Significance
Neuronal morphology is tightly linked to the actin cytoskeleton, which provides stability for synapses while retaining the potential for changes during learning and memory processes (synaptic plasticity). Despite biochemical similarity, isoforms of the small actin-binding protein profilin perform astonishingly diverse tasks in neurons, influencing actin dynamics in an opposing manner. Although the ubiquitous isoform profilin1 supports synaptogenesis, the evolutionary more recent and brain-specific isoform profilin2a mediates synapse function and plasticity. Moreover, we show that in mammalia, only profilin1 is bound by Fragile X mental retardation protein, and therefore is most likely crucial in the cause of and in the devastating outcome of Fragile X syndrome, one of the most common forms of inherited cognitive impairment.
Keywords: actin, spinogenesis, FXS, FMRP, profilin
Abstract
Learning and memory, to a large extent, depend on functional changes at synapses. Actin dynamics orchestrate the formation of synapses, as well as their stabilization, and the ability to undergo plastic changes. Hence, profilins are of key interest as they bind to G-actin and enhance actin polymerization. However, profilins also compete with actin nucleators, thereby restricting filament formation. Here, we provide evidence that the two brain isoforms, profilin1 (PFN1) and PFN2a, regulate spine actin dynamics in an opposing fashion, and that whereas both profilins are needed during synaptogenesis, only PFN2a is crucial for adult spine plasticity. This finding suggests that PFN1 is the juvenile isoform important during development, whereas PFN2a is mandatory for spine stability and plasticity in mature neurons. In line with this finding, only PFN1 levels are altered in the mouse model of the developmental neurological disorder Fragile X syndrome. This finding is of high relevance because Fragile X syndrome is the most common monogenetic cause for autism spectrum disorder. Indeed, the expression of recombinant profilins rescued the impairment in spinogenesis, a hallmark in Fragile X syndrome, thereby linking the regulation of actin dynamics to synapse development and possible dysfunction.
The immense computational power of the central nervous system depends on the formation of functional neuronal networks, which are further refined and adapted to environmental changes by processes of neuronal plasticity throughout the entire life span of an individual. The majority of synapses in highly plastic regions, such as the neocortex and hippocampus, are located at dendritic spines, tiny protoplasmatic membrane protrusions that build the postsynaptic compartment. Changes in spine shape are directly associated with the dynamic actin cytoskeleton, which is highly enriched in dendritic spines (1–6). In fact, up to 80% of actin filaments turn over in less than 2 min in the spine head (7). Hence, an understanding of the detailed molecular machinery and identification of key molecules that control actin polymerization in space and time will help to reveal details of spine function and plasticity, and might eventually also provide a better understanding of neurological disorders characterized by defects in spinogenesis and spine maintenance (8, 9).
The small actin-binding protein profilin—present in the mammalian CNS in two different isoforms, profilin1 (PFN1) and profilin2a (PFN2a) (10)—has been described as such a promising candidate because its activity-dependent translocation into dendritic spines could be shown both in vitro and in vivo (11–13). However, recent studies exploiting knockout animals for either PFN1 or PFN2a demonstrated a surprising lack of a spine phenotype for both isoforms (14, 15). One explanation might reside in the crucial importance of tightly restricted actin dynamics for virtually all aspects of neuronal function that might be preserved in knockout animals by means of compensational effects acting on the expression or regulation of other actin-binding molecules. This theory is supported by work from our group showing that an acute knockdown of PFN2a actually revealed an important function in dendritic spines (16).
In this study, we took advantage of an acute interference RNA (RNAi)-mediated loss-of-function approach, which allowed us to provide evidence that despite the fact that profilins are biochemically very similar, the two brain isoforms perform astonishingly diverse functions. Our results indicate that the ubiquitous isoform PFN1 is of great importance for spine formation. Furthermore, we can show that the expression of PFN1 is developmentally down-regulated in the hippocampus. In contrast to this, we found the evolutionary most-recent and brain-specific isoform PFN2a to be involved in synapse function, spine stabilization, and activity-dependent structural plasticity. Most notably, both isoforms were differentially engaged in regulating actin dynamics in dendritic spines. In line with a role of PFN1 for spine formation during development, we provide evidence that, of the brain profilin isoforms, only the mRNA of PFN1, comparable to the Drosophila homolog chickadee (17), is bound by the Fragile X mental retardation protein (FMRP). Similarly, PFN1 but not PFN2a levels were altered in the mouse model of the neurodevelopmental disorder Fragile X syndrome (FXS), a hallmark of which is an apparent defect in spine formation and maturation (18–20).
Our results therefore point toward intriguingly different functions of profilin isoforms in the brain with a juvenile expression profile, indicating a major role of PFN1 during spinogenesis and a mature expression profile favoring PFN2a as the predominant isoform crucial for spine stabilization, synaptic function, and spine plasticity.
Results
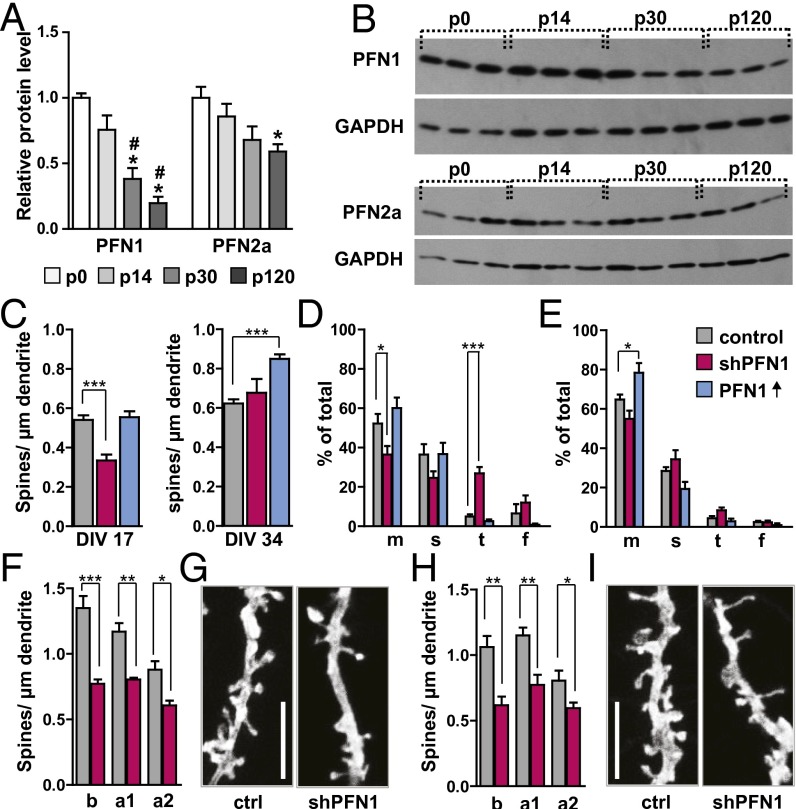
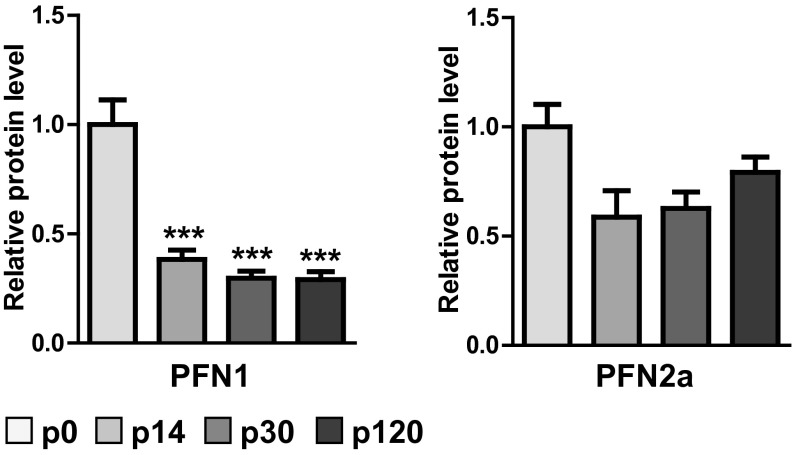
It is well known that both profilins are expressed throughout the entire CNS, leaving no brain region with only one isoform (21). However, to our knowledge it has not been described before whether the relative expression levels of PFN1 and PFN2a are developmentally regulated in a differential manner. Thus, we performed a Western blot analysis of profilin isoform expression levels in the mouse hippocampus, a brain region known to be highly plastic (Fig. 1 A and B). We were especially interested in addressing whether the expression levels would change before, during, and after the peak of synaptogenesis (22, 23). Indeed, our results showed that PFN1 levels started to decrease significantly after postnatal day (P) 14. At 4 mo of age, PFN1 levels were only 20% of those detected at birth, indicating that this isoform might be of special importance during development (Fig. 1 A and B). In contrast, PFN2a levels started to decrease only mildly after the peak of synaptogenesis and became significantly lower (60% of those at birth) only at 4 mo of age (Fig. 1 A and B).
Fig. 1.
PFN1 expression is developmentally regulated in the hippocampus and important for spine formation. (A) Western blot analysis of hippocampal profilin levels revealed a significant reduction of PFN1 levels throughout development, whereas PFN2a levels are only altered mildly. (B) Examples for Western blots of PFN1 and PFN2a levels, three samples for each time point. (C) Spine numbers in primary embryonic cultures were significantly reduced following RNAi-mediated knockdown of PFN1 at 17 DIV but not at 34 DIV, the overexpression of PFN1 only affected 34 DIV neurons. (D and E) Spine subtype composition of neurons analyzed in C. Abbreviations: m, mushroom; s, stubby; t, thin; f, filopodia. (D) Neurons at 17 DIV; (E) neurons at 34 DIV, PFN ↑: overexpression of PFN1. (F–I) PFN1 knockdown led to a significant reduction in spine density in CA1 neurons (F and G) and CA3 neurons (H and I); example image of CA1 (G) or CA3 (I) apical dendrites of control or shPFN1-expressing cells. (Scale bars, 5 µm.) a1 proximal apical (50–200 µm from soma), a2 distal apical tufts, b basal dendrites. Means, SEM, and P values, as well as statistic tests used are depicted in Table S1. *P < 0.05; **P < 0.01; ***P < 0.001 (all compared to ctrl/P0); #P < 0.05 (compared to P14).
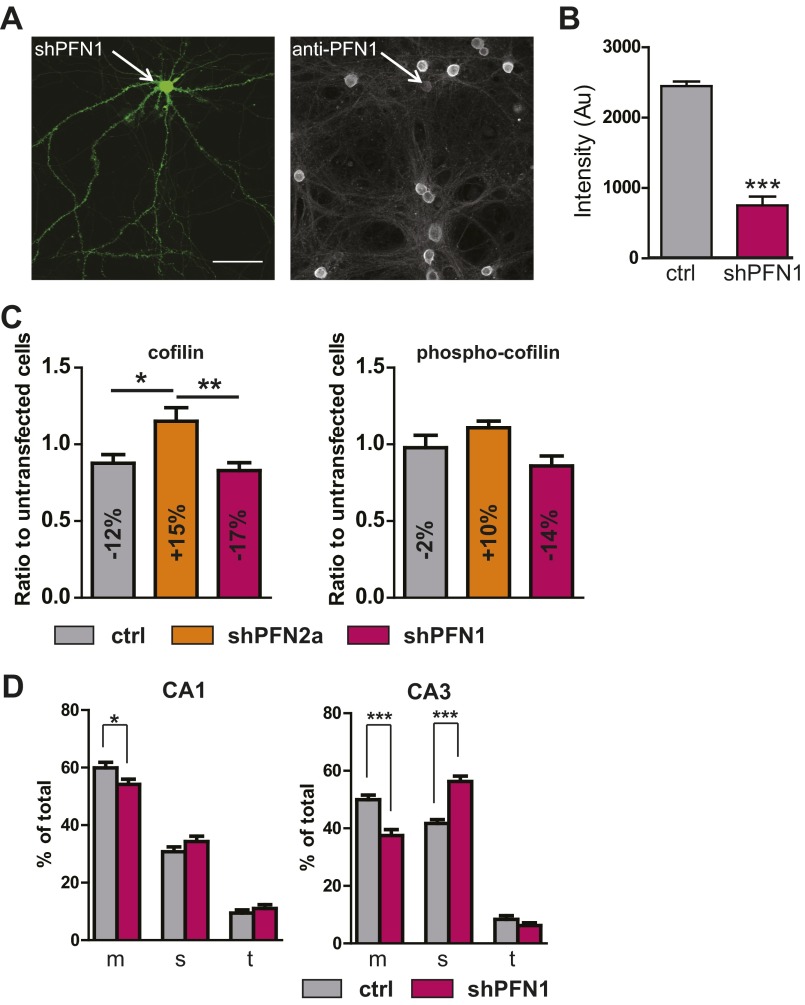
Based on our in vivo findings, we were interested to further elucidate the role of PFN1 during and after the time point of synaptogenesis, as we had shown previously that, indeed, both profilins seem to be important in dendritic spines (16). We took advantage of an acute loss of PFN1 by exploiting an RNAi-based knockdown system to circumvent compensational alterations in the expression of other actin-binding molecules or upstream signaling cascades, because this might be the case in knockout animals that were described as showing no alterations in spine number (14). Primary embryonic hippocampal cultures were transfected at 14 d in vitro (DIV) and 31 DIV with an shRNA expression vector directed against PFN1 (Fig. 1C and Fig. S1 A and B) (we previously used the same vector system to describe an acute loss of PFN2a in hippocampal neurons). Transfection of the vector resulted in a 75% loss of PFN1 compared with untransfected neighboring cells (Fig. S1 A and B). We were interested to learn whether other actin-binding proteins might be affected by the loss of profilin isoforms, and analyzed the most likely candidate cofilin and phospho-cofilin levels in transfected neurons. Only small alterations in cofilin and no change in phospho-cofilin levels could be detected (Fig. S1C).
Fig. S1.
(A) primary embryonic neuron transfected with shPFN1 + eGFP-F, immunostaining using anti-PFN1 reveals loss of PFN1 in the transfected neurons compared with neighboring cells. (Scale bar, 20 µm.) (B) Quantification of knockdown by intensity measurement of the immunostaining seen in A; 20 shPFN1-transfected neurons were compared with neighboring control cells. (C) Cofilin and phospho-cofilin levels were analyzed via immunohistochemistry in primary embryonic hippocampal cultures transfected with the control vector shLuciferase, shPFN1, or shPFN2a (all n = 10); fluorescence intensity was compared with neighboring untransfected neurons. The quantification revealed a mild increase in cofilin expression for PFN2a transfected cells compared with shLuciferase expressing neurons, whereas the phospho-cofilin levels were unaltered upon knockdown of both profilin isoforms compared with control transfected neurons. (D) Spine-type quantification for the organotypic slice cultures described in Fig. 1 F–I. Mushroom (m), stubby (s), thin (t) showed a decrease in the relative number of mushroom spines accompanied by an increase in the number of stubby spines. *P < 0.05; **P < 0.01; ***P < 0.001.
We compared spine density between PFN1-deficient cells, neurons overexpressing recombinant PFN1, and control neurons transfected with the empty vector, as we previously showed that the shRNA control vector system had no adverse side effects on spine number (16) (Fig. 1C). When cells were fixed and analyzed 3 d after transfection, a comparison of spine numbers at DIV 17 revealed a significant reduction upon the loss of PFN1 at this stage, whereas the overexpression had no impact. For cells transfected after the peak of synaptogenesis and analyzed at DIV 34, we found that PFN1 knockdown had no effect on spine numbers, whereas expression of recombinant PFN1 led to a significant increase in spine number compared with controls (Fig. 1C). Moreover, when we analyzed spine subtypes classified by using the same criteria described previously (24), we found that an early reduction of PFN1 at DIV 17 led to an increase in the number of thin spines accompanied by a reduction in mushroom spines; however, no influence of recombinant PFN1 at this stage on spine morphology could be detected (Fig. 1D). After the peak of synaptogenesis was over, we found that the knockdown had no influence on spine subtype composition, whereas the expression of recombinant PFN1 induced an increase in the abundance of mushroom spines (Fig. 1E).
In a second step, we wanted to confirm the importance of PFN1 for spine formation using a system in which the in vivo circuitry of the hippocampus would be better preserved, and therefore used the organotypic slice-culture method. Neurons of the CA1 and CA3 region were transfected at DIV 11 with shPFN1 or the empty-vector backbone. A comparison of spine numbers at DIV 14 revealed that in both subfields the loss of PFN1 led to a robust and significant reduction in spine number (CA1 in Fig. 1 F and G; CA3 in Fig. 1 H and I). In addition, spine-subtype composition was altered in CA1 and CA3 principal neurons, with a reduction in the proportion of mushroom-shaped spines (Fig. S1D). Unless specified otherwise for all further experiments, results of CA1 and CA3 pyramidal neurons were therefore combined.
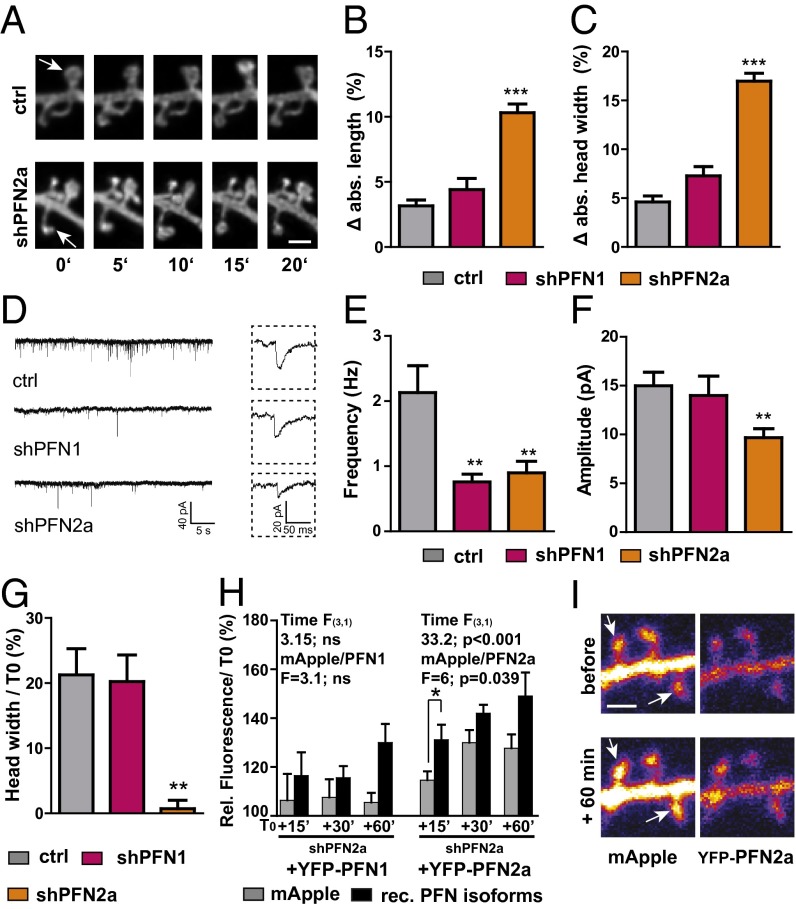
To further elucidate the isoform-specific functions of profilins in dendritic spines, we performed time-lapse imaging experiments in PFN1- and PFN2a-deficient pyramidal neurons in organotypic slice cultures. We started by exploring the stability of dendritic spines in the presence or absence of profilins because spine motility (small changes in length and head width over short periods of time) directly depends on the actin cytoskeleton, and can therefore be used as a read-out for alterations in actin dynamics already under baseline conditions (25). The acquisition of image stacks of dendritic segments using 5-min intervals over an imaging period of 20 min in PFN2a-deficient cells revealed a strongly apparent phenotype of abnormally motile spines (Fig. 2A and Fig. S2 A and B), whereas spines of PFN1-deficient neurons were indistinguishable from those of control cells (Fig. 2 B and C). Absolute changes in length (Fig. 2B) and head width (Fig. 2C) were analyzed between consecutive time points and compared between control as well as shPFN1- and shPFN2a-transfected cells (Fig. 2 B and C and Fig. S2 A and B).
Fig. 2.
PFN2a but not PFN1 is important for spine stability, synapse function, and spine plasticity. (A) Motility was increased in the absence of PFN2a; example spines for each imaging time point of a control cell and a PFN2a-deficient neuron. (Scale bar, 1 µm.) White arrows indicate an example spine. Absolute changes in spine length (B) or head width (C) were significantly increased when averaged for 5-min imaging intervals in PFN2a-deficient pyramidal neurons. (D) Example traces of mEPSCs in control cells or PFN isoform deficient pyramidal cells. (E) The frequency of mEPSCs was significantly reduced upon knockdown of profilin isoforms; however, a significant decrease in amplitude (F) could only be found in PFN2a-deficient neurons. (G) Changes in spine head width 60 min after cLTP induction normalized to before stimulation; activity-dependent structural plasticity was virtually absent in PFN2a-deficient pyramidal neurons. (H) Rescue experiments overexpressing recombinant profilin isoforms in shPFN2a mApple-transfected cells. Only PFN2a was able to rescue the defect in spine plasticity; only recombinant PFN2a was significantly enriched in spines upon stimulation. ns, not significant. (I) Example images of a cell transfected with shPFN2a mApple + YFP-PFN2a before and after stimulation. (Scale bar, 1 µm.) Means, SEM, and P values, as well as statistic tests used are depicted in Table S1. *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. S2.
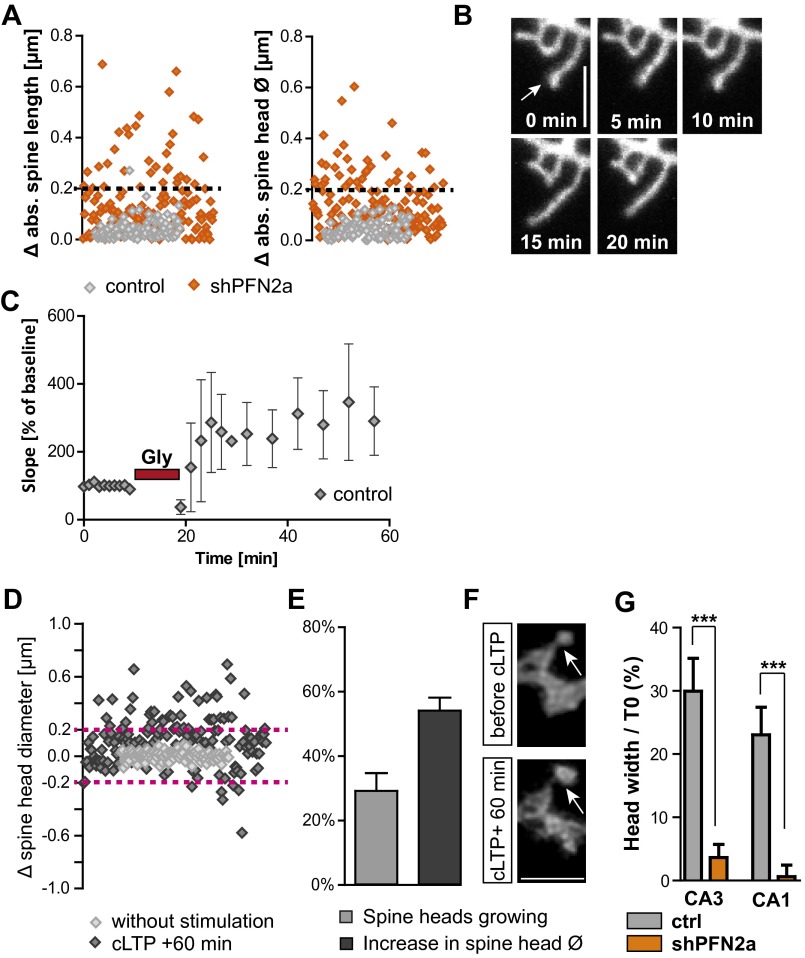
(A) Absolute changes in spine length or head diameter for a 5-min imaging interval in shPFN2a- (n = 7) or control transfected cells (n = 6), dashed line indicates the optical resolution limit. Note that only PFN2a-deficient cells show changes above the resolution limit of 200 nm. (B) Example images of PFN2a-deficient cells showing the increase in spine length motility. (Scale bar, 2 µm.) (C) Electrophysiological recording of the Schaffer collaterals in four organotypic slice cultures using the cLTP induction protocol; after 10 min of baseline recording, 10 mM glycine was applied for 10 min. Recording during the stimulation was stopped and resumed afterward with recordings every 2 min. (D) Changes in the diameter of spine heads of control transfected CA1 neurons with (n = 9) or without stimulation (n = 4); analyzed were the time point directly before stimulation compared with 60 min after cLTP induction or the changes within a 60-min time window without stimulation. Dashed lines indicate the optical resolution limit. Note that only stimulated spines display changes above the resolution limit of 200 nm. (E) Depicted is the fraction of spines of control transfected neurons showing changes in head diameter above the resolution limit and the relative amount of growth per single spine compared with before the stimulation. (F) Example confocal microscopic images of a spine before and 60 min after stimulation. (Scale bar, 2 µm.) (G) Comparison of activity-dependent spine head growth in CA1 and CA3 cells revealed no significant difference between pyramidal cell types. In both neuronal cell types PFN2a deficiency let to an almost complete impairment of activity-dependent spine growth. ***P < 0.001.
As we found a reduction in spine number both for the loss of PFN1 described above and in PFN2a-deficient neurons (16), we next asked the question whether basal synaptic function would be affected upon knockdown of profilins. Therefore, miniature excitatory postsynaptic currents (mEPSCs) were recorded in the presence of Tetrodotoxin (TTX; 0.5 µM) and d-(-)-2-amino-5-phosphonopentanoic acid (APV; 10 µM) from control (shRNA vector against firefly luciferase), as well as shPFN1- and shPFN2a-transfected neurons (Fig. 2 D–F). A significant decrease in mean mEPSC frequency was observed in PFN1-deficient cells and PFN2-deficient neurons compared with the respective control (Fig. 2E). Interestingly, the mean amplitude of mEPSCs was significantly reduced only in shPFN2a-transfected cells compared with control neurons, whereas the amplitude in shPFN1 neurons was not altered (Fig. 2F).
Both profilin isoforms have been shown before to be targeted to dendritic spines in an activity-dependent manner (11, 12). Therefore, we tried to elucidate which profilin isoform might be involved in activity-dependent structural plasticity at dendritic spines. To approach this question, we applied 10 mM glycine for 10 min to modulate activity in organotypic slice cultures, a protocol described previously to lead to a significant increase in synaptic efficacy comparable in many aspects to TBS-induced long-term potentiation (26) (chemical LTP, cLTP) (Fig. 2G and Fig. S2 C–G). Spines were monitored at proximal basal and apical dendrites (distance up to 200 µm from soma). Sixty minutes after cLTP induction, control neurons displayed a prominent increase in spine head width above the resolution limit, whereas no changes could be detected in unstimulated cells (Fig. 2G and Fig. S2D). Upon stimulation, 30% of spines displayed an increase in head diameter above 200 nm (Fig. S2E). When we averaged only changes above the resolution limit of 200 nm in control cells, it resulted in a mean head size increase of 50%, compared with baseline conditions for individual spines (Fig. S2 E and F). This finding indicates that our protocol reliably induced processes of structural spine plasticity. To compare the ability of PFN1- and PFN2a-deficient cells to undergo activity-dependent structural plasticity to control neurons, again all changes were averaged (Fig. 2G). Most notably, whereas PFN1-deficient neurons showed normal cLTP-induced structural changes and displayed a comparable increase, as observed for control cells 60 min after cLTP induction, PFN2a-deprived neurons were characterized by an almost complete lack of activity-dependent structural plasticity at dendritic spines (Fig. 2G and Fig. S2G). In a second step, we tried to rescue the lack of structural plasticity displayed in PFN2a-deficient neurons by expressing recombinant PFN1 or PFN2a as well (Fig. 2H). Only cells transfected with shPFN2a and recombinant PFN2a showed a significant increase in relative mApple fluorescence over time, compared with prestimulus conditions [repeated-measures ANOVA, F(3, 1) time = 33.2, P < 0.001], whereas recombinant PFN1 failed to rescue spine plasticity [F(3, 1) time = 3.15]. In addition, we could detect a significant enrichment of recombinant YFP-PFN2a in spines upon stimulation already at 15 min, the earliest time point investigated (Fig. 2 H and I) [repeated-measures ANOVA with time as the dependent variable and mApple/YFP-PFN2a as the independent variable F(independent variable) = 6, P = 0.039].
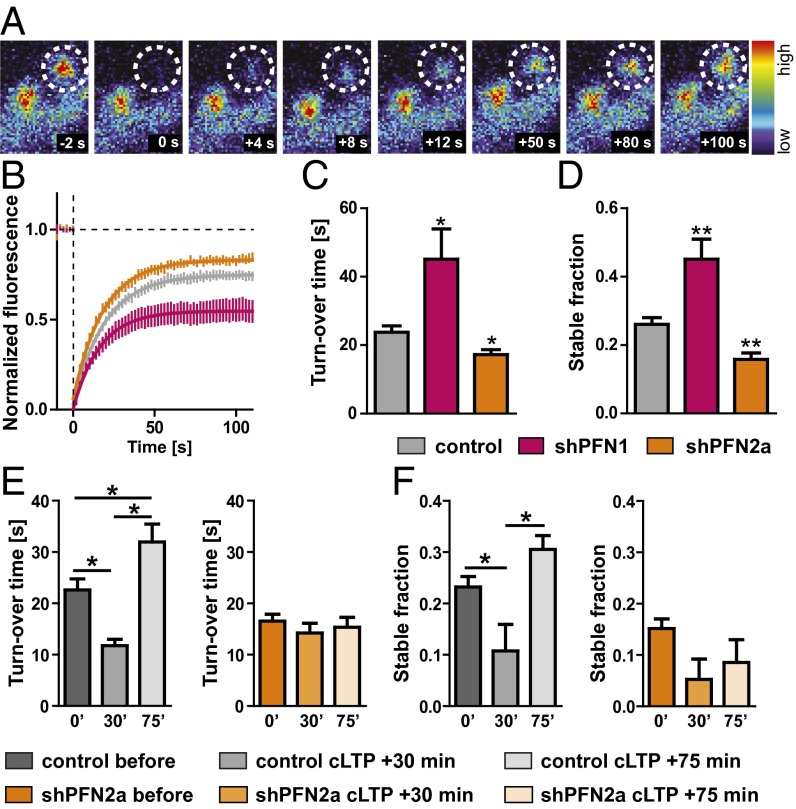
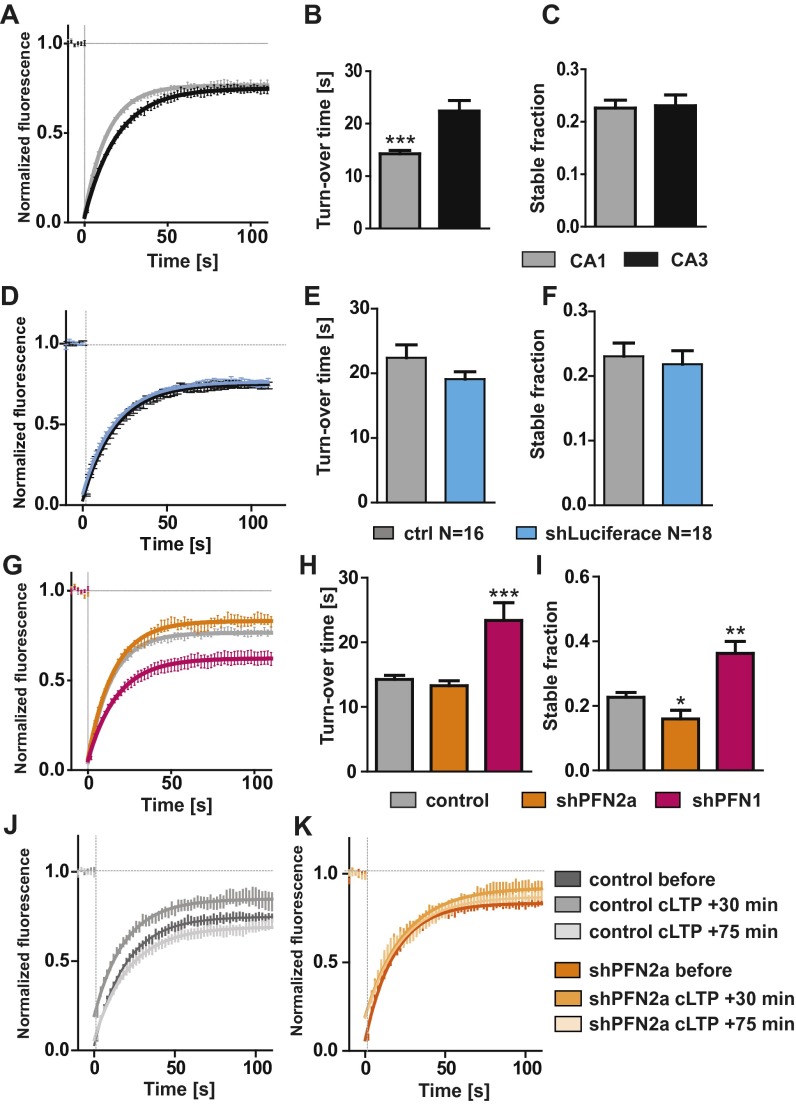
To unravel the underlying cellular mechanisms responsible for the profilin-mediated changes observed in motility and spine plasticity, we used fluorescence recovery after photobleaching (FRAP) experiments to monitor actin dynamics at single spines of hippocampal pyramidal neurons expressing eGFP-actin in the presence or absence of profilin isoforms (Fig. 3A). Surprisingly, control neurons displayed significant differences in actin dynamics within different subregions in hippocampal organotypic slice cultures (i.e., pyramidal neurons of the CA1 versus the CA3 subfield) (Fig. S3 A–C). CA3 neurons displayed slower actin dynamics in single spines indicated by a significantly increased turnover time (Fig. S3B). The stable actin pool (the proportion of eGFP-actin fluorescence that did not recover within the 2-min time window) did not differ between pyramidal neuron subtypes (Fig. S3C).
Fig. 3.
Profilin isoforms regulate spine actin dynamics in an opposing manner. (A) Actin dynamics were analyzed using FRAP expressing eGFP-actin in pyramidal neurons; example spine (dashed circle, diameter: 1 µm) before and at different time points after the 405-nm laser bleaching impulse. (B–D) CA3 control cells were compared with neurons deficient in PFN isoforms, depicted are the recovery curve (B), turnover time (C), and stable actin fraction (D). (E) Quantification of the turnover time and (F) stable actin fraction in control cells (gray) or PFN2a-deficient CA3 neurons (orange) before, at 15–30 min (30′), and at 60–75 min (75′) after cLTP induction. Means, SEM, and P values, as well as statistic tests used are depicted in Table S1. *P < 0.05; **P < 0.01.
Fig. S3.
(A) Fluorescence recovery curve for spines of control (mApple + GFP-actin) expressing CA1 and CA3 pyramidal neurons. (B) The turnover time significantly differed between CA1 and CA3 neurons (CA1 15 ± 0.6 s, CA3 23.7 ± 1.8 s, P < 0.001). (C) The stable fraction indicated by the fluorescence, which did not recover after 112 s, was not different between pyramidal neuron subtypes (CA1 23 ± 1.5%, CA3 26 ± 1.8%). (D) Fluorescence recovery curve for spines of control (mApple + GFP-actin) expressing CA3 neurons and CA3 cells expressing the shRNA control vector directed against firefly luciferase (shLuciferase); no side effects of shRNA transfection could be observed regarding turnover time (E) and stable actin fraction (F). (G–I) CA1 control cells were compared with neurons deficient in PFN isoforms. Depicted are the recovery curve (G), turnover time (H), and stable actin fraction (I). (J and K) Recovery curves of spines before and after induction of cLTP. *P < 0.05; **P < 0.01; ***P < 0.001.
When we compared actin dynamics in single spines derived from PFN1- or PFN2a-deficient neurons within the same subregion to control cells, it became obvious that both isoforms were differentially engaged in regulating actin turnover time and the proportion of the dynamic versus the stable actin fraction in single dendritic spines (CA3 neurons in Fig. 3 B–D; CA1 neurons in Fig. S3 G–I; comparison between mApple-only expressing cells and cells expressing the control shRNA vector in Fig. S3 D–F). In CA3 neurons, the actin turnover time in PFN2a-deprived cells was significantly decreased, indicating a faster polymerization rate, whereas PFN1-deficient neurons showed a significant increase pointing toward slower actin polymerization rates (Fig. 3C). Along this line, the stable actin fraction was significantly reduced in shPFN2a-expressing CA3 neurons and significantly increased in shPFN1-expressing cells (Fig. 3D). Spines of CA1 neurons revealed comparable alterations in actin dynamics (Fig. S3 G–I), except for the decrease in the turnover time shown upon PFN2a ablation in CA3 neurons (Fig. 3F). In a second set of experiments, we were interested whether the lack of activity-dependent structural plasticity in PFN2a-deficient neurons was indeed caused by a dysregulation of actin dynamics. Therefore, we quantified actin polymerization rates before and after cLTP induction. Interestingly, control-transfected neurons showed a significant increase in actin dynamics at 15–30 min after the cLTP stimulus, whereas at 60–75 min after cLTP induction, actin dynamics were significantly reduced compared with basal levels before stimulation (Fig. 3 E and F and Fig. S3 J and K). Most notably, a lack of PFN2a completely prevented these activity-dependent changes (Fig. 3 E and F and Fig. S3 J and K).
Up to this point, we could provide evidence for intriguing differences between profilin isoforms in the brain, especially given the fact that they share a high degree of biochemical similarity. We could show that profilins are regulated differentially throughout development and that they shape actin dynamics in opposing ways in dendritic spines. Moreover, PFN2a specifically could be revealed as the isoform crucial for basal synaptic function, as well as for spine stability and processes of activity-dependent structural plasticity.
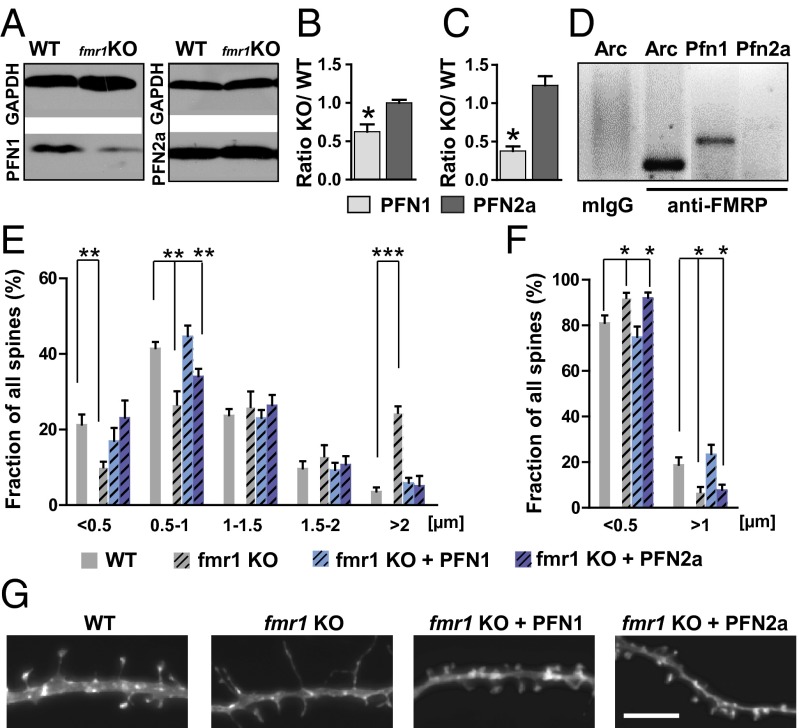
The cytoskeleton is crucial for spine formation and maintenance, and therefore actin dynamics might indeed be tightly linked to the establishment of functional neuronal networks. Evidence from Drosophila connects the profilin homolog chickadee to the neurodevelopmental disorder FXS because the mRNA of chickadee is bound by the Fragile X mental retardation protein, FMRP, and chickadee levels are dysregulated in the FXS Drosophila model (17). In line with the role of PFN1, especially during development and spine formation, the question arose if profilins were actually involved in mediating the spine phenotype in FXS, and if so, whether there would be a difference between the two brain isoforms in the mouse model of FXS (fmr1 KO mice that lack FMRP). Western blot analysis in whole-brain lysates of fmr1 KO mice and WT littermates (Fig. 4 A and B and Fig. S4) revealed that only PFN1 levels were significantly reduced, whereas PFN2a levels were left unaltered. Because we showed previously that PFN1 levels are highest at P0 during postnatal development, we also quantified hippocampal PFN1 and PFN2a levels at this time point and found a significant reduction again only for PFN1 (Fig. 4C). During development, hippocampal levels of PFN1 also decreased drastically in fmr1 KO mice (Fig. S4). Given the fact that FMRP is an RNA-binding protein regulating the localization and expression of its target mRNAs (27, 28), we performed RNA-immunoprecipitation using an anti-FMRP antibody followed by RT-PCR for either Pfn1 or Pfn2a mRNAs (Fig. 4D). In line with our findings described previously (16), only the mRNA of PFN1 could be detected as a target of FMRP, further underlining the fact that the changes in protein expression we observed resulted indeed from a direct interaction of FMRP with the mRNA of PFN1.
Fig. 4.
PFN1 but not PFN2a expression is dysregulated in a mouse model of the FXS. (A) Example Western blots for profilin isoforms in whole-brain lysates derived from adult WT or fmr1 KO mice. (B) Quantification of protein levels revealed a significant reduction for PFN1 in fmr1 KO animals. (C) Hippocampal PFN1 protein levels were reduced as well at P0 in fmr1 KO mice. (D) Representative gel of a RIP using an antibody against FMRP followed by RT-PCR specific for either PFN1 or PFN2a mRNA identified only the mRNA of PFN1 as a target of FMRP (three animals, 2–4 mo of age were analyzed). Arc was used as a positive control; analysis of spine length (E) or head width (F) in WT, fmr1 KO and fmr1 KO neurons expressing recombinant PFN1 or PFN2a revealed an increase in long, thin spines in fmr1 KO neurons that could be only fully rescued by overexpression of PFN1. (G) Example images of WT, fmr1 KO, and fmr1 KO cells expressing recombinant profilin isoforms. (Scale bar, 2 µm.) Means, SEM, and P values as well as statistic tests used are depicted in Table S1. *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. S4.
The developmental regulation of PFN levels was quantified in fmr1 KO animals (three to six animals per time point), whereas the level of PFN1 significantly decreased compared with P0 PFN2a levels remained unaltered, ANOVA with post hoc Bonferroni correction. ***P < 0.001.
A prominent hallmark of FXS is an impairment in the formation of functional neuronal networks (19, 29, 30). Spine numbers are described as being permanently or transiently increased in different brain regions, such as the hippocampus and neocortex, accompanied by alterations in the shape of dendritic spines with an overabundance of long and thin protrusions, indicating impaired synapse formation and maturation (20).
Primary embryonic cultures of hippocampal neurons derived from fmr1 KO animals indeed revealed an increase in spine length (Fig. 4E) and a concomitant decrease in spine head diameter (Fig. 4F), thereby showing an impairment in spine maturation compared with cultures derived from littermate controls. These data also indicate that the spine number was unaltered in these cultures. The immature spine profile, together with the reduction in PFN1 protein levels and its important role for spinogenesis, let us hypothesize whether the recombinant expression of profilins might be able to rescue the spine phenotype in fmr1 KO neurons. Indeed, only the overexpression of PFN1 was able to fully restore spine length as well as spine head diameter to levels indistinguishable from controls, whereas recombinant PFN2a only partially restored spine length and had no effect on spine head width (Fig. 4 E–G).
Discussion
Our present study provides insights into the molecular mechanisms underlying actin-mediated orchestration of spinogenesis and spine stabilization, and further outlines how these mechanisms differ from those used to mediate activity-dependent spine structural plasticity. In addition, we show a link between the dysregulation of actin dynamics and the impairment in spinogenesis observed in one of the most common developmental neurological disorders, FXS. We focused our work on profilins, actin-binding proteins of central importance identified for many years because of their fundamental role during development, where deletion of the ubiquitous isoform PFN1 inevitably results in cell death (31). The expression of tissue-specific isoforms indicates potentially diverse tasks. In this respect, specialized tissues, like the kidney, testis, or CNS express more than one isoform (10), and the question arises to what extent profilins might diverge in their function in the respective tissue. The results we obtained show two different lines of expression pattern and function of profilin isoforms during development (spinogenesis) and in the adult nervous system (activity-dependent structural plasticity). We provide strong evidence that in mature neurons the brain-specific PFN2a is also the profilin isoform mandatory for basal synaptic function, spine stability, and modulation of actin dynamics during processes of structural plasticity.
Given the fact that synaptic plasticity and spine number were reported as normal in PFN2a knockout mice, our phenotype observed in this study was surprisingly strong, as the capacity of spines to undergo activity-dependent structural plasticity was virtually absent. This finding indicates that the function of profilins is in fact crucial for neurons and especially synapses in the CNS. Functional redundancy of actin-binding proteins might be a compensational mechanism to prevent deleterious and otherwise most likely fatal effects for the organism, a fact that may hinder analysis in knockout animals. Aside from clear differences in the relevance of the two isoforms for basal synaptic function and spine plasticity in the mature nervous system, we can also show that the loss of profilins affects actin dynamics in an even opposing manner in dendritic spines. In this respect, PFN1 primarily seems to promote actin polymerization in a prototypical fashion described for profilins. In cooperation with members of the formin family, profilins deliver G-actin to the growing actin filament and can thereby promote the formation of long, unbranched actin filaments that are important for filopodia formation, and thereby might be especially needed during spinogenesis (32). In line with this, the spine phenotype we observed here in PFN1-deficient neurons was much stronger than the phenotype we reported earlier for an acute loss-of-function of PFN2a (16).
Recently, PFN1 has been described as a gatekeeper for actin assembly by competing with the Arp2/3 complex for free-actin monomers in fibroblasts (33). Based on the results presented herein, we conclude that in the mature CNS and in particular within dendritic spines, PFN2a might be the primary isoform restricting the Arp2/3-dependent formation of actin networks by sequestration of actin monomers, because actin dynamics in dendritic spines were even enhanced in cells deprived of PFN2a. This increased actin polymerization rate might also be the reason for the observed increase in spine motility, because enhanced actin dynamics might in fact interfere with spine stabilization. This is further supported by the observation that expression of recombinant PFN2a leads to stabilization of spine structure and rounding of spine shape, the opposite phenotype as observed here for the loss of PFN2a (11). In addition, the reduction in mEPSC amplitude in PFN2a-deficient neurons suggests a reduced number of AMPA receptors, which might point to a function of PFN2a in mediating actin dynamics and thereby receptor clustering directly at the postsynaptic density. Along this line, the lack of activity-dependent spine structural plasticity might not only result from an impairment in actin dynamics, which modulate spine growth but moreover might indicate a role of PFN2a in modulating both functional and structural plasticity. In line with this, we found recombinant YFP-PFN2a significantly enriched in spine heads as soon as 15 min after cLTP induction. Further support for the hypothesis that PFN2a indeed represents the predominant isoform in the mature CNS important for processes of activity-dependent synaptic plasticity underlying learning and memory formation, is provided by our analysis of profilin isoform expression throughout development, which revealed a switch in the ratio of PFN1 versus PFN2a. In this respect, it might be interesting to mention that PFN2a can also be considered as the most recently derived isoform in vertebrate evolution (34, 35). More than being directly responsible for delivery of actin monomers to the growing filament (as might be the case for PFN1, which may indeed fulfill the “traditional” profilin function in neurons), PFN2a might well be a hub molecule mediating the balance between different actin polymerization pools by restricting specific pathways. Based on these observations, one hypothesis is that PFN2a and Arp2/3 could compete for G-actin. In this respect, PFN2a may be even considered as a molecule with attributes suitable to fulfill either the role of a synaptic tag, as it translocates into spines following LTP induction, or it might be used as a plasticity-related protein because it is able to shuttle between active synapses and the nucleus (35).
In line with a role of PFN1 for spinogenesis during development, our results clearly identify this isoform to be linked to the developmental neurological disorder FXS, which shows as its main characteristic an impairment in spine maturation. We show here that only the mRNA of PFN1 is bound by FMRP, which regulates the transport of its target mRNAs to control local translation in dendrites. This finding is in line with the model we propose here, in which PFN1 is the predominant isoform important for spine formation. Similarly, the developmental switch in isoform relevance from the predominance of PFN1, which is regulated by FMRP in juvenile animals, to PFN2a in the mature CNS, which is not bound by FMRP, could in addition provide an explanation for the often-described transient nature of the spine phenotype in the FXS mouse model (20, 29). With an amazing efficacy, the immature spine phenotype with an overabundance of long and thin protrusions detected in fmr1 KO neurons could be rescued by expression of recombinant PFN1. Future experiments determining, on a subcellular level, the localization of PFN1 mRNA within different neuronal subcompartments, will shed light on the detailed role of PFN1 for spine maturation during the course of FXS. A further analysis of synaptic actin dynamics during development might shed light on possible therapeutic treatments of this devastating disease.
Materials and Methods
Mice, Genotyping, Cell Culture, and Transfection.
All procedures concerning animals were approved by the animal welfare representative of the Technische Universität Braunschweig and the LAVES [Oldenburg, Germany, Az. §4 (02.05) TSchB TU BS). Detailed procedures are described in SI Materials and Methods.
DNA Constructs.
The knockdown of PFN1 and PFN2a was achieved using the shRNA expression vector system pRNAT U6.3 vector and insertion of specific oligonucleotides against the respective mRNA. In the case of PFN2a, the knockdown was conducted as previously described (16). To achieve the knockdown for PFN1, the ds oligonucleotide GATCCCGTTGTTGATCAAACCACCGTGGTTGATATCCGCCACGGTGGTTTGATCAACAATTTTTTCCAAA was inserted. The overexpression of PFN1 and PFN2a was achieved using an expression vector carrying YFP-PFN1 under the control of a truncated CMV promoter (16). Knockdown/knockin vectors were previously described (16). For structural analysis of dendritic spines, farnesylated eGFP-F (Clontech) or mApple were used (16, 36, 37).
Western Blot Analysis.
Protein samples were prepared from three animals per time point, except for P0, where hippocampi of three animals had to be pooled together for one sample. Details are described in SI Materials and Methods.
RNA Immunoprecipitation.
RNA immunoprecipitations (RIP) were performed using the MagnaRIPTM Kit (Merk Millipore 17–701) according to the manufacturer’s instructions. FMRP and the associated RNAs were precipitated in hippocampal lysates using an anti-FMRP antibody (clone 1C3, Merk Millipore, MAB2160). For detection of the respective mRNAs, the following primer sequences were used in the RT-PCR: PFN1 forward CACTTGGGGGCCAGAAATGT, reverse ATGGAAAGAAGGGGGTGCAA, PFN2a forward TGTCCACGCAGGCACAATTA, reverse GGAGGGGTGAGAAAGGTGTG.
Imaging and Image Analysis.
Imaging and analysis of fixed neurons was performed as previously described (16). Time-lapse imaging was performed using a Fluoview1000 set-up (Olympus) equipped with a 60× objective (1.0 NA water, pixel size 0.07 µm) as previously described (38). The imaging chamber was heated to 32 °C and perfused with carbogenated artificial cerebrospinal fluid (38). Z-stacks (interval for z-sectioning 0.35 µm) of dendritic segments were captured at 5-min intervals for 20 min. Spine motility was analyzed manually using ImageJ software by quantifying absolute changes in length or head diameter between two consecutive time points. For the cLTP, 10 mM glycine in ACSF was used (26).
Electrophysiology.
Whole-cell patch-clamp recording techniques were used to record mEPSCs from organotypic hippocampal slice cultures (DIV 14–16). The details are described in SI Materials and Methods.
FRAP Experiments.
FRAP experiments were performed as previously described (38); the detailed procedure is described in SI Materials and Methods.
Data Presentation and Statistical Analysis.
Unless otherwise specified, data are depicted as mean ± SEM. An α level of P < 0.05 was used as the criterion to reject the null hypothesis. Means, SEM, and P values as well as statistic tests used are depicted in Table S1.
Table S1.
Means, SEM, P values, and statistic tests used in the figures
| Figure | Treatment | |||
| Fig. 1A | PFN1 P0 | PFN1 P14 | PFN1 P30 | PFN1 P120 |
| 1.0 ± 0.03 n = 3 | 0.75 ± 0.09 n = 3 | 0.38 ± 0.07 n = 3 | 0.20 ± 0.04 n = 3 | |
| P | ANOVA (post hoc Bonferroni) | P0 P = 0.002 | P0 P = 0.0001; P30 P = 0.009 | |
| PFN2a P0 | PFN2a P14 | PFN2a P30 | PFN2a P120 | |
| 1.0 ± 0.08 n = 6 | 0.85 ± 0.09 n = 6 | 0.67 ± 0.1 n = 6 | 0.59 ± 0.06 n = 6 | |
| P | ANOVA (post hoc Bonferroni) | P0 P = 0.01 | ||
| Fig. 1C | Ctrl (DIV 17) | shPFN1 (DIV 17) | PFN1 oe (DIV 17) | |
| 0.54 ± 0.023 n = 10 | 0.33 ± 0.03 n = 10 | 0.55 ± 0.03 n = 10 | ||
| P | Ctrl P < 0.0001 | |||
| Ctrl (DIV 34) | shPFN1 (DIV 34) | PFN1 oe (DIV 34) | ||
| 0.62 ± 0.02 n = 7 | 0.67 ± 0.07 n = 6 | 0.85 ± 0.023 n = 6 | ||
| P | Ctrl P < 0.0001 | |||
| Fig. 1 F and H | CA1 Ctrl | CA1 shPFN1 | CA3 Ctrl | CA3 shPFN1 |
| B: 1.35 ± 0.09 n = 8 | B: 0.77 ± 0.04 n = 5 | B: 1.06 ± 0.083 n = 9 | B: 0.61 ± 0.06 n = 7 | |
| A1: 1.17 ± 0.06 n = 8 | A1: 0.8 ± 0.01 n = 5 | A1: 1.15 ± 0.06 n = 9 | A1: 0.77 ± 0.07 n = 7 | |
| A2:0.88 ± 0.06 n = 8 | A2: 0.60 ± 0.03 n = 5 | A2: 0.81 ± 0.076 n = 9 | A2: 0.6 ± 0.04 n = 7 | |
| P | B: P < 0.0001 A1 P = 0.0011;A2 P = 0.011 | B: P = 0.0014 Mid P = 0.0016 A1 P = 0.04 | ||
| Fig. 2B | Ctrl | shPFN2a | shPFN1 | |
| L 3.2 ± 0.5 n = 6 | L 10.3 ± 0.7 n = 7 | L 4.4 ± 0.9 n = 10 | ||
| W 4.6 ± 0.6 n = 6 | W 17 ± 0.8 n = 7 | W 7.3 ± 0.9 n = 9 | ||
| P | Ctrl L P < 0.0001; Ctrl W P < 0.0001 | |||
| Fig. 2E | Ctrl | shPFN2a | shPFN1 | |
| 2.132 ± 0.4128 n = 7 | 0.89 ± 0.17 n = 8 | 0.76 ± 0.12 n = 10 | ||
| P | Ctrl P = 0.0079 | Ctrl P = 0.0048 | ||
| Fig. 2F | Ctrl | shPFN2a | shPFN1 | |
| 14.98 ± 1.3 n = 7 | 9.672 ± 0.91 n = 8 | 13.99 ± 1.9 n = 10 | ||
| P | Ctrl P = 0.0062 | |||
| Fig. 2G | Ctrl | shPFN2a | shPFN1 | |
| 21.27 ± 4 n = 9 | 0.72 ± 1.3 n = 12 | 20.25 ± 4.7 n = 5 | ||
| P | Ctrl P < 0.0001 | |||
| Fig. 2H | shPFN2a + YFP PFN1 | shPFN2a + YFP PFN2a | ||
| t+15 mApple: 1.06 ± 0.1 n = 4 | t+15 PFN1: 1.16 ± 0.1 n = 4 | t+15 mApple: 1.15 ± 0.03 n = 5 | t+15 PFN1: 1.31 ± 0.06 n = 5 | |
| t+30 mApple: 1.07 ± 0.07 n = 4 | t+30 PFN1: 1.16 ± 0.05 n = 4 | t+30 mApple: 1.3 ± 0.05 n = 5 | t+30 PFN1: 1.42 ± 0.03 n = 5 | |
| t+60 mApple: 1.05 ± 0.04 n = 4 | t+60 PFN1: 1.3 ± 0.08 n = 4 | t+60 mApple: 1.28 ± 0.05 n = 5 | t+60 PFN1: 1.49 ± 0.09 n = 5 | |
| P | n corresponds to the number of cells, average of 10–15 spines per cell; repeated-measures ANOVA with time as the innersubject factor and mApple versus PFN1 or PFN2a as intersubject factor; for F and P values see Fig. 2G | |||
| Fig. 3 C and D | CA3 ctrl | CA3 shPFN2a | CA3 shPFN1 | |
| St. fr. 0.23 ± 0.02 n = 16 | St. fr. 0.15 ± 0.02 n = 20 | St. fr. 0.43 ± 0.058 n = 18 | ||
| t, time 22.40 ± 2 n = 16 | t, time 16.53 ± 1.3 n = 20 | t, time 44.23 ± 8.95 n = 18 | ||
| P | n = spines | Ctrl: st. fr. P = 0.008; t, time P = 0.031 | Ctrl: st. fr. P = 0.008; t, time P = 0.017 | |
| Fig. 3 E and F | CA3 ctrl | shPFN2a | CA3 ctrl | shPFN2a |
| 0.23 ± 0.02 n = 16 | 0.15 ± 0.02 n = 20 | 22.63 ± 2.1 n = 15 | 16.54 ± 1.3 n = 20 | |
| 0.1 ± 0.05 n = 10 | 0.05 ± 0.04 n = 11 | 11.75 ± 1.2 n = 10 | 14.21 ± 1.9 n = 11 | |
| 0.3 ± 0.03 n = 16 | 0.08 ± 0.04 n = 14 | 31.94 ± 3.5 n = 14 | 15.33 ± 1.9 n = 14 | |
| P | ANOVA (post hoc Bonferroni) | Ctrl: 0–30 min P = 0.003; 75 min P = 0.006 | Ctrl: 0–30 min P = 0.026; 30–60 min P < 0.001; 0–75 min P = 0.04 | |
| Fig. 4B | PFN1 | PFN2a | ||
| 0.6254 ± 0.09 n = 3 | 1.0 ± 0.04 n = 3 | |||
| P | Littermate controls P = 0.021 | |||
| Fig. 4C | PFN1 | PFN2a | ||
| 0.37 ± 0.059 n = 3 | 1.23 ± 0.12 n = 3 | |||
| P | Littermate controls P = 0.046, hippocampi of three pups P0 were pooled for one Western blot sample | |||
Unless otherwise specified, means were compared using an unpaired Student´s t test.
SI Materials and Methods
Mice.
All procedures concerning animals were approved by the animal welfare representative of the Technische Universität Braunschweig and the LAVES [Oldenburg, Germany, Az. §4 (02.05) TSchB TU BS]. Unless otherwise specified, C57bl/6 animals were used for slice culture preparation and Western blot analysis of profilin levels. Fmr1 KO mice in an FVB background were obtained from the Jackson Laboratory (Strain 004624).
Genotyping.
Genomic DNA was obtained from tail biopsy and genotyping was performed as described previously (36). The following primer sequences were used: mutant allele CAC GAG ACT AGT GAG ACG TG, WT allele TGT GAT AGA ATA TGC AGC ATG TGA, common CTT CTG GCA CCT CCA GCT T.
Cell Culture.
Organotypic slice cultures and primary embryonic hippocampal cultures were prepared as previously described (16, 37). Primary embryonic hippocampal cultures were plated at a density of 70 × 104 on 12-mm poly-l-lysine–coated coverslips in Neurobasal medium containing 400 µM Glutamine, 2% (vol/vol) N2 supplement, and 2% (vol/vol) B27 (all from Invitrogen).
Transfection.
Hippocampal slice cultures were transfected at the respective ages using the Helios Gene gun method (Bio-Rad), as previously described (16), or single-cell electroporation (38). Primary embryonic hippocampal neurons were transfected to express recombinant PFN2a or PFN1, as well as eGFP-F for morphological analysis using Lipofectamine2000 (Invitrogen) according to the manufacturer’s instructions and to a previously published protocol (16).
Immunhistochemistry and Western Blotting.
Immunhistochemistry on primary hippocampal neurons was performed as previously described (16) using the following antibodies: anit-PFN1 (rabbit, 1:1,000; Catalog no. P7624, Sigma Aldrich), cofilin and phospho-cofilin (rabbit, 1:5,000; Abcam). Western botting was performed on PVDF-membranes using the following antibodies: anit-PFN1 (rabbit, 1:5,000; Catalog No. P7624, Sigma Aldrich), anti-PFN2a (rabbit, 1:20,000; produced by Bioscience Göttingen), anti-GAPDH (rabbit, 1:9,000; Catalog No. AP21839 Acris), and anti-Tubulin DM1a (mouse, 1:10.000; Abcam).
FRAP Experiments.
Actin-GFP at single dendritic spines was bleached using the 405 laser line at a power of 2.3–3 mW (approx. 30%) for 30 ms. Neighboring spines were monitored to ensure that only the spine of interest was bleached. The SIM scanner unit of the Olympus Fluoview1000 set-up allowed simultaneous bleaching at 405 and imaging at 488 nm. For subsequent analysis of actin dynamics, a time-lapse series was acquired with an imaging interval of 2 s for a total imaging time of 120 s. At this time point, a plateau in the fluorescence recovery was reached. The FRAP time series were analyzed using ImageJ software. The background corrected mean intensity values for each time point were calculated per spine and plotted against the time. The fluorescent intensity of each time point was normalized to the average value derived from five prebleaching images (relative fluorescence). Nonlinear curve fitting was performed in GraphPad Prism where the net recovery after photobleaching is provided by the following equation: Y = Y0 + (Plateau − Y0) × (1 − exp(−K × x)), where Y0 is the Y value when time is zero directly after the bleaching impulse, Plateau is the Y value at infinite times, expressed as a fraction of the fluorescence before bleaching and was used to determine the dynamic actin pool (F-actin dynamic). The stable pool (F-actin stable) is the fraction of fluorescence that does not recover within the imaging period of 2 min calculated as 1 − (F-actin dynamic), K is the rate constant, and τ is the time constant, expressed in seconds; it is computed as the reciprocal of K. From this equation, the actin turnover rate was calculated as the time at 50% recovery of prebleaching fluorescence levels.
Electrophysiology.
Cultures were transferred to a recording chamber at 32 °C and bathed continuously with carbogenated ACSF, as described previously (38). Experiments were performed in the presence of 10 μM APV and 0.5 μM TTX. Patch pipettes (tip resistance 5–8 MΩ) contained 126 mM K-gluconate, 4 mM KCl, 4 mM Mg-ATP, 0.3 mM Na-GTP, 10 mM PO-Creatine, 10 mM Hepes (pH = 7.3 with KOH, 290 mOsm with sucrose). Hippocampal neurons were recorded at a holding potential of −60 mV. Signals were amplified with MultiClamp 700B (Axon Instruments) and recorded with pClamp9 (Axon Instruments). Mini Analysis program (Synaptosoft) was used for off-line analysis of recordings.
Acknowledgments
We thank Diane Mundil and Reinhard Huwe for excellent technical assistance, and Marta Zagrebelsky for helpful comments on an earlier version of the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grant KO1674/8-1 (to K.M.-P. and M.K.) and by Grant N-RENNT, Ministry of Science and Culture of Lower Saxony (to G.G. and M.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516697113/-/DCSupplemental.
References
- 1.Fischer M, Kaech S, Knutti D, Matus A. Rapid actin-based plasticity in dendritic spines. Neuron. 1998;20(5):847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- 2.Chen LY, Rex CS, Casale MS, Gall CM, Lynch G. Changes in synaptic morphology accompany actin signaling during LTP. J Neurosci. 2007;27(20):5363–5372. doi: 10.1523/JNEUROSCI.0164-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukazawa Y, et al. Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron. 2003;38(3):447–460. doi: 10.1016/s0896-6273(03)00206-x. [DOI] [PubMed] [Google Scholar]
- 4.Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GC, Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57(5):719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Hotulainen P, et al. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol. 2009;185(2):323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch M, et al. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron. 2014;82(2):444–459. doi: 10.1016/j.neuron.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Star EN, Kwiatkowski DJ, Murthy VN. Rapid turnover of actin in dendritic spines and its regulation by activity. Nat Neurosci. 2002;5(3):239–246. doi: 10.1038/nn811. [DOI] [PubMed] [Google Scholar]
- 8.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14(3):285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanpied TA, Ehlers MD. Microanatomy of dendritic spines: Emerging principles of synaptic pathology in psychiatric and neurological disease. Biol Psychiatry. 2004;55(12):1121–1127. doi: 10.1016/j.biopsych.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Jockusch BM, Murk K, Rothkegel M. The profile of profilins. Rev Physiol Biochem Pharmacol. 2007;159:131–149. doi: 10.1007/112_2007_704. [DOI] [PubMed] [Google Scholar]
- 11.Ackermann M, Matus A. Activity-induced targeting of profilin and stabilization of dendritic spine morphology. Nat Neurosci. 2003;6(11):1194–1200. doi: 10.1038/nn1135. [DOI] [PubMed] [Google Scholar]
- 12.Neuhoff H, et al. The actin-binding protein profilin I is localized at synaptic sites in an activity-regulated manner. Eur J Neurosci. 2005;21(1):15–25. doi: 10.1111/j.1460-9568.2004.03814.x. [DOI] [PubMed] [Google Scholar]
- 13.Lamprecht R, Farb CR, Rodrigues SM, LeDoux JE. Fear conditioning drives profilin into amygdala dendritic spines. Nat Neurosci. 2006;9(4):481–483. doi: 10.1038/nn1672. [DOI] [PubMed] [Google Scholar]
- 14.Görlich A, et al. Preserved morphology and physiology of excitatory synapses in profilin1-deficient mice. PLoS One. 2012;7(1):e30068. doi: 10.1371/journal.pone.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pilo Boyl P, et al. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J. 2007;26(12):2991–3002. doi: 10.1038/sj.emboj.7601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michaelsen K, et al. Fine-tuning of neuronal architecture requires two profilin isoforms. Proc Natl Acad Sci USA. 2010;107(36):15780–15785. doi: 10.1073/pnas.1004406107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeve SP, et al. The Drosophila Fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr Biol. 2005;15(12):1156–1163. doi: 10.1016/j.cub.2005.05.050. [DOI] [PubMed] [Google Scholar]
- 18.Bhakar AL, Dolen G, Bear MF. The pathophysiology of Fragile X (and what it teaches us about synapses) Annu Rev Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Portera-Cailliau C. Which comes first in Fragile X syndrome, dendritic spine dysgenesis or defects in circuit plasticity? Neuroscientist. 2012;18(1):28–44. doi: 10.1177/1073858410395322. [DOI] [PubMed] [Google Scholar]
- 20.He CX, Portera-Cailliau C. The trouble with spines in fragile X syndrome: Density, maturity and plasticity. Neuroscience. 2013;251:120–128. doi: 10.1016/j.neuroscience.2012.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witke W, et al. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998;17(4):967–976. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiala JC, Feinberg M, Popov V, Harris KM. Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J Neurosci. 1998;18(21):8900–8911. doi: 10.1523/JNEUROSCI.18-21-08900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: Implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci. 1992;12(7):2685–2705. doi: 10.1523/JNEUROSCI.12-07-02685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zagrebelsky M, et al. The p75 neurotrophin receptor negatively modulates dendrite complexity and spine density in hippocampal neurons. J Neurosci. 2005;25(43):9989–9999. doi: 10.1523/JNEUROSCI.2492-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halpain S. Actin and the agile spine: How and why do dendritic spines dance? Trends Neurosci. 2000;23(4):141–146. doi: 10.1016/s0166-2236(00)01576-9. [DOI] [PubMed] [Google Scholar]
- 26.Fortin DA, et al. Long-term potentiation-dependent spine enlargement requires synaptic Ca2+-permeable AMPA receptors recruited by CaM-kinase I. J Neurosci. 2010;30(35):11565–11575. doi: 10.1523/JNEUROSCI.1746-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to Fragile X syndrome. Dev Cell. 2008;14(6):926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kao DI, Aldridge GM, Weiler IJ, Greenough WT. Altered mRNA transport, docking, and protein translation in neurons lacking Fragile X mental retardation protein. Proc Natl Acad Sci USA. 2010;107(35):15601–15606. doi: 10.1073/pnas.1010564107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cruz-Martín A, Crespo M, Portera-Cailliau C. Delayed stabilization of dendritic spines in Fragile X mice. J Neurosci. 2010;30(23):7793–7803. doi: 10.1523/JNEUROSCI.0577-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonçalves JT, Anstey JE, Golshani P, Portera-Cailliau C. Circuit level defects in the developing neocortex of Fragile X mice. Nat Neurosci. 2013;16(7):903–909. doi: 10.1038/nn.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkowski DJ. Profilin I is essential for cell survival and cell division in early mouse development. Proc Natl Acad Sci USA. 2001;98(7):3832–3836. doi: 10.1073/pnas.051515498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci. 2013;126(Pt 1):1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rotty JD, et al. Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev Cell. 2015;32(1):54–67. doi: 10.1016/j.devcel.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polet D, Lambrechts A, Vandepoele K, Vandekerckhove J, Ampe C. On the origin and evolution of vertebrate and viral profilins. FEBS Lett. 2007;581(2):211–217. doi: 10.1016/j.febslet.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 35.Birbach A. Profilin, a multi-modal regulator of neuronal plasticity. BioEssays. 2008;30(10):994–1002. doi: 10.1002/bies.20822. [DOI] [PubMed] [Google Scholar]
- 36.Michaelsen K, et al. Neurotrophin receptors TrkB.T1 and p75NTR cooperate in modulating both functional and structural plasticity in mature hippocampal neurons. Eur J Neurosci. 2010;32(11):1854–1865. doi: 10.1111/j.1460-9568.2010.07460.x. [DOI] [PubMed] [Google Scholar]
- 37.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5(6):545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michaelsen-Preusse K, Kellner Y, Korte M, Zagrebelsky M. Analysis of actin turnover and spine dynamics in hippocampal slice cultures. In: Bakota L, Brandt R, editors. Laser Scanning Microscopy and Quantitative Image Analysis of Neuronal Tissue. Springer; New York: 2014. pp. 189–217. [Google Scholar]