Significance
Mutations in cardiac myosin-binding protein C (cMyBP-C) are the leading cause of inherited hypertrophic cardiomyopathy, demonstrating the key role that cMyBP-C plays in the heart’s contractile machinery. Since its discovery >40 years ago, cardiovascular biologists have wondered how cMyBP C’s structure regulates its function. Using atomic force spectroscopy, electron microscopy, in vitro model systems of contractility, and mutant protein expression, we show how phosphorylation affects cMyBP-C’s structure to tune its functions within the contractile apparatus. We also identify a novel molecular mechanism by which calcium ions that trigger each muscle contraction override phosphorylation’s structural and functional impact. This intersection of posttranslational modification and calcium signaling may occur in other biological systems that rely on these pathways to control cellular processes.
Keywords: muscle activation, muscle regulation, cMyBP-C, structure-function
Abstract
During each heartbeat, cardiac contractility results from calcium-activated sliding of actin thin filaments toward the centers of myosin thick filaments to shorten cellular length. Cardiac myosin-binding protein C (cMyBP-C) is a component of the thick filament that appears to tune these mechanochemical interactions by its N-terminal domains transiently interacting with actin and/or the myosin S2 domain, sensitizing thin filaments to calcium and governing maximal sliding velocity. Both functional mechanisms are potentially further tunable by phosphorylation of an intrinsically disordered, extensible region of cMyBP-C’s N terminus, the M-domain. Using atomic force spectroscopy, electron microscopy, and mutant protein expression, we demonstrate that phosphorylation reduced the M-domain’s extensibility and shifted the conformation of the N-terminal domain from an extended structure to a compact configuration. In combination with motility assay data, these structural effects of M-domain phosphorylation suggest a mechanism for diminishing the functional potency of individual cMyBP-C molecules. Interestingly, we found that calcium levels necessary to maximally activate the thin filament mitigated the structural effects of phosphorylation by increasing M-domain extensibility and shifting the phosphorylated N-terminal fragments back to the extended state, as if unphosphorylated. Functionally, the addition of calcium to the motility assays ablated the impact of phosphorylation on maximal sliding velocities, fully restoring cMyBP-C’s inhibitory capacity. We conclude that M-domain phosphorylation may have its greatest effect on tuning cMyBP-C’s calcium-sensitization of thin filaments at the low calcium levels between contractions. Importantly, calcium levels at the peak of contraction would allow cMyBP-C to remain a potent contractile modulator, regardless of cMyBP-C’s phosphorylation state.
Cardiac contractility results from the calcium-dependent sliding of actin-based thin filaments toward the centers of myosin-based thick filaments to shorten the overall length of the sarcomere, the heart’s elementary contractile unit. Filament sliding is turned on and off on a beat-to-beat basis by calcium’s binding to, and release from, the troponin–tropomyosin regulatory complex on the thin filament. These mechanochemical regulatory processes are fine-tuned by proteins within the thick filament, including cardiac myosin-binding protein C (cMyBP-C). Although cMyBP-C is not essential for cardiac contractility, its importance in contractile function is evidenced by mutations in the MYBPC3 gene being a leading cause of inherited hypertrophic cardiomyopathy (1, 2). Because of the prevalence of this disease (affecting 1 in 500 people) and the potential for therapeutic intervention, much work during the last two decades has focused on defining cMyBP-C’s structure and function within the sarcomere (3).
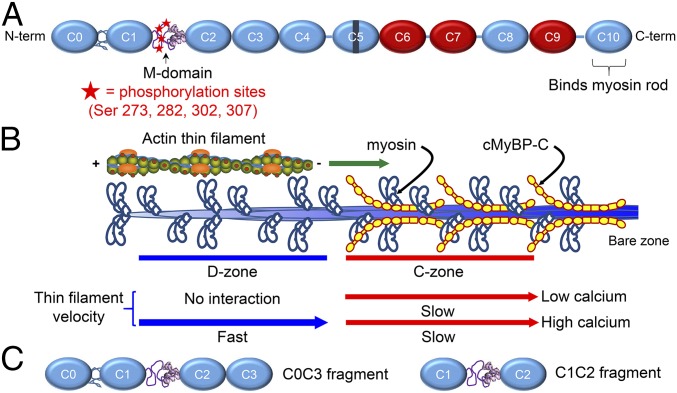
cMyBP-C has an elongated modular structure comprising 11 Ig and fibronectin type III (Fn3) domains, numbered C0–C10 from the N terminus (Fig. 1A) (4, 5). The C-terminal domain (C10) is tightly bound to the thick filament backbone, and the N-terminal domains extend radially from the thick filament (6, 7). Thus cMyBP-C’s N-terminal domains are positioned to bind to neighboring actin filaments and/or the myosin S2 domain to modulate actomyosin activity. However, consensus regarding the actual binding sites and partner(s) in vivo has not yet been reached (8). The N-terminal domains contain long polypeptide linkers between the C0–C1 and C1–C2 domains, the latter termed the “M-domain” or “motif” (Fig. 1A). The M-domain contains four highly conserved serines (S273, S282, S302, and S307; mouse sequence) in an intrinsically disordered region of its N-terminal half and a three-helix bundle (residues 317–351) in its C-terminal region (Fig. 1A) (9, 10). β-Adrenergic–stimulated phosphorylation of these serines is believed to enhance cardiac contractility (11, 12), and a high level of phosphorylation appears to be critical to normal cardiac function, whereas dephosphorylation has been associated with heart failure (13–15). Although the mechanistic role of cMyBP-C phosphorylation in vivo remains unclear, it is known to reduce the extensibility of the M-domain (10, 16), diminish the binding of cMyBP-C to actin (17, 18) and to myosin S2 (19), and to tune cMyBP-C’s ability to modulate actomyosin activity in vitro (20–24).
Fig. 1.
Structure and function of cMyBP-C. (A) Schematic diagram of full-length cMyBP-C. Domains C1 and C2 are connected by the M-domain, containing an intrinsically disordered N-terminal region with four phosphorylatable serines, and a more structured C-terminal half. Ig-like domains are shown in blue, and Fn-like domains are shown in red. (B) Illustration of half of a native thick filament with a native thin filament landing on the tip of the thick filament and being translocated through the D- and C-zones at the different speeds indicated, as observed in the TIRFM experiments. (C) Wild-type C0C3 and C1C2 N-terminal fragments used in motility, AFM, and EM assays. Phosphomimetic counterparts to each fragment were expressed containing aspartic acid substitutions for the four serines highlighted in A.
To gain insight into the function of cMyBP-C in its native structural environment, we recently developed a total internal reflectance microscopy (TIRFM)-based assay to visualize the sliding of actin-based filaments along native cardiac thick (myosin) filaments (21, 23). The actin filaments used in this assay are short 250-nm shards, which allow independent probing of thick filament areas with and without cMyBP-C (the C- and D-zones, respectively) (Fig. 1B). Using this assay, we showed that the sliding velocity of bare F-actin filaments was slowed within the C-zone of thick filaments and that the degree of slowing was reduced by phosphorylation of the four serines within the M-domain (21). Next, we examined the motion of native thin filaments containing the calcium-regulatory troponin–tropomyosin complex over native thick filaments. We found that cMyBP-C in the C-zone activated the native thin filaments at low calcium levels (Fig. 1B) and that the level of cMyBP-C–induced activation was lessened by phosphorylation (23). These findings collectively indicated that phosphorylated cMyBP-C is a less potent modulator of actomyosin interactions, as also has been demonstrated in other in vitro model systems (20, 22).
Here, using in vitro motility assays in combination with atomic force microscopy (AFM) and electron microscopy (EM), we show that phosphorylation attenuates cMyBP-C’s function by multimodal structural ordering within the M-domain, which in turn affects long-range global interactions between cMyBP-C’s N-terminal domains. These phosphorylation-dependent structural changes suggest a molecular basis for cMyBP-C’s ability to modulate cardiac contractility. In addition, we identify another novel modulatory mechanism by which calcium limits these intra- and interdomain structural interactions and the ability of cMyBP-C phosphorylation to affect function. Our findings suggest that phosphorylation and calcium can antagonistically fine-tune cMyBP-C’s modulation of cardiac contractility, so that phosphorylation plays its greatest role when calcium levels within the sarcomere are low. The mechanistic phenomena described here may apply to additional proteins of the contractile machinery that are tuned by phosphorylation and possibly to other biological systems regulated through phosphorylation and changes in intracellular calcium levels.
Results
The Effect of cMyBP-C Phosphorylation on Maximal Thin Filament Sliding Velocity Is Modulated by Calcium.
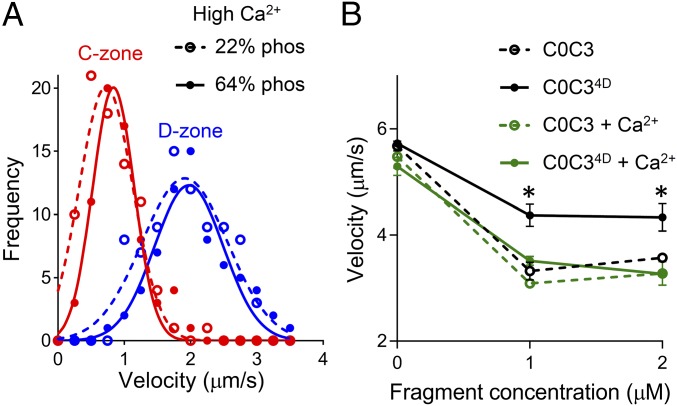
We applied our TIRFM-based assay to visualize the sliding of fluorescently labeled, troponin- and tropomyosin-containing native thin filaments along myosin thick filaments with high temporal (8.3-ms) and spatial (30-nm) resolution (Fig. 1B). The maximal sliding velocity of native thin filaments at high, saturating levels of calcium (0.01 mM free Ca2+, 100 µM ATP, 22 °C) was slowed by 56% within the C-zone of thick filaments containing cMyBP-C with significant levels of endogenous phosphorylation (∼64% at each site) (21) (Fig. 2A). This slowing was similar to that previously described for bare actin filaments in the C-zone of native thick filaments (21). However, in contrast to our prior studies showing the ability of dephosphorylated cMyBP-C to inhibit further the sliding velocity of bare actin filaments in the C-zone (21), the sliding velocity (at high calcium levels) of native thin filaments over thick filaments containing dephosphorylated cMyBP-C (∼22% at each site) (21) was equal to that over thick filaments containing endogenously phosphorylated cMyBP-C (P > 0.05) (Fig. 2A). This unanticipated finding that cMyBP-C phosphorylation did not modulate maximal native thin-filament sliding velocities suggested that either the presence of the troponin and tropomyosin regulatory proteins on the thin filaments or calcium itself limits the ability of phosphorylation to modulate cMyBP-C’s inhibitory function.
Fig. 2.
Effects of cMyBP-C phosphorylation and calcium on actin-filament sliding. (A) Frequency–velocity histograms and Gaussian fits for initial native thin-filament sliding velocity in the D-zone (blue) and C-zone (red) (compare with Fig. 1B). Solid symbols and lines indicate motion on wild-type thick filaments containing highly phosphorylated (∼64%) MyBP-C; open symbols and dashed lines represent motion on thick filaments containing dephosphorylated (∼22%) MyBP-C. Assays were carried out at 100 µM ATP and 22 °C in the presence of 0.01 mM free calcium. (B) Sliding velocities of bare actin filaments on the surface of randomly oriented myosin molecules (skeletal muscle proteins) in the presence of unphosphorylated wild-type C0C3 (open circles, dashed lines) and phosphomimetic C0C34D (closed circles, solid lines), with (green) and without (black) 0.1 mM free calcium. The assay was carried out at 1 mM ATP and 30 °C. *P < 0.01 compared with C0C3, Student’s t test. The impact of cMyBP-C phosphorylation on sliding velocity and its partial reversal by calcium is similar for actomyosin from cardiac (A and Fig. S1) and skeletal (B) muscles, suggesting that calcium acts directly on cMyBP-C.
To test whether calcium was the cause, we used a conventional in vitro motility assay, with a surface of randomly oriented myosin molecules, to examine the inhibitory effect of bacterially expressed cMyBP-C N-terminal fragments (wild-type C0C3, containing domains C0–C3, and a phosphomimetic mutant, C0C34D) on the sliding velocity of native thin filaments and bare actin filaments (lacking regulatory proteins) over a range of calcium concentrations (Fig. S1 and Fig. 2B). These fragments represent the extremes of cMyBP-C phosphorylation, the C0C3 fragment being unphosphorylated and the mutant C0C34D fragment with aspartic acid substitutions for the four phosphorylatable serines in the M-domain representing the fully phosphorylated state. In addition, they have been shown to recapitulate the modulatory effects of intact cMyBP-C both in the context of a native thick filament and in the more simplified in vitro motility assay (21, 24), with the mutant C0C34D fragment both structurally (16) and functionally (18, 21) identical to fragments fully phosphorylated by protein kinase A.
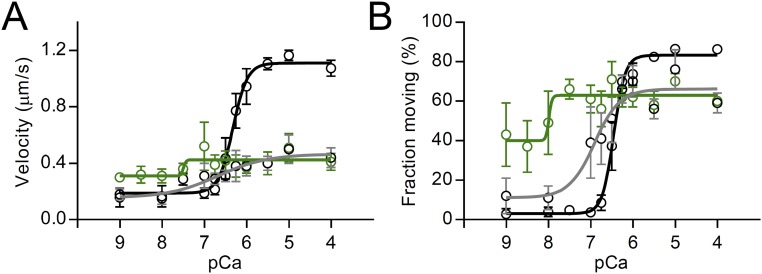
Fig. S1.
The effects of wild-type C0C3 and phosphomimetic C0C34D N-terminal fragments and calcium on mouse cardiac native thin filaments sliding over a surface of randomly oriented mouse cardiac myosin molecules. (A) Velocity and (B) fraction moving versus pCa (−log [Ca2+]) plots (100 µM ATP, 22 °C) for native thin filaments in the absence of cMyBP-C (black symbols and lines) and in the presence of 1 µM wild-type C0C3 (green symbols and lines) and C0C34D (gray symbols and lines). Data were fitted with sigmoidal dose–response curves with variable slopes. Results show that phosphomimetic substitution allows tuning of the activating potential of the C0C3 fragment but has no effect on the modulatory mechanism when the thin filaments are fully activated by calcium.
Over the upper range of physiological calcium levels (0.5–1.2 µM free Ca2+, 100 µM ATP, 22 °C), the addition of 1 µM unphosphorylated wild-type C0C3 and phosphomimetic C0C34D fragments had similar (P > 0.05) effects on the velocity and fraction of native thin filaments sliding over myosin (Fig. S1), as anticipated from the thick filament-based assays containing phosphorylated cMyBP-C (Fig. 2A). To confirm further that calcium was directly affecting the function of the phosphorylated cMyBP-C (Fig. 2A) and phosphomimetic C0C34D fragments (Fig. S1), we repeated the motility assay with non–calcium-regulated F-actin filaments and myosin isolated from chicken skeletal muscle (Fig. 2B). In the absence of calcium, the addition of at least 1 µM of the unphosphorylated wild-type C0C3 fragment inhibited actin sliding velocity (1 mM ATP, 30 °C), and this inhibition was muted in the presence of phosphomimetic C0C34D (24% vs. 40%, P < 0.01) (Fig. 2B). Interestingly, calcium eliminated the phosphorylation-dependent difference (P > 0.05) in the inhibitory capacity between the C0C3 and C0C34D fragments (Fig. 2B) so that the C0C34D was equally inhibitory. In control experiments lacking fragments, calcium had no effect (P > 0.05) on the sliding velocities of bare actin filament (Fig. 2B). We conclude that calcium directly limits the ability of phosphorylation to modulate cMyBP-C’s inhibitory function rather than indirectly affecting the actomyosin system. Based on prior structural (10) and mechanical (16) studies showing that phosphorylation reduces the mechanical extensibility of the M-domain, we hypothesized that calcium must ablate the phosphorylation-induced structural changes, in turn affecting cMyBP-C function.
Calcium Increases the Extensibility of the Phosphorylated M-Domain.
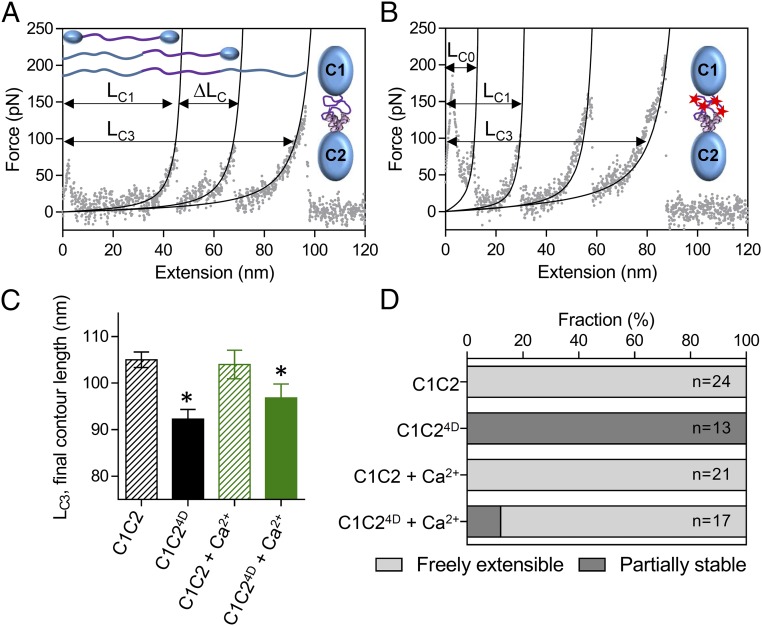
We investigated the effect of calcium on the mechanics of the M-domain using atomic force spectroscopy. To obtain the cleanest data (with the smallest number of force-extension peaks), we examined the extensibility of bacterially expressed wild-type C1C2 (Fig. 1C) and phosphomimetic C1C24D fragments. C1C2 is the shortest fragment of cMyBP-C that maintains the native connections between the M-domain and its flanking Ig domains. With only C1 and C2 serving as attachment handles for pulling on the molecule, the mechanics of the M-domain can be isolated. C1C2 molecules (Fig. 1C) were adsorbed onto a glass coverslip; then one end of the molecule was lifted off the surface by retraction of a nonspecifically attached silicon nitride atomic force microscope (AFM) probe. Force:extension traces of the wild-type C1C2 fragments showed three distinct peaks (Fig. 3A). The rising phase of the first peak (LC1) resulted from the initial lengthening of the freely extensible components within the M-domain and the subsequent drop from the unfolding of one of the Ig-like domains (16). The next peak (LC2) resulted from the lengthening of the now unfolded Ig-like domain and the subsequent drop from the unfolding of the other stable Ig-like domain. The final peak (LC3) was associated with full extension of the unfolded molecule before its detachment from the glass surface or probe (Fig. 3A). The length of LC3 (105.0 ± 8.2 nm; mean ± SD) was consistent with the expected length (107.8 nm) of a fully linearized peptide chain (308 amino acids × 0.35 nm per amino acid), suggesting that all secondary and tertiary structure was removed by pulling with ∼200 pN of force applied by the atomic force microscope. In comparison, force:extension traces of the C1C24D fragments showed an additional peak (LC0) and drop that presumably resulted from the unfolding of a portion of the M-domain (Fig. 3B) that was partially stabilized by phosphorylation, as described previously for longer C0C3 fragments (16). Although phosphomimetic substitution had no effect on the lengths of the unfolded Ig-like (C1 and C2) domains (∆LC) (Table S1), it reduced the final contour length (LC3) of the fully unfolded molecule by 13 nm (Fig. 3C). This reduction was suggestive of highly stable, phosphorylation-dependent structural ordering within the M-domain that could not be extended by the force applied with the atomic force microscope.
Fig. 3.
Effects of phosphorylation and calcium on the structure of the M-domain. (A and B) AFM force:extension curves for single unphosphorylated wild-type C1C2 (A) and phosphomimetic C1C24D (B) molecules in the absence of calcium. The initial contour length of the freely extensible components (LC1) and final contour length of the fully extended molecules (LC3) are indicated, as is the change in contour length associated with the preceding unfolding event (ΔLC). (C) The final contour lengths of the C1C2 and C1C24D fragments (LC3 in A and B) in the absence and presence 0.1 mM free calcium. *P < 0.01, LC3 vs. C1C2, Student’s t test. (D) The relative stability of the M-domain within the single C1C2 and C1C24D molecules in the absence and presence of 0.1 mM free calcium as determined from the number of molecules showing three (freely extensible M-domain) or four (partially stable M-domain) peaks in the force:extension curves.
Table S1.
Contour lengths for C1C2 and C1C24D force:extension curves
| Fragment | LC, nm | ∆LC, nm | |||||
| Peak number | Peaks | ||||||
| 0 | 1 | 2 | 3 | 0–1 | 1–2 | 2–3 | |
| C1C2 | |||||||
| Mean | 45.28 | 71.91 | 104.99 | 26.63 | 33.07 | ||
| SD | 6.91 | 7.75 | 8.16 | 5.95 | 5.04 | ||
| n | 24 | 24 | 24 | 24 | 24 | ||
| C1C24D | |||||||
| Mean | 17.50 | 37.94 | 63.66 | 92.23 | 20.67 | 25.73 | 28.78 |
| SD | 5.16 | 7.73 | 10.24 | 9.15 | 5.73 | 8.20 | 5.35 |
| n | 21 | 21 | 21 | 19 | 19 | 19 | 16 |
| C1C2 + Ca2+ | |||||||
| Mean | 44.56 | 72.47 | 103.98 | 28.03 | 31.51 | ||
| SD | 7.05 | 9.01 | 11.55 | 7.32 | 7.58 | ||
| n | 13 | 14 | 14 | 13 | 14 | ||
| C1C24D + Ca2+ | |||||||
| Mean | 17.22 | 38.47 | 65.28 | 96.78 | 19.27 | 26.81 | 31.50 |
| SD | 1.07 | 6.80 | 8.76 | 12.44 | 3.21 | 5.75 | 8.14 |
| n | 2 | 17 | 17 | 17 | 2 | 17 | 17 |
Shown is the extended contour length (LC) at each numbered peak and the change in contour length between numbered peaks associated with the preceding unfolding event (ΔLC). n, number of molecules.
To determine whether calcium altered the effect of phosphorylation on the development of the highly stable nascent structure and/or the partial stability of the M-domain, 0.1 mM of free calcium was added to the assay. The presence of calcium had no significant effect (P > 0.05) on the final contour length or extensibility of the wild-type C1C2 fragments (Fig. 3 C and D). However, calcium resulted in an 88% reduction in the fraction of C1C24D fragment force:extension traces that exhibited the unique initial unfolding peak, indicating that calcium inhibits the phosphorylation-induced stabilization of the M-domain (Fig. 3D). Despite this change in stability, the final contour lengths for the C1C24D fragments were not significantly different (P > 0.05) in the presence or absence of calcium and still were shorter (P < 0.05) than the wild-type C1C2 fragments (Fig. 3C). This result suggests that phosphorylation has multiple effects on the structure of the M-domain, with calcium affecting only a subset of these changes that are detected in the measurement of partial mechanical stability (Fig. 3D). Could these changes in stability have long-range effects on cMyBP-C’s structure to affect its function?
cMyBP-C Contains a Hinge Point Within the M-Domain.
To test the hypothesis that these local phosphorylation-dependent changes in the M-domain (Fig. 3) affect the overall structure of cMyBP-C’s N terminus, we visualized single cMyBP-C molecules in the absence of calcium using negative staining and rotary shadowing EM. Baculovirus-expressed full-length cMyBP-C molecules imaged by negative staining adopted multiple conformations ranging from elongated flexible molecules with hinge points, similar to chicken cMyBP-C (25), to amorphous structures (Fig. 4A). The elongated molecules comprised globular units 3–5 nm in diameter, consistent with a linear arrangement of individual ∼4-nm Ig and Fn domains (26). Comparable images of Ig and Fn domains have been obtained by EM of titin (27) and fibronectin molecules (28).
Fig. 4.
Localization of a hinge point within the M-domain. (A) Negatively stained EM image of full-length baculovirus-expressed cMyBP-C. (Insets) Single molecules with visible Ig and Fn domains. (B) Rotary-shadowed EM image of full-length cMyBP-C molecules. (Insets) Examples of bent rods with one and two hinge points. (C) Rotary-shadowed EM image of full-length cMyBP-C after treatment with calpain to remove the N-terminal domains. (Insets) Molecules with only a single hinge, resulting from site-specific truncation of the N-terminal segment. (D) Schematic diagram of cMyBP-C showing the location of two hinge points connecting more rigid segments.
Although individual Ig and Fn domains were not as readily detected by rotary shadowing EM (Fig. 4B), the images showed cMyBP-C molecules with similar conformations. These included elongated bent rods with up to two hinge points (50%, n = 200), shorter, more rigid straight rods that were potentially folded molecules (31%), and amorphous structures that could not be classified (19%). Of the elongated bent-rod structures, 60% contained a single central hinge creating a V-like conformation, as previously observed for skeletal MyBP-C (29). The remaining 40% contained a second hinge point near one end of the molecule that created a third short segment that has not been previously reported. The segments were 8 ± 2 nm, 15 ± 3 nm, and 20 ± 2 nm in length (Table 1).
Table 1.
Lengths of rotary shadowed cMyBP-C and C0C3 segments
| Segment 1, nm | Segment 2, nm | Segment 3, nm | |
| cMyBP-C | 8 ± 2 | 15 ± 3 | 20 ± 2 |
| cMyBP-C/calpain | 15 ± 2 | 20 ± 2 | |
| C0C3 | 8 ± 1 | 11 ± 1 |
Segment numbers refer to labels in Fig. 4D. Lengths are mean ± SD; 50 molecules were measured in each case.
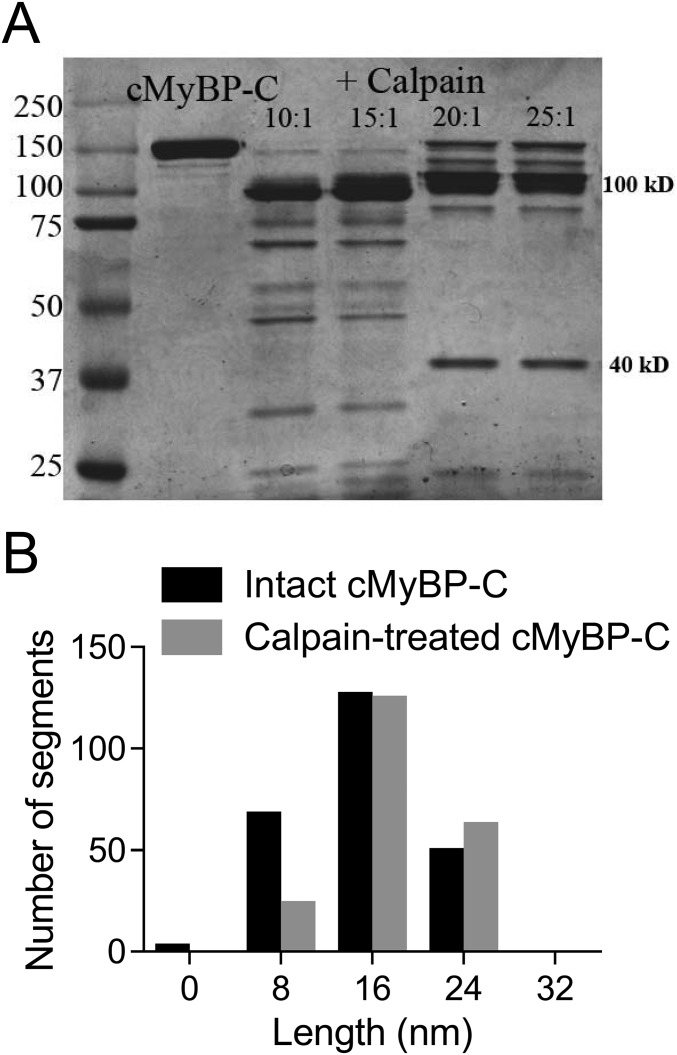
To determine whether this second hinge point was within the M-domain (a possibility suggested by its proximity to one end of the molecule), we digested the full-length cMyBP-C with calpain. Calpain specifically cleaves between R266 and R270, located near the first phosphorylation site (S273) (21), releasing a 29-kDa fragment comprising the C0–C1 domains and the first 17 amino acids of the M-domain (this fragment has also been referred to as a “40-kDa fragment” based on its migration on SDS/PAGE) (Fig. S2). After calpain cleavage (Fig. S2), both elongated bent rods (35 ± 3 nm, n = 100) and short rod-like molecules (9 ± 2 nm, n = 20) were observed in rotary-shadowed EM images (Fig. 4C). The majority (94%) of the elongated molecules contained a single hinge point, creating two segments. The segments on either side of the hinge were asymmetric and when grouped accordingly had lengths of 15 ± 2 nm and 20 ± 2 nm, similar to the lengths of the longer segments observed in intact cMyBP-C (Table 1). The shorter (9-nm) rods were similar in length to the 8-nm segment in intact cMyBP-C (segment 1 in Table 1), suggesting that the hinge point creating this segment is located near the calpain cleavage site within the M-domain (Fig. 4C). With one hinge point located within the M-domain, we can use the lengths of each segment (Table 1) and the ∼4-nm size of the Ig and Fn domains (Fig. 4A) to predict that the central hinge point occurs near the C5 domain (Fig. 4D). Based on our current resolution (±2 nm), we cannot conclusively determine whether this hinge occurs between the C4 and C5 domains, within the disordered ∼30-aa cardiac-specific insert in C5 (30), or between the C5 and C6 domains as previously predicted (31). This central hinge point may be involved with the extension of the molecule from the thick filament, and the N-terminal hinge may function to alter how the N-terminal domains interact with actin and/or the myosin S2 region.
Fig. S2.
Calpain cleavage of full-length cMyBP-C. (A) SDS/PAGE shows that at low calpain concentrations cMyBP-C is cleaved into two major fragments with apparent molecular masses of ∼110 and 40 kDa [confirmed to be 29 kDa by mass spectrometry (1)]. (B) Frequency–length histograms for cMyBP-C segments in elongated wild-type and calpain-cleaved (25:1 ratio) expressed cMyBP-C molecules. Segment lengths for 60 molecules with two hinges and 40 molecules with three hinges were measured for the wild-type molecules (black bars). Segment lengths for 94 calpain-treated cMyBP-C molecules with two hinges and six molecules with three hinges were measured (gray bars). Calpain-treated molecules showed a large reduction in the number with segments ≤8 nm long (segment 1 in Table 1), and only 6% of the calpain-treated molecules showed a terminal bend (three hinges), as compared with 40% of the undigested molecules.
cMyBP-C Phosphorylation Promotes Bending of the N Terminus at the M-Domain.
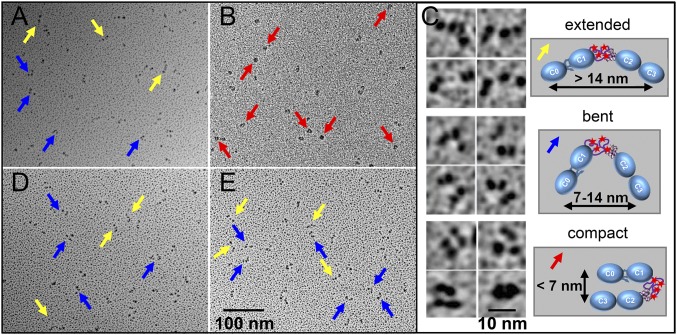
With the presence of a hinge point in the M-domain, it is possible that the phosphorylation-induced partial stability that we observed previously with C0C3 (16) and presently with C1C2 in AFM might promote changes in the structural orientation of the N-terminal domains. Therefore, we examined the structure of bacterially expressed wild-type C0C3, PKA-treated C0C3, and phosphomimetic C0C34D fragments (Fig. 1C) by rotary shadowing EM (Fig. S3A and Fig. 5 A and B). For each fragment, three distinct classes of molecules were observed: extended rods, bent rods with a single hinge point, and compact/closed structures (Fig. 5C). To quantify the relative distribution of these structures in the wild-type and phosphomimetic fragments, we measured the distances between the ends of the molecules in each image (Fig. 5 A–C and Table 2). The majority (85%) of the wild-type C0C3 molecules showed the extended (>14 nm between the two ends) or bent (7–14 nm between ends) conformations (Table 2). The bent rods contained segments of 8 ± 1 nm and 11 ± 1 nm, the 8-nm segment being similar to that observed for segment 1 in intact cMyBP-C (Fig. 4D and Table 1). In contrast, the majority (61%) of the C0C34D molecules exhibited the compact structure rather than the extended or bent conformations (Fig. 5B and Table 2). The phosphorylation-induced closure of the N terminus around this hinge suggests that local changes in the structure of the M-domain (Fig. 3) promote global changes in orientation of the N-terminal domains seen in these rotary shadowed images.
Fig. S3.
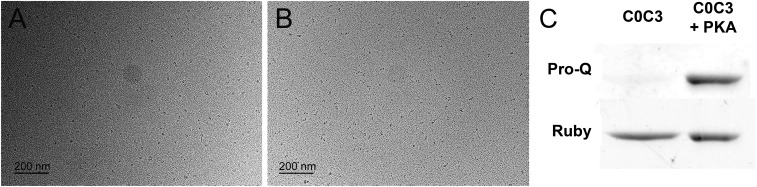
The effects of phosphorylation and calcium on the orientation of the N-terminal domains. (A and B) Rotary-shadowed EM images of PKA-treated C0C3 fragments in the absence (A) and presence (B) of 0.1 mM free Ca2+. (C) Blots from a Pro-Q–stained SDS/PAGE gel confirming the presence of phosphate in the PKA-treated sample. As observed for the phosphomimetic C0C34D fragments (Fig. 5 B and E), PKA treatment resulted in molecules adopting a compact conformation, and the presence of calcium shifted these molecules into bent or extended conformations.
Fig. 5.
Effects of phosphorylation and calcium on the orientation of the N-terminal domains. (A and B) Rotary-shadowed EM images of nonphosphorylated wild-type C0C3 (A) and phosphomimetic C0C34D (B) fragments in the absence of calcium. (C) Representative EM images (Left) and schematic diagrams (Right) of the extended rods (Top), bent rods (Middle), and compact/closed structures (Bottom) classified in each EM image. (D and E) Rotary-shadowed EM images as in A and B for C0C3 (D) and C0C34D (E) fragments in the presence of calcium. Yellow, blue, and red arrows point to molecules with extended, bent, and compact structures, respectively.
Table 2.
Phosphorylation and calcium affect C0C3 structure
| Extended, % | Bent, % | Compact, % | |
| C0C3 | 39 | 46 | 15 |
| C0C34D | 5 | 34 | 61 |
| C0C3 + Ca2+ | 42 | 49 | 9 |
| C0C34D + Ca2+ | 37 | 46 | 17 |
One hundred molecules were measured in each case.
Phosphorylation-Induced Closure of the N-Terminal Domains Is Attenuated by Calcium.
Does calcium’s ability to destabilize the phosphorylated M-domain (Fig. 3D) mitigate the closure of cMyBP-C’s N-terminal domains? If so, this effect could account for the loss of phosphorylation-dependent modulation of thin filament velocities in the motility assays in the presence of calcium (Fig. 2). To address this question, rotary shadowed EM images of the wild-type C0C3, PKA-treated C0C3, and phosphomimetic C0C34D fragments were obtained in the presence of 0.1 mM free calcium (Fig. 5 D and E and Fig. S3B). The addition of calcium resulted in fewer of the PKA-treated and phosphomimetic molecules adopting the compact conformation. When quantified, there was an 89% reduction in the fraction of C0C34D molecules adopting the compact conformation, with the distribution of molecules in the extended and bent rod conformations being similar to that in the wild-type C0C3 fragments regardless of the presence of calcium (Table 2). This calcium-induced reopening of the C0C34D molecules likely results from a disruption in global interactions within the N-terminal domains (Fig. 3D) and could explain how calcium ablated the ability of phosphorylation to modulate the function of cMyBP-C in our motility assays (Fig. 2).
Discussion
Phosphorylation sites are commonly found in intrinsically disordered regions of proteins, with phosphorylation having both local effects on the order of secondary structures and global effects on the orientation of tertiary states (32–34). Such structural rearrangements can result in changes in the affinity for binding partners (33, 35) and/or the mechanics of the phosphoprotein in the bound state (32, 36). Each of these mechanisms allows exquisite tuning of the protein interactions involved in complex biological processes.
Here, we determined that phosphorylation of four residues within an intrinsically disordered region of cMyBP-C’s M-domain significantly impacts cMyBP-C’s structure, in turn tuning cMyBP-C’s ability to modulate actomyosin motility. Phosphorylation-dependent structural changes within the M-domain were evident by a reduction in the C1C2 fragment’s final contour length (Fig. 3C) and a change in mechanical extensibility (Fig. 3D) and were manifest beyond the M-domain by shifts in the conformation of cMyBP-C’s N terminus (Fig. 5 A and B). The decrease in contour length upon phosphorylation suggests the formation of secondary and/or tertiary structures within the M-domain that are so stable that they cannot be unraveled by the AFM pulling forces (∼200 pN). The formation of these nascent structures may partially stabilize the M-domain by their surface charge distribution, allowing more electrostatic interactions with either the three-helix bundle or other neighboring domains. In support of this notion, previous AFM studies showed that the phosphorylation-dependent mechanical stability of the M-domain was sensitive to ionic strength (16). Importantly, these local alterations within the M-domain appear to have long-range, global effects on the equilibrium between the extended and compact conformational states that cMyBP-C’s N terminus adopts (Fig. 5 A and B). Specifically, phosphorylation shifts the N-terminal domains (C0C3) from the extended conformation to a compact conformation as the C0C3 bends in half at a hinge point near the sites of phosphorylation (Fig. 6). Such conformational closure may directly control the accessibility of binding sites for both actin and/or myosin S2, allowing phosphorylation to limit cMyBP-C’s affinity for either partner, as previously described (17–19), and/or directly alter the mechanics of cMyBP-C in the structurally bound state (32, 36). Either scenario could explain the phosphorylation-dependent reduction in cMyBP-C’s inhibition of actomyosin motility (Fig. 2).
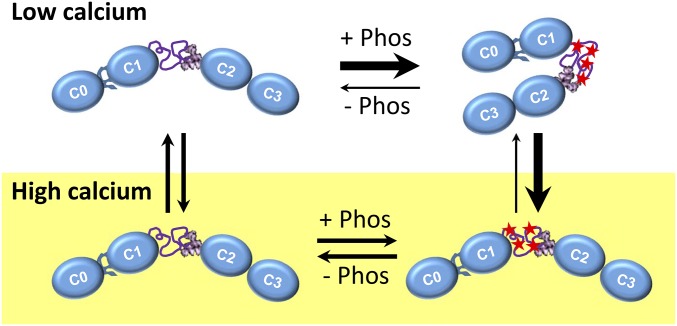
Fig. 6.
Diagrams of the effects of phosphorylation on the overall structure of cMyBP-C’s N terminus at low (Top) and high (Bottom) calcium levels.
Interestingly, the presence of calcium at the concentrations found at the peak of cardiac muscle contraction can effectively reverse the impact of phosphorylation on the equilibrium of the N-terminal conformational state (Figs. 5E and 6), the partial stability of the M-domain (Fig. 3D), and the modulation of actomyosin motility (Fig. 2 and Fig. S1). Calcium ions are positively charged and thus may simply shield negative charges within the phosphorylation-dependent nascent structure that partially stabilizes the M-domain (Fig. 3D). Alternatively, phosphorylation may result in the formation of a calcium-binding motif within the M-domain. By either mechanism, the presence of calcium could shift the C0C34D fragment’s conformational equilibrium back to the extended, rod-like conformation (Fig. 5E and Table 2) favored by C0C3, thus explaining why the C0C34D and C0C3 fragments become functionally indistinguishable in the motility assay when calcium is present.
The complex intersection of posttranslational modification and calcium signaling that we report has broad implications for our understanding of cMyBP-C’s role in modulating cardiac contractility. The interplay between phosphorylation and calcium on cMyBP-C structure and function must be dynamic and allow fine tuning of cMyBP-C’s mechanics within every heartbeat, because calcium ebbs and flows within each cardiac muscle cell. Although cMyBP-C’s in vivo binding partner(s) have yet to be defined, cMyBP-C may serve at least two distinct functional roles: to enhance the activation state of thin filaments at low calcium concentrations and to limit the maximal activity of actomyosin interactions once the sarcomere is fully saturated with calcium (23). We recently have proposed a model based on these roles in which the localization of cMyBP-C to the sarcomeric C-zone serves to sensitize the thin filaments to calcium, helping correct an intrinsic inhomogeneity in calcium activation during the onset of contraction (23). Then, at the peak of contraction, when calcium levels have increased, cMyBP-C serves a second role as a governor that effectively throttles back actomyosin activity. Without the effects of calcium on cMyBP-C’s structure, phosphorylation of cMyBP-C at normal in situ levels would partially mitigate both these modulatory roles (21, 23). However, because of the antagonistic interplay between phosphorylation and calcium, physiologically saturating levels of calcium would, in turn, tend to reverse the impact of phosphorylation on cMyBP-C structure and function so that cMyBP-C remains a potent modulator of contractility, as demonstrated in vitro (Fig. S2). Thus, phosphorylation and calcium together fine-tune cMyBP-C’s modulatory function through changes in cMyBP-C’s structure.
Multiple cardiac proteins, including the myosin regulatory light chain, troponin I, and phospholamban, appear to be regulated by phosphorylation-dependent structural transitions similar to those observed for cMyBP-C (32). It currently is unknown whether the interplay between phosphorylation and calcium is specific to cMyBP-C or is a general phenomenon that would impact the structure and function of these other cardiac contractile proteins by a similar mechanism. In fact, it is possible that such mechanisms may occur in other biological systems, because phosphorylation and calcium are ubiquitous signaling mechanisms critical to protein function.
Materials and Methods
Detailed methods, given in SI Materials and Methods, are summarized here.
Proteins.
Native thick and thin filaments and depolymerized myosin were isolated from FVB mouse hearts (21, 23, 37). cMyBP-C phosphorylation levels were measured by LC-MS, and, where applicable, thick filaments were dephosphorylated with λ-phosphatase (New England BioLabs) (21). Skeletal myosin and F-actin were purified from chicken pectoralis muscle (38, 39). F-actin and native thin filaments were fluorescently labeled with equimolar TRITC (37). Wild-type and phosphomimetic mutant (S273, S282, S302, and S307 to D substitution) C1–C2 (amino acids 151–448) and C0–C3 (amino acids 1–539) mouse cMyBP-C N-terminal fragments were bacterially expressed and purified (40, 41). Full-length mouse cMyBP-C cDNA was expressed in the Baculovirus-insect cell system, purified, and where applicable, truncated with calpain-1 (Calbiochem), as described in SI Materials and Methods.
Motility Assays.
The movement of 250-nm shards of thin filaments on thick filaments was observed (100 µM ATP, 22 °C) by TIRFM (21). The movement of thin filaments (100 µM ATP, 22 °C) and bare actin filaments (1 mM ATP, 30 °C) on randomly oriented surfaces of mouse cardiac and skeletal muscle myosin molecules was observed by epifluorescence microscopy in the absence and presence of wild-type C0C3 and phosphomimetic C0C34D fragments (18, 21, 42). Free calcium concentrations were determined using MaxChelator software (43).
Atomic Force Spectroscopy.
The unfolding of wild-type C1C2 and phosphomimetic C1C24D fragments was observed by pulling (180 nm at 700 nm/s) with an atomic force microscope (MFP-3 Bio; Asylum Research) (16). AFM force:extension traces were identified by a fingerprinting routine (44) and then were fit using a wormlike chain model (45).
EM.
Full-length and truncated cMyBP-C and wild-type, PKA-treated C0C3, and phosphomimetic C0C34D fragments were visualized by negative staining and/or rotary shadowing EM. EM grids were examined using a transmission electron microscope (FEI), and lengths were measured from digital images using the line tool in ImageJ.
SI Materials and Methods
Proteins.
Native thick filaments and thin filaments were isolated from the apex of FVB mouse hearts by mechanochemical means, as previously described (21, 23, 37). A 10-µL drop of 0.2 U/µL calpain-1 from porcine erythrocytes (Calbiochem) in 20 mM imidazole (pH 6.8), 5 µM 2-mercaptoethanol, and 1 mM calcium acetate was used for the final dissociation of the thick filament to limit proteolytic cleavage of cMyBP-C as described (21). The thick filaments were stored in 250 μL of chilled 75 mM NaCl, 5 mM MgCl2, 2 mM EGTA, 1 mM DTT, 7 mM phosphate buffer (pH 7), 10 mM creatine phosphate, and 2.5 mM ATP. Average phosphorylation levels in the wild-type thick filaments were ∼64% on serines 273, 282, 302, and 307 within the M-domain, when measured by LC-MS (21). Where applicable, 50-μL aliquots of thick filament samples were incubated with 5 μL (2,000 U) of λ-phosphatase from Escherichia coli (New England BioLabs) and 6.5 μL of 10 mM MnCl2 (1 h, 30 °C) to decrease the phosphorylation of the endogenous cMyBP-C to 22% (21). Myosin and F-actin were purified from chicken pectoralis muscle (38, 39). Native thin filaments and F-actin filaments were fluorescently labeled with equimolar TRITC (21, 37).
Clones encoding domains C1–C2 (amino acids 151–448) and C0–C3 (amino acids 1–539) of cMyBP-C were inserted into the bacterial expression vector pET28A using the restriction sites Nco1 and Xho1. The cDNA was ligated in-frame with the C-terminal 6His tag, allowing purification of the expressed protein by passage over Talon His-Tag purification resin (Clontech) (40, 41). Clones were modified using a Site-Directed QuikChange Mutagenesis Kit (Stratagene), allowing specific amino acids to be changed to aspartic acid (S273, S282, S302, S307D). For protein production, clones were transformed into BL21(DE3), grown and induced with isopropyl β-d-1-thiogalactopyranoside followed by lysis using HisTALON xTractor Buffer (Clontech). Cell lysates were passed over the resin, aliquots were collected, and samples were pooled and dialyzed against 1× PBS/10 mM DTT. Where applicable, the C0C3 fragment was phosphorylated by incubation with 1 U/µL of PKA (Sigma), and the presence of phosphate was qualitatively verified by Pro-Q Diamond phosphostaining (Invitrogen) as previously described (24).
Full-length cDNA encoding the mouse cMyBP-C protein was cloned into the pIEx/Bac-3 3C/LIC Expression transfer vector (Novagen) by ligation-independent cloning using compatible homologous ends introduced by PCR. Clones were verified by sequencing followed by recombination into a digested baculovirus vector. The virus was propagated in Sf9 insect cells, expanded, and plaque purified. Positive clones were used for further protein expression. Constructs cloned in-frame into the transfer vector produced protein with an N-terminal 6His tag, and the C terminus contained a Strep-tag II, allowing isolation of full-length protein from affinity columns. Tags were removed from the ends of the protein using the proteases HRV3C and thrombin.
Truncation of Full-Length cMyBP-C.
The calpain-induced truncation of cMyBP-C was carried out on 20-µL aliquots of 1 mg/mL cMyBP-C. The aliquots were incubated with 1.2 U (0.45 μL) calpain-1 (Calbiochem) in 200 µL of 25 mM KCl buffer for 20 min at 30 °C in the presence of 1 mM CaCl2 to ensure proteolytic cleavage. The fragments were diluted in 25 mM KCl buffer with 1 mM EGTA.
Fluorescence Image Acquisition.
Fluorescently labeled actin-based filament sliding was visualized using a Nikon Eclipse Ti-U microscope equipped with a Plan Apo oil immersion objective lens (100×, 1.40 N.A.) for epifluorescence or through-the-objective TIRFM using a custom Micro Optic Fiber Launch total internal reflectance microscope (21). A Lumen 200-W metal arc lamp (Prior Scientific) and 532-nm diode-pumped solid-state, 50-mW laser (Lasever Inc.) were used for excitation, respectively. An intensified high-resolution Mega Z 10-bit digital camera running Piper Control v2.6.09 software (Stanford Photonics) was used for imaging without pixel binning (93 nm per pixel).
Thick and Thin Filament Assays.
The movement of 250-nm shards of TRITC-labeled thin filaments was observed on thick filaments from three separate preparations by TIRFM at 22 °C as previously described (21). Briefly, native thick filaments were incubated on a Sigmacote (Sigma Aldrich)-coated flow cell in 75 mM KCl, 1 mM EGTA, 40 mM DTT, 25 mM imidazole, 4 mM MgCl2, adjusted to pH 7.4 (20 min, 30 °C). The surface was blocked with BSA and was washed with 1 mM ATP. Next, short, 250-nm sonicated TRITC-labeled native thin filaments in motility buffer [25 mM KCl, 1 mM EGTA, 40 mM DTT, 25 mM imidazole, 4 mM MgCl2, adjusted to pH 7.4, containing an oxygen scavenging system (0.1 µg/mL glucose oxidase, 0.018 µg/mL catalase, 2.3 µg/mL glucose)] were incubated on the surface for 1 min. The flow cell was rinsed three times with motility buffer containing 100 µM ATP, CaCl2, and short TRITC-labeled native thin filaments. The CaCl2 concentration was adjusted to achieve 0.01 mM free calcium in solution as determined using MaxChelator software (43) to account for the 1 mM EGTA in the buffer.
Thin filament motion was recorded at 120 frames/s for 1,000 frames in multiple areas of each flow cell. The image stacks were opened and inspected for the movement of actin shards. Image substacks were created for each shard, and the position was tracked in the x,y visual plane using ImageJ SpotTracker 2D (21). The fitted positions were converted to nanometers and plotted using Excel 2013 (Microsoft). The centroid position for each linear track was used to calculate the displacement from the origin with respect to time and was fitted with a segmental linear regression using GraphPad Prism 6 for Windows (GraphPad Software) to calculate velocities, as described (21). The resultant initial and final velocity distributions, being motion on the tip and within the C-zone each thick filament, were fitted with a Gaussian using GraphPad. The statistical significance for velocity comparisons was determined by the Mann–Whitney–Wilcox test.
Bare Actin Motility Assays.
The sliding of bare actin filaments on a randomly oriented surface of skeletal muscle myosin molecules was observed at 30 °C in the absence and presence of wild-type C0C3 and phosphomimetic C0C34D fragments (1 or 2 µM) using an in vitro motility assay as previously described (18, 21, 42). Briefly, 100 µg/mL of myosin was added to a nitrocellulose-coated flow cell; then the surface was blocked with BSA. Next, unlabeled actin filaments were incubated on the surface for 1 min and then were washed with 1 mM ATP. TRITC-labeled actin filaments were incubated on the surface for 1 min in motility buffer lacking ATP. The flow cell was rinsed, and motility buffer containing 1 mM ATP and 0.5% methyl cellulose was added to the flow cell. CaCl2 was added to the motility buffer to achieve a concentration of 0.1 mM free calcium, and 1 or 2 µM wild-type C0C3 or phosphomimetic C0C34D was added where applicable.
For motility, 150 images were collected at 10 frames/s; the velocity of each moving filament was determined from 25 frames of each image stack using DiaTrack 3.03 software for Windows (Semasopht). The mean velocities ± SEM were determined from four fields of view for each experimental condition, from four independent experimental preparations. Data were plotted using GraphPad Prism 6 for Windows (GraphPad Software), and statistical significance between velocities was determined using a Student’s t test.
Thin Filament Motility Assays.
The sliding of native cardiac thin filaments on a randomly oriented surface of cardiac muscle myosin molecules was observed at 22 °C in the absence and presence of 1 µM wild-type C0C3 and phosphomimetic C0C34D fragments using an in vitro motility assay as previously described (37). The solutions were added to the flow cells as described above for the bare actin motility assays, but native thin filaments and 100 µg/mL cardiac muscle myosin were substituted, and the final ATP concentration was 100 µM. The CaCl2 concentration was adjusted to achieve free calcium concentrations in solution as determined using MaxChelator software (43). Data collection and analysis were identical to those for the bare actin motility assays, but the movies were down-sampled to two frames/s using ImageJ software to avoid oversampling. The mean velocities ± SEM were determined from four fields of view for each experimental condition from three independent experimental preparations. Data were plotted using GraphPad Prism 6 for Windows (GraphPad Software) and fitted with sigmoidal dose–response curves with variable slopes.
Atomic Force Spectroscopy.
The unfolding of wild-type C1C2 and phosphomimetic C1C24D fragments was observed by AFM as previously described (16). Briefly, N-terminal fragments were diluted in 25 mM KCl buffer to an approximate concentration of 10 nM. A 50-µL droplet of the diluted protein was placed on a precleaned glass slide (Gold Seal) and incubated for 10 min. Unadsorbed protein was removed by washing three times with 50 µL of dilution buffer. A 150-µL droplet of buffer was left on the slide over the adsorbed fragments. For experiments examining the effects of calcium, CaCl2 was added to the 25-mM KCl buffer to achieve final concentrations of 0.1 mM free calcium.
An MFP-3 Bio atomic force microscope (Asylum Research) fitted with an uncoated silicon nitride probe (Budget Sensors) was used to pull on the adsorbed molecules. Before testing, the spring constant of the probe (average 52 pN/nm) was measured by equipartition using the Asylum Research software. Force:extension traces were acquired by scanning square grids (5–20 µm) with 400–1,600 points. At each point, the probe of the atomic force microscope was brought into contact with the glass slide and, after a 1-s dwell, was retracted to 180 nm (farther than the 3∼100-nm expected length of the unfolded C1C2 fragment) at 700 nm/s. Probe position and deflection were sampled at 8 kHz during retraction.
AFM traces consisting of three force peaks resulting from unfolding of the C1 and C2 Ig domains and detachment were identified using a molecular fingerprinting routine (45) implemented in Matlab (MathWorks, Inc.) after pulling on the C1C2 fragments. Briefly, this technique is carried out by ranking all force:extension traces collected based on how well they correlate with a template trace. Based on prior experiments (16), force extension traces for C1C2 fragments were expected to have either three force peaks (unfolding of the two IG domains and detachment from the atomic force microscope probe) or four peaks (including an additional peak arising from a stable M-domain). A pair of theoretical force:extension traces representing these two states (extensible and nonextensible M-domain) were created and used as templates in the fingerprinting routine. To assess the relative fractions of fragments with extensible and nonextensible M-domains accurately, traces from all experimental conditions were compared against both templates. Force:extension traces identified by the fingerprinting routine then were fit using a wormlike chain model (44) of contour lengths, LC0-3, at each force peak. To assess the effect of calcium on cMyBP-C N-terminal fragment mechanics, C1C2 and C1C24D were tested in the presence of 0.1 mM free calcium by the addition of CaCl2 to achieve the desired concentration. Data were plotted using GraphPad Prism 6 for Windows (GraphPad Software), and statistical significance for LC3 was determined using a Student’s t test.
EM.
Full-length and truncated baculovirus-expressed cMyBP-C, bacterially expressed wild-type C0C3, and phosphomimetic C0C34D N-terminal fragments were visualized by negative staining and/or rotary shadowing EM. For negative staining, full-length protein was diluted to 50 nM in 25 mM KCl buffer. A 5-μL droplet was applied to a carbon-coated grid that had been freshly (within 30 min) irradiated with UV light using an R-52G ozone-producing Mineralight lamp (UVP) for 40 min (46), and the grid was stained with 1% uranyl acetate. For rotary shadowing, full-length cMyBP-C and expressed C0C3/C0C34D molecules were diluted to 0.7 and 0.9 µM in 25 mM KCl buffer. Proteins then were mixed with an equal volume of glycerol and sprayed onto freshly cleaved mica. The droplets were allowed to dry for 40 min; then the mica was placed in a Denton (DV-502) vacuum evaporator and pumped to <10−7 Torr. Ten micrograms of platinum wire was evaporated over a 1-min period from a distance of 9.5 cm at a 6° angle onto the rotating samples, and then thin carbon was deposited over the platinum replica. The carbon-supported platinum replica was floated off the mica onto water and adsorbed onto 400 mesh copper grids. For high-calcium conditions, CaCl2 was added to the dilution buffer to achieve 0.1 mM free calcium.
Grids were examined at 80 kV using an FEI Tecnai 12 or Philips CM120 transmission electron microscope, and digital images were recorded with an FEI Eagle 4K × 4K CCD camera (Tecnai) or a TVIPS F224HD 2K × 2K CCD camera (CM120). In all experiments single molecules on each grid were initially classified by eye. In the rotary-shadowed images, length measurements were made using the line tool in ImageJ software. The lengths were corrected for shadow thickness based on the apparent thickness of the myosin tail, known to be 2 nm in diameter, in control images of single myosin molecules. All segment length measurements were reported as mean ± SD.
For intact full-length cMyBP-C, 200 molecules in the images were determined to be elongated bent rods (with up to two hinge points), shorter straight rigid rods, or amorphous structures that could not be classified. The lengths of the segments from the elongated bent rods were measured. Additional images were evaluated, and the three segment lengths of 50 molecules with elongated bent-rod structures containing two hinges were measured. For calpain-treated cMyBP-C, segment lengths were measured from the images for 100 elongated molecules (with up to two hinge points) and 20 short fragments. Additional images were evaluated, and the two-segment lengths of 50 elongated molecules with only a single hinge were measured. The two segment lengths in each molecule were asymmetric and were grouped accordingly for averaging. For C0C3 and C0C34D molecules in the absence and presence of calcium, molecular structures were classified as extended rods, bent rods, and compact/closed by measuring the distance between the ends of each molecule. A total of 100 molecules were measured for each condition, with lengths >14 nm being considered extended rods, lengths between 7 and 14 nm considered bent rods, and lengths <7 nm considered compacted/closed conformations. For C0C3 in the absence of calcium, the segment lengths of 50 molecules having either the extended or bent conformation were measured.
Acknowledgments
We thank G. Kennedy from the University of Vermont Instrumentation and Model Facility for TIRFM imaging expertise and A. Quinn at the Microscopy Imaging Center of the University of Vermont (supported by NIH Grant S10RR025498) for assistance with AFM. EM was carried out in the Core Electron Microscopy Facility at the University of Massachusetts Medical School. M.J.P., J.Y.M., A.J.M., S.B.P., J.G., J.R., D.M.W., and R.C. were supported by NIH Grant HL059408; M.J.P was supported by NIH Grant HL124041; A.J.M. was supported by NIH Grants HL007944 and HL007647; J.R. was supported by NIH Grant HL069779; D.M.W. was supported by NIH Grant HL126909; and R.C. was supported by NIH Grant AR034711.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 3133.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522236113/-/DCSupplemental.
References
- 1.Harris SP, Lyons RG, Bezold KL. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circ Res. 2011;108(6):751–764. doi: 10.1161/CIRCRESAHA.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sequeira V, Witjas-Paalberends ER, Kuster DW, van der Velden J. Cardiac myosin-binding protein C: Hypertrophic cardiomyopathy mutations and structure-function relationships. Pflugers Arch. 2014;466(2):201–206. doi: 10.1007/s00424-013-1400-3. [DOI] [PubMed] [Google Scholar]
- 3.Sadayappan S, de Tombe PP. Cardiac myosin binding protein-C as a central target of cardiac sarcomere signaling: A special mini review series. Pflugers Arch. 2014;466(2):195–200. doi: 10.1007/s00424-013-1396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flashman E, Redwood C, Moolman-Smook J, Watkins H. Cardiac myosin binding protein C: Its role in physiology and disease. Circ Res. 2004;94(10):1279–1289. doi: 10.1161/01.RES.0000127175.21818.C2. [DOI] [PubMed] [Google Scholar]
- 5.Craig R, Lee KH, Mun JY, Torre I, Luther PK. Structure, sarcomeric organization, and thin filament binding of cardiac myosin-binding protein-C. Pflugers Arch. 2014;466(3):425–431. doi: 10.1007/s00424-013-1426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luther PK, et al. Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proc Natl Acad Sci USA. 2011;108(28):11423–11428. doi: 10.1073/pnas.1103216108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee K, Harris SP, Sadayappan S, Craig R. Orientation of myosin binding protein C in the cardiac muscle sarcomere determined by domain-specific immuno-EM. J Mol Biol. 2015;427(2):274–286. doi: 10.1016/j.jmb.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfuhl M, Gautel M. Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): Who does what, with what, and to whom? J Muscle Res Cell Motil. 2012;33(1):83–94. doi: 10.1007/s10974-012-9291-z. [DOI] [PubMed] [Google Scholar]
- 9.Jia W, Shaffer JF, Harris SP, Leary JA. Identification of novel protein kinase A phosphorylation sites in the M-domain of human and murine cardiac myosin binding protein-C using mass spectrometry analysis. J Proteome Res. 2010;9(4):1843–1853. doi: 10.1021/pr901006h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howarth JW, Ramisetti S, Nolan K, Sadayappan S, Rosevear PR. Structural insight into unique cardiac myosin-binding protein-C motif: A partially folded domain. J Biol Chem. 2012;287(11):8254–8262. doi: 10.1074/jbc.M111.309591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stelzer JE, Patel JR, Moss RL. Protein kinase A-mediated acceleration of the stretch activation response in murine skinned myocardium is eliminated by ablation of cMyBP-C. Circ Res. 2006;99(8):884–890. doi: 10.1161/01.RES.0000245191.34690.66. [DOI] [PubMed] [Google Scholar]
- 12.Sadayappan S, et al. Cardiac myosin binding protein-C phosphorylation in a beta-myosin heavy chain background. Circulation. 2009;119(9):1253–1262. doi: 10.1161/CIRCULATIONAHA.108.798983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadayappan S, et al. Cardiac myosin binding protein C phosphorylation is cardioprotective. Proc Natl Acad Sci USA. 2006;103(45):16918–16923. doi: 10.1073/pnas.0607069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacques AM, et al. Myosin binding protein C phosphorylation in normal, hypertrophic and failing human heart muscle. J Mol Cell Cardiol. 2008;45(2):209–216. doi: 10.1016/j.yjmcc.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 15.Tong CW, et al. Phosphoregulation of cardiac inotropy via myosin binding protein-C during increased pacing frequency or beta1-adrenergic stimulation. Circ Heart Fail. 2015;8(3):595–604. doi: 10.1161/CIRCHEARTFAILURE.114.001585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalek AJ, et al. Phosphorylation modulates the mechanical stability of the cardiac myosin-binding protein C motif. Biophys J. 2013;104(2):442–452. doi: 10.1016/j.bpj.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaffer JF, Kensler RW, Harris SP. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J Biol Chem. 2009;284(18):12318–12327. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weith AE, et al. The extent of cardiac myosin binding protein-C phosphorylation modulates actomyosin function in a graded manner. J Muscle Res Cell Motil. 2012;33(6):449–459. doi: 10.1007/s10974-012-9312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: A modulator of cardiac contraction? EMBO J. 1995;14(9):1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coulton AT, Stelzer JE. Cardiac myosin binding protein C and its phosphorylation regulate multiple steps in the cross-bridge cycle of muscle contraction. Biochemistry. 2012;51(15):3292–3301. doi: 10.1021/bi300085x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Previs MJ, Beck Previs S, Gulick J, Robbins J, Warshaw DM. Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science. 2012;337(6099):1215–1218. doi: 10.1126/science.1223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belknap B, Harris SP, White HD. Modulation of thin filament activation of myosin ATP hydrolysis by N-terminal domains of cardiac myosin binding protein-C. Biochemistry. 2014;53(42):6717–6724. doi: 10.1021/bi500787f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Previs MJ, et al. Myosin-binding protein C corrects an intrinsic inhomogeneity in cardiac excitation-contraction coupling. Sci Adv. 2015;1(1):e1400205. doi: 10.1126/sciadv.1400205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weith A, et al. Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. J Mol Cell Cardiol. 2012;52(1):219–227. doi: 10.1016/j.yjmcc.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartzell HC, Sale WS. Structure of C protein purified from cardiac muscle. J Cell Biol. 1985;100(1):208–215. doi: 10.1083/jcb.100.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Govada L, et al. Crystal structure of the C1 domain of cardiac myosin binding protein-C: Implications for hypertrophic cardiomyopathy. J Mol Biol. 2008;378(2):387–397. doi: 10.1016/j.jmb.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 27.Tskhovrebova L, et al. Shape and flexibility in the titin 11-domain super-repeat. J Mol Biol. 2010;397(4):1092–1105. doi: 10.1016/j.jmb.2010.01.073. [DOI] [PubMed] [Google Scholar]
- 28.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett P, Starr R, Elliott A, Offer G. The structure of C-protein and X-protein molecules and a polymer of X-protein. J Mol Biol. 1985;184(2):297–309. doi: 10.1016/0022-2836(85)90381-x. [DOI] [PubMed] [Google Scholar]
- 30.Idowu SM, Gautel M, Perkins SJ, Pfuhl M. Structure, stability and dynamics of the central domain of cardiac myosin binding protein C (MyBP-C): Implications for multidomain assembly and causes for cardiomyopathy. J Mol Biol. 2003;329(4):745–761. doi: 10.1016/s0022-2836(03)00425-x. [DOI] [PubMed] [Google Scholar]
- 31.Jeffries CM, et al. Human cardiac myosin binding protein C: Structural flexibility within an extended modular architecture. J Mol Biol. 2011;414(5):735–748. doi: 10.1016/j.jmb.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 32.Colson BA, Gruber SJ, Thomas DD. Structural dynamics of muscle protein phosphorylation. J Muscle Res Cell Motil. 2012;33(6):419–429. doi: 10.1007/s10974-012-9317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bah A, et al. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015;519(7541):106–109. doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- 34.Forbes JG, et al. Titin PEVK segment: Charge-driven elasticity of the open and flexible polyampholyte. J Muscle Res Cell Motil. 2005;26(6-8):291–301. doi: 10.1007/s10974-005-9035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16(1):18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colson BA, Rybakova IN, Prochniewicz E, Moss RL, Thomas DD. Cardiac myosin binding protein-C restricts intrafilament torsional dynamics of actin in a phosphorylation-dependent manner. Proc Natl Acad Sci USA. 2012;109(50):20437–20442. doi: 10.1073/pnas.1213027109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mun JY, et al. Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc Natl Acad Sci USA. 2014;111(6):2170–2175. doi: 10.1073/pnas.1316001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85(Pt B):164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 39.Margossian SS, Lowey S. Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 1982;85(Pt B):55–71. doi: 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
- 40.Sadayappan S, et al. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circ Res. 2005;97(11):1156–1163. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mun JY, et al. Electron microscopy and 3D reconstruction of F-actin decorated with cardiac myosin-binding protein C (cMyBP-C) J Mol Biol. 2011;410(2):214–225. doi: 10.1016/j.jmb.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmiter KA, et al. R403Q and L908V mutant beta-cardiac myosin from patients with familial hypertrophic cardiomyopathy exhibit enhanced mechanical performance at the single molecule level. J Muscle Res Cell Motil. 2000;21(7):609–620. doi: 10.1023/a:1005678905119. [DOI] [PubMed] [Google Scholar]
- 43.Patton C, Thompson S, Epel D. Some precautions in using chelators to buffer metals in biological solutions. Cell Calcium. 2004;35(5):427–431. doi: 10.1016/j.ceca.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 44.Dietz H, Rief M. Detecting Molecular Fingerprints in Single Molecule Force Spectroscopy Using Pattern Recognition. Jpn J Appl Phys. 2007;46(8B):5540–5542. [Google Scholar]
- 45.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265(5178):1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 46.Burgess SA, Walker ML, Thirumurugan K, Trinick J, Knight PJ. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J Struct Biol. 2004;147(3):247–258. doi: 10.1016/j.jsb.2004.04.004. [DOI] [PubMed] [Google Scholar]