuations. To address this question, we examined temporal changes in the mitochondrial proteome by applying MS-based quantitative proteomics on isolated mitochondria from mouse liver. We obtained the first, to our knowledge, comprehensive mitochondrial proteome around the clock and found that ∼38% of mitochondrial proteins oscillate in abundance throughout the day, with a predominant peak during the early light phase. Remarkably, several rate-limiting mitochondrial enzymes that process different nutrients accumulate in a diurnal manner and are dependent on the clock proteins PER1/2. Concurrently, we uncovered daily oscillations in mitochondrial respiration that are substrate-specific and peak during different times of the day. We propose that PERIOD proteins regulate the diurnal utilization of different nutrients by the mitochondria and thus, optimize mitochondrial function to daily changes in energy supply/demand.
Results
Quantitative Proteomics Identify a Predominant Daily Phase for Mitochondrial Protein Accumulation.
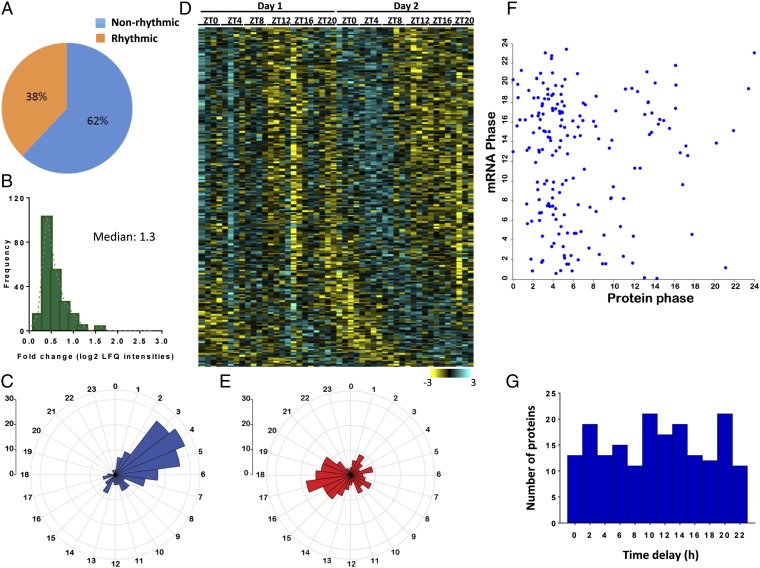
To examine the daily regulation of mitochondrial function, we commenced with a MS-based quantitative proteomics approach. Mice were killed at 4-h intervals over 2 d, livers were harvested, and mitochondria were isolated by biochemical fractionation. For each time point, mitochondria from four individual animals were independently analyzed. Proteomic analysis identified 1,537 proteins with label-free intensities (22) in at least three of four biological replicates for each measured time point. As expected, the quantified proteome was statistically enriched with mitochondrial-annotated proteins (Fig. S1A and Dataset S1). In addition, we identified some endoplasmic reticulum- and peroxisome-associated proteins that were likely to be copurified with mitochondria. Cycling analysis using a 24-h periodicity revealed that, of 1,537 proteins, 452 proteins (29%) exhibited statistically significant daily oscillations [q value (permutation-based false discovery rate) < 0.15] (Fig. S1B and Dataset S2A), with median amplitude of ∼1.4-fold (Fig. S1C). Oscillating proteins exhibited two prominent phases 12 h apart: Zeitgeber time (ZT) ∼4 and ∼ZT16 (Fig. S1 D and E). Interestingly, when the analysis was limited to exclusively mitochondrial-annotated proteins (Uniprot annotation), a larger fraction oscillated (223 of 590; 38%; q value < 0.15) (Fig. 1A and Dataset S2B), with median amplitude of 1.3-fold (Fig. 1B). Remarkably, the majority of cycling proteins peaked at ∼ZT4 (Fig. 1 C and D). Phase enrichment analysis (using a less stringent cutoff q value < 0.5) showed that proteins from key mitochondrial functions cycle with phases statistically enriched (false discovery rate < 0.1) during the light phase (Fig. S2).
Fig. S1.
Analysis of diurnal protein oscillations in the mitochondrial fraction. (A) Scatter plot showing protein categories (UniProt keywords) that were statistically enriched (Fisher exact test P < 0.05) in the quantified mitochondrial fraction proteome compared with an in silico mouse proteome. Highlighted categories are the most statistically enriched. Categories statistically overrepresented (enrichment factor >1) are indicated in red, and categories statistically underrepresented (enrichment factor <1) are indicated in blue. (B) The percentage of proteins that exhibit a daily pattern of accumulation in the mitochondrial fraction prepared from mouse liver. Of 1,537 identified and quantified proteins, 452 exhibited statistically significant (q value < 0.15) daily oscillations (29%; 12 time points; n = 4 for each). (C) Fold change distribution of rhythmic proteins calculated using the median of the log2 label-free (LFQ) intensities for each time point. The median fold change of the population was 1.41-fold. (D) Radar plot shows the distribution of phases for rhythmic proteins. The y axis on the upper left side indicates the scale of the histogram bins. (E) Hierarchical clustering of rhythmic proteins based on the phase of the cycle. Each row corresponds to a protein group, and each column corresponds to the analyzed sample. The color scale of the values (Z-scored normalized log2 label-free intensities) is shown in the bottom bar [high (light blue) and low (yellow)].
Fig. 1.
Daily oscillations in the mitochondrial proteome. (A) The percentage of annotated mitochondrial proteins that exhibit a diurnal pattern of accumulation. Of 590 quantified and annotated as mitochondrial proteins, 223 proteins exhibited a diurnal pattern of accumulation (38%; 12 time points; n = 4 for each; q value < 0.15). (B) Fold change of rhythmic proteins annotated as mitochondria calculated using the median of the log2 label-free (LFQ) intensities for each time point. The median fold change of the population was 1.3-fold. (C) Phases distribution of the rhythmic proteins annotated as mitochondrial proteins (223 proteins). The y axis on the upper left side indicates the scale of the histogram bins. (D) Hierarchical clustering of rhythmic proteins annotated as mitochondria based on the phase of their maximal expression. Each row corresponds to a protein group entry, and each column indicates the intensities for all of the biological replicates at each time point. The color scale of the intensity values (Z-scored normalized log2 intensities) is shown in the bottom bar [high (light blue) and low (yellow)]. (E) Phase distribution of rhythmic transcripts corresponding to the cycling mitochondrial proteins (185 transcripts). The y axis on the upper left side indicates the scale of the histogram bins. (F) Scatter plot showing the phases for every cycling mitochondrial protein and its corresponding rhythmic mRNA (Pearson correlation r = 0.01). (G) Distribution of the time delays between the peak of the cycling protein and the peak of the corresponding oscillating mRNA.
Fig. S2.
Cycling mitochondrial proteins involved in key processes show high abundance during the light phase. Scatter plot showing the result of cycling annotation distribution using UniProt keywords as protein categories. The indicated categories show statistically significant (FDR < 0.1) enrichment in a phase-dependent manner for the mitochondrial-annotated dataset. Phase (ZT) is indicated in the x axis, and the P value (−log10) of the statistical analysis is indicated in the y axis.
To examine the relation between transcript and protein levels, we compared our cycling mitochondrial-annotated proteome with a widely used mouse liver transcriptome dataset (23). We applied the same statistical algorithm that was used for the proteome analysis to identify oscillations in the corresponding transcripts. Of 223 cycling mitochondrial proteins (216 had matched transcript data), 185 showed rhythms at the mRNA level (86%); however, the phase distribution of cycling proteins and transcripts differed (compare Fig. 1C with Fig. 1E). The majority of cycling transcripts encoding for oscillating mitochondrial proteins peaked at ∼ZT17. The phase correlation between transcript and their corresponding protein was very low (Pearson’s r = 0.01) (Fig. 1F). Hence, we observed a wide distribution in the phase delay between the peak in protein accumulation and the peak in its respective transcript (Fig. 1G). Some proteins followed their transcript levels within a reasonable timeframe, whereas others were significantly delayed. About two-thirds of proteins accumulated in mitochondria more than 6 h later than the peak in their transcript levels, suggesting that posttranscriptional mechanisms shape the phase of protein accumulation in mitochondria.
Our comprehensive quantitative proteomics analysis thus revealed daily oscillations in a large fraction of the mitochondrial proteome. Cycling mitochondrial proteins mostly reached their zenith levels during the early light phase. Although the transcript levels of the majority of oscillating proteins cycled, the transcript to protein phases differed, pointing toward the involvement of posttranscriptional control. Conceivably, the diurnal oscillations in mitochondrial proteins are expected to affect mitochondrial function throughout the day.
Key Mitochondrial Metabolic Enzymes Accumulate in a Daily Manner.
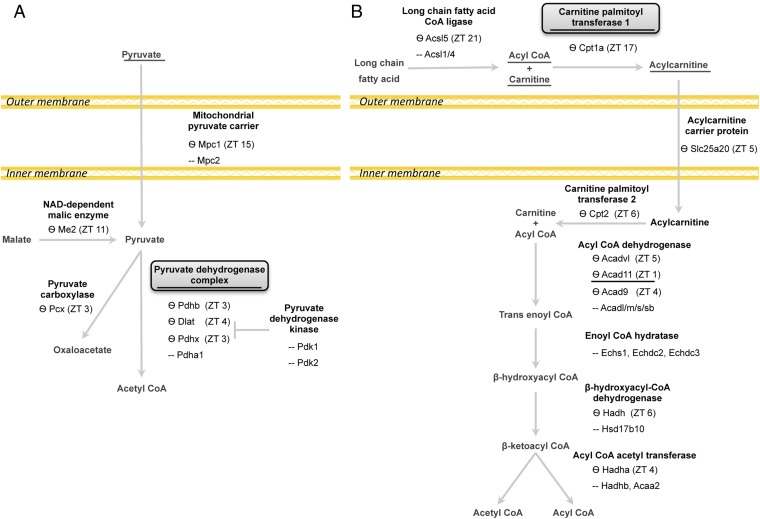
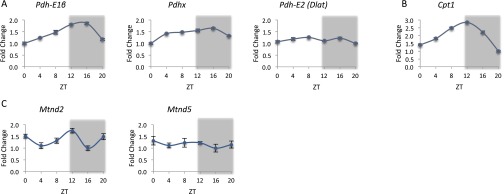
To pinpoint mitochondrial processes that might be under circadian control, we generated flow charts of several central mitochondrial metabolic pathways (i.e., carbohydrate metabolism, fatty acid uptake and FAO, the Krebs cycle, and electron transport chain). In each pathway, we marked the enzymes that accumulate in a daily manner together with their peak abundance time according to our proteomics analysis, and we highlighted the rate-limiting steps (Figs. 2 and 3). The rate-limiting step in mitochondrial carbohydrate metabolism is carried out by the PDC, a multiprotein complex that catalyzes the oxidative decarboxylation of pyruvate (24). We found that several components of the PDC, namely the catalytic pyruvate dehydrogenase PDH-E1β (Pdhb), PDH-E2 (Dlat), and the regulatory subunit PDHX (Pdhx), accumulate in a daily manner with maximum levels at ∼ZT3 (Fig. 2A and Dataset S3). The mRNA levels of Pdh-E1β and Pdhx also cycled but reached their zenith levels at ∼ZT16, whereas Pdh-E2 (Dlat) was relatively constant throughout the day (Fig. S3A). The rate-limiting step for the entry of long-chain fatty acids into the mitochondrial matrix is the synthesis of acylcarnitine from acyl CoA and carnitine, which is mediated by carnitine palmitoyl-transferase 1 (CPT1) (25). We identified oscillations in CPT1 (Cpt1a) protein levels with zenith levels at ∼ZT17 (Fig. 2B and Dataset S4). The mRNA levels of Cpt1 cycled throughout the day, with peak levels at ∼ZT12 (Fig. S3B). After fatty acids enter the mitochondria, they are oxidized. Several enzymes within the FAO pathway cycled in a daily manner and peaked at ∼ZT4 [e.g., acyl CoA dehydrogenase VL (ACADVL), ACAD11, ACAD9, HADH, and HADHa] (Fig. 2B and Dataset S4). Moreover, our analysis identified daily rhythms in several enzymes of the Krebs cycle and various proteins within the respiratory complexes (Fig. 3 and Datasets S5 and S6). Finally, we found that several members of the mitochondrial protein translocation machinery, namely the TIM/TOM complex, accumulate in a daily manner, reaching their peak levels predominantly during the early light phase (Dataset S7), which might suggest that protein entry to the mitochondria is temporally gated.
Fig. 2.
Diurnal oscillations of enzymes in pyruvate metabolism and fatty acid uptake and oxidation. Schematic depiction of the following principal mitochondrial pathways: (A) pyruvate metabolism and (B) fatty acid uptake and FAO. Metabolites are marked in gray, and enzymes are in black; known rate-limiting enzymes are shown as squares. Oscillating enzymes according to the proteomics analysis are marked with a wave sign (Ѳ) together with their peak time indicated by ZT. Metabolites used as substrates for mitochondrial respiration assays in Figs. 4 and 5 and relevant enzymes are underlined.
Fig. 3.
Diurnal oscillations of enzymes in the Krebs cycle and the respiratory chain. Schematic depiction of the following principal mitochondrial pathways: (A) Krebs cycle and (B) respiratory chain complexes. Metabolites are marked in gray, and enzymes are in black. Oscillating enzymes according to the proteomics analysis are marked with a wave sign (Ѳ) together with their peak time indicated by ZT.
Fig. S3.
Daily expression profiles of enzymes participating in mitochondrial nutrient processing. WT mice fed ad libitum were killed at 4-h intervals throughout the day. Total RNA was prepared from liver, and mRNA expression levels of enzymes participating in mitochondrial nutrient processing were determined by quantitative real-time PCR and presented as fold change relative to the lowest value, with means ± SDs of four mice per time point. (A) Pdh-E1β, Pdhx, and Pdh-E2 (Dlat). (B) Cpt1. (C) Mtnd2 and Mtnd5. Gray shading represents the dark phase.
We also quantified 2 mitochondrial-encoded proteins of the known 14, namely NADH-ubiquinone oxidoreductase chain 2 (MTND2) and MTND5. MTND2 was found to be rhythmic, with peak levels at ∼ZT13 (q value = 0.041), whereas MTND5 did not show statistically significant oscillations (q value > 0.15). In accordance, the transcript levels of Mtnd2 were rhythmic with zenith levels at ∼ZT12, whereas Mtnd5 transcript levels were relatively constant throughout the day (Fig. S3C).
In summary, our quantitative proteomics analysis evinced that key enzymes in principal mitochondrial metabolic pathways oscillate throughout the day.
Diurnal Oscillations in Mitochondrial Respiration in Response to Different Nutrients.
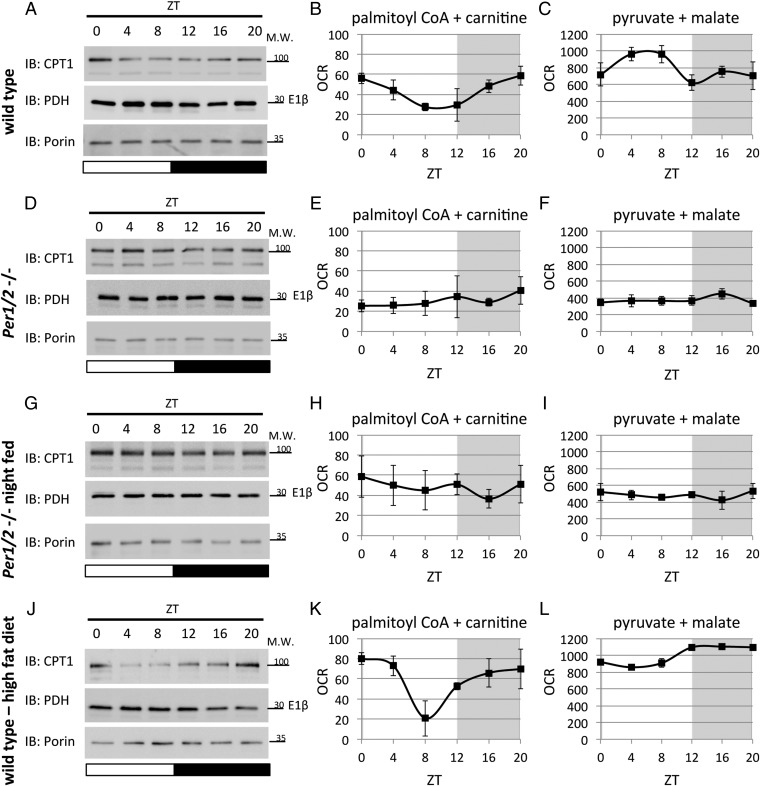
To corroborate the diurnal accumulation of mitochondrial enzymes identified in our proteomics analysis, mitochondrial protein extracts were analyzed by SDS/PAGE and immunoblots. We centered our analysis on the following key rate-limiting enzymes, each related to a distinct mitochondrial pathway: (i) CPT1 (i.e., CPT1α), the rate-limiting enzyme for the entry of long-chain fatty acids to the inner mitochondrial matrix; and (ii) PDH (i.e., PDH-E1β), which catalyzes the rate-limiting step in mitochondrial carbohydrate metabolism as part of the PDC. Immunoblot analysis showed that the protein levels of CPT1 and PDH oscillate in mitochondrial extracts prepared from WT mice (Fig. 4A and Fig. S4A). In line with our proteomics analysis, CPT1 reached zenith levels at ∼ZT20, whereas PDH accumulated at ∼ZT4 (Fig. 4A and Fig. S4A).
Fig. 4.
Daily oscillations in accumulation of rate-limiting mitochondrial enzymes and mitochondrial respiration are PER1/2-dependent. (A) Mitochondrial protein extracts of WT mice fed ad libitum were analyzed by SDS/PAGE and immunoblot (IB) with indicated antibodies. Oxygen consumption rates (OCRs) of isolated mitochondria prepared from WT mice fed ad libitum were quantified using the Seahorse Flux Analyzer in the presence of the indicated substrates: (B) palmitoyl CoA and carnitine and (C) pyruvate and malate. (D) Mitochondrial protein extracts of PER1/2 null mice fed ad libitum were analyzed by SDS/PAGE and IB. OCRs of isolated mitochondria prepared from PER1/2 null mice fed ad libitum were quantified in the presence of the indicated substrates: (E) palmitoyl CoA and carnitine and (F) pyruvate and malate. (G) Mitochondrial protein extracts of night-fed PER1/2 null mice were analyzed by SDS/PAGE and IB. OCRs of isolated mitochondria prepared from night-fed PER1/2 null mice were quantified in the presence of the indicated substrates: (H) palmitoyl CoA and carnitine and (I) pyruvate and malate. (J) Mitochondrial protein extracts of WT mice fed with a high-fat diet for 3 d were analyzed by SDS/PAGE and IB. OCRs of isolated mitochondria prepared from WT mice fed with a high-fat diet for 3 d were quantified in the presence of the indicated substrates: (K) palmitoyl CoA and carnitine and (L) pyruvate and malate. Carbonilcyanide p-trifluoromethoxyphenylhydrazone (FCCP) was specifically added in the case of pyruvate and malate according to standard protocols as detailed in Materials and Methods. For SDS/PAGE and IB, porin levels were used as a loading control, and each time point consists of a mix of mitochondria isolated from three to four individual mice (Fig. S4 shows quantification of the different IBs). OCR measurements are presented in picomoles per minute as means ± SEMs, with individual measurements of three to five animals per time point. Gray shading represents the dark phase. Molecular mass (M.W.) is indicated in kilodaltons.
Fig. S4.
Immunoblot quantifications of CPT1, PDH, and ACAD11 protein levels. Quantifications of CPT1, PDH-E1β, and ACAD11 relative to porin levels from three to four independent experiments; each comprises of a single mouse per time point at six time points throughout the day. (A) WT mice fed ad libitum. (B) PER1/2 null mice fed ad libitum. (C) Night-fed PER1/2 null mice. (D) WT mice fed with a high-fat diet for 3 d. Data are presented as fold change relative to the lowest value, with means ± SEMs of three to four different immunoblots. *P value < 0.05; **P value < 0.01; ***P value < 0.001.
Because these key enzymes accumulated at specific times of the day, we posited that mitochondria might display a distinct diurnal metabolic response in the presence of their corresponding substrates. To test this hypothesis, we monitored the respiration rate of isolated mitochondria prepared from mice killed throughout the day using the Seahorse Flux Analyzer. CPT1 function was examined in the presence of palmitoyl CoA and carnitine, and PDH activity was tested with pyruvate and malate. Our working premise was that, for any substrate provided in excess to isolated mitochondria, the respiration rate would primarily reflect the activity of the first rate-limiting enzyme (26). On incubation with palmitoyl CoA and carnitine, mitochondrial respiration exhibited a diurnal response, with peak levels at ∼ZT20 (Fig. 4B), which corresponded to the oscillations in CPT1 protein levels (Fig. 4A). In the presence of pyruvate and malate, mitochondrial respiration showed diurnal oscillations with maximum levels at ∼ZT4 (Fig. 4C), concomitant with the accumulation profile observed for PDH protein (Fig. 4A).
We also examined whether mitochondrial respiration is rhythmic in isolated mitochondria from cultured cells that exhibit circadian rhythmicity (i.e., NIH 3T3) (27) (Fig. S5A). In contrast to mitochondria prepared from mouse liver, respiration of isolated mitochondria from NIH 3T3 cells in the presence of palmitoyl CoA and carnitine or pyruvate and malate was relatively even throughout the circadian cycle (Fig. S5B). NIH 3T3 exhibits rhythmic gene expression of core clock and several output genes; however, the amplitude is often shallow compared with liver, likely because of absence of systemic timing cues. Thus, we cannot exclude the possibility that we failed to detect rhythmic respiration in isolated mitochondria from NIH 3T3 because of their low amplitude.
Fig. S5.
Mitochondrial respiration of isolated mitochondria from synchronized NIH 3T3 throughout the circadian cycle. NIH 3T3 cells were synchronized with a Dexamethasone (Dex) shock as previously described (27) and harvested at 4-h intervals from 24 to 44 h after the Dex shock. (A) Total RNA was prepared, and mRNA expression levels of Bmal1 and Per2 were determined by quantitative real-time PCR. Data are presented as relative expressions, with means ± SEMs of three independent experiments. (B) Mitochondria were isolated from NIH 3T3 cells as described in Materials and Methods. Oxygen consumption rates (OCRs) of isolated mitochondria were quantified using the Seahorse Flux Analyzer in the presence of palmitoyl CoA and carnitine or pyruvate and malate as indicated. OCR measurements are presented in picomoles per minute as means ± SEMs of thee independent experiments.
To address the potential dependency of CPT1 and PDH daily accumulation and their respective mitochondrial function on circadian clocks and more specifically, the clock proteins PER1/2, we isolated mitochondria from PER1/2 null mice throughout the day. The circadian expression of core clock and clock-controlled genes is largely abolished in mice lacking both PER1 and PER2 (28). Immunoblot analysis revealed that mitochondria isolated from PER1/2 null mice exhibit relatively constant CPT1 and PDH protein levels throughout the day (Fig. 4D and Fig. S4B). Likewise, we did not observe significant diurnal oscillations in mitochondrial respiration in response to their respective substrates (Fig. 4 E and F). Thus, the equal daily levels of CPT1 and PDH and the constant mitochondrial respiration in Per1/2−/− mice suggested that the oscillations in these pathways are dependent on the circadian clock PERIOD proteins.
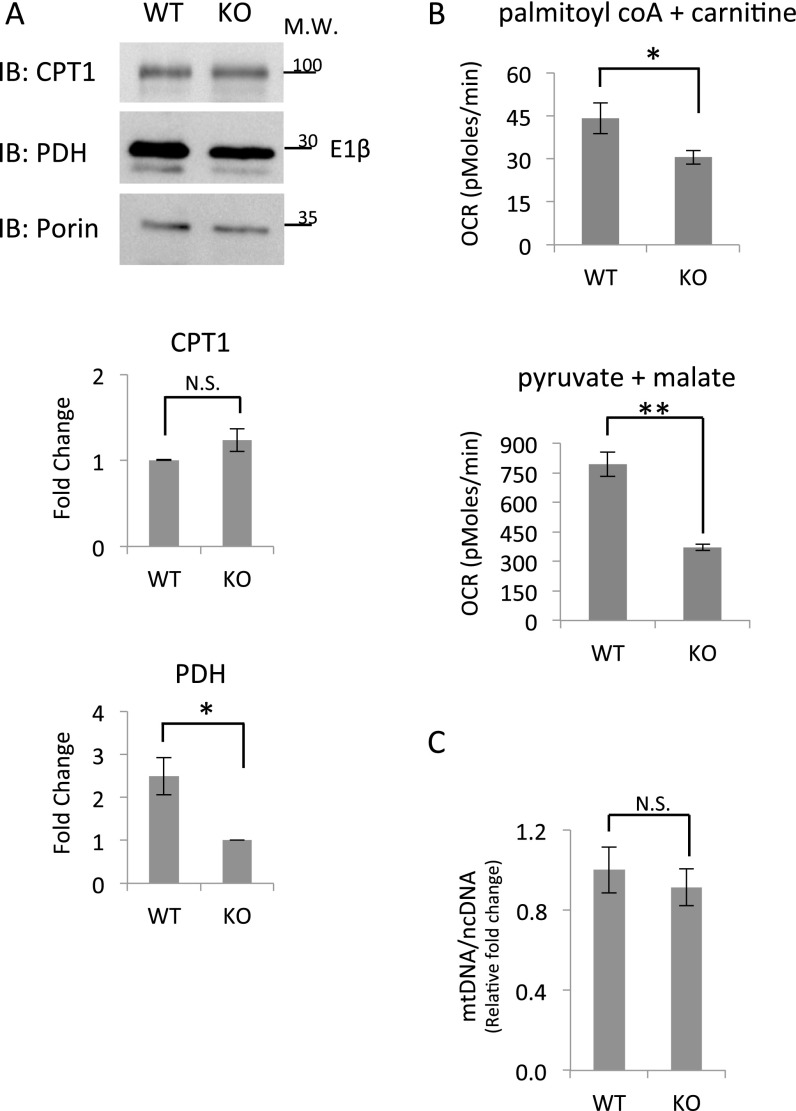
Comparison of the overall daily levels of CPT1 and PDH between WT and Per1/2−/− mice fed ad libitum revealed similar CPT1 levels but lower PDH levels in Per1/2−/− mitochondria (Fig. S6A). The daily mean oxygen consumption rates in the presence of palmitoyl CoA and carnitine or pyruvate and malate were significantly lower in Per1/2−/− compared with WT mitochondria (Fig. S6B). Thus, the lower PDH levels in Per1/2−/− mitochondria corresponded to the overall reduction in their oxygen consumption rate with pyruvate and malate. By contrast, the decline in mean oxygen consumption rates in the presence of palmitoyl CoA and carnitine could not be attributed to overall changes in CPT1 levels in Per1/2−/− compared with in WT mitochondria. It is possible that overall CPT levels are similar but that its enzymatic activity in general is reduced in Per1/2−/− mice. In this respect, leptin was shown to increase liver CPT1 activity (29), and recently, it was found to be disregulated in PER1/2 null mice (30). In addition, potential differences in the levels of malonyl CoA that inhibit CPT1 activity (25) and posttranslational modifications of CPT1 (31) may also account for differences in its overall activity levels. We also examined whether the number of mitochondria differs between the two mouse strains. Hence, we quantified the number of mitochondria in WT and Per1/2−/− mice by measuring the ratio between the mtDNA and nuclear DNA and did not observe a significant difference (Fig. S6C).
Fig. S6.
Comparison of daily mean CPT1 and PDH protein levels, mitochondrial respiration, and the number of mitochondria in WT and Per1/2−/− mice. (A) Mitochondrial protein extracts of WT and Per1/2−/− mice (KO) fed ad libitum were analyzed by SDS/PAGE and immunoblot (IB) with indicated antibodies. Porin levels were used as a loading control, and each condition consists of a mixture of all six time points throughout the day, with mitochondria isolated from three to four individual mice for each time point. CPT1 and PDH-E1β levels were quantified relative to porin levels. Data are presented as fold change relative to the lowest value, with means ± SEMs of three independent experiments. (B) Oxygen consumption rates (OCRs) of isolated mitochondria from WT and Per1/2−/− mice (KO) fed ad libitum were measured in the presence of palmitoyl CoA and carnitine or pyruvate and malate. Carbonilcyanide p-trifluoromethoxyphenylhydrazone (FCCP) was added in the case of pyruvate and malate according to standard protocols as detailed in Materials and Methods. For each condition, data are presented in picomoles per minute as means ± SEMs of all six time points throughout the day, with three to four individual mice for each time point. (C) The average ratio of mtDNA to nuclear DNA (ncDNA) in WT and Per1/2−/− mice (KO) fed ad libitum was quantified by real-time PCR as described in Materials and Methods. Data are presented as fold change relative to the highest value, with means ± SEMs of n = 4. Molecular mass (M.W.) is indicated in kilodaltons. N.S., nonsignificant. *P value < 0.05; **P value < 0.001.
Mice normally ingest most of their food during the dark phase. Clock-deficient mice (e.g., Clock mutant and Cry1/2 and Per1/2 double-KO mice) exhibit greatly attenuated diurnal feeding rhythms, because they consume more food during the light phase compared with WT mice (11, 16, 32, 33). To examine whether feeding rhythms might play a role in the diurnal accumulation of CPT1 and PDH and consequently, mitochondrial respiration, we applied a nighttime-restricted feeding regimen on PER1/2 null mice. Immunoblot analysis of mitochondria isolated from night-fed Per1/2−/− mice showed that CPT1 and PDH protein levels are relatively constant throughout the day (Fig. 4G and Fig. S4C), similar to those in Per1/2−/− mice fed ad libitum (Fig. 4D). Likewise, the daily profile of mitochondrial respiration in response to their respective substrates was relatively constant (Fig. 4 H and I).
Next, we asked whether the diet composition might play a role in the diurnal accumulation of CPT1 and PDH and consequently, mitochondrial respiration. Thus, WT mice were fed with a high-fat diet for 3 days. Immunoblot analysis of mitochondria isolated from high-fat diet-fed mice showed that CPT1 oscillated throughout the day, with trough levels at ∼ZT8 (Fig. 4J and Fig. S4D), similar to mice fed with regular chow (Fig. 4A). Mitochondrial respiration in the presence of CPT1 substrates, namely palmitoyl CoA and carnitine, cycled and exhibited a similar pattern under both diets (Fig. 4 B and K). By contrast, mitochondria isolated from high-fat diet-fed mice showed relatively constant levels of PDH protein (Fig. 4J and Fig. S4D), and their respiration in the presence of pyruvate and malate was fairly even throughout the day (Fig. 4L).
Taken together, we detected daily oscillations in key mitochondrial enzymes and observed concomitant diurnal changes in mitochondrial respiration in the presence of their respective substrates. Thus, the response to palmitoyl CoA and carnitine as well as pyruvate and malate elicited daily cycles in mitochondrial respiration with different phases (∼ZT20 and ∼ZT4, respectively), concurrent with the oscillating levels of their rate-limiting enzyme (i.e., CPT1 and PDH). These oscillations were abolished in PER1/2 null mice irrespective of the feeding schedule, suggesting that they are dependent on the clock PERIOD proteins independent of feeding time. Remarkably, a high-fat diet specifically affected the daily accumulation of PDH and related mitochondrial respiration but did not alter the diurnal oscillations in CPT1 levels and its respective mitochondrial function.
Daily Rhythms in FAO and Related Enzymes.
Long-chain fatty acids are transported into mitochondria through the carnitine shuttle, in which the rate-limiting step is catalyzed by CPT1. After fatty acids enter the mitochondria, they are catabolized through FAO generating acetyl CoA. Our proteomics analysis evinced that several enzymes within the FAO pathway oscillate with zenith levels at ∼ZT4 (Fig. 2B and Dataset S4), among them several ACADs. ACADs are a class of enzymes that catalyzes the initial step in each cycle of FAO. They differ in their specificity for different chain lengths of fatty acid acyl CoA substrates (34). As a proof of principle, we examined ACAD11, which specifically metabolizes long-chain fatty acid substrates (e.g., palmitoyl CoA) (34).
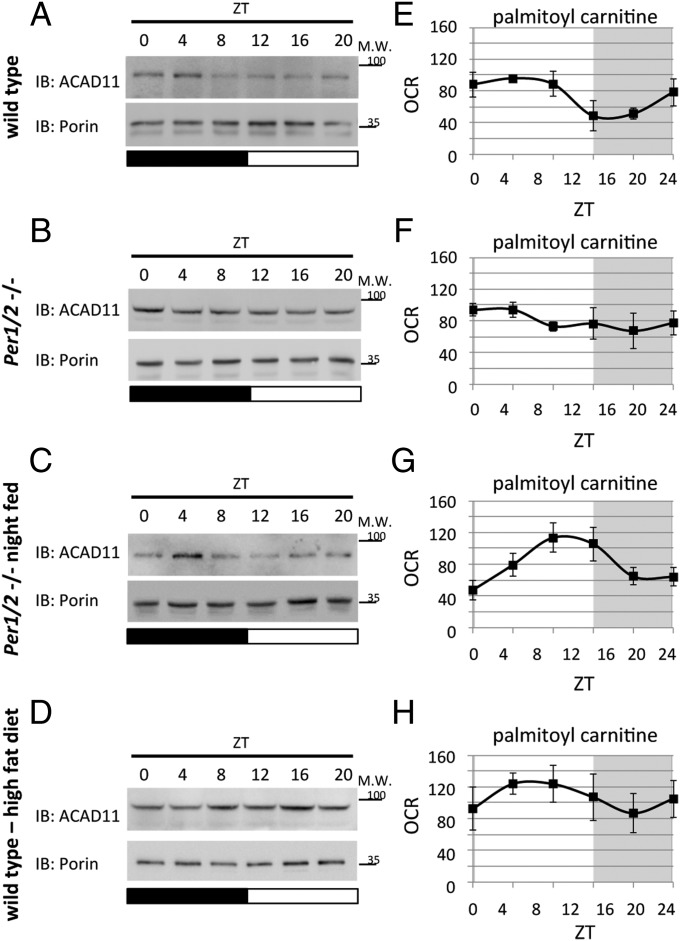
Analysis of mitochondrial protein levels of ACAD11 throughout the day by SDS/PAGE and immunoblot showed, in accordance with our proteomics analysis, that they oscillate, with peak levels at ∼ZT0 (Fig. 5A and Fig. S4A). The rhythmic accumulation of ACAD11 was lost in mitochondria purified from PER1/2 null mice fed ad libitum (Fig. 5B and Fig. S4B). However, when we tested mitochondria that were isolated from night-fed PER1/2 null mice, the daily oscillations in ACAD11 accumulation were restored (Fig. 5C and Fig. S4C), albeit with a phase delay compared with that in WT mice (i.e., ∼ZT4). Next, we asked whether the food composition might also play a role in the daily accumulation of ACAD11. Remarkably, 3 d of high-fat diet were sufficient to damp the oscillations of ACAD11 accumulation in mitochondria isolated from WT mice (Fig. 5D and Fig. S4D).
Fig. 5.
Analysis of FAO in WT and PER1/2 null mice under different feeding regimens. Mitochondrial protein extracts were analyzed by SDS/PAGE and immunoblot (IB) with indicated antibodies. (A) WT mice fed ad libitum. (B) PER1/2 null mice fed ad libitum. (C) Night-fed PER1/2 null mice. (D) WT mice fed with a high-fat diet for 3 d. Oxygen consumption rates (OCRs) of isolated mitochondria in the presence of palmitoyl carnitine and malate were quantified using the Seahorse Flux Analyzer. (E) WT mice fed ad libitum. (F) PER1/2 null mice fed ad libitum. (G) Night-fed PER1/2 null mice. (H) WT mice fed with a high-fat diet for 3 d. For SDS/PAGE and IB, porin levels were used as a loading control, and each time point consists of a mix of mitochondria isolated from three to five individual mice (Fig. S4 shows quantification of the different IBs). OCR measurements are presented in picomoles per minute as means ± SEMs, with individual measurements of three to five animals per time point. Gray shading represents the dark phase. Molecular mass (M.W.) is indicated in kilodaltons.
These findings incited us to examine the daily changes in FAO by monitoring the oxygen consumption of isolated mitochondria in the presence of palmitoyl carnitine, the product of CPT1 enzymatic activity. In line with a recent study that identified circadian oscillations in FAO in cells and mice (35), isolated mitochondria from WT mice exhibited daily oscillations in oxygen consumption in the presence of palmitoyl carnitine and malate, with peak levels at ∼ZT4 (Fig. 5E). By contrast, mitochondria from PER1/2 null mice fed ad libitum did not exhibit significant daily changes in oxygen consumption (Fig. 5F). Nighttime-restricted feeding restored the daily oscillations in mitochondrial respiration of PER1/2 null mice (Fig. 5G), but we observed a phase delay compared with that in WT mice. Finally, we did not detect significant daily oscillations in respiration of mitochondria isolated from WT mice that were fed high-fat diet for 3 d (Fig. 5H).
The above-described experiments showed that the daily changes in ACAD11 levels correspond to oscillations in FAO as monitored by mitochondrial respiration in the presence of palmitoyl carnitine. Both were rhythmic in WT mice with zenith levels at ∼ZT4, and the rhythmicity was lost on high-fat diet or in PER1/2 null mice and restored in nighttime-fed PER1/2 null mice.
Analysis of Respiratory Exchange Ratio, Feeding, and Locomotor Activity.
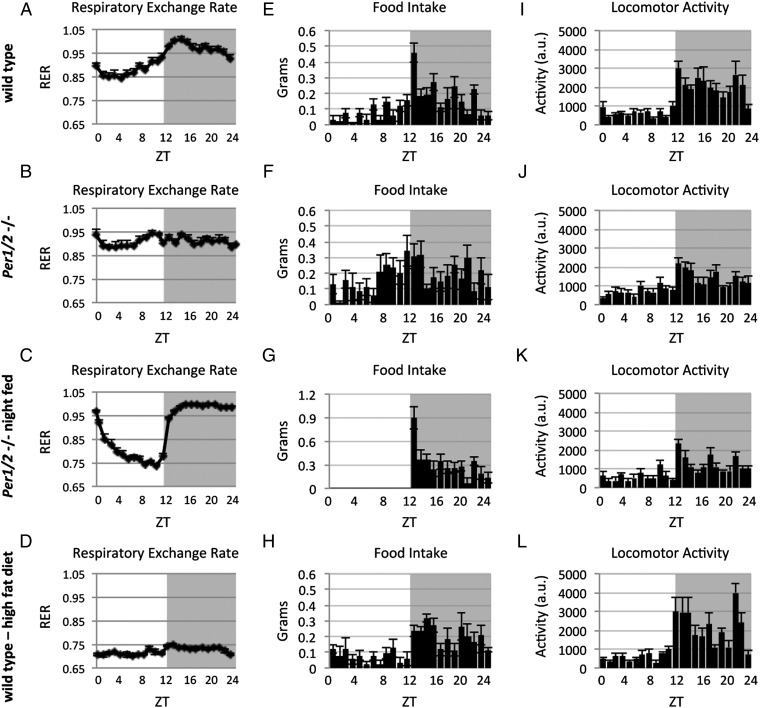
Hitherto, we analyzed mitochondrial respiration using isolated mitochondria in response to different nutrients. To assess these parameters in living animals we measured the respiratory exchange ratio (RER) using metabolic cages. The RER reflects the respiratory quotient, an indicator of which fuel (carbohydrate or fat) is primarily metabolized as an energy source. An RER of 0.70 is indicative of fat being the predominant fuel, whereas a value of 1.00 or above points toward carbohydrate as the major fuel (36). WT mice exhibited diurnal oscillations in RER values ranging from 0.85 to 1, with nadir levels at ∼ZT4 and zenith levels at ∼ZT16 (Fig. 6A). In PER1/2 null mice fed ad libitum, the RER was relatively constant throughout the day (Fig. 6B). Nighttime-restricted feeding restored the diurnal oscillations in RER of PER1/2 null mice, with high-amplitude oscillations that peaked during the dark phase (Fig. 6C). Similar effects of nighttime-restricted feeding were previously reported for WT mice (37). Notably, on 3 d of high-fat diet, WT mice exhibited low (∼0.7) and constant RER levels (Fig. 6D).
Fig. 6.
Analysis of RER, feeding behavior, and locomotor activity of WT and PER1/2 null mice under different feeding regimens. RER, food consumption, and voluntary locomotor activity were recorded using metabolic cages. (A) RER of WT mice fed ad libitum. (B) RER of PER1/2 null mice fed ad libitum. (C) RER of night-fed PER1/2 null mice. (D) RER of WT mice fed with a high-fat diet for 3 d. (E) Food consumption of WT mice fed ad libitum (29.3% and 70.7% during the light and dark phases, respectively; n = 7; P value = 1E-05). (F) Food consumption of PER1/2 null mice fed ad libitum (44.8% and 55.2% during the light and dark phases, respectively; n = 8; P value = 0.01). (G) Food consumption of night-fed PER1/2 null mice. (H) Food consumption of WT mice fed with a high-fat diet for 3 d (26.7% and 73.3% during the light and dark phases, respectively; n = 4; P value = 6E-06). (I) Locomotor activity of WT mice fed ad libitum (23.6% and 76.4% during the light and dark phases, respectively; n = 7; P value = 1E-07). (J) Locomotor activity of PER1/2 null mice fed ad libitum (33.1% and 66.9% during the light and dark phases, respectively; n = 8; P value = 7E-05). (K) Locomotor activity of night-fed PER1/2 null mice (31% and 69% during the light and dark phases, respectively; n = 8; P value = 2E-04). (L) Locomotor activity of WT mice fed with a high-fat diet for 3 d (22.7% and 77.3% during the light and dark phases, respectively; n = 4; P value = 1E-04). Data are presented as means ± SEMs, with individual measurements of four to eight animals per time point. Locomotor activity is presented in arbitrary units (a.u.). Gray shading represents the dark phase.
Concomitantly, we monitored daily food consumption (Fig. 6 E–H) and voluntary locomotor activity (Fig. 6 I–L) of WT and PER1/2 null mice under the above-described conditions. In agreement with previous reports on clock-disrupted mice (11, 16, 32, 33), Per1/2−/− mice exhibited attenuated diurnal feeding rhythms and consumed more food during the light phase compared with WT mice (∼45% vs. ∼29% during the day and ∼55% vs. ∼71% during the night in PER1/2 null and WT mice, respectively). Short duration (i.e., 3 d) of high-fat diet in contrast to 6 wk of high-fat diet (38) did not significantly alter the daily feeding habits of WT mice. As expected, mice were mostly active during the dark phase, but PER1/2 null mice were more active during the light phase and less during the dark phase compared with WT mice (∼33% vs. ∼24% during the day and ∼67% vs. ∼76% during the night in PER1/2 null and WT mice, respectively). This effect was maintained under different feeding regimens, namely nighttime-restricted feeding and high-fat diet.
Thus, our findings suggested that WT mice mostly use lipids as an energy source during the rest phase when they ingest little food. PER1/2 null mice seem to use both carbohydrates and lipids equally throughout the day, likely because of their attenuated feeding rhythms. A high-fat diet shifted the balance to lipids use throughout the entire day. These changes matched the oscillations in mitochondrial FAO as reflected by respiration measurements of isolated mitochondria in the presence of palmitoyl carnitine and malate (Fig. 5 E–H).
Discussion
In this study, we examined the temporal changes in the mitochondrial proteome by applying MS-based quantitative proteomics on isolated mitochondria from mouse liver. We obtained the first, to our knowledge, comprehensive mitochondrial proteome around the clock and found that ∼38% of mitochondrial proteins oscillate throughout the day. Unexpectedly, we discovered that the majority of cycling proteins in mitochondria reach their zenith levels at ∼ZT4. Hence, mitochondrial proteins are predominantly gated to accumulate during the early light phase, raising the question of how this temporal coordination is achieved at the molecular level. When we tested the possibility that transcription regulation plays a role in this process, we found that, although the transcript levels of the vast majority of cycling mitochondrial proteins do oscillate, the phase of oscillating mitochondrial proteins poorly correlated with the phase of their cycling transcript levels. It should be noted that our proteomics analysis was done from mice housed under a 12-h light–dark regimen, whereas the transcriptome dataset (23) was obtained under constant darkness. The difference in the experimental setup may account for some differences in peak time of abundance for RNA and protein. Recent proteomics studies on whole-liver samples already showed that the oscillations of proteins encoded by rhythmically expressed mRNAs greatly differ in their cycling phases (13, 15). Thus, although rhythmic transcription is at the core of circadian regulation, it seems that posttranscriptional mechanisms, such as translational control (39, 40) and temporal regulation of protein synthesis/degradation (41), play a significant role in shaping the circadian proteome landscape. In the case of the mitochondrial proteome, another mechanism that comes to mind is the translocation of protein from the cytoplasm, where the vast majority of mitochondrial proteins are synthesized, to the mitochondria. This process is mediated through the TIM/TOM complex and tightly regulated, for example, by cytosolic kinases, such as casein kinase 2 (42). Interestingly, casein kinases have been previously implicated in regulation of circadian rhythmicity (7). Along this line, our proteomics analysis also evinced that several members of the mitochondrial protein translocation machinery, namely the TIM/TOM complexes, accumulate in a daily manner during the early light phase, which might suggest that protein entry to the mitochondria is temporally gated by them. Lastly, it was recently shown that mitochondrial dynamics through fusion, fission, and mitophagy occur in a daily manner under the control of the liver clock (43). Hence, it is plausible that these alterations might also shape the phase of protein accumulation in mitochondria.
In contrast to previous circadian proteomics that were done on whole-liver samples (13, 15), we performed herein the proteomics analysis on isolated mitochondria from liver. After stringent filtering, we precisely quantified 590 different mitochondrial-annotated proteins of ∼1,000 known mitochondrial proteins (44). Comparison of our mitochondrial proteome with formerly reported whole-liver proteome (15) revealed that mitochondrial isolation before the proteomics analysis increases the identification and quantification rate. Of the total of 590 mitochondrial-annotated proteins quantified in this study, only 412 were previously quantified in whole-liver samples. Interestingly, although 157 of 223 cycling mitochondrial proteins (based on this study) were also quantified in the whole-liver proteome, only 6 of them were considered to be cycling. This striking difference further highlights the critical importance of mitochondrial fractionation when examining rhythmic protein accumulation in mitochondria and is likely to reflect the temporal and spatial dynamics in protein accumulation in subcellular compartments.
We detected daily oscillations in key mitochondrial enzymes and observed concomitant diurnal mitochondrial respiration profiles in the presence of their respective substrates. Thus, the response to palmitoyl CoA and carnitine as well as pyruvate and malate elicited daily cycles in mitochondrial respiration with different phases (∼ZT20 and ∼ZT4) concurrent with the oscillating levels of their rate-limiting enzyme (i.e., CPT1 and PDH). The oscillations in mitochondrial respiration and the abundances of the rate-limiting enzymes were abolished in PER1/2 null mice irrespective of feeding schedule, suggesting that they are dependent on the clock proteins PER1/2 independent of feeding time. Remarkably, high-fat diet specifically affected the daily accumulation of PDH and related mitochondrial respiration but did not alter the diurnal oscillations in CPT1 levels and its respective mitochondrial function. Based on the correlation between the oscillations in the protein levels of the aforementioned rate-limiting enzymes and the diurnal oscillation in mitochondrial respiration in the presence of their respective substrates, we proposed that daily rhythmicity in mitochondrial function is governed, at least in part, by the cycling level of these enzymes. However, it is conceivable that additional mechanisms, such as covalent modification (e.g., phosphorylation and acetylation), of the abovementioned rate-limiting enzymes (i.e., CPT1 and PDH) might also participate in the circadian control of mitochondrial respiration. Recent studies identified circadian changes in the mitochondrial acetylome (35, 45); however, neither CPT1 nor PDH was reported to undergo rhythmic changes in their acetylation status.
In this study, we assessed whole-body energy utilization using RER measurements. We found that they correspond to oscillations in diurnal capacity of isolated mitochondria from mouse liver to oxidize fatty acids under different experimental setups. Energy expenditure rate consists of integration of multiple parameters, such as activity, food intake, and body temperature. Furthermore, it represents the contribution of different organs, such as skeletal muscle, smooth muscle in the gastrointestinal tract, adipose tissue, liver, and others. Of relevance is also the environmental temperature that strongly affects the overall energy expenditure, and even at vivarium (i.e., 22 °C), thermogenesis consists of a major part of animals’ total energy expenditure (46). In view of the above information, our correlation between whole-body energy utilization and liver mitochondria activity should be interpreted cautiously and further explored. Nevertheless, in view of previous work identifying rhythmic FAO in muscle cells (35), it is likely that our rhythmic measurements of liver mitochondrial FAO are also relevant to mitochondrial activity in other organs.
We show that CPT1 and PDH accumulate in a daily manner and that their diurnal oscillations are dependent on the clock PERIOD proteins. Both PER1 and PER2 are considered as core clock components that are also systemically regulated, most likely by feeding, and thus, serve as a conduit for systemic input to the clock (47). Hence, the diminished circadian rhythmicity of CPT1 and PDH and their related mitochondrial respiration in Per1/2−/− mice might be caused by the lack of sensory input mediated through PER1/2 function and/or the nonfunctional core clock in these mice.
FAO exhibited daily oscillations with zenith levels during the light phase that are PER1/2-dependent. In accordance, liver metabolomics and lipidomics studies reported that free fatty acids and triglycerides accumulate in liver around the same time of the day in a clock-dependent manner (16, 19, 48). Furthermore, feeding time defines the phase of lipid accumulation in liver (16, 37). Indeed, we found that nighttime-restricted feeding restored the oscillations in FAO. Therefore, it is likely that the lack of diurnal oscillations in FAO in PER1/2 null mice were because of attenuated feeding–fasting cycles. Intriguingly, short-term high-fat diet hindered the temporal control of mitochondrial FAO without altering feeding–fasting cycles, suggesting that an additional mechanism plays a role in the temporal regulation of FAO. Because substrates are provided in excess, the respiration assay with isolated mitochondria reflects inherent changes in mitochondrial structure/function. It, thus, seems that changes in feeding time or food composition are sufficient to alter mitochondrial composition and hence, function.
In summary, we found that utilization of long-chain fatty acids by mitochondria through the carnitine shuttle and mitochondrial pyruvate metabolism is dependent on the clock proteins PER1/2, irrespective of feeding time. The former peaks late at night, whereas the latter rises at the middle of the day. By contrast, FAO per se is optimized to the beginning of the day; it is also controlled by PER1/2 but responds to feeding time. In this respect, it should be noted that FAO uses not only long-chain fatty acids but also, short and medium chains, which do not require the carnitine shuttle, because they freely diffuse into the mitochondria (49). We conclude that PERIOD proteins and potentially, the circadian clock regulate the diurnal utilization of different nutrients by the mitochondria and thus, optimize mitochondrial response to daily changes in energy supply/demand.
Materials and Methods
Animals.
All animal experiments and procedures were conducted in conformity with the Weizmann Institute Animal Care and Use Committee guidelines. For mitochondria isolation, we used 3-mo-old WT and Per1/2−/− males as previously described (28). Mice were kept under a 12-h light–dark regimen and fed either ad libitum or exclusively during the dark phase for 2 wk. Animals were fed either regular chow or a high-fat diet (60% of kilocalories as fat; D12492; Open Source Diets). Mice were killed at 4-h intervals throughout the day; ZT0 corresponded to the time that lights were turned on in the animal facility, and ZT12 corresponded to the time that lights were turned off in the animal facility. The temperature in the housing cages and metabolic cages was maintained at 22 °C.
Mitochondrial Preparation.
Mice were killed, and livers were isolated and minced in ∼10 vol Mitochondrial Isolation Buffer (MIB; 70 mM sucrose, 210 mM mannitol, 10 mM Hepes, 1 mM EDTA, pH 7.5) supplemented with 0.2% BSA at 4 °C. All subsequent steps of the preparation were performed on ice. The material was rinsed with MIB several times to remove blood. The tissue was then homogenized using a Teflon dounce homogenizer. Homogenate was centrifuged at 600 × g for 10 min at 4 °C. After centrifugation, fat/lipids were carefully aspirated, and the remaining supernatant was centrifuged at 7,000 × g for 15 min. The pellet was resuspended in MIB supplemented with 0.5 mM EGTA and centrifuged for 10 min at 7,000 × g. Subsequently, the pellet was resuspended in MIB and centrifuged for 5 min at 7,000 × g. The final pellet was collected. Total protein (milligrams per milliliter) was determined using Bradford Assay Reagent (Bio-Rad). In the case of mitochondria isolation from NIH 3T3 cells, cells were collected and washed with PBS, homogenized in MIB buffer using a syringe, and isolated by subsequent centrifugations as described above.
Mitochondrial Respiration Assay.
Mitochondrial oxygen consumption rate experiments were conducted using the Seahorse Bioscience XFe24 Extracellular Flux Analyzer; 5 μg per well mouse liver-isolated mitochondria or 10 μg per well mitochondria purified from NIH 3T3 cells were seeded in Mitochondrial Assay Solution [70 mM sucrose, 220 mM mannitol, 10 mM KH2PO4, 5 mM MgCl2, 2 mM Hepes, 1.0 mM EGTA, 0.2% (wt/vol) fatty acid-free BSA, pH 7.2]. Mitochondrial Assay Solution was supplemented according to standard protocols (50) with (i) 40 μM palmitoyl CoA and 40 μM carntine; (ii) 10 mM pyruvate, 2 mM malate, and 5 μM carbonilcyanide p-trifluoromethoxyphenylhydrazone (FCCP) or (iii) 40 μM palmitoyl-carnitine and 0.5 mM malate. FCCP was added in the case of pyruvate and malate according to established protocols to enable accurate measurements of oxygen consumption rate within the dynamic range in response to these specific substrates.
Metabolic Cages Analysis.
The voluntary locomotor activity, food consumption, and RER were monitored using Phenomaster Metabolic Cages (TSE Systems).
Liquid Chromatography and MS.
Protein quantification was done using the label-free algorithm (22). Samples were prepared, and data were analyzed as detailed in SI Materials and Methods.
Protein Extraction and Immunoblotting.
SDS/PAGE and immunoblot were performed according to standard procedures (51). Each time point included a mix of mitochondria isolated from three to four individual mice. Antibodies used were mouse anti-CPT1 (ab128568; abcam), PDH antibody (ab110416; abcam), ACAD11 antibody (sc514027; SANTA CRUZ), and porin (ab14734; abcam).
RNA Analysis by Real-Time Quantitative PCR.
RNA extraction and transcript quantification by real-time PCR technology were performed as previously described (16). Real-time PCR measurements were performed using SYBR green or Taqman probes with a LightCycler II Machine (Roche). Normalization was performed relative to the geometrical mean of five housekeeping genes: Tbp, Hprt, Actin, Rplp0, and Gapdh. Primers and probes are listed in SI Materials and Methods.
mtDNA Quantification.
Total DNA was isolated from liver using TRI Reagent (SIGMA) according to the manufacturer’s protocol, and quantitative real-time PCR was performed with mitochondrial- (ChrM and Cox1) and genomic-specific (Hprt and Glucagon) primers as listed below. The relative number of mitochondria was determined as the ratio between the average of mtDNA and the average genomic DNA based on the real-time PCR with the different specific primers:
ChrM fw 5′-AATCAACTCGTCTATGTGGCAAAA-3′;
ChrM rev 5′-CCAGCTATCACCAAGCTCGTT-3′;
Cox1 fw 5′-AGCCCACTTCGCCATCATAT-3′;
Cox1 rev 5′-GCGTCGTGGTATTCCTGAAAG-3′;
Glucagon fw 5′-CAGGGCCATCTCAGAACC-3′;
Glucagon rev 5′-GCTATTGGAAAGCCTCTTGC-3′;
Hprt fw 5′-TCGTAATTTGACCCGACTGATG-3′; and
Hprt rev 5′-AAGTTCCAGTTCTAAGGACGTCTGTAC-3′.
SI Materials and Methods
Sample Preparation for MS.
Isolated mitochondrion pellets were lysed with a buffer containing 100 mM Tris⋅HCl, pH 8, 4% (vol/vol) SDS, and 50 mM DTT. Samples were incubated at 95 °C for 5 min followed by sonication in a bioruptor (Diagenode) for 15 min. Proteins were alkylated with Iodoacetamide (55 mM final concentration) for 30 min at 4 °C in the dark, and after that, samples were cleared by centrifugation at 16,100 × g for 10 min at room temperature. Proteins were precipitated at −20 °C overnight with 5 vol ethanol/acetate. Protein pellets were washed twice with ice-cold ethanol/acetate and finally, resuspended with sonication in a buffer containing 6 M guanidinium hypochloride. For protein digestion, samples were diluted twofold with 6 M urea/2 M thiourea in 10 mM Hepes, and subsequently, 1:100 LysC endopeptidase (Wako) was added and incubated for 3 h at 37 °C. Samples were then diluted to 0.6 M Guanidinium hypochloride with 10% acetonitrile before digestion with 1:100 Trypsin (Sigma-Aldrich) overnight at 37 °C. The digestion was stopped by the addition of 1 μL trifluoroacetic acid. The acidified peptides were desalted and purified by loading on Stage Tips (self-made pipette tips containing three layers of SDB-RPS; Sigma-Aldrich) as previously described (52).
Liquid Chromatography and MS.
Approximately 2 µg digested peptides were analyzed by a nanoflow liquid chromatograph on an EASY-nLC 1000 Ultrahigh-Pressure System (Thermo Fisher Scientific), which was coupled to the Q Exactive HF Mass Spectrometer (Thermo Fisher Scientific) by a nanoelectrospray source (Thermo Fisher Scientific). For the chromatography, a 50-cm, 75-µm i.d. column was packed in house with 1.9-µm reverse-phase silica beads (ReprosSil-Pur C18-AQ; Dr. Maisch GmbH). Peptides were separated with a 120-min gradient from 5% to 50% of 80% acetonitrile and 0.1% formic acid at a constant flow of 250 nL/min. Column temperature was kept at 50 °C by an in house-designed oven with a Peltier Element. MS data were acquired with an automatic switch between one full scan and subsequent MS/MS scans of the 15 most abundant peaks (top 15 method). Target value for the full-scan MS spectra was 3 × 106 charges in the 300−1,650 m/z range, with a maximum injection time of 25 ms and a resolution of 120,000 at 400 m/z. Isolation of precursors was performed with a 1.4-m/z window. Precursors were fragmented by higher-energy C-trap dissociation, with a normalized collision energy of 27 eV. MS/MS scans were acquired at a resolution of 15,000 at m/z 400, with an ion target value of 1 × 105 and a maximum injection time of 50 ms. Repeat sequencing of peptides was minimized by excluding the selected peptide candidates for 20 s.
Data Analysis.
Raw MS files were processed with MaxQuant (version 1.5.3.2), a freely available software suite. Peak list files were searched by the ANDROMEDA search engine incorporated into the MaxQuant framework (53) against the Mus musculus Uniprot FASTA Database (version May 29, 2013) and a file containing commonly observed contaminants. The search included variable modifications for oxidation of methionine, protein N-terminal acetylation, and carbamidomethylation as a fixed modification. Peptides with at least seven amino acids were considered for identification, specifying as enzyme Trypsin with cleavage C-terminal to K or R and allowing two missed cleavages. The false discovery rate (FDR), determined by searching a decoy database, was set at 0.01 FDR for both peptides and proteins. Identification across different replicates and adjacent fractions was achieved by enabling the matching between runs option in MaxQuant within a time window of 0.7 min and an alignment time window of 20 min. During the main search, parent masses were allowed an initial mass deviation of 4.5 ppm, and fragment ions were allowed a mass deviation of 0.5 Da. Label-free quantification (LFQ) of proteins was done using the MaxLFQ algorithm (22) integrated into MaxQuant. The parameters were as follows: minimum ratio count was set to two, the FastLFQ option was enabled, LFQ minimum number of neighbors was set to three, and the LFQ average number of neighbors was set to six as per default.
Bioinformatic Analysis.
All bioinformatic analyses were performed with the Perseus software (www.coxdocs.org/doku.php?id=perseus:start) as previously described (15). Briefly, to determine the subset of cycling proteins, each protein expression profile based on the label-free intensities is fitted to a cosine with a fixed period of 24 h and the amplitude and phase as free parameters. We used a cutoff of q value (permutation-based FDR) < 0.15. There are no clear guidelines regarding the appropriate q value for this type of analysis. Previous circadian proteomics studies used a more permissive q value of 0.33 (15) or 0.25 (13), whereas circadian transcriptome studies often use a more stringent q value of 0.05. We believe that a cutoff of 0.15 is a good compromise between the two approaches. For transcriptome data, the period was fixed to 23.6 h, because samples were collected in constant darkness. Profiles are ranked by their variance ratio, which is the part of the variance explained by the fit divided by the contribution to the variance that is not accounted for by the fit. Based on this ranking, we determine a permutation-based FDR (q value) by repeating the same procedure 1,000 times on the same profiles but with scrambled time labels. Hierarchical clustering was done in a phase-preserving way by restricting the order of elements to the one determined by the output of the cosine model-based fitting. During the growth of the tree in hierarchical clustering, only those links that conserve this order were permitted. Although obviously, the order of the terminal branches is not an outcome of the algorithm, the cluster structure is still a nontrivial result of the clustering.
Cycling annotation enrichments were performed as previously described (15). Briefly, phase enrichment analysis of categorical annotations was done with a wider set of cycling candidates (405) obtained after using a less stringent q-value cutoff (q < 0.5). The significance of the cycling annotation distribution was determined by using a Fisher exact test cutoff (FDR < 0.1). The test detects statistically significant clustering of protein categories at specific times of the circadian cycle.
The MS proteomics data have been deposited to the ProteomeXchange Consortium (proteomecentral.proteomexchange.org) through the PRIDE partner repository (54) with the dataset identifier PXD001732.
List of primers and probes used for transcript analysis
| Primer name | Primer sequence |
| Hprt fw | 5′-GGCCAGACTTTGTTGGATTTG-3′ |
| Hprt rev | 5′-TGCGCTCATCTTAGGCTTTGT-3′ |
| Tbp fw | 5′-CCCTATCACTCCTGCCACACCAGC-3′ |
| Tbp rev | 5′-GTGCAATGGTCTTTAGGTCAAGTTTACAGCC-3′ |
| Gapdh fw | 5′-AGGTCGGTGTGAACGGATTTG-3′ |
| Gapdh rev | 5′-TGTAGACCATGTAGTTGAGGTCA-3′ |
| Actin fw | 5′-GGCTGTATTCCCCTCCATCG-3′ |
| Actin rev | 5′-CCAGTTGGTAACAATGCCATGT-3′ |
| Rplp0 fw | 5′-AGATTCGGGATATGCTGTTGGC-3′ |
| Rplp0 rev | 5′-TCGGGTCCTAGACCAGTGTTC-3′ |
| Cpt1 fw | 5′-CTCCGCCTGAGCCATGAAG-3′ |
| Cpt1 rev | 5′-CACCAGTGATGATGCCATTCT-3′ |
| Pdh E1β fw | 5′-AGGAGGGAATTGAATGTGAGGT-3′ |
| Pdh E1β rev | 5′-ACTGGCTTCTATGGCTTCGAT-3′ |
| Pdhx fw | 5′-GCTTCACTGTAACCAGCCG-3′ |
| Pdhx rev | 5′-CCCTTGCTCCATCGTAGGAGA-3′ |
| Pdh E2 fw | 5′-GACCAGCTTAAAGCCACAGG-3′ |
| Pdh E2 rev | 5′-AGAGGACTAACGAACACCCTT-3′ |
| Mtnd2 fw | 5′-ATACTAGCAATTACTTCTATTTTCATAGGG-3′ |
| Mtnd2 rev | 5′-GAGGGATGGGTTGTAAGGAAG-3′ |
| Mtnd5 fw | 5′-ACGAAAATGACCCAGACCTC-3′ |
| Mtnd5 rev | 5′-GAGATGACAAATCCTGCAAAGATG-3′ |
| Bmal1 fw | 5′-CCAAGAAAGTATGGACACAGACAAA-3′ |
| Bmal1 rev | 5′-GCATTCTTGATCCTTCCTTGGT-3′ |
| Bmal1 probe | FAM-TGACCCTCATGGAAGGTTAGAATATGCAGAA-TAMRA |
| Per2 rev | 5′-ATGCTCGCCATCCACAAGA-3′ |
| Per2 fw | 5′-GCGGAATCGAATGGGAGAAT-3′ |
| Per2 probe | FAM-ATCCTACAGGCCGGTGGACAGCC-TAMRA |
Supplementary Material
Acknowledgments
We thank K. Mayr, I. Paron, and G. Sowa for their assistance with the MS measurements and D. Wischnewski and M. Elimelech for technical help. We also thank A. Gross and H. Reinke for their valuable comments on the manuscript. A.N.-C. and Y.A. received a postdoctoral fellowship from the Feinberg Graduate School, Weizmann Institute of Science. The work performed in the laboratory of G.A. was supported by Israel Science Foundation Grant ISF 138/12, the Abish-Frenkel Foundation, Human Frontier Science Program (HFSP) Career Development Award HFSP CDA00014/2012, and European Research Council Grant ERC-2011 METACYCLES 310320.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The MS proteomics data have been deposited in the ProteomeXchange Consortium (proteomecentral.proteomexchange.org) through the Proteomics Identifications (PRIDE) partner repository (dataset identifier PXD001732).
See Commentary on page 3127.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519650113/-/DCSupplemental.
References
- 1.Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17(4):491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nunnari J, Suomalainen A. Mitochondria: In sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13(2):125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Bass J. Circadian topology of metabolism. Nature. 2012;491(7424):348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 5.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134(5):728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asher G, Sassone-Corsi P. Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161(1):84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 8.McCarthy JJ, et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31(1):86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 10.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 11.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106(50):21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci USA. 2014;111(45):16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mauvoisin D, et al. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci USA. 2014;111(1):167–172. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 15.Robles MS, Cox J, Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10(1):e1004047. doi: 10.1371/journal.pgen.1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adamovich Y, et al. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014;19(2):319–330. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ang JE, et al. Identification of human plasma metabolites exhibiting time-of-day variation using an untargeted liquid chromatography-mass spectrometry metabolomic approach. Chronobiol Int. 2012;29(7):868–881. doi: 10.3109/07420528.2012.699122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci USA. 2012;109(7):2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckel-Mahan KL, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci USA. 2012;109(14):5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasukawa T, et al. Human blood metabolite timetable indicates internal body time. Proc Natl Acad Sci USA. 2012;109(37):15036–15041. doi: 10.1073/pnas.1207768109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minami Y, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci USA. 2009;106(24):9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel MS, Nemeria NS, Furey W, Jordan F. The pyruvate dehydrogenase complexes: Structure-based function and regulation. J Biol Chem. 2014;289(24):16615–16623. doi: 10.1074/jbc.R114.563148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schreurs M, Kuipers F, van der Leij FR. Regulatory enzymes of mitochondrial beta-oxidation as targets for treatment of the metabolic syndrome. Obes Rev. 2010;11(5):380–388. doi: 10.1111/j.1467-789X.2009.00642.x. [DOI] [PubMed] [Google Scholar]
- 26.Lanza IR, Nair KS. Functional assessment of isolated mitochondria in vitro. Methods Enzymol. 2009;457:349–372. doi: 10.1016/S0076-6879(09)05020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagoshi E, et al. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119(5):693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Zheng B, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105(5):683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 29.Wein S, Ukropec J, Gasperíková D, Klimes I, Seböková E. Concerted action of leptin in regulation of fatty acid oxidation in skeletal muscle and liver. Exp Clin Endocrinol Diabetes. 2007;115(4):244–251. doi: 10.1055/s-2007-956166. [DOI] [PubMed] [Google Scholar]
- 30.Kettner NM, et al. Circadian dysfunction induces leptin resistance in mice. Cell Metab. 2015;22(3):448–459. doi: 10.1016/j.cmet.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerner J, Lee K, Hoppel CL. Post-translational modifications of mitochondrial outer membrane proteins. Free Radic Res. 2011;45(1):16–28. doi: 10.3109/10715762.2010.515218. [DOI] [PubMed] [Google Scholar]
- 32.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2(11):e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He M, et al. Identification and characterization of new long chain acyl-CoA dehydrogenases. Mol Genet Metab. 2011;102(4):418–429. doi: 10.1016/j.ymgme.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peek CB, et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. 2013;342(6158):1243417. doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flatt JP. Body composition, respiratory quotient, and weight maintenance. Am J Clin Nutr. 1995;62(5) Suppl:1107S–1117S. doi: 10.1093/ajcn/62.5.1107S. [DOI] [PubMed] [Google Scholar]
- 37.Hatori M, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15(6):848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6(5):414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Atger F, et al. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci USA. 2015;112(47):E6579–E6588. doi: 10.1073/pnas.1515308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janich P, Arpat AB, Castelo-Szekely V, Lopes M, Gatfield D. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res. 2015;25(12):1848–1859. doi: 10.1101/gr.195404.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J Cell Sci. 2011;124(Pt 3):311–320. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt O, et al. Regulation of mitochondrial protein import by cytosolic kinases. Cell. 2011;144(2):227–239. doi: 10.1016/j.cell.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 43.Jacobi D, et al. Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab. 2015;22(4):709–720. doi: 10.1016/j.cmet.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masri S, et al. Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc Natl Acad Sci USA. 2013;110(9):3339–3344. doi: 10.1073/pnas.1217632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abreu-Vieira G, Xiao C, Gavrilova O, Reitman ML. Integration of body temperature into the analysis of energy expenditure in the mouse. Mol Metab. 2015;4(6):461–470. doi: 10.1016/j.molmet.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5(2):e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gachon F, et al. Proline- and acidic amino acid-rich basic leucine zipper proteins modulate peroxisome proliferator-activated receptor alpha (PPARalpha) activity. Proc Natl Acad Sci USA. 2011;108(12):4794–4799. doi: 10.1073/pnas.1002862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papamandjaris AA, MacDougall DE, Jones PJ. Medium chain fatty acid metabolism and energy expenditure: Obesity treatment implications. Life Sci. 1998;62(14):1203–1215. doi: 10.1016/s0024-3205(97)01143-0. [DOI] [PubMed] [Google Scholar]
- 50.Rogers GW, et al. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One. 2011;6(7):e21746. doi: 10.1371/journal.pone.0021746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shalev M, et al. The PXDLS linear motif regulates circadian rhythmicity through protein-protein interactions. Nucleic Acids Res. 2014;42(19):11879–11890. doi: 10.1093/nar/gku873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11(3):319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 53.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 54.Vizcaíno JA, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2013;41(Database issue):D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.