Significance
Mammals including humans use highly specific pathways for tissue differentiation. One such pathway is operative in reticulocytes and involves the programmed destruction of the cell’s organellar complement by 15-lipoxygenase (15-LOX), which oxygenates polyunsaturated membrane fatty acids and provokes organelle leakage. As we report here, plants make use of a similar LOX pathway to degrade their chloroplasts during leaf senescence. The enzyme involved is a 13-LOX with unique positional specificity and molecular terms. Because 15-LOX and 13-LOX pathway products likewise operate in biological defense, a mechanism of cross-kingdom conservation of pathway regulation and function was uncovered for multicellular eukaryotes.
Keywords: chloroplast envelope, membrane destruction, oxylipins, green leaf volatiles, herbivore deterrence
Abstract
Leaf senescence is the terminal stage in the development of perennial plants. Massive physiological changes occur that lead to the shut down of photosynthesis and a cessation of growth. Leaf senescence involves the selective destruction of the chloroplast as the site of photosynthesis. Here, we show that 13-lipoxygenase (13-LOX) accomplishes a key role in the destruction of chloroplasts in senescing plants and propose a critical role of its NH2-terminal chloroplast transit peptide. The 13-LOX enzyme identified here accumulated in the plastid envelope and catalyzed the dioxygenation of unsaturated membrane fatty acids, leading to a selective destruction of the chloroplast and the release of stromal constituents. Because 13-LOX pathway products comprise compounds involved in insect deterrence and pathogen defense (volatile aldehydes and oxylipins), a mechanism of unmolested nitrogen and carbon relocation is suggested that occurs from leaves to seeds and roots during fall.
Despite considerable progress made over the last few years, little is still known about the molecular mechanism governing plant senescence. Up-regulation of thousands of different genes has been reported (1–3). These so-called senescence‐associated genes (SAGs) encode components active in signal perception, transduction, and execution, as well as hormone homoeostasis (1–3). Genetic and biochemical studies identified several receptor‐like kinases, MAP kinase cascades, as well as NAC and WRKY transcription factors as regulating the senescence program (4, 5).
During leaf senescence, chloroplasts as sites of photosynthesis undergo massive destruction and finally collapse. Symptoms similar to those observed during natural senescence can be induced by treating leaf tissues with jasmonic acid (JA) and its methyl ester, methyl jasmonate (MeJA), which are widespread plant cyclopentanone compounds with similarities to prostaglandins (see ref. 6 for review). During natural and MeJA-induced senescence, common decreases in the photosynthetic capacity and chlorophyll content have been observed (1–3). Carbon and nitrogen relocation is of fundamental importance for seed filling, and thus mass degradation of photosynthetic constituents needs to be tightly controlled in time and space (7, 8). In fact, more than one billion tons of chlorophyll, coming from the disassembled photosynthetic apparatus, needs to be turned over every year (9). Similarly, the key enzyme of photosynthesis, ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo), has to be degraded (10). Turnover of these key macromolecules is thought to proceed through a tight interaction between senescing chloroplasts, the cytosol, and lytic vacuoles. Several stromal proteases are implicated in the early steps of RuBisCo breakdown (11–13). Degradation of chlorophyll involves numerous, well-characterized steps and involves plastid and nonplastid reactions (14). Lytic vacuoles have been implicated in chloroplast breakdown, and autophagy is supposed to provide a major step in final chloroplast dispersal (see ref. 15 for review).
The question of how the initial steps of chloroplast breakdown may proceed has not been answered yet. Previous work suggested some leakage of plastid constituents from senescing chloroplasts (see ref. 16 for review). In the present study, we provide evidence for a mechanism of controlled chloroplast destruction that involves a 13-lipoxygenase (13-LOX), the molecular properties of which match perfectly those of an enzyme attacking unsaturated membrane fatty acids of the plastid envelope and thereby introducing holes for mass export of stromal constituents. The expression and localization of 13-LOX explain how the chloroplast compartment is deconstructed in naturally senescing plants and in plants senescing artificially as a result of MeJA treatment. Structural modeling of 13-LOX provides insights into the molecular mechanisms underlying membrane dispersal.
Results
Chloroplast Destruction During Natural and MeJA-Induced Leaf Senescence.
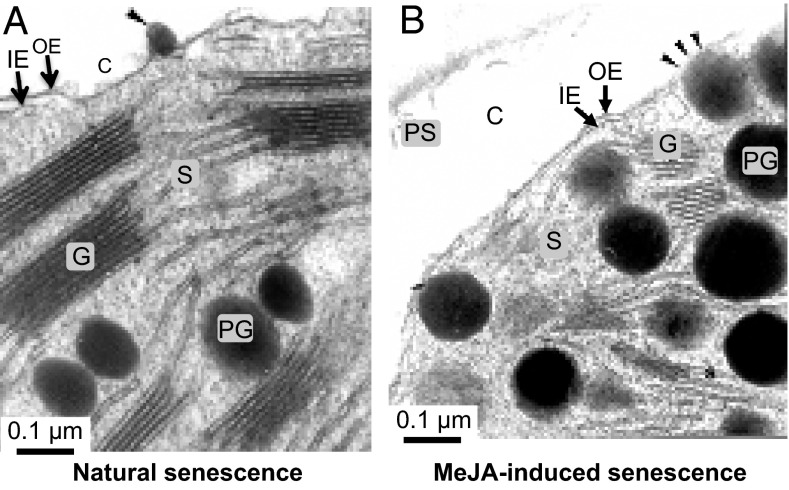
Barley was used as a model to study natural and artificially induced, MeJA-triggered leaf senescence. Electron microscopy of ultrathin leaf sections of naturally senescing and artificially senescing, MeJA-treated plants revealed similar decreases to occur in the number and size of chloroplasts per cell as well as massive changes in chloroplast ultrastructure. A hallmark event in either case was the disintegration of thylakoids, which was accompanied by a mass exodus of plastoglobules to the cytosol, occurring through small holes appearing in the plastid envelope (Fig. 1 A and B). The entire process of programmed organelle destruction was reminiscent of that triggered by 15-LOX in mammalian cells. van Leyen et al. (17) reported on a 15-LOX (i) integrating into the limiting membranes of various cell organelles, including those of the endoplasmic reticulum, Golgi apparatus, peroxisomes, and mitochondria; (ii) introducing holes by catalyzing the regio- and stereo-specific oxidation of arachidonic acid (ARA); and (iii) permitting the release of proteins from the organelle lumen and the access of proteases to both luminal and integral membrane proteins.
Fig. 1.
Plastid leakage during natural senescence (A) and MeJA-induced leaf senescence (B). Scale bars are indicated. Arrowheads indicate extrusion sites of organic matter from chloroplasts. C, cytosol; G, grana thylakoids; IE, inner plastid envelope membrane; OE, outer plastid envelope membrane; PG, plastoglobules; PS, protoplasmic stream; S, stroma.
Identification and Characterization of Plastid Envelope 13-LOX.
A database search was performed to identify lipoxygenases that could contribute to the observed chloroplast dissolution in senescing plants. Special emphasis was placed on LOX enzymes accumulating in the plastid envelope. By searching public plastid proteome databases (18–20), indeed, one enzyme was identified that is encoded by At3g45140 in Arabidopsis (21). The closest equivalent enzyme in barley was LOX2:Hv:1 (22). A search for similar amino acid sequences in the Protein Data Bank (PDB) using BLAST revealed that vegetative soybean (Glycine max) lipoxygenase VLX-B (PDB ID code 2IUJ) (23) showed the highest score (615 bits), with 60% amino acid similarity to the barley 13-LOX (Fig. S1), followed by LOX-3, VLX-D, and LOX-1, which are all from soybean, with scores of 607 (58%), 606 (58%), and 597 (58%) bits, respectively.
Fig. S1.
Amino acid sequence alignment of barley 13-LOX and soybean VLX-B generated using the Needleman–Wunsch alignment algorithm, performed with European Molecular Biology Open Software Suite Neelde Protein Alignment web-based software (52). NH2-terminal regions where no predictable 3D structure was obtained for either 13-LOX or VLX-B are marked with a bracket. Five Fe-coordinating residues are highlighted in red, of which H587, H592, H777, and N781 were replaced by in vitro mutagenesis (22).
Catalytic Activity of 13-LOX.
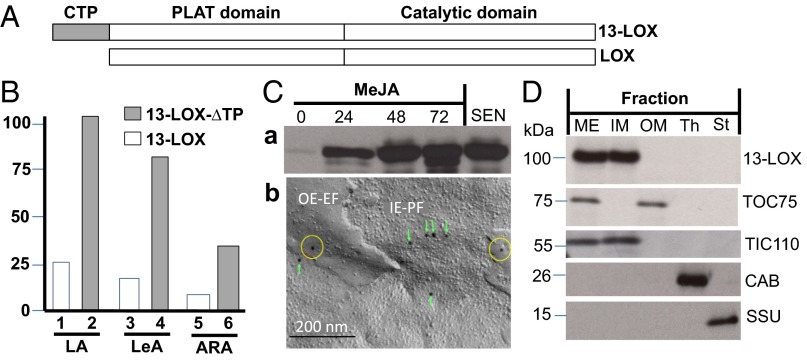
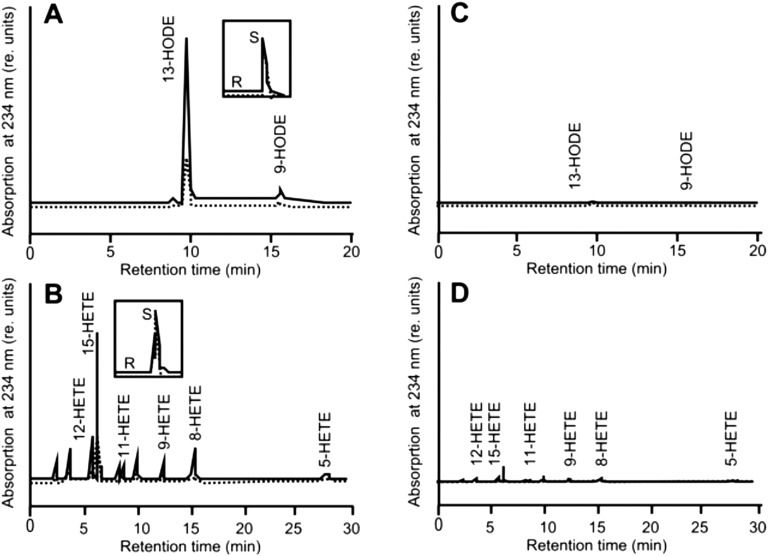
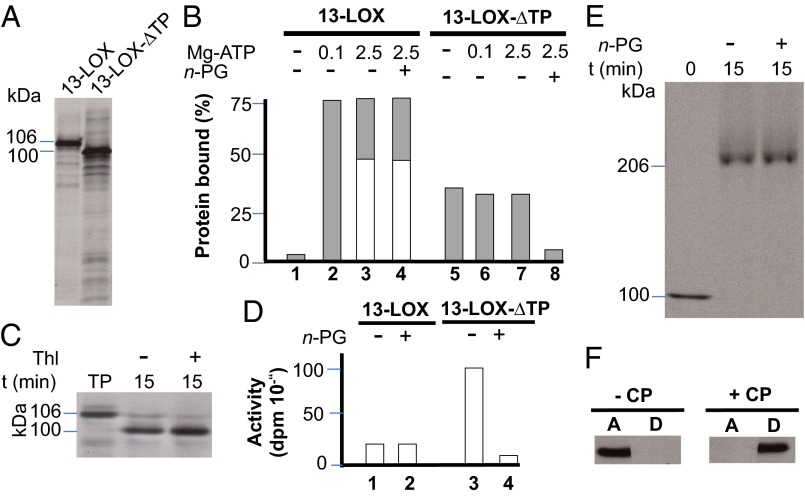
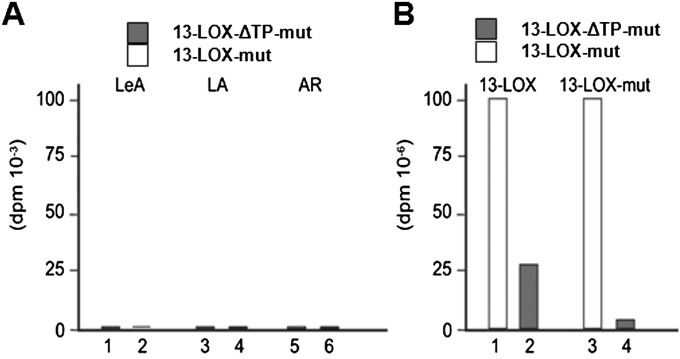
LOX2:Hv:1, henceforth designated barley 13-LOX, consists of an NH2-terminal chloroplast transit peptide (CTP), PLAT domain (eight stranded β-barrel), and catalytic domain (Fig. 2A). cDNAs were constructed for 13-LOX containing or lacking the predicted CTP (13-LOX and 13-LOX-ΔTP, respectively) and bearing either NH2-terminal or COOH-terminal (His)6 tags. Protein was expressed in Escherichia coli, purified to apparent homogeneity by Ni-NTA agarose chromatography, and used for activity measurements (22–26). In fact, all lipoxygenases including mammalian 15-LOX and plant 13-LOX catalyze the stereo- and regio-specific dioxygenation of polyunsaturated membrane fatty acids containing a (Z,Z)-1,4-pentadiene such as hexadecadienoic (linoleic) acid (LA), octadecatrienoic acid [linolenic acid (α-LeA)], as well as ARA (24) into the lipoxygenases’ respective 13(S)-hydroperoxy products. Pilot experiments showed that the presence of the COOH-terminal (His)6 tag completely abolished the enzyme activity, as expected from the Fe-coordination role of carboxyl terminus in other LOX enzymes (Fig. S2). Therefore, all activity measurements were conducted with NH2-terminally (His)6-tagged 13-LOX and 13-LOX-ΔTP enzymes. HPLC and GC-MS analyses used to identify and quantify the substrates and products of the 13-LOX reaction are shown in Fig. 2B and Fig. S2. Consistent with previous results (22, 26), 13-LOX converted α-LeA, LA, and, to a lesser extent, ARA into their respective 13(S)-hydroperoxy products (Fig. 2B and Fig. S2). Interestingly, the presence of the CTP significantly lowered 13-LOX activity on all tested substrates (Fig. S2).
Fig. 2.
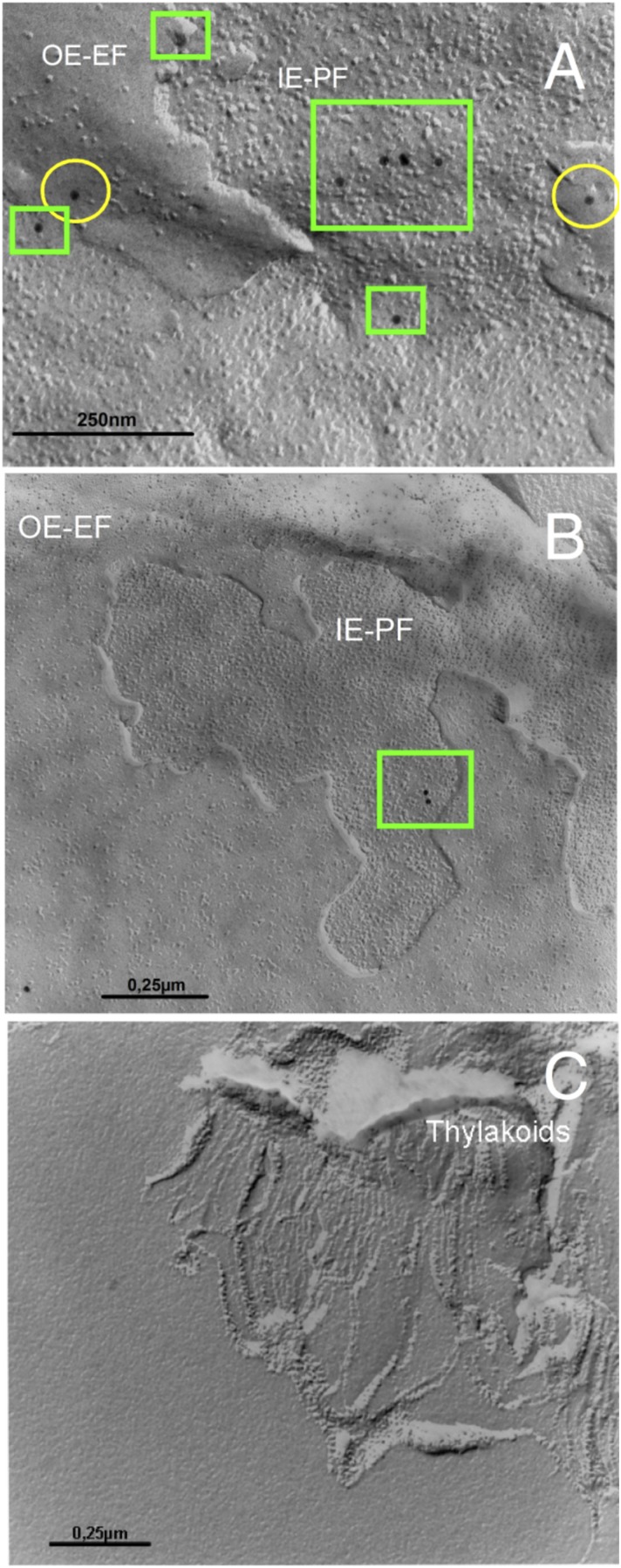
Structure, activity, expression, and localization of 13-LOX in chloroplasts. (A) Domain structure of 13-LOX and 13-LOX-ΔTP. (B) Activity of 13-LOX and 13-LOX-ΔTP on LA, LeA, and ARA. (C) Expression of 13-LOX in MeJA-treated plants and during natural senescence (C, a) and immunodetection of the enzyme in outer envelope exoplasmic faces (OE-EF) (yellow circles) and inner envelope protoplasmic faces (IE-PF) (green boxes) obtained after freeze-fracturing chloroplasts of MeJA-treated leaves using 13-LOX antibodies and 10-nm gold-conjugated secondary antibodies (C, b). (D) Immunolocalization of 13-LOX in the inner plastid envelope. Controls show markers for outer envelope membranes (OM), inner envelope membranes (IM), and mixed outer and inner envelope membranes (ME), as well as thylakoids (Th) and stroma (St).
Fig. S2.
HPLC identification of reaction products formed by NH2-terminally (His)6-tagged (A and B) and COOH-terminally (His)6-tagged (C and D) 13-LOX-ΔTP (solid line) and 13-LOX (dotted line) using LA (A and C) and ARA (B and D) as substrates. Note that 13-LOX converts its substrates exclusively into the corresponding S-isomers of 13-HODE (13:9-HODE ratio: 89:11; enantiomeric (S/R) ratio of 13-HODE: 93:7) and 15-HETE (15:12/11:5-HETE ratio: 92: 3.2:4.8; S/R ratio of 15-HETE: 92:8).
Expression of 13-LOX During Natural and JA-Induced Leaf Senescence.
If 13-LOX played a key role in programmed chloroplast destruction, it should be expressed in the plastid envelope of senescing chloroplasts. Indeed, strong 13-LOX signals were obtained in leaf extracts of naturally senescing and MeJA-treated plants (Fig. 2 C, a and Fig. S3). Electron microscopy of freeze-fractured chloroplasts from MeJA-treated plants revealed the presence of immune-reactive 13-LOX signals at outer envelope exoplasmic faces (Fig. 2 C, b: OE-EF,yellow circles) and inner envelope protoplasmic faces (Fig. 2 C, b: IE-PF, green boxes; and Fig. S4). In organelle fraction studies, 13-LOX was detectable in the inner plastid envelope membrane (Fig. 2D).
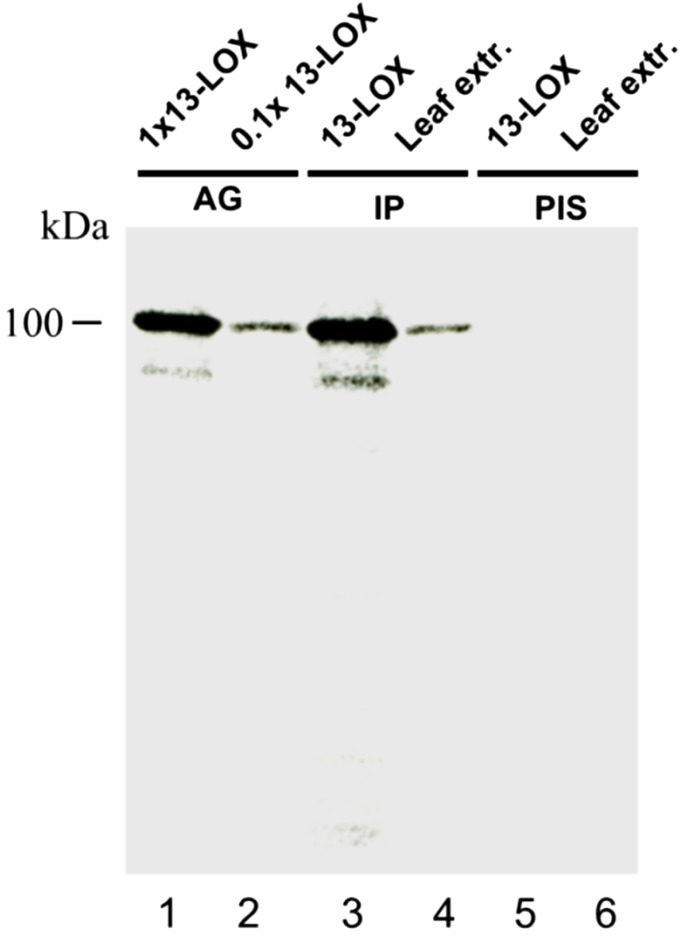
Fig. S3.
Characterization of 13-LOX antiserum. Two hundred nanograms of chemically pure recombinant 13-LOX-ΔTP protein [antigen (AG), lane 1; lane 2 contains a 10-fold dilution] were used to immunize rabbits. A respective polyclonal antiserum was obtained and tested by immunoprecipitation (IP) for its specificity toward the antigen (lane 3, corresponding to lane 1) and 25 µg of total leaf protein from 10-d-old barley plants that had been treated with a 45 µM solution of MeJA for 24 h (lane 4). The respective preimmune serum (PIS) was tested for comparison with recombinant protein (lane 5, corresponding to lane 1) and leaf protein (lane 6, corresponding to lane 4).
Fig. S4.
Detection of 13-LOX in freeze-fractured chloroplasts and respective subfractions by electron microscopy. Detection of 13-LOX occurred in leaf tissues of plants that had been exposed to MeJA (A and C) or water (B) for 24 h. Whereas A and B show the plastid envelope regions of chloroplasts, C depicts an image of the thylakoid membrane fraction. Note that 13-LOX is detectable in outer envelope exoplasmic faces (OE-EF) (yellow circles) and inner envelope protoplasmic faces (IE-PF) (green boxes) (A and B) but not in the thylakoids (C). Immunodetection was made using anti-LOX plus 10-nm gold-conjugated secondary antibodies. Bars indicate scale markers.
Role of 13-LOX in Programmed Chloroplast Destruction.
We next tested the binding of 13-LOX and 13-LOX-ΔTP to isolated chloroplasts. Sedimentation analyses were conducted essentially as described by van Leyen et al. (17). Equimolar amounts of [35S]methionine-labeled protein (Fig. 3A) were incubated with isolated chloroplasts, which had been prepared from 7-d-old freshly harvested barley plants and energy-depleted. Incubations were carried out either in the absence of Mg-ATP or in the presence of 0.1 mM Mg-ATP or 2.5 mM Mg-ATP. These Mg-ATP concentrations were used to distinguish between CTP-dependent binding of 13-LOX to the chloroplast (0.1 mM Mg-ATP) from translocation of 13-LOX across the envelope membranes and import into the inner envelope membrane (2.5 mM Mg-ATP). In addition, incubations were performed in the absence of Mg-ATP and Mg-GTP to detect whether a spontaneous binding of 13-LOX and 13-LOX-ΔTP to the lipid bilayers occurs through the PLAT and catalytic domains. After incubation, the plastids were collected by centrifugation, and the binding of 13-LOX and 13-LOX-ΔTP was determined (27).
Fig. 3.
Binding and import of 13-LOX and 13-LOX-ΔTP to chloroplasts. (A) SDS/PAGE and autoradiography of [35S]Met-labeled 13-LOX and 13-LOX-ΔTP used for the binding and import assays. (B) Chloroplast binding of [35S]13-LOX and [35S]13-LOX-ΔTP at different Mg-ATP concentrations (in mM) tested in the presence (+) or absence (−) of n-propyl gallate (n-PG) as a LOX inhibitor. Light gray columns show precursor levels bound to resedimented chloroplasts, whereas white bars give mature [35S]protein levels after import. (C) Import of 106-kDa [35S]13-LOX (pLOX) and conversion into a smaller, ∼100-kDa product (LOX) corresponding to [35S]13-LOX-ΔTP in chloroplasts at 2.5 mM Mg-ATP. Thl, thermolysin; TP, translation product. (D) Activity of bacterially expressed and purified 13-LOX and 13-LOX-ΔTP tested with α-LeA in the presence and absence of n-propyl gallate (n-PG). (E) Detection of higher-molecular-weight complexes containing imported [35S]13-LOX in the inner envelope membranes of barley chloroplasts after solubilization with 1.3% (vol/vol) decylmaltoside and nondenaturing PAGE. (F) Phase partitioning of imported and processed 13-LOX, as assessed with Triton X-114. After centrifugation of assays containing chloroplasts (+CP) or lacking chloroplasts (−CP), [35S]LOX was detected in the aqueous phase (A) and detergent phase (D) by SDS/PAGE and autoradiography.
Fig. 3B illustrates that 13-LOX and 13-LOX-ΔTP exhibited different plastid-binding capabilities. Whereas a significant fraction (ca. 35–40%) of added 13-LOX-ΔTP bound to the plastids under all tested conditions, binding of 13-LOX containing the CTP was energy-dependent and did not occur in the absence of Mg-ATP. In the presence of 0.1 mM Mg-ATP, ca. 75–80% of added precursor was recovered in a plastid-bound form (Fig. 3B). In the presence of 2.5 mM Mg-ATP, a major fraction of 13-LOX was imported and converted into a smaller, mature-size ∼100-kDa product that was resistant to added thermolysin (Fig. 3C), a protease that does not penetrate chloroplasts but degrades surface-exposed plastid envelope proteins (28). Activity measurements confirmed that the CTP partially blocked the catalytic activity of 13-LOX on α-LeA (Fig. 3D). Tests with n-propyl gallate (PG), a known LOX inhibitor (29), underscored the different roles of the catalytic domain and CTP in catalysis and membrane binding (Fig. 3D). Whereas the CTP is indispensable for the specific import of 13-LOX into the plastid envelope membranes and is removed, the catalytic and PLAT domains are needed for subsequent fatty acid binding and conversion. The imported, mature 13-LOX established larger complexes in the inner envelope membrane (Fig. 3E) and behaved as an integral membrane protein, as revealed by phase partitioning with Triton X-114 (Fig. 3F).
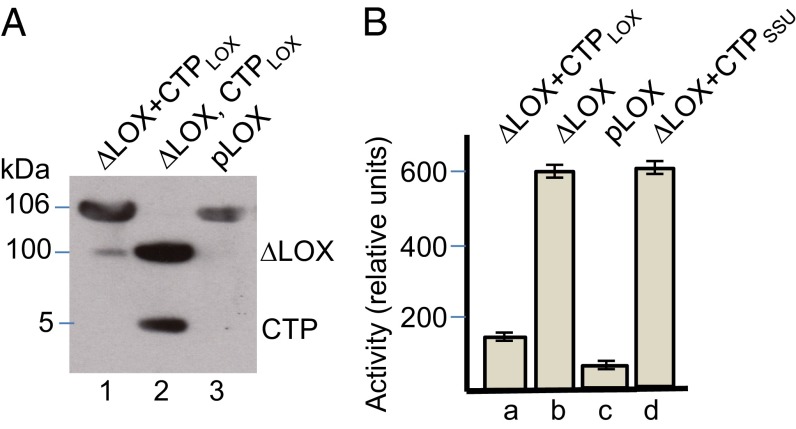
Thus far, the data suggested a crucial role of the CTP in regulating 13-LOX activity. To deepen this point, we expressed the CTP of 13-LOX comprising amino acids 1–47 in vitro, purified it, and carried out reconstitution experiments with the mature 13-LOX. CTP interactions with the mature 13-LOX were assessed by nondenaturing PAGE as well as activity measurements. Fig. 4A shows tight binding of the CTP to the mature 13-LOX, giving rise to a protein band that migrated identically to that of the full-length 13-LOX protein containing the covalently linked CTP. Activity measurements revealed a highly specific inhibition of 13-LOX activity by added 13-LOX CTP, while showing no effect of the CTP of the small subunit of RuBisCo (Fig. 4B). On the basis of these results, we concluded that the CTP of 13-LOX blocks the enzyme activity. It will be interesting to see whether the CTP-dependent inhibition of 13-LOX activity may also block the enzyme’s binding to mitochondria and other cell organelles.
Fig. 4.
Inhibition of 13-LOX activity by its in vitro-expressed CTP. (A) Expression and reconstitution of pseudo full-length 13-LOX from ∆13-LOX and added CTP. Lane 1 shows pseudo full-length 13-LOX reconstituted from ∆13-LOX and the CTP, whereas lane 2 shows each component individually after detergent treatment. Lane 3 depicts the native full-length 13-LOX expressed from the cDNA, referred to as pLOX, and containing the covalently linked CTP, used as reference. (B) Activity measurements carried out for the reconstituted pseudo full-length 13-LOX (∆LOX+CTPLOX) (B, a), mature LOX lacking the CTP (∆LOX) (B, b), and full-length native 13-LOX (pLOX) (B, c). For comparison, 13-LOX activity was measured in assay mixtures containing ∆LOX and the CTP of the small subunit of RuBisCo (B, d). Error bars refer to three independent experiments.
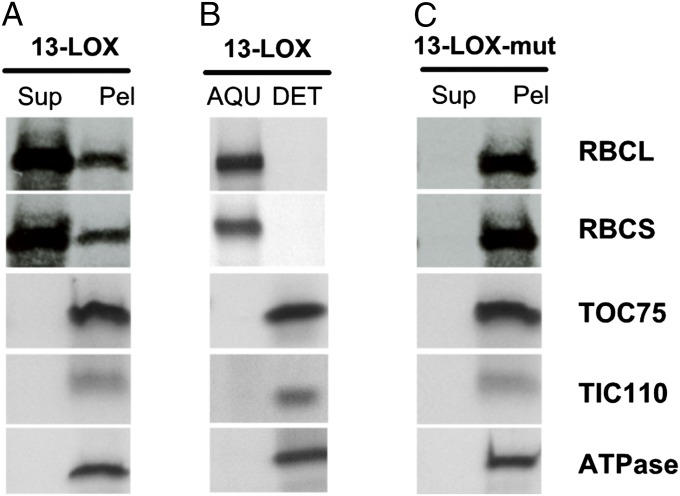
Interestingly, changing polypeptide patterns were noted after prolonged incubations of chloroplasts containing imported and processed 13-LOX (Fig. 5A). Stromal proteins such as the large and small subunits of RuBisCo (RBCL and RBCS) were released into the supernatant obtained after sedimentation of chloroplasts containing imported 13-LOX. This effect was not attributable to a general solubilization of the outer and inner envelope membranes, as evidenced by the absence of membrane proteins, such as TOC75 and TIC110, as components of the standard protein import machinery of chloroplasts (30) and the thylakoid ATPase 52-kDa subunit in the supernatant of sedimented chloroplasts (Fig. 5A). Phase partitioning with Triton X-114 proved the membrane localization of TOC75, TIC110, and ATPase (Fig. 5B).
Fig. 5.
Sedimentation analysis on chloroplasts containing imported 13-LOX (A and B) or 13-LOXmut bearing His/Asn-to-Ala substitutions at positions 587, 592, 777, and 781 (C). (A and C) Levels of the RuBisCo large subunit (RBCL) and RuBisCo small subunit (RBCS), the 52-kDa subunit of the ATPase, as well as TOC75 and TIC110 in the supernatant (Sup) and plastid pellets (Pel) obtained after plastid sedimentation, as assessed by Western blotting using specific antisera. (B) As in A but showing protein levels in the aqueous phase (AQU) and detergent phase (DET) after Triton X-114 partitioning of chloroplasts bearing imported 13-LOX.
Together, our results were strongly supportive of a function of 13-LOX in selective envelope membrane destruction during plant senescence. To corroborate this conclusion, sedimentation analyses were conducted with chloroplasts bearing a catalytically inactive mutant form of 13-LOX (13-LOX-mut). As shown previously (22, 23), all plant 13-LOX isoforms contain highly conserved His and Asn residues, with H587, H592, H777, and N781 establishing the active site in the case of the barley 13-LOX studied here. Replacing H587, H592, H777, and N781 by Ala residues rendered the mature 13-LOX catalytically inactive and partly hampered its spontaneous membrane binding (Fig. S5). Chloroplast transit peptide-dependent binding of the 13-LOX mutant precursor to chloroplasts was unaffected, however (Fig. S5). Chloroplasts bearing the imported and processed mature mutant 13-LOX were intact and retained stromal proteins, such as RBCS and RBCL, in respective sedimentation analyses (Fig. 5C). These results proved the essential role of 13-LOX activity for selective plastid envelope destruction occurring during senescence.
Fig. S5.
Catalytic activity and chloroplast binding properties of mutant 13-LOX containing His/Asn-to-Ala substitutions at positions H587, H592, H777, and N781. (A) Activity measurements carried out with LeA, LA, and ARA (AR). Quantification refers to respective 13(S)-hydroperoxy products identified by HPLC and shows no detectable activity on any tested substrate. White and gray bars refer to reactions containing the full-length His/Asn-to-Ala-13-LOX (lanes 2, 4, and 6) and respective transit peptide-less 13-LOX-ΔTP mutant (lanes 1, 3, and 5) proteins. (B) Membrane-binding assays carried out with chloroplasts isolated from 7-d-old, freshly harvested barley plants. White and gray bars refer to the amount of chloroplast-bound full-length 13-LOX and 13-LOX-ΔTP that was determined for the wild-type 13-LOX (lanes 1 and 2) and the His/Asn-to-Ala-13-LOX mutant protein (lanes 3 and 4).
Discussion
Role of 13-LOX During Plant Senescence.
Leaf senescence is an active, highly regulated process that involves the disassembly of the photosynthetic apparatus and ends with the full dissolution of the chloroplast (1–3). The first steps of chloroplast breakdown involve the local destruction of the plastid envelope, leading to holes as exit sites for organic matter (16). In the present study, we asked whether a LOX-mediated mechanism could account for this effect in naturally senescing and artificially senescing, MeJA-treated barley plants. Database searches identified several plastid lipoxygenases, of which the one encoded by At3g45140 in Arabidopsis is orthologous to the barley 13-LOX studied here. This 13-LOX accumulated in chloroplasts of both naturally senescing and artificially senescing, MeJA-treated plants and caused local membrane destruction and leakage of stromal constituents. Our data are similar to findings reported for mammalian 15-LOX, which programs selective membrane destruction and organelle fate in reticulocytes and presumably also in central fiber lens cells (17, 31). In contrast to these mammalian cells, which remain viable, although unable to divide and proliferate, leaf mesophyll cells undergo further destructions and finally die during the senescence program (1, 2).
Regulation of 13-LOX Function.
How can the chloroplast 13-LOX identified in this work operate so specifically during the senescence process? Firstly, the expression of 13-LOX is confined to senescent plants. No 13-LOX protein was found in 7-d-old, freshly harvested plant tissues (Fig. 2 C, a). Leaf senescence and 15-LOX expression in Arabidopsis thaliana are regulated by the miR319-regulated clade of TCP (TEOSINTE BRANCHED/CYCLOIDEA/PCF) transcription factor genes (32). Similarly, expression of many genes during the differentiation of animal eye lens cells is under the control of miRNAs (33), and that of 13-LOX peaks shortly before organelle destruction (17), suggesting a cross-kingdom conservation of LOX pathway regulation to determine organelle fate.
Secondly, unique structural features of 13-LOX’s NH2 terminus appear to ensure the proper targeting to the chloroplast but not to other cell organelles. Once bound to the plastid envelope, 13-LOX was first imported into the chloroplast, then processed, and finally sorted out of the import channel, before 13-LOX became operational (Fig. 3 B and C). The CTP of 13-LOX hereby directed the precursor to the respective protein import machinery and prevented the unspecific access of the catalytic and PLAT domains to other membranes. Once imported into chloroplasts, the mature 13-LOX oxidized the membrane fatty acids α-LeA and LA and, presumably by virtue of this lipid modification, introduced holes in the plastid envelope (Fig. 3D) that permitted the release of plastoglobules and stromal proteins, such as the RuBisCo large and small subunits (RBCL and RBCS), to the cytosol. Both our microscopic studies (Fig. 1) and sedimentation analyses (Fig. 5) are consistent with this view and confirm and extend previous work by Sharma et al. (34) who, using transgenic expression, demonstrated a link between the occurrence of 13-LOX100 and senescence.
Substrate Recognition and Conversion by 13-LOX.
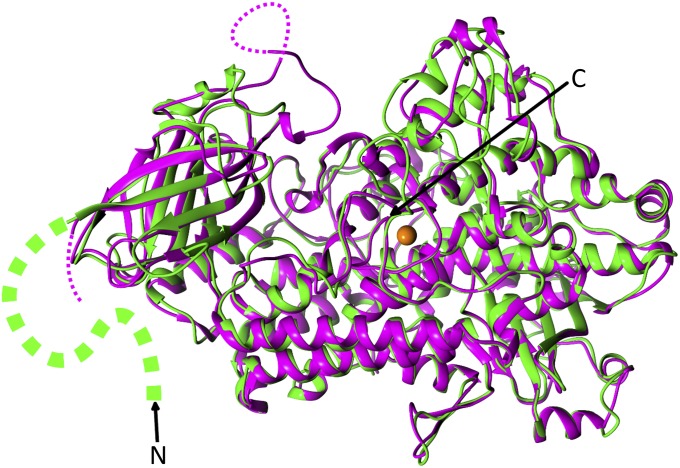
A 3D structural modeling was performed for understanding 13-LOX’s obvious capability for binding and attacking membrane-bound fatty acids. Given the high overall sequence similarity of barley 13-LOX to soybean lipoxygenase VLX-B, for which X-ray data have been obtained (23), such modeling was expected to provide insights into the catalytic mechanism of barley 13-LOX and how this may be affected by the CTP. Although there are a few areas of short insertion or deletion, most of those insertions and deletions are located in the exposed loop areas, and thus the overall 3D structure of barley 13-LOX (green in Fig. 6) was very similar to that of VLX-B (violet in Fig. 6). None of the corresponding amino acid substitutions in 13-LOX versus VLX-B produced any major high-energy van der Waals contacts. However, there is a heterogeneity among those lipoxygenases in the first ∼150 residues, which cover the CTP and a part of the NH2-terminal PLAT domain, and the corresponding region of 13-LOX is larger than any of those compared lipoxygenases (Fig. 6). A sequence alignment of the 13-LOX with the sequences of those lipoxygenases revealed several insertions and deletions, indicating that the PLAT domain of 13-LOX has different secondary structural features compared with other known lipoxygenases (Fig. 6). In particular, the side of the PLAT domain facing the substrate entrance site of 13-LOX is quite different compared with that of VLX-B (Fig. 6).
Fig. 6.
Superimposed views of Cα positions of 13-LOX and VLX-B are depicted in green and violet color, respectively. Dotted violet lines indicate disordered areas in VLX-B, and the segment green line indicates the structurally unresolved part of PLAT domain and CTP region in 13-LOX. N and C indicate N and C termini, respectively. This figure was generated with the program PyMOL (version 0.99). Molecular images were produced using the Chimera graphics package (University of California, San Francisco; National Institutes of Health Grant P41 RR-01081).
Similar to other lipoxygenases, 13-LOX uses nonheme iron for activity, as confirmed by our mutagenesis study. The five residues coordinating this nonheme iron are H587H513,H499, H592H518,H504, H777H704,H690, N781N708,N694, and I936I853,I839, which are conserved in 13-LOX, VLX-B, and LOX-1 (residue numbering refers to 13-LOX, with the corresponding numbers in VLX-B and LOX-1 given in superscript). Three residues of the second-coordination sphere (35) around the iron coordination sphere, Q784Q711,Q697 and L842L768,L754, are also conserved, which is known to promote a productive conformation of substrate (35–37). In addition, Q583Q509,Q495 and I626I552,I538, which are located near the Fe2+ site and C-14 and C-8 atoms of substrate such as LA, are conserved and superimposable with those of VLX-B and LOX-1. Likewise, most of the residues constituting the hydrophobic substrate-binding pocket, so-called subcavity IIa, are highly conserved among 13-LOX, VLX-B, and LOX-1. The internal cavity of known lipoxygenases is roughly composed of three branches: the extended pocket, O2 cavity, and entrance site. Among 11 residues constituting the extended pocket (A582A505,S491, S644T570,T556, F645F571,F557, A789G715,G701, I791I718,I704, R794R721,R707, T796T723,T709, V835D761,S747, L836L762,L748, I838I764,I750, L839I765,I751), 6 residues are completely conserved and 4 are conservative changes; however, the corresponding residue of V835 D761,S747 are polar amino acids in both VLX-B and LOX-1. Among the nine residues constituting the O2 cavity branch (Q583Q509,Q495, L584I510,L496, W588W514,W500, A630A556,A542, L634L560,L546, I635V561,I547, I641I567,I553, L652L578,V564, L842L768,L754), six residues are completely conserved among 13-LOX, VLX-B, and LOX-1, and the remaining three are conservative changes. L634L560,L546, I641I567,I553, and L842L768,L754, which have been proposed to have a guiding role for O2 access by blocking access to the C-9 of LA (38), are conserved and superimposable. In addition, A630A556,A542 that are completely conserved among S-lipoxygenases and have been proposed to determine the stereochemistry of products, are conserved and superimposable (39, 40). As shown in Fig. 6, the substrate entrance site of our 13-LOX model has a comparable size to that of VLX-B. However, long insertion in the NH2 terminus and the noticeable structural difference in PLAT domain near the substrate entrance site may restrict (or regulate) the entry port for 13-LOX. In summary, the molecular model of 13-LOX based on the crystal structures of VLX-B, VLX-D, and LOX-1 shows that the subcavities of 13-LOX involved in substrate binding and O2 access are very similar to the subcavities of VLX-B, VLX-D, and LOX-1, supporting the similar enzymatic character among all of these lipoxygenases in terms of positional specificity. Because of the presence of several unique structural motifs including long insertion in the PLAT domain, 13-LOX is obviously rendered capable of binding both membrane and free polyunsaturated fatty acids and converting them into their respective 13(S)-hydroperoxy products.
Biological Significance of the 13-LOX Pathway in Insect Deterrence and Microbial Defense.
Pathway products of 13-LOX that accumulate in planta after release from the membrane comprise bioactive aldehydes and oxylipins (24, 41). Whereas volatile aldehydes are involved in direct and indirect defenses against herbivores (42–44), oxylipins, including JA, regulate defense gene activation and cell death in response to microbial pathogens (6, 41, 45). The whole array of 13-LOX pathway products permits the bearing relocation of carbon and nitrogen resources from source to sink tissues needed for optimal seed filling and thus is of pivotal importance for plant viability in the ecosystem.
Materials and Methods
Molecular Modeling.
The homology model of 13-LOX was constructed using SWISS-MODEL (46). Soybean lipoxygenase-B (PDB ID 2IUJ) was used as the template, yielding a model with a global quality estimation score (GMQE) of 0.67.
Plant Materials.
For studying natural senescence, 56-d-old barley plants (Hordeum vulgare L. Scarlett) were used that had been grown under continuous white light illumination provided by fluorescent bulbs (30 W/m2). For studying prematurely induced artificial senescence, primary leaves were cut from 7-d-old seedlings and floated on 45 µM aqueous solution of MeJA for appropriate periods.
Activity Measurements of 13-LOX.
Activity measurements were conducted according to Vörös et al. (22), using n-propyl gallate (PG) as a LOX inhibitor (29). Identification and quantification of substrates and products of the 13-LOX reaction was done by HPLC and GC-MS (22, 26) (SI Text).
Chloroplast Binding and Import of 13-LOX.
Binding and import assays were conducted as described, using Percoll-purified, energy-depleted barley chloroplasts and bacterially expressed, chemically pure [35S]Met-labeled 13-LOX and 13-LOX-ΔTP (47). Incubations were carried out either in the absence of Mg-ATP or in the presence of 0.1 mM Mg-ATP (used to study binding) or 2.5 mM Mg-ATP (used to study import). Binding was assessed as described by Friedman and Keegstra (27). After import, intact plastids were rapidly reisolated on sucrose/Percoll. Postimport protease treatment of plastids with thermolysin and extraction of membranes with sodium carbonate, pH 11, or 1 M NaCl were carried out according to Cline et al. (28). Plastid subfractionation into envelopes, stroma, and thylakoids was achieved according to Li et al. (48). For the experiment described in Fig. 5, plastids were pretreated with an excess of bacterially expressed and purified 13-LOX or mutant 13-LOX containing His/Asn-to-Ala substitutions at positions H587, H592, H777, and N781 for 2 h before further analysis. Phase partitioning with Triton X-114 was carried out according to Bordier (49).
Immunoblotting.
Protein was extracted and run by denaturing or nondenaturing PAGE and blotted onto nitrocellulose membranes (50, 51). Western blotting was carried out with a specific antiserum raised against the bacterially expressed and purified barley 13-LOX (Fig. S3). For sedimentation assays used to explore the lytic activity of 13-LOX on isolated chloroplasts, antisera against the large and small subunits of RuBisCo (RBCL and RBCS), the thylakoid ATPase 52-kDa subunit, as well as TOC75 and TIC110 were used. Protein detection was made using either an enhanced chemiluminescence (ECL) system (Amersham) or an anti-rabbit, anti-goat alkaline phosphatase system.
SI Text
Activity measurements were carried out according to Vörös et al. (22) and Bachmann et al. (26). Bacterially expressed, purified enzymes were incubated with 300 mM LA or ARA in 0.1 M phosphate buffer, pH 7.3, for 15 min. The hydroperoxy derivatives then were reduced with sodium borohydride. Subsequently, the hydroxyl derivatives of LA (HODEs) and the corresponding hydroxyl eicosatetraenoic acids (HETEs) were extracted, isolated by reversed-phase HPLC on a Nucleosil C-18 column (250 × 4.6 mm; 5-μm particle size; KS-system; Macherey-Nagel) with a solvent system consisting of methanol/water/acetic acid (85:15:0.1 vol/vol) and a flow rate of 1 mL/min, essentially as described (22, 26). The insets in Fig. S2 show separation of positional isomers of 13-HODE and 15-HETE, respectively, by chiral phase HPLC (the curves were normalized). Note that only the amino-terminally tagged 13-LOX-ΔTP and 13-LOX enzymes are active, although to extents that are fivefold different.
Acknowledgments
We thank G. Acker (formerly of the University Bayreuth) for help with immunogold electron microscopy. Antisera against LOX were kind gifts from J. Lehmann (formerly Leibnitz Institute of Plant Biochemistry) and H. Sanchez-Serano (University of Madrid), and those against TOC75 and TIC110 were gifts from F. Kessler (Université Neuchatel) and D. J. Schnell (University of Massachusetts). Antiserum against the thylakoid ATPase was kindly provided by K. Apel (formerly Institute for Plant Sciences, ETH Zurich). We thank C. Wasternack (formerly of the Leibnitz Institute of Plant Biochemistry) for critical reading of the manuscript and helpful comments. This work was supported by Marie-Curie Actions Program from the European Community, FP7-PEOPLE-CIG-2011-303744 (to S.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525747113/-/DCSupplemental.
References
- 1.Guo Y, Gan S. Leaf senescence: Signals, execution, and regulation. Curr Top Dev Biol. 2005;71:83–112. doi: 10.1016/S0070-2153(05)71003-6. [DOI] [PubMed] [Google Scholar]
- 2.Reinbothe C, Reinbothe S. 2005. Regulation of photosynthetic gene expression by the environment: From seedling de-etiolation to leaf senescence, Photoprotection, Photoinhibition, Gene Regulation, and Environment, eds Demmig-Adams B, Adams W, III, Mattoo A, Advances in Photosynthesis and Respiration, series ed. Govidjee B (Springer, Dordrecht, The Netherlands), Vol 21, pp 333–365.
- 3.Sarwat M, Naqvi AR, Ahmad P, Ashraf M, Akram NA. Phytohormones and microRNAs as sensors and regulators of leaf senescence: Assigning macro roles to small molecules. Biotechnol Adv. 2013;31(8):1153–1171. doi: 10.1016/j.biotechadv.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Zentgraf U, Laun T, Miao Y. The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur J Cell Biol. 2010;89(2-3):133–137. doi: 10.1016/j.ejcb.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Christiansen MW, Gregersen PL. Members of the barley NAC transcription factor gene family show differential co-regulation with senescence-associated genes during senescence of flag leaves. J Exp Bot. 2014;65(14):4009–4022. doi: 10.1093/jxb/eru046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinbothe C, Pollmann S, Reinbothe S. Singlet oxygen signaling links photosynthesis to translation and plant growth. Trends Plant Sci. 2010;15(9):499–506. doi: 10.1016/j.tplants.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Distelfeld A, Avni R, Fischer AM. Senescence, nutrient remobilization, and yield in wheat and barley. J Exp Bot. 2014;65(14):3783–3798. doi: 10.1093/jxb/ert477. [DOI] [PubMed] [Google Scholar]
- 8.Gregersen PL, Culetic A, Boschian L, Krupinska K. Plant senescence and crop productivity. Plant Mol Biol. 2013;82(6):603–622. doi: 10.1007/s11103-013-0013-8. [DOI] [PubMed] [Google Scholar]
- 9.Brown SB, Houghton JD, Hendry GF. 1991. Chlorophyll breakdown, Chlorophylls, Scheer H (CRC Press, Boca Raton, FL), pp 465–492.
- 10.Feller U, Anders I, Mae T. Rubiscolytics: Fate of Rubisco after its enzymatic function in a cell is terminated. J Exp Bot. 2008;59(7):1615–1624. doi: 10.1093/jxb/erm242. [DOI] [PubMed] [Google Scholar]
- 11.Prins A, van Heerden PD, Olmos E, Kunert KJ, Foyer CH. Cysteine proteinases regulate chloroplast protein content and composition in tobacco leaves: A model for dynamic interactions with ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) vesicular bodies. J Exp Bot. 2008;59(7):1935–1950. doi: 10.1093/jxb/ern086. [DOI] [PubMed] [Google Scholar]
- 12.Parrott DL, Martin JM, Fischer AM. Analysis of barley (Hordeum vulgare) leaf senescence and protease gene expression: A family C1A cysteine protease is specifically induced under conditions characterized by high carbohydrate, but low to moderate nitrogen levels. New Phytol. 2010;187(2):313–331. doi: 10.1111/j.1469-8137.2010.03278.x. [DOI] [PubMed] [Google Scholar]
- 13.Lee TA, van de Wetering SW, Brusslan JA. Stromal protein degradation is incomplete in Arabidopsis thaliana autophagy mutants undergoing natural senescence. BMC Res Notes. 2013;6:17. doi: 10.1186/1756-0500-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hörtensteiner S, Kräutler B. Chlorophyll breakdown in higher plants. Biochim Biophys Acta. 2011;1807(8):977–988. doi: 10.1016/j.bbabio.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Ishida H, Izumi M, Wada S, Makino A. Roles of autophagy in chloroplast recycling. Biochim Biophys Acta. 2014;1837(4):512–521. doi: 10.1016/j.bbabio.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Krupinska K. Fate and activities of plastids during leaf senescence. In: Wise RR, Hoober JK, editors. The Structure and Function of Plastids. Springer; Dordrecht, The Netherlands: 2007. pp. 433–449. [Google Scholar]
- 17.van Leyen K, Duvoisin RM, Engelhardt H, Wiedmann M. A function for lipoxygenase in programmed organelle degradation. Nature. 1998;395(6700):392–395. doi: 10.1038/26500. [DOI] [PubMed] [Google Scholar]
- 18.Bräutigam A, Weber AP. Proteomic analysis of the proplastid envelope membrane provides novel insights into small molecule and protein transport across proplastid membranes. Mol Plant. 2009;2(6):1247–1261. doi: 10.1093/mp/ssp070. [DOI] [PubMed] [Google Scholar]
- 19.Bruley C, Dupierris V, Salvi D, Rolland N, Ferro M. AT_CHLORO: A chloroplast protein database dedicated to sub-plastidial localization. Front Plant Sci. 2012;3:205. doi: 10.3389/fpls.2012.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang M, et al. Construction of plastid reference proteomes for maize and Arabidopsis and evaluation of their orthologous relationships; The concept of orthoproteomics. J Proteome Res. 2013;12(1):491–504. doi: 10.1021/pr300952g. [DOI] [PubMed] [Google Scholar]
- 21.Joyard J, et al. Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism. Prog Lipid Res. 2010;49(2):128–158. doi: 10.1016/j.plipres.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Vörös K, et al. Characterization of a methyljasmonate-inducible lipoxygenase from barley (Hordeum vulgare cv. Salome) leaves. Eur J Biochem. 1998;251(1-2):36–44. doi: 10.1046/j.1432-1327.1998.2510036.x. [DOI] [PubMed] [Google Scholar]
- 23.Youn B, et al. Crystal structures of vegetative soybean lipoxygenase VLX-B and VLX-D, and comparisons with seed isoforms LOX-1 and LOX-3. Proteins. 2006;65(4):1008–1020. doi: 10.1002/prot.21182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feussner I, Wasternack C. The lipoxygenase pathway. Annu Rev Plant Biol. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- 25.Feussner I, Wasternack C, Kindl H, Kühn H. Lipoxygenase-catalyzed oxygenation of storage lipids is implicated in lipid mobilization during germination. Proc Natl Acad Sci USA. 1995;92(25):11849–11853. doi: 10.1073/pnas.92.25.11849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachmann A, et al. Jasmonate-induced lipid peroxidation in barley leaves initiated by distinct 13-LOX forms of chloroplasts. Biol Chem. 2002;383(10):1645–1657. doi: 10.1515/BC.2002.185. [DOI] [PubMed] [Google Scholar]
- 27.Friedman AL, Keegstra K. Chloroplast protein import : Quantitative analysis of precursor binding. Plant Physiol. 1989;89(3):993–999. doi: 10.1104/pp.89.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cline K, Werner-Washburne M, Andrews J, Keegstra K. Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol. 1984;75(3):675–678. doi: 10.1104/pp.75.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walther M, Anton M, Wiedmann M, Fletterick R, Kuhn H. The N-terminal domain of the reticulocyte-type 15-lipoxygenase is not essential for enzymatic activity but contains determinants for membrane binding. J Biol Chem. 2002;277(30):27360–27366. doi: 10.1074/jbc.M203234200. [DOI] [PubMed] [Google Scholar]
- 30.Paila YD, Richardson LG, Schnell DJ. New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J Mol Biol. 2014;427(5):1038–1060. doi: 10.1016/j.jmb.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schewe T, Halangk W, Hiebsch C, Rapoport SM. A lipoxygenase in rabbit reticulocytes which attacks phospholipids and intact mitochondria. FEBS Lett. 1975;60(1):149–152. doi: 10.1016/0014-5793(75)80439-x. [DOI] [PubMed] [Google Scholar]
- 32.Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008;6(9):e230. doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frederikse PH, Donnelly R, Partyka LM. miRNA and Dicer in the mammalian lens: Expression of brain-specific miRNAs in the lens. Histochem Cell Biol. 2006;126(1):1–8. doi: 10.1007/s00418-005-0139-0. [DOI] [PubMed] [Google Scholar]
- 34.Sharma VK, et al. Transgenic barley plants overexpressing a 13-lipoxygenase to modify oxylipin signature. Phytochemistry. 2006;67(3):264–276. doi: 10.1016/j.phytochem.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Schenk G, Neidig ML, Zhou J, Holman TR, Solomon EI. Spectroscopic characterization of soybean lipoxygenase-1 mutants: The role of second coordination sphere residues in the regulation of enzyme activity. Biochemistry. 2003;42(24):7294–7302. doi: 10.1021/bi027380g. [DOI] [PubMed] [Google Scholar]
- 36.Gillmor SA, Villaseñor A, Fletterick R, Sigal E, Browner MF. The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity. Nat Struct Biol. 1997;4(12):1003–1009. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 37.Tomchick DR, Phan P, Cymborowski M, Minor W, Holman TR. Structural and functional characterization of second-coordination sphere mutants of soybean lipoxygenase-1. Biochemistry. 2001;40(25):7509–7517. doi: 10.1021/bi002893d. [DOI] [PubMed] [Google Scholar]
- 38.Knapp MJ, Seebeck FP, Klinman JP. Steric control of oxygenation regiochemistry in soybean lipoxygenase-1. J Am Chem Soc. 2001;123(12):2931–2932. doi: 10.1021/ja003855k. [DOI] [PubMed] [Google Scholar]
- 39.Coffa G, et al. On the relationships of substrate orientation, hydrogen abstraction, and product stereochemistry in single and double dioxygenations by soybean lipoxygenase-1 and its Ala542Gly mutant. J Biol Chem. 2005;280(46):38756–38766. doi: 10.1074/jbc.M504870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oldham ML, Brash AR, Newcomer ME. Insights from the X-ray crystal structure of coral 8R-lipoxygenase: Calcium activation via a C2-like domain and a structural basis of product chirality. J Biol Chem. 2005;280(47):39545–39552. doi: 10.1074/jbc.M506675200. [DOI] [PubMed] [Google Scholar]
- 41.Wasternack C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 2007;100(4):681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howe GA, Jander G. Plant immunity to insect herbivores. Annu Rev Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- 43.Wu J, Baldwin IT. New insights into plant responses to the attack from insect herbivores. Annu Rev Genet. 2010;44:1–24. doi: 10.1146/annurev-genet-102209-163500. [DOI] [PubMed] [Google Scholar]
- 44.Dicke M, Baldwin IT. The evolutionary context for herbivore-induced plant volatiles: Beyond the ‘cry for help’. Trends Plant Sci. 2010;15(3):167–175. doi: 10.1016/j.tplants.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Reinbothe C, Springer A, Samol I, Reinbothe S. Plant oxylipins: Role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 2009;276(17):4666–4681. doi: 10.1111/j.1742-4658.2009.07193.x. [DOI] [PubMed] [Google Scholar]
- 46.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reinbothe S, Mache R, Reinbothe C. A second, substrate-dependent site of protein import into chloroplasts. Proc Natl Acad Sci USA. 2000;97(17):9795–9800. doi: 10.1073/pnas.160242597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li HM, Moore T, Keegstra K. Targeting of proteins to the outer envelope membrane uses a different pathway than transport into chloroplasts. Plant Cell. 1991;3(7):709–717. doi: 10.1105/tpc.3.7.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256(4):1604–1607. [PubMed] [Google Scholar]
- 50.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 51.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA. 1979;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]