Abstract
Dissolved organic matter (DOM) in the oceans is one of the largest pools of reduced carbon on Earth, comparable in size to the atmospheric CO2 reservoir. A vast number of compounds are present in DOM, and they play important roles in all major element cycles, contribute to the storage of atmospheric CO2 in the ocean, support marine ecosystems, and facilitate interactions between organisms. At the heart of the DOM cycle lie molecular-level relationships between the individual compounds in DOM and the members of the ocean microbiome that produce and consume them. In the past, these connections have eluded clear definition because of the sheer numerical complexity of both DOM molecules and microorganisms. Emerging tools in analytical chemistry, microbiology, and informatics are breaking down the barriers to a fuller appreciation of these connections. Here we highlight questions being addressed using recent methodological and technological developments in those fields and consider how these advances are transforming our understanding of some of the most important reactions of the marine carbon cycle.
Keywords: dissolved organic matter, marine microbes, cyberinfrastructure
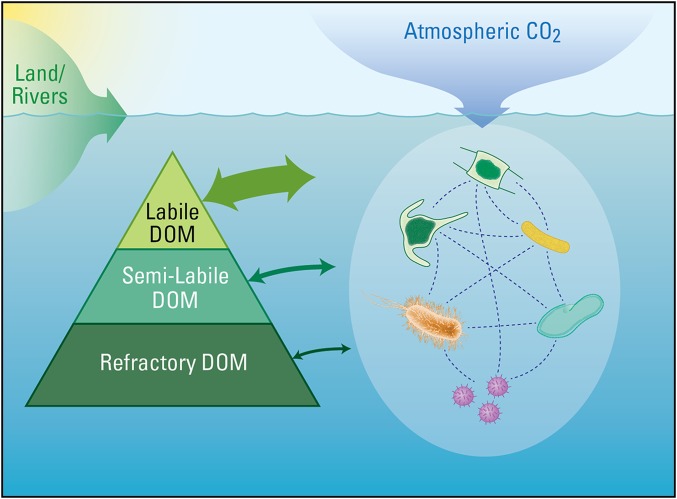
The global cycling of carbon supports life on Earth and affects the state of the biosphere within which humans reside. Industrial processes are now altering the balance of this natural cycle by adding fossil carbon to the contemporary atmosphere and changing our climate (1). Marine dissolved organic matter (DOM) is central to the current and future global cycle, storing as much carbon as the current atmospheric CO2 reservoir (2) (Fig. 1).
Fig. 1.
Oceanic DOM is a complex mixture of molecules that are produced and consumed by billions of heterotrophic and autotrophic microbes in each liter of seawater. These heterogeneous molecules have varied reactivities toward microbial metabolism, including high reactivity (labile DOM, wide arrows) and minimal reactivity (refractory DOM, narrow arrows). Microbe−DOM interactions affect the concentration and fate of atmospheric CO2, the accumulation of refractory carbon in the deep ocean, and flux of carbon through the ocean's food webs.
Flux of carbon through the marine DOM pool is mediated largely by microbial activity. However, the intertwined relationships between the molecules making up the DOM pool and the ocean microbes that process them remain poorly characterized. The complexity of each has defied easy characterization, and fundamental interactions have been necessarily oversimplified to yield a scientifically tractable framework. The principles of organization and interactions between ocean microbial communities and DOM have parallels in other complex ecosystems such as mammalian microbiomes, soils, rhizospheres, extreme environments, and the built environment. Thus, progress in mapping microbe−DOM interactions in the oceans will enhance knowledge across seemingly disparate fields, culminating in a better understanding of element cycling in Earth’s varied ecosystems.
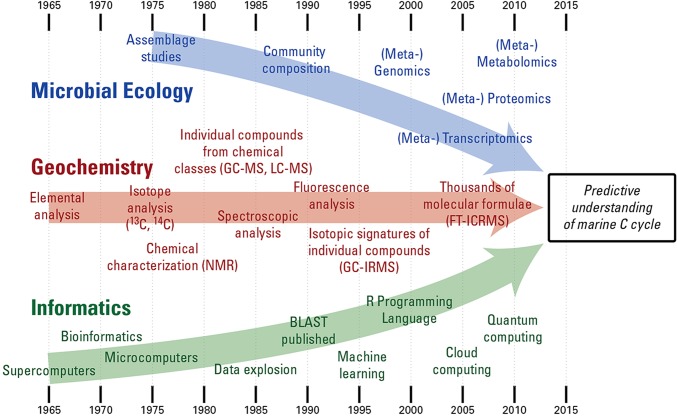
Recent advances in chemistry, microbiology, and data science have directly addressed the complexity of DOM cycling in marine environments and led to a reexamination of basic concepts. A revolution in DNA sequencing technology (3), advances in mass spectrometry (4–6), new approaches to identify metabolites from genome sequences (7), the growth of informatics (8, 9), and the building of knowledge and analysis cyberinfrastructures (10–12) are key tools already in place or in development. As a result, the DOM pool is now known to conservatively consist of tens to hundreds of thousands of different organic molecules (13), for which formulas are rapidly emerging (14). Meanwhile, the ocean microbiome has been estimated to consist of more than a hundred thousand different bacterial, archaeal, and eukaryotic taxa (15, 16) with diverse ecological and metabolic strategies for producing and consuming fixed carbon (17–19). Until recently, major gains in understanding ocean carbon cycling have moved largely along independent lines within the fields of biology and chemistry. Now, it is at the confluence of these disciplines, enabled through innovative data science, that transformative advances are being made (Fig. 2).
Fig. 2.
Significant advances that have occurred independently in three fields—microbial ecology, geochemistry, and informatics—have positioned oceanographers for a deeper understanding of the ocean's carbon cycle. The integration of these three fields is yielding insights into the reactions at the foundation of the global carbon cycle. BLAST, basic local alignment search tool; FT-ICRMS, Fourier transform ion cyclotron resonance mass spectrometry; GC-IRMS, gas chromatography isotope ratio mass spectrometry; GC-MS, gas chromatography mass spectrometry; LC-MS, liquid chromatography mass spectrometry; NMR, nuclear magnetic resonance spectroscopy.
Here we present six fundamental questions in marine biogeochemistry that are benefiting from integrated research strategies. The questions are organized along a general gradient in apparent DOM reactivity that is based on persistence under typical ocean conditions (2). “Labile” DOM refers to the molecules that are consumed by microbes within hours or days of production (Fig. 1). “Semilabile” DOM is less reactive and persists in the surface ocean for weeks to years. “Refractory” DOM is the least biologically reactive and circulates through the major ocean basins on millennial time scales. Although all three operational categories occur throughout the ocean, their relative importance loosely corresponds to a depth gradient. In the surface ocean, the photosynthesis of organic molecules from CO2 by phytoplankton (i.e., primary production) is the source of most of the ocean's labile and semilabile DOM. Semilabile DOM persists long enough to be transported to moderate ocean depths (hundreds of meters below the surface) before it is metabolized (20). Refractory DOM has its strongest signature in the deep ocean (depths greater than 1,000 m) (2, 21). The linkages among individual molecules and microbes that culminate in the global carbon cycle and give rise to the DOM reactivity spectrum lie at the foundation of the questions posed here.
Which Compounds Represent the Largest Conduits of Carbon Flux Through the Labile Marine DOM Pool?
Each year in the surface ocean, ∼20 Gt of carbon recently fixed into organic matter by phytoplankton photosynthesis is rapidly taken up by heterotrophic bacteria (1 Gt = one gigaton or 1 × 1015 g). For perspective, the current annual increase in the atmospheric CO2 pool is 4 Gt C (22), and annual processing of refractory marine DOM is <0.2 Gt C. Many of the labile compounds mediating this brisk and quantitatively important carbon flux into the microbial food web are thought to have half-lives on the order of minutes and concentrations in the picomolar range (23). The very characteristics that define highly labile compounds make their study extremely challenging.
Because phytoplankton cells are rich in proteins and carbohydrates, and these polymers are typically degraded extracellularly into oligomers or monomers before transport into bacterial cells, early research on biologically labile DOM focused primarily on rates and kinetics of amino acid and sugar uptake (24–26). Today, a wealth of new data coming largely but not exclusively from the “'omics” tools (genomics, transcriptomics, proteomics, and metabolomics) suggests that a much wider variety of molecules participate in the rapid heterotrophic DOM flux. For example, gene expression studies in both the ocean and laboratory indicate that labile DOM can take the form of monocarboxylic and dicarboxylic acids (27, 28), glycerols and fatty acids (27, 29), and the nitrogen-containing metabolites taurine, choline, sarcosine, polyamines, methylamines, and ectoine (27, 30, 31). One-carbon compounds such as methanol (27, 29, 31), as well as several sulfonates (32), have recently been added to the list. Chemical analyses concur: Photosynthate released directly from phytoplankton is highly complex, consisting of hundreds of different compounds (33, 34). This “dissolved primary production”—the material released from living phytoplankton—supports a major fraction of labile carbon flux in the surface ocean (35).
Complexity in the composition and concentration of labile DOM presents an ecological opportunity for microbes but an analytical challenge for chemists. Substrates used by heterotrophic bacteria will not accumulate if their demand is higher than their supply; therefore, the most important biologically labile molecules are inherently difficult to recognize against the chemical background of organic compounds in seawater. For example, monomeric amino acids and sugars have concentrations below one billionth of a gram per liter, which is at or below the limits of quantification in marine waters (36, 37). However, detecting low-concentration high-flux compounds has recently become more tractable with methodological advances in chemistry [e.g., better separation methodologies, sensitivity, accuracy, and resolving power (5, 38, 39)], biology [e.g., deducing key substrates from transcriptome analysis (28, 29, 40)], and cyberinfrastructure [e.g., determining patterns of DOM−bacterial interaction networks (41)]. The complementarity of these research fields is key to identifying this massive yet all but invisible flux in the ocean’s active carbon cycle (Fig. 2).
How Are Element Cycles Linked Through Marine DOM?
In addition to driving major fluxes of carbon, microbial production and consumption of DOM in the surface ocean also plays a central role in the cycling of nitrogen (N), phosphorus (P), and sulfur (S), along with micronutrients such as iron, cobalt, nickel, and zinc. Molecules within the marine DOM pool that contain N, P, or S include amino acids and proteins (42), nucleotides and nucleic acids (43), various osmolytes (44), siderophores (45), vitamins (46), and primary metabolites.
Advances in understanding the fate of these diverse components of DOM have occurred despite the fact that extraction of element-specific compound classes quantitatively from seawater remains a challenge, and that characterization of the myriad biological systems that support uptake and transformation of N-, P-, and S-containing organic compounds is daunting. Two main lines of scientific inquiry have motivated progress. The first is a long-standing question in oceanography on the role of organic forms of N, P, and S in alleviating nutrient stress for ocean microbes. This question is particularly relevant in oligotrophic oceans where inorganic nutrients (that is, nitrate, nitrite, ammonium, phosphate, and sulfate) are perennially limiting or energetically expensive to reduce to biologically active forms. Microbes that use organic nutrients may have a significant advantage over those that cannot. With 'omics data as an influential driver, substantial progress is being made in understanding the microbial production and use of organic nutrients. Phosphorylated organic compounds, for example, provide a source of inorganic phosphate after cleavage by phosphatases and nucleotidases (47, 48). Phosphonates are synthesized by marine cyanobacteria, archaea, and other microbes (49–52) and subsequently consumed by both microbial autotrophs (53, 54) and heterotrophs (55). Nitrogen stress is lessened in the oligotrophic ocean by use of urea and cyanate (56, 57). The fate of these dissolved organic N, P, and S molecules in seawater represents a confluence of Earth’s element cycles.
The second issue stimulating research into organic N-, P-, and S-containing compounds is their use as biochemical intermediates and cofactors by auxotrophic microbes (i.e., those unable to synthesize metabolites critical for their own growth). Auxotrophy in the ocean is thought to reflect a microorganism's evolutionary positioning along the trade-off between the expense of biosynthesis of complex molecules, on the one hand, and the risk of relying on neighbors on the other (58). For instance, bacteria in the Pelagibacterales are missing the genetic capability for using extremely abundant sulfate and instead scavenge organic sulfur from seawater (59, 60); this is truly remarkable for what is arguably the most successful heterotrophic microbial group in the ocean. Many marine phytoplankton with critical roles in global carbon fixation have lost biosynthetic pathways for N-, P-, and S-rich vitamins such as B1 and B12 (61) and must scavenge them from the DOM pool. Genomic data have been ideal for learning which marine microbes depend on the DOM pool for energetically expensive biomolecules, whereas metabolomics advances have detected dilute components of DOM that were previously not measurable (39, 46).
How Do Microbe−Microbe Relationships Influence DOM?
Early studies of the marine microbial food web revealed a major role for trophic interactions in the formation and flux of DOM. A surprising 20–50% of microbial biomass is turned over each day in the ocean by viral infection (62), releasing intracellular organic matter into surrounding seawater (63–65). Similarly, protistan grazing on bacteria and phytoplankton converts up to 30% of ingested carbon to dissolved form (66). Protists also directly consume DOM (67) in this intricate network of microbial predation.
Our knowledge of the DOM molecules that arise from or facilitate interactions between marine microbes is growing. Metabolomics approaches have revealed that viral infection increases the concentrations of N-rich metabolites in infected bacteria (38). In this cycle within a cycle, a portion of the organic matter initially assimilated from the DOM pool into bacterial biomass is returned to the DOM pool as viral lysate (62) but is enhanced in N relative to metabolites of noninfected cells (38). New categories of nonpredatory microbial alliances that release organic compounds into seawater are also being recognized. These include molecules in microbial cytosols and exudates (39, 68) that serve as substrates, signaling molecules, and allelochemicals to neighboring microbes (69, 70). Genes able to mediate microbial interactions have also been uncovered (71, 72), including a high prevalence of virulence gene homologs in marine bacteria and archaea that could facilitate direct contact with eukaryotic plankton (73). The conditions under which genes mediating microbial interactions in the ocean are expressed are now better understood because of metatranscriptomic surveys (30, 32). Examples include the alteration of marine phytoplankton growth rates by bacterial release of phenylacetic acid (74) and indole acetic acid (72), and the modulation of bacterial quorum-sensing molecules (75) and antibiotic production (76) by phytoplankton. Global patterns of marine plankton cooccurrences can be better explained by factors involving microbial interactions (such as grazing, viral infection, and parasitic relationships) than by environmental conditions (77). Thus, it has become clear that compounds released into the DOM pool by ocean microbes are considerably more chemically diverse than predicted from the composition of plankton biomass, at least in part because many are synthesized for roles occurring beyond the cell wall.
The full consequences of microbe−microbe interactions depend to a large extent on factors such as cell encounter frequencies in seawater and life history traits of the microbial participants (78). Thus, their prediction must also rely on modeling approaches that consider small- and large-scale dynamics and feedbacks in ocean waters. As examples, a heterotrophic bacterium will experience higher DOM concentrations when associated with a particle compared with when it is free-living (78, 79), and viral−host interactions can, at the same time, kill individual cells while stimulating overall ecosystem productivity (80). New generations of biogeochemical models are explicitly incorporating 'omics-derived data (81, 82) to more directly link microbes and the fate of marine DOM.
How Many Metabolic Pathways Are Required for the Bacterial Transformation of Marine DOM?
Although there is currently no way to know the full biochemical diversity behind microbial processing of DOM, headway is being made. Early genomic studies addressing this question typically focused on transporter systems because they mediate the first essential step in utilization of DOM by heterotrophic bacteria. Transporters involved in organic compound uptake have been reported to account for 13% of expressed genes (28) and 35% of expressed peptides (83) in marine bacterial communities. Over 100 protein families predicted to function in the uptake of organic compounds from seawater have been described in metagenomic, metatranscriptomic, and metaproteomic datasets (28, 84, 85).
However, assigning an exact function to microbial genes remains a stubborn obstacle to more effective use of genomic databases to address DOM transformations. In the case of transporter genes, many are poorly annotated with regard to substrate specificity, and consequently are assigned only to broad categories (such as “branched chain amino acid” or “carboxylate” transporter) based on homology to a limited number of experimentally characterized genes. Making matters worse, transporters classified into the same protein family may mediate uptake of different substrates (86); a single transporter can have multiple substrates (87); and higher molecular weight DOM is assimilated through generic, and therefore uninformative, transporter systems following hydrolysis at the cell surface (88). In the case of catabolic genes, those encoding conserved central metabolic pathways are generally well characterized, but pathways for upstream reactions that ultimately feed into central metabolism, the linchpins of many essential biogeochemical transformations, are poorly known. When we are ultimately successful in identifying a new biogeochemically relevant gene, it is often the case that it was initially annotated with a misleading or uninformative function (89–91).
Substantive improvement in characterization of microbial genes is widely acknowledged as a central goal in biology. One approach to annotation of genes relevant to ocean carbon cycling is to use model microbial systems amenable to experimental manipulation and genetic modification. For example, the first marine bacterial gene mediating catabolism of dimethylsulfoniopropionate (DMSP), a compound known for 50 y to be critical in the global sulfur cycle, was found in 2006 by generating transposon mutants of a marine bacterium (92), and the first phytoplankton gene mediating synthesis of DMSP was found in 2015 by shotgun proteomics of phytoplankton isolates (93). A full suite of DMSP gene discoveries is now enabling studies of the dominant transformation pathways and their regulation in the ocean (94, 95). Similarly, the genes mediating marine bacterial transport and metabolism of organic compounds such as sulfonates (90, 96), ectoine and hydroxyectoine (97), and methylamines and choline (31, 98, 99) have been recently elucidated through model organism systems.
This type of characterization work is slow due to both the small fraction of marine bacteria amenable to culturing and the challenges of developing genetic systems for them. A culture-independent twist involves cloning DNA from marine environments into laboratory strains and performing screens to identify genes conferring a function of interest. Examples include an early effort that identified chitin degradation genes in marine microbial community DNA (100) and a recent study that discovered genes for use of a novel phosphonate (55).
The expansion of cyberinfrastructure capabilities has opened up possibilities of using pattern mining of combinatorial datasets (DOM and metabolite composition, or microbial genes and transcript inventories) to generate hypotheses regarding gene function. This approach is already having success in secondary metabolite research (7, 101). Such efforts will be particularly informative when guided by knowledge of which unknown protein families are ubiquitous in genomes of ocean microbes, or demonstrate phylogenetic coherence, or show biogeographic patterns. Answering the question of how many metabolic pathways are required for the bacterial transformation of marine DOM is perhaps an impossible task, but identification of a subset of pathways that mediate important fluxes of dissolved compounds through the oceanic carbon reservoir is steadily pushing understanding forward.
Why Does Semilabile DOM Accumulate in the Surface Ocean?
Semilabile DOM is operationally defined as the dissolved organic compounds that accumulate in surface waters over time frames of weeks to years but then disappear once exported to depth (20). Why, exactly, these molecules resist degradation in the surface ocean where heterotrophic microbes are often limited by substrates and nutrients remains a mystery. As a substantial and temporally stable component of DOM, the semilabile pool affects the overall rate of carbon turnover in the oceans (Fig. 1). Therefore, illuminating its composition and identifying the metabolic pathways that can degrade it are important for predictive understanding of carbon sequestration (that is, the transfer of excess carbon from the atmosphere into long-term storage in the ocean).
New data are beginning to untangle the factors that covary with semilabile DOM and depth in the ocean, and thereby helping in understanding its fate. Microbial diversity is lower in surface than in deep waters, which suggests that a limited genetic repertoire in surface heterotrophs might restrict degradation of certain compounds (15). At the species level, oligotrophic bacteria such as Pelagibacterales dominate open ocean gyres where semilabile DOM accumulates, and these cells typically have small genomes with fewer and less varied transporters and catabolic pathways (102). Recent experiments with the marine bacterium Alteromonas, harboring a substantially larger genome than the Pelagibacterales, showed that, although this one strain can degrade the labile fraction of marine DOM in a period of days, an amount of DOM equivalent to the semilabile pool remained untouched. Instead, the full microbial community was needed to degrade the semilabile DOM (103). Earlier studies showed that the addition of both labile DOM and inorganic nutrients is needed to degrade semilabile DOM (104), signifying a complex relationship between DOM accumulation, microbial diversity, and the availability of nutrients and cometabolites.
The chemical and optical signatures of seawater also differ from the deep ocean background in locations where semilabile DOM accumulates, indicating that this material is compositionally distinct from labile or refractory DOM. For instance, fluorescence signals indicative of dissolved proteins are elevated and signatures of carbohydrates and aliphatic material are enriched in semilabile DOM relative to the signatures of deep ocean DOM (105, 106). Nevertheless, extraction protocols are insufficient and chemical understanding is too limited to physically isolate semilabile DOM from seawater at this time. Instead, indirect experiments such as time series studies (107), long-term incubations (103), and the isolation of representative microbes (108) are being used to address first-order questions regarding the molecular composition of this enigmatic DOM pool and the metabolic pathways by which it is degraded. New data analysis methods are also helping to parse small but crucial biological signals from these complex data sets.
How Refractory Is Deep Ocean DOM and Why Does It Persist?
The deep ocean represents a challenging ecosystem to study because of its remoteness, the low concentrations of organic molecules, and the slow rates of microbial metabolism at high pressure and low temperature. However, this is the repository for over 70% of the carbon sequestered in DOM and is a major reservoir in the global carbon cycle (Fig. 1). Bulk radiocarbon dating indicates that deep ocean DOM has an average age of 6,000 y (109). More recent radiocarbon techniques showed that the apparent ages of individual molecules are not normally distributed around this average. Instead, different reactivity pools were identified representing both semilabile (radiocarbon enriched) and refractory (radiocarbon deplete) pools within the DOM, with the most deplete fraction having a radiocarbon age of ∼12,000 y (21).
That these energy-rich molecules exist for millennia in the deep ocean is a paradox that seems to contradict the laws of thermodynamics: Why, in a marine environment rich in other necessities for life, would microbes fail to use such a large reservoir of organic carbon? One line of reasoning posits that this pool contains inherently biologically recalcitrant molecules. For example, condensed polycyclic aromatics generated by processes such as wildfire and biomass burning on land accumulate throughout the deep ocean (110, 111) and have radiocarbon ages exceeding those of other DOM pools (112). These molecules are susceptible to photodegradation because of their aromatic functional groups, suggesting that upwelling of deep waters to the surface during ocean circulation may determine the half-life of photochemically active yet biologically refractory compounds (113–115). The majority of deep ocean refractory DOM, however, likely represents the accumulation of metabolic products of ocean microbes that are refractory to further biological degradation (116), a phenomenon termed the “microbial carbon pump” (117). It is not clear if the microbial carbon pump generates inherently refractory molecules from labile forms, or if labile DOM is diversified by the pump until each molecule is present at vanishingly low concentrations. In the former case, refractory DOM would consist of a pool of survivor molecules that are biologically intractable and enriched at depth. In the latter case, refractory DOM would represent a highly diverse suite of compounds, each at its limiting concentration of metabolic utility (118–121).
Conducting the laboratory and field experiments to test current theories of the nature of refractory deep ocean DOM is proving to be both enlightening and challenging. Incubation experiments seeking to measure changes in DOM concentrations and chemistry under conditions that mimic the deep ocean are hampered by inherently low rates of net carbon turnover and analytical techniques that provide limited structural resolution of resistant molecules (121–123). Further, refractory organic matter is defined based on its lifetime in the ocean (2) rather than on inherent chemical structures, making for an elusive experimental target. Extraction of DOM from seawater, a prerequisite to most analytical methods, does not presently yield all of the compounds dissolved in seawater (124). Thus, our view of the molecular composition of DOM remains restricted to the fraction that can be physically isolated and analyzed. On the biological side, the percentage of microbial genes with no known function increases with depth in the ocean (15). The metabolic pathways that degrade refractory molecules may be hidden within these unannotated genes with no analogs in known metabolic pathways. When the question of why deep ocean DOM persists is finally resolved, the answer is likely to be a combination of concentration, chemical structure, bioenergetics, and microbial diversity.
A final notable aspect of the deep ocean ecosystem is that, although it is home to the largest reservoir of refractory DOM, it also harbors labile and semilabile molecules. Multiple lines of evidence have recently revealed labile DOM−microbe interactions far below the photic zone (21, 125–128). For example, carbon isotopic analysis of microbial DNA confirms the incorporation of modern organic matter into microbial biomass in the ocean depths (129). Release from sinking particles is the primary recognized source of modern DOM at depth. Indeed, the dissolved organic compounds liberated by microbes from sinking particles are now thought to fuel up to 90% of carbon cycling in the deep ocean (125, 129–133).
The Next Step: Prototypical Molecules of the Marine Carbon Cycle
An opportunity to identify a broader range of molecular currencies of the marine carbon cycle can be found at the intersection of marine chemistry and ‘omics methodologies, in the context of developments in informatics. Admittedly, successful identification of even several hundred new molecules seems a trivial advance stacked against the enormous chemical diversity of seawater organic matter. However, microbial ecologists would nearly unanimously agree that genome sequences of just 175 marine bacteria (134) out of the hundred thousand taxa present in seawater fueled a revolution in our understanding of element cycling in the ocean. Indeed, many of the recent DOM advances discussed here were directly enabled by foundational data on the genomes of marine plankton (28, 29, 31–33, 38, 39, 70). A corresponding suite of model organic compounds representative of those cycling through the world’s oceans will provide tools for unraveling pathways of carbon flux.
Two categories of prototypical molecules are of particular interest in this endeavor. The first is molecules rapidly produced and metabolized by marine microbes in the fast loop of labile DOM, including biogeochemical intermediates and signaling compounds relevant to organic matter flux. The second is molecules from less labile components of marine DOM that will improve understanding of why molecules are biologically refractory and what characteristics determine their half-life in the ocean. Identification of prototypical compounds will then lead to methodologies for their analysis in bulk seawater and microbial metabolomes, and to synthesis and labeling for flux studies.
Already, modern targeted chemical workflows are enabling quantification of intermediates of biogeochemical cycling. Correspondingly, nontargeted workflows are helping us to discover new molecules we didn’t know to look for (33, 39, 70, 105, 135). Newly developed informatics approaches are supporting data mining across multiple studies and systems (136, 137) (see Box 1). ‘Omics data are allowing us to use microbes as biosensors for the compounds being synthesized, assimilated, and metabolized in the ocean microbiome (27, 28, 138–140). Genetic systems are assigning substrates to uncharacterized genes through knockouts and heterologous expression (31, 76, 92). All of these tools, and others on the horizon, will expand our knowledge of the organic compounds produced and transformed by microbes of the ocean.
Box 1: Cyberinfrastructure
The merging of chemical and microbiological data for resolving microbe−DOM interactions in the ocean is being enabled by advances in data management capabilities and systems, collectively referred to as cyberinfrastructure. In genomics research, the core cyberinfrastructure method is well established: Sequence databases are searched for homology using tools such as basic local alignment search tool (BLAST), and then analyzed for taxonomy and function. Analogous datasets and cyberinfrastructure are now emerging that can be applied to investigate DOM chemistry, for example MetaboLights (www.ebi.ac.uk/metabolights/) and Global Natural Products Social Molecular Networking (GNPS; gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp), which emphasize mass spectrometry knowledge capture and dissemination using social networking. In concert with data accessibility (141), new or existing infrastructure can be specifically dedicated to the growing needs of the DOM community.
Well-engineered data systems adopted by collaborating scientists are the key to keeping up with the burgeoning capacity to generate data. For microbe−DOM research, such systems will require coordinated cyberinfrastructure elements that include descriptions of chemical composition; inventories and interpretation of transcripts, genes, and proteins; and data that are curated and searchable. Publication of open-access datasets must be easy and rewarded. Open source tools for data reduction and intercomparison, such as parallel factor analysis (PARAFAC) for fluorescence data (142), must be developed. By this approach, processing tasks once considered difficult can be automated. Validation, provisions for searchable metadata, provenance, repeatability, and archiving are all considerations that weave into robust cyberinfrastructure development.
Two related elements of cyberinfrastructure design are managing data volume and making data more available. This latter refers both to researchers not directly involved in acquisition and to questions that are not yet anticipated. For example, a field scientist could generate synoptic assays of the near-surface microbiome–DOM systems at a rate of one snapshot every few minutes over a period of weeks, actively tracking the biogeochemical pathways of the ocean. Well-designed cyberinfrastructure would make the resulting data discoverable, explorable, and queryable by other scientists, in addition to performing data reduction and organizational tasks at the many-terabyte scale. As we envision scientists being rewarded for proliferating public data and software, so too should cyberinfrastructure developers be rewarded for building data systems that reduce analysis times from months to minutes and for coordinating with data discovery mechanisms in the scientific community.
Conclusions
Exciting discoveries have been moving the needle on our understanding of the marine microbe−DOM network over the past decade. Successes include improved knowledge of the organic compounds through which nearly a quarter of net global photosynthesis passes within days of fixation, a grasp of the chemical formulas of compounds that persist for tens of thousands of years in the ocean, knowledge of how organic forms of limiting nutrients take part in element cycles, and realization of the crucial roles of marine microbial interactions in Earth's biogeochemical cycles. More discoveries are in the pipeline, helped by innovation in high-throughput methodologies and effective cyberinfrastructures. The next decade will continue this period of rapid learning, both in ways that we glimpse already (through growing accessibility of metabolomics, the speed and lowered cost of next-generation sequencing, and the development of efficient screening tools for gene function) and from directions not yet predictable.
The DOM−microbe complexity challenge has synergies with other areas of science where the chemical foundations of microbial community function are crucial. Microbiome studies, for example, have the same scientific goals of discovering, identifying, and quantifying molecules that link a genome-encoded potential with a realized metabolic and ecological function. Annotation of gene function likewise cuts across many fields and organisms. A compelling example is the recent discovery of the genetic basis of bacterial degradation of the sulfolipid component of photosynthetic membranes, based on studies conducted with bacteria from soil (90), coastal seawater (32), and the human gut (89). Another example is the development of cross-discipline databases for metabolite annotation, including the use of crowdsourcing to solve common problems in compound identification (gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp). The new classes of data and types of methodologies being developed to explore both molecules and microbes will be necessary to predict carbon cycle response to challenges ranging from oil spills to climate change (see Box 2).
Box 2: Microbe−DOM Climate Responses
Climate change effects on ocean systems are being manifested as global shifts in temperature, seawater pH, sea level, circulation patterns, oxygen content, and nutrient and DOM loading from land. Marine ecosystems are also being affected regionally by coastal eutrophication, invasive species, and habitat degradation. Today, oligotrophic subtropical gyres are regions of DOM accumulation (143) and export (144). The predicted growth in the areal extent of gyres in the future, evidenced by a 56% increase of the North Atlantic gyre wintertime area between 1998 and 2006 (145, 146), may therefore increase net oceanic DOM production. However, experimental studies suggest that rising temperatures and ocean acidification will increase bacterial DOM consumption (147, 148), whereas the same drivers may reduce formation of colloids and microgels from DOM (149). Thus, whether or not the future ocean will experience greater accumulation of DOM or alterations in its chemical composition (150) is still unclear.
Climate change is also predicted to alter the distribution and composition of marine phytoplankton communities and create new physical regimes that shift longstanding chemical distributions, throwing together microbes and carbon forms with limited evolutionary history. The emergence of new high-temperature oceanic biomes, currently rare regions where mean sea surface temperatures exceed 31 °C, is projected to establish more than 25 million square kilometers of altered ocean by 2100 (145). Whether microbes inhabiting these and other new niches will interact with DOM as analogs of current assemblages is unknown. Discovery and prediction of microbe−DOM linkages as they react to and shape the future ocean will rely heavily on the tools and concepts discussed in this perspective.
Acknowledgments
We thank Jack Cook for graphics expertise. Formative discussions for this perspective occurred at a workshop entitled “Linking Marine Microbes and the Molecules of Dissolved Organic Matter,” held in New York City in November 2014. The workshop was supported by the Gordon and Betty Moore Foundation and Microsoft Research Corporation. Additional support was provided by National Science Foundation Grants OCE1356010, OCE1154320, and OCE1356890, and by Gordon and Betty Moore Foundation Grant 3304.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Doney SC, et al. Climate change impacts on marine ecosystems. Annu Rev Mar Sci. 2012;4:11–37. doi: 10.1146/annurev-marine-041911-111611. [DOI] [PubMed] [Google Scholar]
- 2.Hansell DA. Recalcitrant dissolved organic carbon fractions. Annu Rev Mar Sci. 2013;5:421–445. doi: 10.1146/annurev-marine-120710-100757. [DOI] [PubMed] [Google Scholar]
- 3.DeLong EF, Karl DM. Genomic perspectives in microbial oceanography. Nature. 2005;437(7057):336–342. doi: 10.1038/nature04157. [DOI] [PubMed] [Google Scholar]
- 4.Koch BP, Ludwichowski KU, Kattner G, Dittmar T, Witt M. Advanced characterization of marine dissolved organic matter by combining reversed-phase liquid chromatography and FT-ICR-MS. Mar Chem. 2008;111(3-4):233–241. [Google Scholar]
- 5.Bouslimani A, Sanchez LM, Garg N, Dorrestein PC. Mass spectrometry of natural products: Current, emerging and future technologies. Nat Prod Rep. 2014;31(6):718–729. doi: 10.1039/c4np00044g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kujawinski EB, Hatcher PG, Freitas MA. High-resolution Fourier transform ion cyclotron resonance mass spectrometry of humic and fulvic acids: Improvements and comparisons. Anal Chem. 2002;74(2):413–419. doi: 10.1021/ac0108313. [DOI] [PubMed] [Google Scholar]
- 7.Cimermancic P, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158(2):412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dührkop K, Shen H, Meusel M, Rousu J, Böcker S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc Natl Acad Sci USA. 2015;112(41):12580–12585. doi: 10.1073/pnas.1509788112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watrous J, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA. 2012;109(26):E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amaral-Zettler L, et al. A global census of marine microbes. In: McIntyre AD, editor. Life in the World’s Oceans: Diversity, Distribution and Abundance. Wiley-Blackwell; Oxford: 2010. pp. 223–245. [Google Scholar]
- 11.Haug K, et al. MetaboLights—An open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 2013;41(Database issue):D781–D786. doi: 10.1093/nar/gks1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun S, et al. Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: The CAMERA resource. Nucleic Acids Res. 2011;39(Database issue):D546–D551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S, Kramer RW, Hatcher PG. Graphical method for analysis of ultrahigh-resolution broadband mass spectra of natural organic matter, the van Krevelen diagram. Anal Chem. 2003;75(20):5336–5344. doi: 10.1021/ac034415p. [DOI] [PubMed] [Google Scholar]
- 14.Hertkorn N, et al. Characterization of a major refractory component of marine dissolved organic matter. Geochim Cosmochim Acta. 2006;70(12):2990–3010. [Google Scholar]
- 15.Sunagawa S, et al. Tara Oceans coordinators Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348(6237):1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 16.Gibbons SM, et al. Evidence for a persistent microbial seed bank throughout the global ocean. Proc Natl Acad Sci USA. 2013;110(12):4651–4655. doi: 10.1073/pnas.1217767110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganesh S, Parris DJ, DeLong EF, Stewart FJ. Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J. 2014;8(1):187–211. doi: 10.1038/ismej.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall KT, Morris RM. Isolation of an aerobic sulfur oxidizer from the SUP05/Arctic96BD-19 clade. ISME J. 2013;7(2):452–455. doi: 10.1038/ismej.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran MA, et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature. 2004;432(7019):910–913. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- 20.Hansell DA, Carlson CA. Net community production of dissolved organic carbon. Global Biogeochem Cycles. 1998;12(3):443–453. [Google Scholar]
- 21.Follett CL, Repeta DJ, Rothman DH, Xu L, Santinelli C. Hidden cycle of dissolved organic carbon in the deep ocean. Proc Natl Acad Sci USA. 2014;111(47):16706–16711. doi: 10.1073/pnas.1407445111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciais P, et al. Carbon and other biogeochemical cycles. In: Stocker T, et al., editors. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge Univ Press; New York: 2014. pp. 465–570. [Google Scholar]
- 23.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5(10):782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 24.Ferguson RL, Sunda WG. Utilization of amino acids by planktonic marine bacteria: Importance of clean technique and low substrate additions. Limnol Oceanogr. 1984;29(2):258–274. [Google Scholar]
- 25.Hodson RE, et al. Microbial uptake of dissolved organic matter in McMurdo Sound, Antarctica. Mar Biol. 1981;61(2-3):89–94. [Google Scholar]
- 26.Hollibaugh JT, Azam F. Microbial degradation of dissolved proteins in seawater. Limnol Oceanogr. 1983;28(6):1104–1116. [Google Scholar]
- 27.Gifford SM, Sharma S, Booth M, Moran MA. Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J. 2013;7(2):281–298. doi: 10.1038/ismej.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poretsky RS, Sun S, Mou X, Moran MA. Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ Microbiol. 2010;12(3):616–627. doi: 10.1111/j.1462-2920.2009.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarren J, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci USA. 2010;107(38):16420–16427. doi: 10.1073/pnas.1010732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q, Lu X, Tolar BB, Mou X, Hollibaugh JT. Concentrations, turnover rates and fluxes of polyamines in coastal waters of the South Atlantic Bight. Biogeochemistry. 2015;123(1-2):117–133. [Google Scholar]
- 31.Lidbury I, Murrell JC, Chen Y. Trimethylamine N-oxide metabolism by abundant marine heterotrophic bacteria. Proc Natl Acad Sci USA. 2014;111(7):2710–2715. doi: 10.1073/pnas.1317834111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durham BP, et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci USA. 2015;112(2):453–457. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becker JW, et al. Closely related phytoplankton species produce similar suites of dissolved organic matter. Front Microbiol. 2014;5:111. doi: 10.3389/fmicb.2014.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bittar TB, Vieira AAH, Stubbins A, Mopper K. Competition between photochemical and biological degradation of dissolved organic matter from the cyanobacteria Microcystis aeruginosa. Limnol Oceanogr. 2015;60(4):1172–1194. [Google Scholar]
- 35.Morán XAG, Ducklow HW, Erickson M. Carbon fluxes through estuarine bacteria reflect coupling with phytoplankton. Mar Ecol Prog Ser. 2013;489:75–85. [Google Scholar]
- 36.Kaiser K, Benner R. Organic matter transformations in the upper mesopelagic zone of the North Pacific: Chemical composition and linkages to microbial community structure. J Geophys Res. 2012;117(C1):C01023. [Google Scholar]
- 37.Mopper K, et al. Determination of sugars in unconcentrated seawater and other natural waters by liquid chromatography and pulsed amperometric detection. Environ Sci Technol. 1992;26(1):133–138. [Google Scholar]
- 38.Ankrah NY, et al. Phage infection of an environmentally relevant marine bacterium alters host metabolism and lysate composition. ISME J. 2014;8(5):1089–1100. doi: 10.1038/ismej.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiore CL, Longnecker K, Kido Soule MC, Kujawinski EB. Release of ecologically relevant metabolites by the cyanobacterium Synechococcus elongates CCMP 1631. Environ Microbiol. 2015;17(10):3949–3963. doi: 10.1111/1462-2920.12899. [DOI] [PubMed] [Google Scholar]
- 40.Beier S, Rivers AR, Moran MA, Obernosterer I. The transcriptional response of prokaryotes to phytoplankton-derived dissolved organic matter in seawater. Environ Microbiol. 2015;17(10):3466–3480. doi: 10.1111/1462-2920.12434. [DOI] [PubMed] [Google Scholar]
- 41.Raes J, Foerstner KU, Bork P. Get the most out of your metagenome: Computational analysis of environmental sequence data. Curr Opin Microbiol. 2007;10(5):490–498. doi: 10.1016/j.mib.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 42.Keil RG, Kirchman DL. Dissolved combined amino acids: Chemical form and utilization by marine bacteria. Limnol Oceanogr. 1993;38(6):1256–1270. [Google Scholar]
- 43.Björkman KM, Karl DM. Presence of dissolved nucleotides in the North Pacific Subtropical Gyre and their role in cycling of dissolved organic phosphorus. Aquat Microb Ecol. 2005;39(2):193–203. [Google Scholar]
- 44.Diaz MR, Visscher PT, Taylor BF. Metabolism of dimethylsulfoniopropionate and glycine betaine by a marine bacterium. FEMS Microbiol Lett. 1992;96(1):61–65. [Google Scholar]
- 45.Gledhill M, Buck KN. The organic complexation of iron in the marine environment: A review. Front Microbiol. 2012;3:69. doi: 10.3389/fmicb.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sañudo-Wilhelmy SA, et al. Multiple B-vitamin depletion in large areas of the coastal ocean. Proc Natl Acad Sci USA. 2012;109(35):14041–14045. doi: 10.1073/pnas.1208755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dyhrman ST, et al. The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS One. 2012;7(3):e33768. doi: 10.1371/journal.pone.0033768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sebastian M, Ammerman JW. The alkaline phosphatase PhoX is more widely distributed in marine bacteria than the classical PhoA. ISME J. 2009;3(5):563–572. doi: 10.1038/ismej.2009.10. [DOI] [PubMed] [Google Scholar]
- 49.Cade-Menun BJ, Benitez-Nelson CR, Pellechia P, Paytan A. Refining 31P nuclear magnetic resonance spectroscopy for marine particulate samples: Storage conditions and extraction recovery. Mar Chem. 2005;97(3-4):293–306. [Google Scholar]
- 50.Dyhrman ST, Benitez-Nelson CR, Orchard ED, Haley ST, Pellechia PJ. A microbial source of phosphonates in oligotrophic marine systems. Nat Geosci. 2009;2(10):696–699. [Google Scholar]
- 51.Metcalf WW, et al. Synthesis of methylphosphonic acid by marine microbes: A source for methane in the aerobic ocean. Science. 2012;337(6098):1104–1107. doi: 10.1126/science.1219875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Mooy BA, et al. Phosphorus cycling. Major role of planktonic phosphate reduction in the marine phosphorus redox cycle. Science. 2015;348(6236):783–785. doi: 10.1126/science.aaa8181. [DOI] [PubMed] [Google Scholar]
- 53.Feingersch R, et al. Potential for phosphite and phosphonate utilization by Prochlorococcus. ISME J. 2012;6(4):827–834. doi: 10.1038/ismej.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karl DM, et al. Aerobic production of methane in the sea. Nat Geosci. 2008;1(7):473–478. [Google Scholar]
- 55.Martínez A, Ventouras LA, Wilson ST, Karl DM, Delong EF. Metatranscriptomic and functional metagenomic analysis of methylphosphonate utilization by marine bacteria. Front Microbiol. 2013;4:340. doi: 10.3389/fmicb.2013.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saito MA, et al. Multiple nutrient stresses at intersecting Pacific Ocean biomes detected by protein biomarkers. Science. 2014;345(6201):1173–1177. doi: 10.1126/science.1256450. [DOI] [PubMed] [Google Scholar]
- 57.Glibert PM, et al. Evidence for dissolved organic nitrogen and phosphorus uptake during a cyanobacterial bloom in Florida Bay. Mar Ecol Prog Ser. 2004;280:73–83. [Google Scholar]
- 58.Morris JJ, Lenski RE, Zinser ER. The Black Queen Hypothesis: Evolution of dependencies through adaptive gene loss. MBio. 2012;3(2):e00036-12. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carini P, Steindler L, Beszteri S, Giovannoni SJ. Nutrient requirements for growth of the extreme oligotroph ‘Candidatus Pelagibacter ubique’ HTCC1062 on a defined medium. ISME J. 2013;7(3):592–602. doi: 10.1038/ismej.2012.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tripp HJ, et al. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452(7188):741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- 61.Bertrand EM, Allen AE. Influence of vitamin B auxotrophy on nitrogen metabolism in eukaryotic phytoplankton. Front Microbiol. 2012;3:375. doi: 10.3389/fmicb.2012.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suttle CA. Marine viruses—Major players in the global ecosystem. Nat Rev Microbiol. 2007;5(10):801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 63.Haaber J, Middelboe M. Viral lysis of Phaeocystis pouchetii: Implications for algal population dynamics and heterotrophic C, N and P cycling. ISME J. 2009;3(4):430–441. doi: 10.1038/ismej.2008.125. [DOI] [PubMed] [Google Scholar]
- 64.Lønborg C, Middelboe M, Brussaard CPD. Viral lysis of Micromonas pusilla: Impacts on dissolved organic matter production and composition. Biogeochemistry. 2013;116(1-3):231–240. [Google Scholar]
- 65.Weinbauer MG, Chen F, Wilhelm SW. Virus-mediated redistribution and partitioning of carbon in the global oceans. In: Jiao N, Azam F, Sanders S, editors. Microbial Carbon Pump in the Ocean. Science/AAAS; Washington, DC: 2011. pp. 54–56. [Google Scholar]
- 66.Sherr EB, Sherr BF. Bacterivory and herbivory: Key roles of phagotrophic protists in pelagic food webs. Microb Ecol. 1994;28(2):223–235. doi: 10.1007/BF00166812. [DOI] [PubMed] [Google Scholar]
- 67.Sherr EB. Direct use of high molecular weight polysaccharide by heterotrophic flagellates. Nature. 1988;335(6188):348–351. [Google Scholar]
- 68.Paul C, Barofsky A, Vidoudez C, Pohnert G. Diatom exudates influence metabolism and cell growth of co-cultured diatom species. Mar Ecol Prog Ser. 2009;389:61–70. [Google Scholar]
- 69.Zelezniak A, et al. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci USA. 2015;112(20):6449–6454. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson WM, Kido Soule MC, Kujawinski EB. Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. ISME J. 2016 doi: 10.1038/ismej.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rinta-Kanto JM, et al. Analysis of sulfur-related transcription by Roseobacter communities using a taxon-specific functional gene microarray. Environ Microbiol. 2011;13(2):453–467. doi: 10.1111/j.1462-2920.2010.02350.x. [DOI] [PubMed] [Google Scholar]
- 72.Amin SA, et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522(7554):98–101. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 73.Persson OP, et al. High abundance of virulence gene homologues in marine bacteria. Environ Microbiol. 2009;11(6):1348–1357. doi: 10.1111/j.1462-2920.2008.01861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem. 2011;3(4):331–335. doi: 10.1038/nchem.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schaefer AL, et al. A new class of homoserine lactone quorum-sensing signals. Nature. 2008;454(7204):595–599. doi: 10.1038/nature07088. [DOI] [PubMed] [Google Scholar]
- 76.Geng H, Belas R. Molecular mechanisms underlying Roseobacter-phytoplankton symbioses. Curr Opin Biotechnol. 2010;21(3):332–338. doi: 10.1016/j.copbio.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 77.Lima-Mendez G, et al. Tara Oceans coordinators Ocean plankton. Determinants of community structure in the global plankton interactome. Science. 2015;348(6237):1262073. doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- 78.Stocker R. Marine microbes see a sea of gradients. Science. 2012;338(6107):628–633. doi: 10.1126/science.1208929. [DOI] [PubMed] [Google Scholar]
- 79.Kiørboe T, Jackson GA. Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol Oceanogr. 2001;46(6):1309–1318. [Google Scholar]
- 80.Weitz JS, et al. A multitrophic model to quantify the effects of marine viruses on microbial food webs and ecosystem processes. ISME J. 2015;9(6):1352–1364. doi: 10.1038/ismej.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Follows MJ, Dutkiewicz S, Grant S, Chisholm SW. Emergent biogeography of microbial communities in a model ocean. Science. 2007;315(5820):1843–1846. doi: 10.1126/science.1138544. [DOI] [PubMed] [Google Scholar]
- 82.Reed DC, Algar CK, Huber JA, Dick GJ. Gene-centric approach to integrating environmental genomics and biogeochemical models. Proc Natl Acad Sci USA. 2014;111(5):1879–1884. doi: 10.1073/pnas.1313713111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sowell SM, et al. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 2009;3(1):93–105. doi: 10.1038/ismej.2008.83. [DOI] [PubMed] [Google Scholar]
- 84.Morris RM, et al. Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME J. 2010;4(5):673–685. doi: 10.1038/ismej.2010.4. [DOI] [PubMed] [Google Scholar]
- 85.Shi Y, Tyson GW, Eppley JM, DeLong EF. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2011;5(6):999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saier MH., Jr Families of transmembrane sugar transport proteins. Mol Microbiol. 2000;35(4):699–710. doi: 10.1046/j.1365-2958.2000.01759.x. [DOI] [PubMed] [Google Scholar]
- 87.van der Heide T, Poolman B. ABC transporters: One, two or four extracytoplasmic substrate-binding sites? EMBO Rep. 2002;3(10):938–943. doi: 10.1093/embo-reports/kvf201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arnosti C. Microbial extracellular enzymes and the marine carbon cycle. Annu Rev Mar Sci. 2011;3:401–425. doi: 10.1146/annurev-marine-120709-142731. [DOI] [PubMed] [Google Scholar]
- 89.Denger K, et al. Sulphoglycolysis in Escherichia coli K-12 closes a gap in the biogeochemical sulphur cycle. Nature. 2014;507(7490):114–117. doi: 10.1038/nature12947. [DOI] [PubMed] [Google Scholar]
- 90.Mayer J, et al. 2,3-Dihydroxypropane-1-sulfonate degraded by Cupriavidus pinatubonensis JMP134: Purification of dihydroxypropanesulfonate 3-dehydrogenase. Microbiology. 2010;156(Pt 5):1556–1564. doi: 10.1099/mic.0.037580-0. [DOI] [PubMed] [Google Scholar]
- 91.Todd JD, et al. Molecular dissection of bacterial acrylate catabolism—Unexpected links with dimethylsulfoniopropionate catabolism and dimethyl sulfide production. Environ Microbiol. 2010;12(2):327–343. doi: 10.1111/j.1462-2920.2009.02071.x. [DOI] [PubMed] [Google Scholar]
- 92.Howard EC, et al. Bacterial taxa that limit sulfur flux from the ocean. Science. 2006;314(5799):649–652. doi: 10.1126/science.1130657. [DOI] [PubMed] [Google Scholar]
- 93.Alcolombri U, et al. MARINE SULFUR CYCLE. Identification of the algal dimethyl sulfide-releasing enzyme: A missing link in the marine sulfur cycle. Science. 2015;348(6242):1466–1469. doi: 10.1126/science.aab1586. [DOI] [PubMed] [Google Scholar]
- 94.Levine NM, et al. Environmental, biochemical and genetic drivers of DMSP degradation and DMS production in the Sargasso Sea. Environ Microbiol. 2012;14(5):1210–1223. doi: 10.1111/j.1462-2920.2012.02700.x. [DOI] [PubMed] [Google Scholar]
- 95.Varaljay VA, et al. Single-taxon field measurements of bacterial gene regulation controlling DMSP fate. ISME J. 2015;9(7):1677–1686. doi: 10.1038/ismej.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Denger K, et al. Bifurcated degradative pathway of 3-sulfolactate in Roseovarius nubinhibens ISM via sulfoacetaldehyde acetyltransferase and (S)-cysteate sulfolyase. J Bacteriol. 2009;191(18):5648–5656. doi: 10.1128/JB.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lecher J, et al. The crystal structure of UehA in complex with ectoine-A comparison with other TRAP-T binding proteins. J Mol Biol. 2009;389(1):58–73. doi: 10.1016/j.jmb.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 98.Chen Y, McAleer KL, Murrell JC. Monomethylamine as a nitrogen source for a nonmethylotrophic bacterium, Agrobacterium tumefaciens. Appl Environ Microbiol. 2010;76(12):4102–4104. doi: 10.1128/AEM.00469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lidbury I, Kimberley G, Scanlan DJ, Murrell JC, Chen Y. Comparative genomics and mutagenesis analyses of choline metabolism in the marine Roseobacter clade. Environ Microbiol. 2015;17(12):5048–5062. doi: 10.1111/1462-2920.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cottrell MT, Moore JA, Kirchman DL. Chitinases from uncultured marine microorganisms. Appl Environ Microbiol. 1999;65(6):2553–2557. doi: 10.1128/aem.65.6.2553-2557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wilson MC, Piel J. Metagenomic approaches for exploiting uncultivated bacteria as a resource for novel biosynthetic enzymology. Chem Biol. 2013;20(5):636–647. doi: 10.1016/j.chembiol.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 102.Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309(5738):1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 103.Pedler BE, Aluwihare LI, Azam F. Single bacterial strain capable of significant contribution to carbon cycling in the surface ocean. Proc Natl Acad Sci USA. 2014;111(20):7202–7207. doi: 10.1073/pnas.1401887111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Carlson CA, et al. Effect of nutrient amendments on bacterioplankton production, community structure, and DOC utilization in the northwestern Sargasso Sea. Aquat Microb Ecol. 2002;30(1):19–36. [Google Scholar]
- 105.Hertkorn N, Harir M, Koch BP, Michalke B, Schmitt-Kopplin P. High-field NMR spectroscopy and FTICR mass spectrometry: Powerful discovery tools for the molecular level characterization of marine dissolved organic matter. Biogeosciences. 2013;10(3):1583–1624. [Google Scholar]
- 106.Jørgensen L, et al. Global trends in the fluorescence characteristics and distribution of marine dissolved organic matter. Mar Chem. 2011;126(1):139–148. [Google Scholar]
- 107.Treusch AH, et al. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 2009;3(10):1148–1163. doi: 10.1038/ismej.2009.60. [DOI] [PubMed] [Google Scholar]
- 108.Sosa OA, Gifford SM, Repeta DJ, DeLong EF. High molecular weight dissolved organic matter enrichment selects for methylotrophs in dilution to extinction cultures. ISME J. 2015;9(12):2725–2739. doi: 10.1038/ismej.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Williams PM, Druffel ERM. Radiocarbon in dissolved organic matter in the central North Pacific Ocean. Nature. 1987;330(6145):246–248. [Google Scholar]
- 110.Dittmar T, Koch BP. Thermogenic organic matter dissolved in the abyssal ocean. Mar Chem. 2006;102(3):208–217. [Google Scholar]
- 111.Dittmar T, Paeng J. A heat-induced molecular signature in marine dissolved organic matter. Nat Geosci. 2009;2(3):175–179. [Google Scholar]
- 112.Ziolkowski LA, Druffel ERM. Aged black carbon identified in marine dissolved organic carbon. Geophys Res Lett. 2010;37:L16601. [Google Scholar]
- 113.Mopper K, Kieber DJ, Stubbins A, Hansell D, Carlson C. Marine photochemistry of organic matter: Processes and impacts. In: Hansell DA, Carlson CA, editors. Biogeochemistry of Marine Dissolved Organic Matter. 2nd Ed. Academic; Boston: 2015. pp. 389–450. [Google Scholar]
- 114.Stubbins A, Niggemann J, Dittmar T. Photo-lability of deep ocean dissolved black carbon. Biogeosciences. 2012;9(5):1661–1670. [Google Scholar]
- 115.Medeiros PM, et al. Dissolved organic matter composition and photochemical transformations in the northern North Pacific Ocean. Geophys Res Lett. 2015;42(3):863–870. [Google Scholar]
- 116.Lechtenfeld OJ, Hertkorn N, Shen Y, Witt M, Benner R. Marine sequestration of carbon in bacterial metabolites. Nat Commun. 2015;6:6711. doi: 10.1038/ncomms7711. [DOI] [PubMed] [Google Scholar]
- 117.Jiao N, et al. Microbial production of recalcitrant dissolved organic matter: Long-term carbon storage in the global ocean. Nat Rev Microbiol. 2010;8(8):593–599. doi: 10.1038/nrmicro2386. [DOI] [PubMed] [Google Scholar]
- 118.Hertkorn N, et al. Natural organic matter and the event horizon of mass spectrometry. Anal Chem. 2008;80(23):8908–8919. doi: 10.1021/ac800464g. [DOI] [PubMed] [Google Scholar]
- 119.Arrieta JM, et al. Ocean chemistry. Dilution limits dissolved organic carbon utilization in the deep ocean. Science. 2015;348(6232):331–333. doi: 10.1126/science.1258955. [DOI] [PubMed] [Google Scholar]
- 120.Dittmar T. Reasons behind the long-term stability of dissolved organic matter. In: Hansell DA, Carlson CA, editors. Biogeochemistry of Marine Dissolved Organic Matter. 2nd Ed. Academic; Boston: 2015. pp. 369–388. [Google Scholar]
- 121.Osterholz H, Niggemann J, Giebel H-A, Simon M, Dittmar T. Inefficient microbial production of refractory dissolved organic matter in the ocean. Nat Commun. 2015;6:7422. doi: 10.1038/ncomms8422. [DOI] [PubMed] [Google Scholar]
- 122.Aluwihare LI, Repeta DJ, Chen RF. A major biopolymeric component to dissolved organic carbon in surface sea water. Nature. 1997;387(6629):166–169. [Google Scholar]
- 123.Carlson CA, et al. Interactions among dissolved organic carbon, microbial processes, and community structure in the mesopelagic zone of the northwestern Sargasso Sea. Limnol Oceanogr. 2004;49(4):1073–1083. [Google Scholar]
- 124.Green NW, et al. An intercomparison of three methods for the large-scale isolation of oceanic dissolved organic matter. Mar Chem. 2014;161:14–19. [Google Scholar]
- 125.Arístegui J, Gasol JM, Duarte CM, Herndl GJ. Microbial oceanography of the dark ocean’s pelagic realm. Limnol Oceanogr. 2009;54(5):1501–1529. [Google Scholar]
- 126.Beaupré SR, Aluwihare L. Constraining the 2-component model of marine dissolved organic radiocarbon. Deep Sea Res Part II. 2010;57(16):1494–1503. [Google Scholar]
- 127.DeLong EF, et al. Community genomics among stratified microbial assemblages in the ocean’s interior. Science. 2006;311(5760):496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 128.Repeta DJ, Aluwihare LI. Radiocarbon analysis of neutral sugars in high-molecular-weight dissolved organic carbon: Implications for organic carbon cycling. Limnol Oceanogr. 2006;51(2):1045–1053. [Google Scholar]
- 129.Hansman RL, et al. The radiocarbon signature of microorganisms in the mesopelagic ocean. Proc Natl Acad Sci USA. 2009;106(16):6513–6518. doi: 10.1073/pnas.0810871106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cho BC, Azam F. Major role of bacteria in biogeochemical fluxes in the ocean’s interior. Nature. 1988;332(6163):441–443. [Google Scholar]
- 131.Ingalls AE, et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA. 2006;103(17):6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nagata T. Production mechanisms of dissolved organic matter. In: Kirchman DL, editor. Microbial Ecology of the Oceans, Wiley Series in Ecological and Applied Microbiology. Wiley; New York: 2000. pp. 121–152. [Google Scholar]
- 133.Swan BK, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science. 2011;333(6047):1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- 134.Yooseph S, et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 2010;468(7320):60–66. doi: 10.1038/nature09530. [DOI] [PubMed] [Google Scholar]
- 135.Koch BP, Witt MR, Engbrodt R, Dittmar T, Kattner G. Molecular formulae of marine and terrigenous dissolved organic matter detected by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Geochim Cosmochim Acta. 2005;69(13):3299–3308. [Google Scholar]
- 136.Duncan KR, et al. Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species. Chem Biol. 2015;22(4):460–471. doi: 10.1016/j.chembiol.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Johnson CH, Ivanisevic J, Benton HP, Siuzdak G. Bioinformatics: The next frontier of metabolomics. Anal Chem. 2015;87(1):147–156. doi: 10.1021/ac5040693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ferrera I, Sebastian M, Acinas SG, Gasol JM. Prokaryotic functional gene diversity in the sunlit ocean: Stumbling in the dark. Curr Opin Microbiol. 2015;25:33–39. doi: 10.1016/j.mib.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 139.Lau WW, Armbrust EV. Detection of glycolate oxidase gene glcD diversity among cultured and environmental marine bacteria. Environ Microbiol. 2006;8(10):1688–1702. doi: 10.1111/j.1462-2920.2006.01092.x. [DOI] [PubMed] [Google Scholar]
- 140.Teeling H, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336(6081):608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 141.da Silva RR, Dorrestein PC, Quinn RA. Illuminating the dark matter in metabolomics. Proc Natl Acad Sci USA. 2015;112(41):12549–12550. doi: 10.1073/pnas.1516878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stedmon CA, Bro R. Characterizing dissolved organic matter fluorescence with parallel factor analysis: A tutorial. Limnol Oceanogr Methods. 2008;6(11):572–579. [Google Scholar]
- 143.Church MJ, Ducklow HW, Karl DM. Multi-year increases in dissolved organic matter inventories at Station ALOHA in the North Pacific Subtropical Gyre. Limnol Oceanogr. 2002;47(1):1–10. [Google Scholar]
- 144.Carlson CA, Ducklow HW, Michaels AF. Annual flux of dissolved organic carbon from the euphotic zone in the Northwestern Sargasso Sea. Nature. 1994;371(6496):405–408. [Google Scholar]
- 145.Polovina JJ, Dunne JP, Woodworth PA, Howell EA. Projected expansion of the subtropical biome and contraction of the temperate and equatorial upwelling biomes in the North Pacific under global warming. ICES J Mar Sci. 2011;68(6):986–995. [Google Scholar]
- 146.Polovina JJ, Howell EA, Abecassis M. Ocean’s least productive waters are expanding. Geophys Res Lett. 2008;35(3):L03618. [Google Scholar]
- 147.Endres S, Galgani L, Riebesell U, Schulz KG, Engel A. Stimulated bacterial growth under elevated p CO₂: Results from an off-shore mesocosm study. PLoS One. 2014;9(6):e99228. doi: 10.1371/journal.pone.0099228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Engel A, et al. Impact of CO2 enrichment on organic matter dynamics during nutrient induced coastal phytoplankton blooms. J Plankton Res. 2014;36(3):641–657. [Google Scholar]
- 149.Chen CS, Anaya JM, Chen EY, Farr E, Chin WC. Ocean warming-acidification synergism undermines dissolved organic matter assembly. PLoS One. 2015;10(2):e0118300. doi: 10.1371/journal.pone.0118300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zark M, Riebesell U, Dittmar T. Effects of ocean acidification on marine dissolved organic matter are not detectable over the succession of phytoplankton blooms. Sci Adv. 2015;1(9):e1500531. doi: 10.1126/sciadv.1500531. [DOI] [PMC free article] [PubMed] [Google Scholar]