Significance
Contractility in cardiac muscle is finely tuned in response to changing circulatory demands, and phosphorylation of cardiac myosin-binding protein C (cMyBP-C) plays an important role in this regulation of function. The molecular mechanism of this regulation is not well understood because the N-terminal region [domains C0–C2 (C0C2)] containing the phosphorylatable region of cMyBP-C is highly dynamic and disordered, thus inaccessible to standard structural techniques. Therefore, we used site-directed spectroscopic probes (fluorescence and magnetic resonance) and computational simulations that are optimized for analyzing dynamic disorder. We found that phosphorylation makes C0C2 more compact and less disordered in the linker region and phosphorylation motif, and a new binding site simultaneously emerges. Our results provide insight into muscle contraction with implications for heart failure.
Keywords: muscle, protein kinase A, fluorescence resonance energy transfer, DEER, molecular dynamics simulation
Abstract
We have used the site-directed spectroscopies of time-resolved fluorescence resonance energy transfer (TR-FRET) and double electron–electron resonance (DEER), combined with complementary molecular dynamics (MD) simulations, to resolve the structure and dynamics of cardiac myosin-binding protein C (cMyBP-C), focusing on the N-terminal region. The results have implications for the role of this protein in myocardial contraction, with particular relevance to β-adrenergic signaling, heart failure, and hypertrophic cardiomyopathy. N-terminal cMyBP-C domains C0–C2 (C0C2) contain binding regions for potential interactions with both thick and thin filaments. Phosphorylation by PKA in the MyBP-C motif regulates these binding interactions. Our spectroscopic assays detect distances between pairs of site-directed probes on cMyBP-C. We engineered intramolecular pairs of labeling sites within cMyBP-C to measure, with high resolution, the distance and disorder in the protein’s flexible regions using TR-FRET and DEER. Phosphorylation reduced the level of molecular disorder and the distribution of C0C2 intramolecular distances became more compact, with probes flanking either the motif between C1 and C2 or the Pro/Ala-rich linker (PAL) between C0 and C1. Further insight was obtained from microsecond MD simulations, which revealed a large structural change in the disordered motif region in which phosphorylation unmasks the surface of a series of residues on a stable α-helix within the motif with high potential as a protein–protein interaction site. These experimental and computational findings elucidate structural transitions in the flexible and dynamic portions of cMyBP-C, providing previously unidentified molecular insight into the modulatory role of this protein in cardiac muscle contractility.
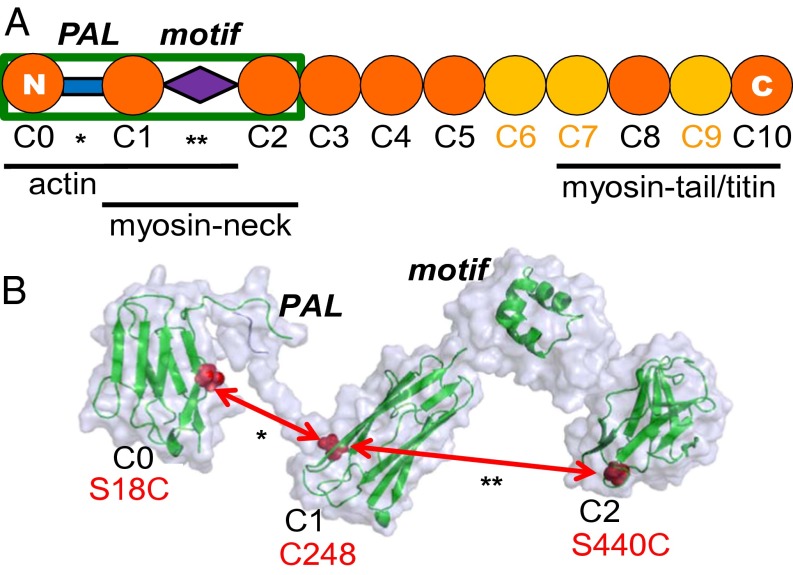
Phosphorylation of cardiac myosin-binding protein C (cMyBP-C) accelerates contraction kinetics in skinned myocardium and plays a critical role in modulating the strength and kinetics of force development and relaxation in the heart via β-adrenergic stimulus (1–3). cMyBP-C is uniquely situated in the sarcomere to influence contractility and dynamic interactions between myosin and actin, anchored at its C-terminal end to the myosin tail that forms the thick filament (4–6) and available to interact at its N-terminal end with both the myosin neck region subfragment-2 (S2) (2, 4, 7) and actin (8–11) (Fig. 1A). In vitro analyses have shown that the interactions of cMyBP-C with myosin–S2 and the regulated actin filament are subject to modulatory tuning with respect to affinity, stoichiometry, and function (12). Thus, with varied cardiovascular load or adrenergic tone, cMyBP-C regulates the probability of interaction between myosin and actin to help govern cross-bridge binding to actin and the activation of the thin filament (13). Similar to the molecular building blocks of titin (14), cMyBP-C is composed of eight Ig I and three fibronectin type III domains (Fig. 1A) and is phosphorylated by PKA, as well as by Ca2+/calmodulin-dependent kinase II and other kinases, at a cluster of three serine residues in the MyBP-C motif (also called the “M-domain”). The cardiac isoform, as compared with the two skeletal isoforms, has an additional Ig domain, C0, at its N terminus, which is linked to the neighboring C1 domain by a Pro/Ala-rich linker (PAL) (Fig. 1).
Fig. 1.
(A) Organization of cardiac MyBP-C from the N terminus to the C terminus (C0–C10). Regions proposed to interact with myofilament-binding partners are indicated below the schematic. (B) Proposed structure of C0C2 (enclosed in the green box in A) (modified from refs. 9, 16, 20, 42), showing the Ig-like domains and the intervening PAL (*) and MyBP-C motif domains (**). Site pairs used to measure interdomain distances across the PAL and motif are labeled in red.
The N-terminal region of cMyBP-C [domains C0–C2 (C0C2)] plays a crucial role in modulating contractile kinetics in a phosphorylation-dependent manner. The MyBP-C motif regulatory domain, situated between domains C1 and C2, is an intrinsically disordered motif composed of ∼100 residues, approximately the size of an Ig domain, that contains the cardiac PKA phosphorylation sites (2). Unlike the rest of this primarily disordered domain, a segment at the motif’s C-terminal end is a well-ordered triple-helix bundle (Fig. 1B) (15–17). We hypothesize that phosphorylation-dependent structural changes in this region are critical for the function of cMyBP-C.
The PAL, positioned between domains C0 and C1 (Fig. 1), contains a high content of Pro and Ala, which probably cause this segment to be flexible, extended, and capable of influencing the interactions of its flanking domains with actin and myosin. The MyBP-C motif and PAL regions within C0C2 contain apparent binding sites for myosin-S2 and actin (Fig. 1A) and play important regulatory roles, but their intrinsic structural disorder has made them largely inaccessible to conventional techniques of determining protein structure, such as crystallography and NMR.
Therefore, in the present study, we used site-directed spectroscopic and computational techniques that are capable of resolving structural transitions, even in the presence of substantial disorder, to define the phosphorylation-dependent structural transitions of the N-terminal portion of cMyBP-C, i.e., C0C2 (Fig. 1). We have shown previously that cMyBP-C alters the torsional dynamics of actin and that this restriction of the actin filament’s twisting motion varies with the phosphorylation state of cMyBP-C (18). Here, we performed time-resolved fluorescence resonance energy transfer (TR-FRET) and dipolar electron paramagnetic resonance (EPR; DEER) on purified recombinant cMyBP-C C0C2 from mouse, with pairs of engineered labeling sites (Figs. 1 and 2), to quantitate the distribution of interprobe distances sampled in this region of cMyBP-C. The protein was labeled with pairs of fluorescent dyes for FRET or spin labels for DEER. We assessed these distance distributions before and after PKA-catalyzed phosphorylation of cMyBP-C. To obtain atomistic models, we performed all-atom molecular dynamics (MD) simulations of the motif in the presence and absence of phosphorylation at each of three sites. This combination of site-directed spectroscopy and MD simulations provides previously unidentified insight into the structural dynamics of cMyBP-C, with important implications for the regulation of cardiac contractility in health and disease.
Fig. 2.
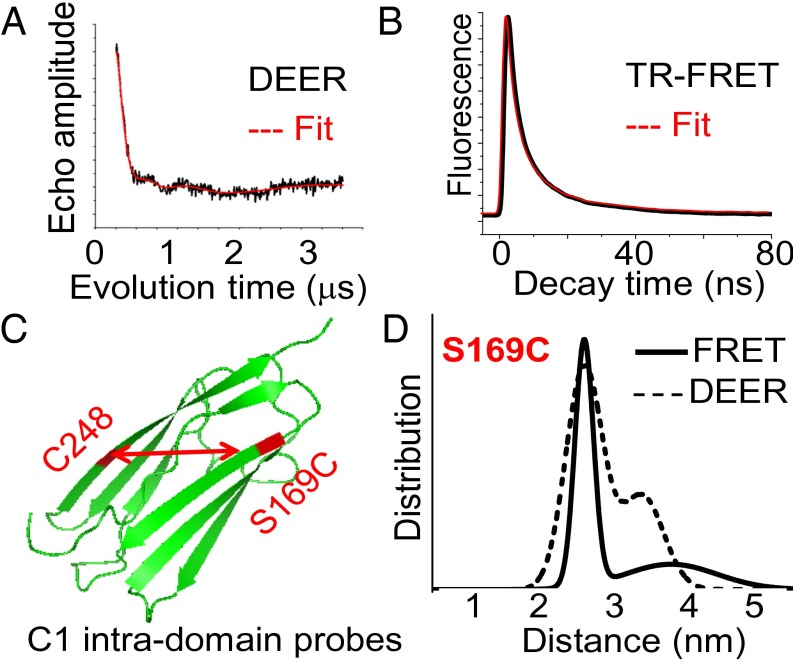
(A and B) Representative traces (black) and fit (red) of intramolecular DEER (A) and TR-FRET decays (B) with probes placed on C0C2 at S169C and C248. (C) High-resolution structure of the Ig-like C1 domain (20), showing the location of the labeling sites (red) used to measure intradomain distance controls. (D) Distance distributions from the data in A and B. The best fit was to a two-Gaussian model.
Results
Validation of Distance Measurement Methodology Using cMyBP-C.
We first aimed to validate our spectroscopic methods of measuring interprobe distances in cMyBP-C by comparing our results with data from crystal and NMR structures of the C1 domain (Fig. 2 and SI Appendix, Table S2) (19, 20). In the FRET and DEER measurements, the major peak mean distance between C1 probes was determined to be 2.55 nm, very near the 2.3-nm distance predicted from the crystal structure (19). Thus, our fluorescent and spin probes can reliably determine the molecular distances and dynamics in C0C2 (Fig. 2 and SI Appendix, Table S2). Using continuous wave EPR, we found that there are no contributions from distances shorter than the DEER detection range (<2 nm) (SI Appendix, Fig. S1 and Table S1). Our spectroscopic distance measurements within C1 reveal significant disorder (∼0.5 nm FWHM) in the distribution of distances that exists within a single Ig-domain (Fig. 2 and SI Appendix, Table S2). This disorder probably arises from varying conformations in solution, which could not be resolved by static protein structural approaches such as X-ray crystallography or electron microscopy. As expected, PKA treatment did not affect the intra-C1 domain probes in C0C2.
Allosteric Effect of Motif Phosphorylation in Altering Structural States of the PAL.
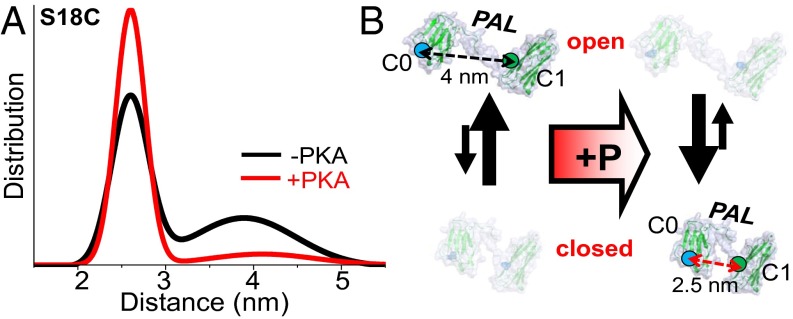
We examined the flexible portion of cMyBP-C at its linker domain, which connects C0 to C1 in the cardiac isoform of MyBP-C (and ends the skeletal isoforms that lack the C0 domain) (Fig. 1). The probe sites in the C0 and C1 domains flanking the PAL (S18C.C248 distances are shown in Fig. 3) determined by both FRET and DEER exhibit markedly increased disorder relative to the other two regions we studied here (C1 probes in Fig. 2 and the motif-flanking probes in Fig. 4, discussed below). This finding is consistent with previous structural studies showing that the C0–C1 region is extended and flexible (16, 21). In both unphosphorylated and phosphorylated C0C2, the distribution of the linker probes reveals two distances in each phosphorylation state: a short distance that is more ordered (closed conformation) and a long distance that is more disordered (open conformation). The main effect of PKA phosphorylation is to increase the mole fraction of the closed conformation. The observation that a structural change occurs in the PAL region, making it very compact upon phosphorylation of the MyBP-C motif region, suggests that allosteric changes propagate from the motif across the C1 domain to the PAL (Fig. 1). This model is consistent with in vitro observations that contacts N-terminal of the motif region (i.e., C1, the PAL, and/or C0) are capable of influencing function (10, 15, 21).
Fig. 3.
Effect of PKA phosphorylation on C0C2 linker (PAL) structure and disorder, detected by TR-FRET (shown) and DEER. (A) Distance distributions of probes flanking the PAL region (S18C–C248). (B) Cartoon of PKA effects on PAL mole fraction shift in structural states (open-to-closed transition upon phosphorylation in the linker region).
Fig. 4.
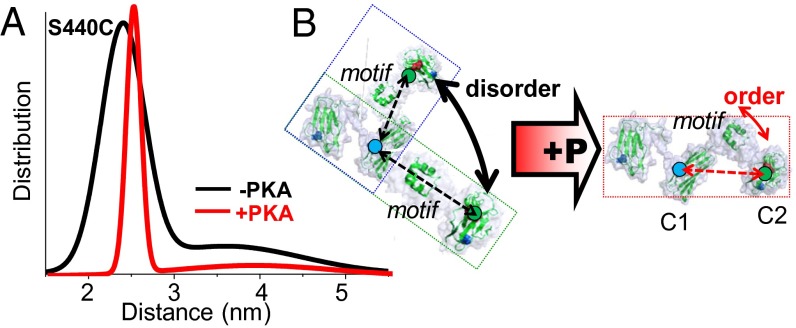
Effect of PKA phosphorylation on C0C2 motif structure and disorder, detected by TR-FRET (shown) and DEER (see SI Appendix, Fig. S2, dashed lines). (A) Distance distributions of probes flanking the cMyBP-C motif region (C248–S440C). (B) Illustration of PKA effects on motif tertiary order. Unphosphorylated C0C2 exhibits an ensemble of conformations, such as bent or extended C0C2 structures (blue and green boxes). Increased tertiary order or a molecular collapse in motif would result in sampling a much narrower distribution of dynamic conformations, depicted here as a single structure (red box).
Probes Flanking the MyBP-C Motif Reveal Phosphorylation-Dependent Changes in Disorder.
PKA treatment of N-terminal cMyBP-C with probes flanking the motif substantially affects both FRET and DEER decays for the labeled C0C2 mutant C248.S440C, indicating that the major effect of phosphorylation in the vicinity of MyBP-C motif is to reduce the level of molecular disorder, as indicated by a much narrower distance distribution (Fig. 4 and SI Appendix, Fig. S2). This finding also is consistent with the alternative interpretation that a disordered region collapses without acquiring ordered structure. Thus, the addition of negatively charged phosphates to the serines within the motif elicits a structural change resulting in a more compact distribution of distances in C0C2, indicating either less disorder or the collapse of a disordered MyBP-C motif region. This effect of phosphorylation on motif conformation probably is important for enhancing cMyBP-C’s mechanical stability in binding its myofilament interaction partners in the cardiac sarcomere. Although both the PAL and motif regions of C0C2 become compact with phosphorylation, they differ in that the PAL undergoes a population shift toward the more compact structural state (Fig. 3), whereas phosphorylation in MyBP-C motif leads to a drastic reduction in tertiary molecular disorder (Fig. 4 and SI Appendix, Fig. S5). These findings are consistent with the model that C0C2 is a very dynamic region of cMyBP-C with high potential for numerous sarcomeric binding partners. These results also are consistent with findings that the MyBP-C motif is intrinsically disordered (15, 16) and that phosphorylation elicits a structural change to a more compact tertiary protein structure (22). To confirm further the nature of the motif’s intrinsic disorder and the specific structural changes associated with the unphosphorylated and phosphorylated states, we next harnessed recent advances in MD simulations (23, 24) and applied these approaches to cMyBP-C.
MD Simulations Reveal Unique Structural Details of Regulation.
To complement our spectroscopic studies and obtain models with atomistic detail, we used microsecond MD to investigate the structural changes associated with PKA phosphorylation across the motif. We performed two sets of 2-µs-long MD simulations of the MyBp-C motif (residues His255–Lys356): unphosphorylated and phosphorylated at serines 273, 282, and 302. A complete description of the construction of the motif and MD methods is supplied in the SI Appendix, SI Text. This in silico approach has been useful in previous studies of muscle protein structural dynamics to complement studies of muscle contraction (23) and of other intrinsically disordered proteins (24). Here, we performed two independent MD simulations to determine the localized effects of cMyBP-C phosphorylation (Figs. 5 and 6).
Fig. 5.
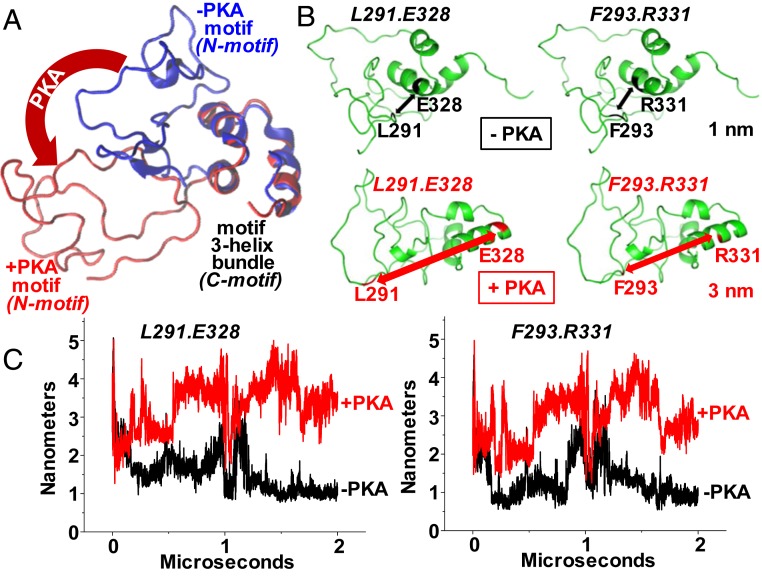
(A) Microsecond-long MD simulations were performed to determine the effect of phosphorylation on the MyBP-C motif (residues 259–353). The accessibility of the three α-helix bundles changes as the disordered region of the motif rotates away from the helices (blue to red conformation). (B) Distance measurements were simulated between three-helix bundle residues (E328 or R331) and disordered region residues (L291 or F293) with (red) and without (black) PKA phosphorylation. (C) Trajectories of the distances between these pairs with or without phosphorylation.
Fig. 6.
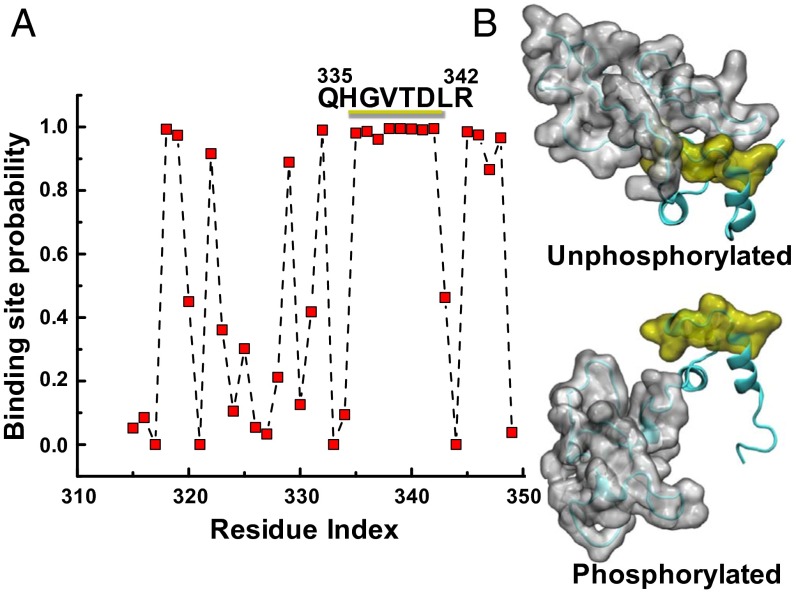
(A) A phosphorylated MyBP-C motif (residues 259–353) protein–protein binding site is predicted to be at residues 335–342 using cons-PPISP, in which a score of 0 is improbable and 1 is highly probable. See SI Appendix, SI Results and Table S3 for residue-specific values. (B) MD simulations suggest that this region is accessible for binding only when phosphorylated: A series of eight residues (yellow) is uncovered as the disordered N-motif (gray) shifts from the unphosphorylated (Upper) to the phosphorylated (Lower) conformation.
Analysis of the secondary structure revealed that the N-terminal segment of the motif (the N-motif region, His255–Glu315) is unstructured on the microsecond time scale (Figs. 5 and 6). Although a transient helix forms near the phosphorylation site Ser282, this disordered region remains unstructured upon phosphorylation (Figs. 5 and 6).
We also found that the triple α-helix bundle (the C-motif region) remains folded in the trajectories of both the unphosphorylated and phosphorylated MyBP-C motif (Figs. 5 and 6). Moreover, phosphorylation of the N terminus has no effect on the folding or structural stability of the α-helix bundle, or on nearby residues, as also depicted in time-dependent plots of the motif secondary structure (SI Appendix, Fig. S8). Despite this lack of change in the secondary structure of the motif, we observed that phosphorylation of the three PKA sites in the MyBP-C motif, which are known to impact function, induced a large rotation of the disordered N-motif region away from residues Q335–R342 in the ordered C-motif region (i.e., the triple-helix bundle). We found that the region Q335–R342 is buried by the N-terminal segment and that phosphorylation-induced rotation of the N terminus unmasks this segment (Figs. 5A and 6A). Remarkably, the region Q335–R342 is predicted to be a protein–protein interaction site by the consensus neural network method of protein–protein interaction site prediction (cons-PPISP) (25).
We next measured the changes in distance upon in silico phosphorylation by selecting two pairs of sites within the motif, with one site in each pair in the disordered N-motif region (L291 or F293) and the other site in the ordered C-motif region (E328 or R331). In both cases, these sites were in very close proximity (∼1 nm apart) when unphosphorylated, and phosphorylation induced a large structural change, with the sites separated by ∼3 nm by the rotation of the N-motif away from the C-motif (Fig. 5 B and C). These results suggest that a local unfolding or extension within the motif, promoted by phosphorylation (Fig. 5), allows the concomitant collapsing or compacting of the overall C0C2 region of cMyBP-C (Figs. 3 and 4).
Further analysis of electrostatic charge shows that the helix region that emerges in MD simulations upon phosphorylation comprises a clustered series of eight residues that are likely to be protein–protein interaction sites (Figs. 5 and 6 and SI Appendix, Table S3), and that are accessible for binding only in the phosphorylated state. cMyBP-C is likely to be highly phosphorylated in healthy mouse and human hearts as compared with failing hearts (26). Previous atomic force microscopy studies have suggested that structural changes occur within the motif upon phosphorylation (22).
Discussion
We used time-resolved FRET and DEER spectroscopies in complementary studies to resolve protein distance distributions, molecular disorder, and the mole fractions of cMyBP-C structural states across its dynamic N-terminal regions. The C0C2 portion of the cardiac accessory protein cMyBP-C is critical for modulating contractility and mediating diastolic function (27, 28). We aimed to define the relationship between phosphorylation-mediated enhancement of contractility and structural dynamics to understand better the mechanistically relevant changes that occur upon phosphorylation. This approach is ideal for resolving the details of the dynamic C0C2 regions—such as the proline/alanine-rich linker PAL or the MyBP-C motif (sometimes called the M-domain) containing the cardiac MyBP-C phosphoserines (Fig. 1)—which are important for modulating contractile function but for which defined structural information determined by other biophysical approaches is lacking. We further complemented our FRET and DEER studies of C0C2 phosphorylation with all-atom MD simulations of the MyBP-C motif and probed the structural changes in this intrinsically disordered motif upon in silico phosphorylation; this approach revealed additional mechanistic molecular detail relevant to the enhancement of cardiac muscle contractility by cMyBP-C phosphorylation.
N-Terminal cMyBP-C Becomes More Compact and Less Disordered upon Phosphorylation.
Using spectroscopy, we found that C0C2 becomes much more compact and less disordered upon PKA phosphorylation (Figs. 3 and 4), and using MD simulation we found that phosphorylation simultaneously leads to an altered binding site in the motif that is not present in unphosphorylated C0C2 (Fig. 5). Because we did not detect increased structural content (e.g., newly formed α-helices or β-sheets) within the phosphorylated motif in our MD simulations, an alternative explanation for the narrowed distance distribution in our spectroscopic studies (Fig. 4) is that cMyBP-C phosphorylation involves the collapsing of N-terminal domains as the disordered motif structure rearranges intradomain contacts upon phosphorylation. In both unphosphorylated and phosphorylated conditions, FRET and DEER indicate that cMyBP-C domains are distributed over two distinct structural states for each set of probe pairs in the motif and the PAL. These two conformations persist in each phosphorylation state, but phosphorylation changes their distribution by altering either the level of disorder or the mole fraction occupied by each distance center. Here we refer to the shorter distance as the “closed structural state” and the longer distance as the “open state.” Strikingly, phosphorylation influences the structural dynamics of both the motif and the PAL by making these N-terminal regions much more compact and less disordered. Specifically, these distance distributions are modified by phosphorylation in unique ways. In the MyBP-C motif region, the level of structural disorder is markedly decreased and/or the motif collapses, because the width of the Gaussian distribution was reduced. In the PAL, the relative populations of the two states shifted from the open structural state toward the closed conformation (Fig. 3). Intradomain spectroscopic probes showed that the C1 domain exhibits some degree of folding-related dynamics, as is typical of Ig domains in muscle and similar to those found in titin (29).
Phosphorylation Unmasks the Charged Binding Strut of the Triple-Helix Bundle.
Our MD simulations coupled with a consensus prediction program (25) suggest that phosphorylation within the motif domain uncovers a series of residues in the ordered C-motif bundle (within the MyBP-C motif) that forms a binding site for another protein. It is likely that the N-motif region shields this region from binding when cMyBP-C is unphosphorylated and that the N-motif rotates away to expose this C-motif helix region upon phosphorylation of the cMyBP-C motif. We found that the triple α-helix bundle remains folded upon phosphorylation (Fig. 5A), that is to say, we did not detect evidence of helix bundle destabilization or rearrangement (SI Appendix, Fig. S8), in contrast to a previously proposed hypothetical model (17). As our spectroscopic results suggested, we found that when this predicted protein–protein interaction site (the α-helical binding strut) is uncovered upon phosphorylation (Fig. 5), the overall order (stability) of C0C2, which includes the motif, is increased (Fig. 4). This increased stability could enhance the probability of binding this newly exposed region of the motif. It remains unknown which myofilament-binding interactions would be present during either low or high phosphorylation and how the altered binding modifies the contractile function of the intact sarcomere. However, this region certainly would be available for binding interactions in phosphorylated cMyBP-C. Thus, this single α-helical binding strut is especially relevant to both physiological and pathophysiological states, because the level of phosphorylation is typically high in healthy donor hearts and low in failing myocardium (30). Further, how [Ca2+]in would affect MyBP-C motif structural dynamics is not known, because PKA influences contractility at submaximal but not maximal levels of Ca2+ activation (31).
Allosteric and Propagating Effects of cMyBP-C Phosphorylation on Actomyosin.
How does motif phosphorylation influence contractility and alter N-terminal cMyBP-C binding with its myofilament binding partners in the cardiac sarcomere? The effects of phosphorylation on all three probe sets within MyBP-C’s N-terminal domains on cMyBP-C’s structural dynamics could have profound physiological and pathological significance. Our major finding is that PKA phosphorylation leads to a more compact and ordered cMyBP-C N terminus with robust structural changes in both the motif and PAL regions. The changes observed—the reduced level of disorder in MyBP-C motif and the allosteric change in the mole fraction favoring the closed structural state of the PAL upon phosphorylation—may be crucial for optimal function of the myosin–actin interaction in the sarcomere. Whether these transitions occur in myocardium has not been determined, but such an allosteric change in the PAL upon phosphorylation of the motif linker could allow the propagation of alterations in binding interactions of the full-length cMyBP-C molecule or could amplify the influence of neighboring domains, such as the motif region, in cMyBP-C’s regulation of cardiac contractility. It is even possible that the C0 domain interacts directly with the motif under certain conditions, such as after phosphorylation, although this idea has not yet been tested. The necessity of the C0 domain extending beyond the linker only in cardiac muscle also remains unclear, but these results suggest it may be important in phosphorylation, given the allosteric structural change. It has been shown previously that the C0C2 end of cMyBP-C interacts with actin and myosin S2, with similar changes detected in Bmax and Kd, respectively (2, 10). These changes in dynamics and exposed binding sites probably play a significant role in the mechanism by which cMyBP-C modulates cardiac contractility via modified myofilament interactions. We conclude that changes in the dynamic motif and PAL of C0C2 are important in modulating contractility, conceivably by switching the accessibility of exposed binding sites and the level of protein disorder and dynamics to alter cMyBP-C binding and/or binding partners in the myofilament.
Tuning Contractility via cMyBP-C Phosphorylation: From Residues to Myocardium.
What is the specific change in motif structure that occurs with the addition of phosphate groups to cMyBP-C’s PKA sites? Our structural measurements suggest that eight residues on the α-helix of the triple-helix bundle that are buried when the MyBP-C motif is unphosphorylated become exposed, and overall motif dynamics become more stable, upon phosphorylation. Because cMyBP-C is likely to be highly phosphorylated under resting physiological conditions in healthy humans and studied mouse models of human heart disease, this uncovered helix may be an important binding region critical to normal cardiac function (26, 28, 32, 33) and would certainly be the dominant cMyBP-C site during β-adrenergic stimulation with high phosphorylation levels, as occurs with the fight-or-flight acute stress response. Furthermore, because cMyBP-C phosphorylation decreases in patients with heart failure and hypertrophic cardiomyopathy (30, 32, 34), the loss of this exposed binding site could have markedly detrimental effects on the contractility response in failing hearts.
Conclusion
We used TR-FRET, DEER, and all-atom MD simulations to characterize the structural dynamics of the flexible and disordered regions of the cMyBP-C N-terminal fragment C0C2 and assayed the structural changes associated with PKA phosphorylation. We detected structural changes in the probability distribution of conformations sampled by cMyBP-C with phosphorylation via probes placed on the C1 and C2 domains to flank the MyBP-C motif region or probes placed on the C0 and C1 domains to flank the flexible linker. The effect on FWHM to reduce disorder and increase compactness in the motif was expected. The allosteric structural change in which the mole fraction of conformations across the PAL linker region become more compact upon phosphorylation of the motif was a more surprising result. Phosphorylation of MyBP-C produces states of cMyBP-C dynamics that are likely to impact myosin cross-bridge kinetics, allowing the tuning of the sarcomere’s response to myocardial inotropy and other physiological factors, primarily through compaction of the N terminus and exposure of phosphorylation-specific binding sites. With this knowledge of the tertiary structural dynamics, we further investigated the changes in secondary structure associated with phosphorylation. The MD simulations of motif led us to the model in which cMyBP-C phosphorylation tunes cardiac muscle contractility by altering the N-terminal surfaces available for binding in the sarcomere. Our combined results reveal that phosphorylation induces specific structural changes, leading to a well-ordered and compact cMyBP-C N terminus, including the exposure of a region of the motif that is likely to be ideal for myofilament interactions.
Methods
Methods for protein preparations, labeling, thiophosphorylation, mass spectrometry, and detailed descriptions of spectroscopic waveform analyses and modeling are given in SI Appendix, SI Text.
TR-FRET.
The protein was labeled with IAEDANS (1,5-IAEDANS, 5-[([(2-iodoacetyl)amino]ethyl)amino]naphthalene-1-sulfonic acid; molecular probes) and/or FMal (fluorescein-5-maleimide; molecular probes) for TR-FRET experiments. IAEDANS-FMal C0C2 donor-only and donor-acceptor FRET samples were excited using a passively Q-switched microchip YAG laser (NanoUV-532; JDS Uniphase) at 355 nm with a pulse repetition frequency of 10 kHz. The high-energy (1 μJ per pulse) and narrow (∼1 ns FWHM) laser pulses are highly uniform in shape and intensity. Emitted photons were passed through a polarizer set at a magic angle of 54.7°, followed by an interference band-pass filter (Semrock 600/15 nm and 470/20 nm), and detected with a photomultiplier tube module (H5774-20; Hamamatsu) and digitizer (Acqiris DC252; time resolution, 0.125 ns). All FRET experiments were performed at 25 °C in cMyBP-C buffer (SI Appendix, SI Text) with 1 μM labeled C0C2 with or without PKA treatment. TR-FRET waveforms were analyzed globally as described previously (35) and as described in SI Appendix, SI Methods. Data were best-fit to three exponentials (SI Appendix, Fig. S3) and a two-Gaussian distance distribution (SI Appendix, Fig. S4) characterized by a center distance and a FWHM (SI Appendix, Fig. S5).
EPR Spectroscopy.
We performed DEER on C0C2 samples doubly labeled with Maleimide spin label [MSL: N-(1-oxyl-2,2,5,5-tetramethyl pyrrolidinyl)maleimide; Toronto Research Chemicals]. Samples were prepared for spectroscopy by dialyzing 150 μM spin-labeled C0C2 into cMyBP-C buffer with or without PKA treatment, followed by the addition of 10% (vol/vol) glycerol for cryoprotection. Samples were loaded into quartz capillaries (1.1-mm i.d., 1.6-mm o.d., 20-μL sample volume) (Wilmad Lab Glass), flash-frozen in liquid nitrogen, and stored at −80 °C until use. A Bruker E580 spectrometer was used operating at Q-band (34 GHz) with an EN5107 resonator, using a four-pulse DEER protocol (36). The π/2 pulse width was 12 ns, and the electron double resonance (ELDOR) pulse width was 24 ns. The ELDOR frequency (pump position) was assigned to the maximum of the nitroxide absorption spectrum, with the observe position (chosen by selection of the static magnetic field) being placed at a 24 Gauss higher magnetic field strength on the field swept absorption spectrum. Temperature was maintained at 65 K during acquisition, which lasted 4–24 h. The background-corrected DEER decay was analyzed using the Tikhonov regularization method provided in the software DeerAnalysis2013.2 (37) to determine the distribution of distances encoded in the spectrum, followed by Gaussian analysis of the distribution assuming a model of discrete conformational states (38). In all cases the dominant distance distributions reported by Tikhonov regularization were well fit by two Gaussians (SI Appendix, Figs. S6 and S7).
MD Simulations.
A starting structure of the M-domain of mouse cMyBP-C, corresponding to residues 259–353 with four additional residues from C1 and C2 on each end of the motif to simulate all-atomic trajectories for residues 255–357 (SI Appendix, Fig. S8), was generated previously using NMR (16). The structure of the MyBP-C fragment was solvated using TIP3P water molecules with a minimum margin of 2 nm between the protein and the edges of the periodic box. Na+ and Cl− ions were added to the system to neutralize the charge of the system and to produce an ion concentration of ∼150 mM. The details of the preparation of the motif structure for modeling are given in SI Appendix, SI Text. We performed MD simulations using the program NAMD 2.9 (39). Periodic boundary conditions (40), particle mesh Ewald summation (41), and a nonbonded cutoff of 0.9 nm and a 2-fs time step were used. The isothermal–isobaric ensemble was maintained with a Langevin thermostat (310 K) and a Langevin piston barostat (1 atm). The system was first subjected to energy minimization for 1,000 steps, followed by a warming up period of 200 ps. This procedure was followed by equilibration for 10 ns with backbone atoms harmonically restrained using a force constant of 100.0 kcal⋅mol−1⋅nm−2. Unrestrained production MD simulations of the unphosphorylated and phosphorylated MyBP-C motif were continued for 2 μs each. Binding-site predictions (Fig. 6 and SI Appendix, Table S3) were based on a computerized cons-PPISP (25).
Supplementary Material
Acknowledgments
Experiments reported in this paper were performed at the Biophysical Technology Center, University of Minnesota Department of Biochemistry, Molecular Biology, and Biophysics. Norma Jimenez Ramirez and Benjamin Zeman provided technical support. Richard Moss (University of Wisconsin, Madison) provided mouse C0C2 cDNA. This study was supported by NIH Grant AR032961 (to D.D.T.), American Heart Association (AHA) Grant 12SDG12060656 (to L.M.E.-F.), and AHA Postdoctoral Fellowship 13POST17250009 and NIH Grant R00 HL122397 (to B.A.C.). This project made use of the outstanding high-performance computing facilities at the Minnesota Supercomputing Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 3133.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521281113/-/DCSupplemental.
References
- 1.Hartzell HC. Phosphorylation of C-protein in intact amphibian cardiac muscle. Correlation between 32P incorporation and twitch relaxation. J Gen Physiol. 1984;83(4):563–588. doi: 10.1085/jgp.83.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: A modulator of cardiac contraction? EMBO J. 1995;14(9):1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moss RL, Fitzsimons DP, Ralphe JC. Cardiac MyBP-C regulates the rate and force of contraction in mammalian myocardium. Circ Res. 2015;116(1):183–192. doi: 10.1161/CIRCRESAHA.116.300561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moos C, Offer G, Starr R, Bennett P. Interaction of C-protein with myosin, myosin rod and light meromyosin. J Mol Biol. 1975;97(1):1–9. doi: 10.1016/s0022-2836(75)80017-9. [DOI] [PubMed] [Google Scholar]
- 5.Weisberg A, Winegrad S. Alteration of myosin cross bridges by phosphorylation of myosin-binding protein C in cardiac muscle. Proc Natl Acad Sci USA. 1996;93(17):8999–9003. doi: 10.1073/pnas.93.17.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci USA. 2008;105(7):2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunst G, et al. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circ Res. 2000;86(1):51–58. doi: 10.1161/01.res.86.1.51. [DOI] [PubMed] [Google Scholar]
- 8.Moos C, Mason CM, Besterman JM, Feng IN, Dubin JH. The binding of skeletal muscle C-protein to F-actin, and its relation to the interaction of actin with myosin subfragment-1. J Mol Biol. 1978;124(4):571–586. doi: 10.1016/0022-2836(78)90172-9. [DOI] [PubMed] [Google Scholar]
- 9.Jeffries CM, Whitten AE, Harris SP, Trewhella J. Small-angle X-ray scattering reveals the N-terminal domain organization of cardiac myosin binding protein C. J Mol Biol. 2008;377(4):1186–1199. doi: 10.1016/j.jmb.2008.01.080. [DOI] [PubMed] [Google Scholar]
- 10.Shaffer JF, Kensler RW, Harris SP. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J Biol Chem. 2009;284(18):12318–12327. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mun JY, et al. Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc Natl Acad Sci USA. 2014;111(6):2170–2175. doi: 10.1073/pnas.1316001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfuhl M, Gautel M. Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): Who does what, with what, and to whom? J Muscle Res Cell Motil. 2012;33(1):83–94. doi: 10.1007/s10974-012-9291-z. [DOI] [PubMed] [Google Scholar]
- 13.Stelzer JE, Patel JR, Walker JW, Moss RL. Differential roles of cardiac myosin-binding protein C and cardiac troponin I in the myofibrillar force responses to protein kinase A phosphorylation. Circ Res. 2007;101(5):503–511. doi: 10.1161/CIRCRESAHA.107.153650. [DOI] [PubMed] [Google Scholar]
- 14.Tskhovrebova L, Trinick J. Properties of titin immunoglobulin and fibronectin-3 domains. J Biol Chem. 2004;279(45):46351–46354. doi: 10.1074/jbc.R400023200. [DOI] [PubMed] [Google Scholar]
- 15.Lu Y, Kwan AH, Jeffries CM, Guss JM, Trewhella J. The motif of human cardiac myosin-binding protein C is required for its Ca2+-dependent interaction with calmodulin. J Biol Chem. 2012;287(37):31596–31607. doi: 10.1074/jbc.M112.383299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howarth JW, Ramisetti S, Nolan K, Sadayappan S, Rosevear PR. Structural insight into unique cardiac myosin-binding protein-C motif: A partially folded domain. J Biol Chem. 2012;287(11):8254–8262. doi: 10.1074/jbc.M111.309591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bezold KL, Shaffer JF, Khosa JK, Hoye ER, Harris SP. A gain-of-function mutation in the M-domain of cardiac myosin-binding protein-C increases binding to actin. J Biol Chem. 2013;288(30):21496–21505. doi: 10.1074/jbc.M113.474346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colson BA, Rybakova IN, Prochniewicz E, Moss RL, Thomas DD. Cardiac myosin binding protein-C restricts intrafilament torsional dynamics of actin in a phosphorylation-dependent manner. Proc Natl Acad Sci USA. 2012;109(50):20437–20442. doi: 10.1073/pnas.1213027109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher SJ, et al. An investigation into the protonation states of the C1 domain of cardiac myosin-binding protein C. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 6):658–664. doi: 10.1107/S0907444908008792. [DOI] [PubMed] [Google Scholar]
- 20.Ababou A, et al. Myosin binding protein C positioned to play a key role in regulation of muscle contraction: Structure and interactions of domain C1. J Mol Biol. 2008;384(3):615–630. doi: 10.1016/j.jmb.2008.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Y, Kwan AH, Trewhella J, Jeffries CM. The C0C1 fragment of human cardiac myosin binding protein C has common binding determinants for both actin and myosin. J Mol Biol. 2011;413(5):908–913. doi: 10.1016/j.jmb.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 22.Michalek AJ, et al. Phosphorylation modulates the mechanical stability of the cardiac myosin-binding protein C motif. Biophys J. 2013;104(2):442–452. doi: 10.1016/j.bpj.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Espinoza-Fonseca LM, Colson BA, Thomas DD. Effects of pseudophosphorylation mutants on the structural dynamics of smooth muscle myosin regulatory light chain. Mol Biosyst. 2014;10(10):2693–2698. doi: 10.1039/c4mb00364k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espinoza-Fonseca LM, Kelekar A. High-resolution structural characterization of Noxa, an intrinsically disordered protein, by microsecond molecular dynamics simulations. Mol Biosyst. 2015;11(7):1850–1856. doi: 10.1039/c5mb00170f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Zhou HX. Prediction of interface residues in protein-protein complexes by a consensus neural network method: Test against NMR data. Proteins. 2005;61(1):21–35. doi: 10.1002/prot.20514. [DOI] [PubMed] [Google Scholar]
- 26.Copeland O, et al. Analysis of cardiac myosin binding protein-C phosphorylation in human heart muscle. J Mol Cell Cardiol. 2010;49(6):1003–1011. doi: 10.1016/j.yjmcc.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Colson BA, et al. Myosin binding protein-C phosphorylation is the principal mediator of protein kinase A effects on thick filament structure in myocardium. J Mol Cell Cardiol. 2012;53(5):609–616. doi: 10.1016/j.yjmcc.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosas PC, et al. Phosphorylation of cardiac Myosin-binding protein-C is a critical mediator of diastolic function. Circ Heart Fail. 2015;8(3):582–594. doi: 10.1161/CIRCHEARTFAILURE.114.001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alegre-Cebollada J, et al. S-glutathionylation of cryptic cysteines enhances titin elasticity by blocking protein folding. Cell. 2014;156(6):1235–1246. doi: 10.1016/j.cell.2014.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Armouche A, et al. Decreased phosphorylation levels of cardiac myosin-binding protein-C in human and experimental heart failure. J Mol Cell Cardiol. 2007;43(2):223–229. doi: 10.1016/j.yjmcc.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Colson BA, et al. Protein kinase A-mediated phosphorylation of cMyBP-C increases proximity of myosin heads to actin in resting myocardium. Circ Res. 2008;103(3):244–251. doi: 10.1161/CIRCRESAHA.108.178996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kooij V, Holewinski RJ, Murphy AM, Van Eyk JE. Characterization of the cardiac myosin binding protein-C phosphoproteome in healthy and failing human hearts. J Mol Cell Cardiol. 2013;60:116–120. doi: 10.1016/j.yjmcc.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong CW, Nair NA, Doersch KM, Liu Y, Rosas PC. Cardiac myosin-binding protein-C is a critical mediator of diastolic function. Pflugers Arch. 2014;466(3):451–457. doi: 10.1007/s00424-014-1442-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barefield D, Sadayappan S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J Mol Cell Cardiol. 2010;48(5):866–875. doi: 10.1016/j.yjmcc.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, James ZM, Dong X, Karim CB, Thomas DD. Structural and functional dynamics of an integral membrane protein complex modulated by lipid headgroup charge. J Mol Biol. 2012;418(5):379–389. doi: 10.1016/j.jmb.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 37.Jeschke G, Koch A, Jonas U, Godt A. Direct conversion of EPR dipolar time evolution data to distance distributions. J Magn Reson. 2002;155(1):72–82. doi: 10.1006/jmre.2001.2498. [DOI] [PubMed] [Google Scholar]
- 38.Blackburn ME, Veloro AM, Fanucci GE. Monitoring inhibitor-induced conformational population shifts in HIV-1 protease by pulsed EPR spectroscopy. Biochemistry. 2009;48(37):8765–8767. doi: 10.1021/bi901201q. [DOI] [PubMed] [Google Scholar]
- 39.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber W, Hünenberger PH, McCammon JA. Molecular dynamics simulations of a polyalanine octapeptide under Ewald Boundary conditions: Influence of artificial periodicity on peptide conformation. J Phys Chem B. 2000;104(15):3668–3675. [Google Scholar]
- 41.Essmann U, Perera L, Berkowitz ML. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 42.Ababou A, Gautel M, Pfuhl M. Dissecting the N-terminal myosin binding site of human cardiac myosin-binding protein C. Structure and myosin binding of domain C2. J Biol Chem. 2007;282(12):9204–9215. doi: 10.1074/jbc.M610899200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.