Significance
Genome biology aims to gain insight into nuclear function through the study of genome architecture. Analysis of the completed sequences of various eukaryotic genomes indicates that genes are nonrandomly distributed along chromosomes. Recent molecular approaches based on chromosome confirmation capture have identified topologically associated domains (TADs) as a unifying structural model for chromatin organization; however, whether linear gene order and TADs intersect to affect nuclear organization remains to be resolved. Using human myogenesis as a model, we found that a population of TADs have significant enrichment for myogenic-specific genes that results in changes in their subnuclear and intrachromosomal territory structure. We found that these changes in organization impact biallelic transcription and require cell division to be established.
Keywords: cell differentiation, gene regulation, nuclear organization, transcriptional noise, 3D image analysis
Abstract
The linear distribution of genes across chromosomes and the spatial localization of genes within the nucleus are related to their transcriptional regulation. The mechanistic consequences of linear gene order, and how it may relate to the functional output of genome organization, remain to be fully resolved, however. Here we tested the relationship between linear and 3D organization of gene regulation during myogenesis. Our analysis has identified a subset of topologically associated domains (TADs) that are significantly enriched for muscle-specific genes. These lineage-enriched TADs demonstrate an expression-dependent pattern of nuclear organization that influences the positioning of adjacent nonenriched TADs. Therefore, lineage-enriched TADs inform cell-specific genome organization during myogenesis. The reduction of allelic spatial distance of one of these domains, which contains Myogenin, correlates with reduced transcriptional variability, identifying a potential role for lineage-specific nuclear topology. Using a fusion-based strategy to decouple mitosis and myotube formation, we demonstrate that the cell-specific topology of syncytial nuclei is dependent on cell division. We propose that the effects of linear and spatial organization of gene loci on gene regulation are linked through TAD architecture, and that mitosis is critical for establishing nuclear topologies during cellular differentiation.
Vertebrate genomes are innately organized, with defined regions of gene density and paucity distributed along chromosomes. Beyond this basic level of organization, there are gene clusters that facilitate coordinate regulation; for example, the β-globin locus, epidermal differentiation complex, and Hox gene cluster exhibit a critical dependence on both the linear proximity and order of their genes for appropriate temporal regulation during development (1–3). The combination of genomic sequences and genome-wide expression studies have revealed another form of nonrandom gene distribution, termed tandem gene arrays (TGAs), which represent contiguous stretches of gene loci that display differential regulation during cellular development and/or function (4, 5). Importantly, gene clusters and TGAs also demonstrate expression-dependent nuclear localization patterns (6–9); therefore, gene loci are both deterministically arrayed along chromosomes and positioned within the nucleus (10–15).
Of course, genomes are not organized within the interphase nucleus as strands of DNA, but rather exist as discrete, irregularly shaped structures known as chromosome territories (CTs) (16, 17). 5C and Hi-C, derivatives of chromosome conformation capture analysis, have identified topologically associated domains (TADs) as a feature of subchromosomal structural organization (18, 19). TADs are molecularly defined linear regions of intrachromosomal association that display high frequencies of physical interaction within a given domain but lower frequencies outside of these domains. Thus, TADs represent a potential link between linear chromosomal organization and 3D genome structure (18).
Intriguingly, TAD boundaries are relatively stable through the dynamic changes in genome architecture associated with both cell division and differentiation (18, 19). This stability may be due in part to the enrichment of architectural proteins at TAD boundaries (20). Boundaries appear to be intransient according to cellular identity, yet gene loci have demonstrated cell-specific nuclear localization patterns. Thus, it is possible that changes observed in subnuclear positioning of a single gene locus affect adjacent loci within the TAD or even adjacent TADs. However, the distribution of lineage-specific genes during differentiation relative to TADs, and the possible effect of this distribution on genome structure, have yet to be tested.
Although there is ample evidence for the relationship between nuclear organization and gene expression, how this correlation mechanistically impacts transcriptional regulation remains incompletely understood. An intriguing possibility for proximity influencing regulation has been observed in various model systems; in isogenic backgrounds, homologous alleles do not necessarily behave identically (21). This breakdown in expression homogeneity occurs because transcriptional activation requires the stochastic binding of transcription factors to cognate sites, and also because cells exist in a milieu with varying concentrations of external cues (22, 23).
The outcome of these probabilistic conditions is defined as intrinsic (transcription factor binding) and extrinsic (cellular context) transcriptional noise. Importantly, transcriptional noise has been implicated in establishing patterns of expression that can lead to cellular diversity (24–26). Therefore, a mechanistic output of linear gene distribution and the spatial proximity of coregulated alleles may be attenuation of the stochastic fluctuations of regulatory proteins that underlie intrinsic transcriptional noise (27). Specifically, the proximity of loci would lead to an increase in the local concentration of the DNA elements that direct the expression of these genes, resulting in a concomitant increase in the concentrations of the regulatory proteins that bind them (27–29). This hypothesis has yet to be tested with endogenous loci during cellular differentiation.
Another central unresolved question in the study of genome organization is how patterns of nonrandom nuclear positioning of gene loci are established. Specific genes, generally those involved in a rapid cellular response to a stimulus such as stress, can dynamically change positions (13), and these changes can alter CT positioning (30, 31); however, interphase chromatin demonstrates constrained diffusion for the most part, consistent with the Brownian motion of an object tethered in space (32, 33). During mitosis, the relationship between gene expression and organization is lost on dissolution of the nucleus (34). Importantly, it has been observed that in a stably cycling population of cells, the interphase organization of the genome can reemerge as a facsimile in daughter nuclei (35). Although this behavior may be probabilistic and dependent on methodology (36, 37), it is likely that a given cell type “remembers” its genomic organization (38) to reestablish a cell-specific expression profile. Proliferation is a hallmark of cellular differentiation and has been suggested to play a role in self-organizing genome topologies (30, 39). We suggest that during cellular development, relevant transcription factors are up-regulated, and a latent landscape for regulation emerges as a result of their binding with cognate binding sites. The transcription factor-bound, euchromatic status of these genes would thereby facilitate genome organization on nuclear reconstitution. Testing this hypothesis has been an intractable problem, because it is difficult to uncouple differentiation from proliferation (40–42).
Myogenesis is a process of differentiation mediated by the master regulatory factor MyoD, in which adherent cells initiate Myogenin expression, drop out of the cell cycle, and fuse to form multinucleated syncytia, known as myotubes (43). A myotube is in a terminally differentiated state and contains a population of nuclei arrested in G0. Here we used myogenesis as a model system of cellular differentiation to gain insight into how linear features of genome organization manifest 3D nuclear topologies. Using data from a whole genome microarray expression set, we have identified TADs that are significantly enriched for muscle-specific genes. We examine the effect of these lineage-enriched TADs (LE-TADs) on nuclear organization in the context of human chromosome 1 (HSA1). Our analysis indicates that LE-TADs drive local changes in HSA1 organization, as well as general alterations to nuclear topology.
Importantly, we provide evidence that proximal localization of alleles leads to a reduction in transcriptional noise, suggesting a unique functional role for patterns of nuclear organization. Finally, taking advantage of the properties of cell–cell fusion during myogenesis to uncouple differentiation and cell division, we demonstrate that differentiation-mediated changes in nuclear topology require mitosis. Our findings suggest that the linear clustering of lineage-restricted genes into chromosomal domains serves to shape cell-specific functional nuclear topologies, and that the emergence of these topologies is dependent on cell division.
Results
Genes Coregulated During Myogenesis Are Enriched in Specific TADs.
To analyze the relationship between linear gene order and 3D genome organization, we first defined a myogenic gene set by interrogating a single robust whole genome gene expression array of primary human myoblasts differentiated to myotubes (44). Within this dataset, we identified 2,275 genes with a >1.75-fold change in expression as the myogenic gene set. Having determined their linear genomic positions, we then generated a simulated gene set for statistical comparison (Materials and Methods). As has been reported for various model systems (5, 15, 45), we identified a significant enrichment of TGAs in the myogenic set (P < 0.001), with certain chromosomes contributing disproportionately to this bias (SI Appendix, Fig. S1 and Table S1). Thus, as anticipated, genes coregulated during myogenesis are nonrandomly distributed in the human genome.
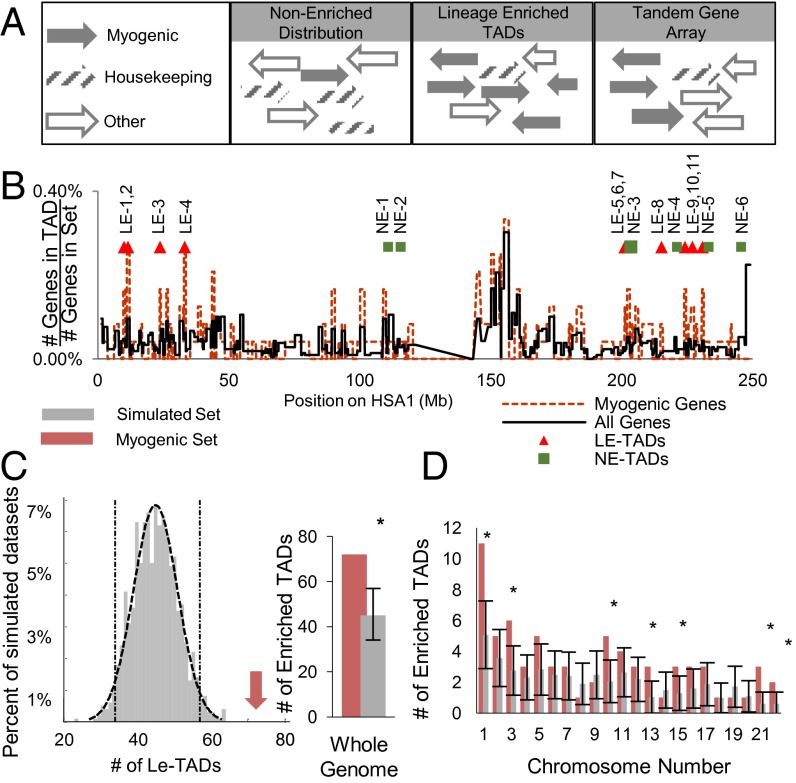
Because TADs represent patterns of chromosome organization, or folding, they compose the vast majority of the human genome. Nevertheless, given the nonrandom distribution of myogenic-specific genes, we investigated whether there are TADs enriched for these genes (Fig. 1A). Using previously defined IMR-90 TAD boundaries (18), we generated gene density maps, on a per-TAD basis, of the entire genome and compared them with the maps derived from the myogenic gene subset (Fig. 1B). By testing the mapped occupancies against a cumulative binomial distribution, we identified 73 TADs significantly enriched for myogenic genes, known as LE-TADs. Importantly, this identification of myogenic LE-TADs uncovered a greater-than-expected frequency, assuming a distribution of genes representative of the total genome (P < 0.001) (Fig. 1C).
Fig. 1.
Myogenic genes have a nonrandom distribution along linear chromosomes. (A) Schematic illustrating a representative LE-TAD arrangement in comparison with TGAs and more typical gene arrangements. (B) Gene density map, on a per TAD basis, of HSA1. Triangles denote locations of LE-TADs. (C) Histogram of the number of LE-TADs observed in the 1,000 simulated gene sets (gray). Dashed lines show the fit of a normal curve and the SD of the mean. The red arrow denotes the 73 LE-TADs observed in the myogenic set (P < 0.001). The data are also shown as a bar graph to highlight the relationship between C and D. (D) LE-TAD distribution by chromosome. Gray bars are the average number of LE-TADs per chromosome among the simulated gene sets with the SD of the random simulation shown in error bars. Red bars indicate the number of LE-TADs per chromosome in the myogenic set. *Significant enrichments of LE-TADs; P < 0.05.
In addition, seven chromosomes were themselves significantly enriched for LE-TADs (Fig. 1D and SI Appendix, Fig. S2; a complete list of TADs and their resident genes is provided in Dataset S1). Out of these seven chromosomes, we chose HSA1 owing to its level of complexity and enrichment, to test whether linear gene distributions affect the spatial organization of the genome during myogenesis.
The Chromosome Territory Morphology of HSA1 Changes During Myogenesis.
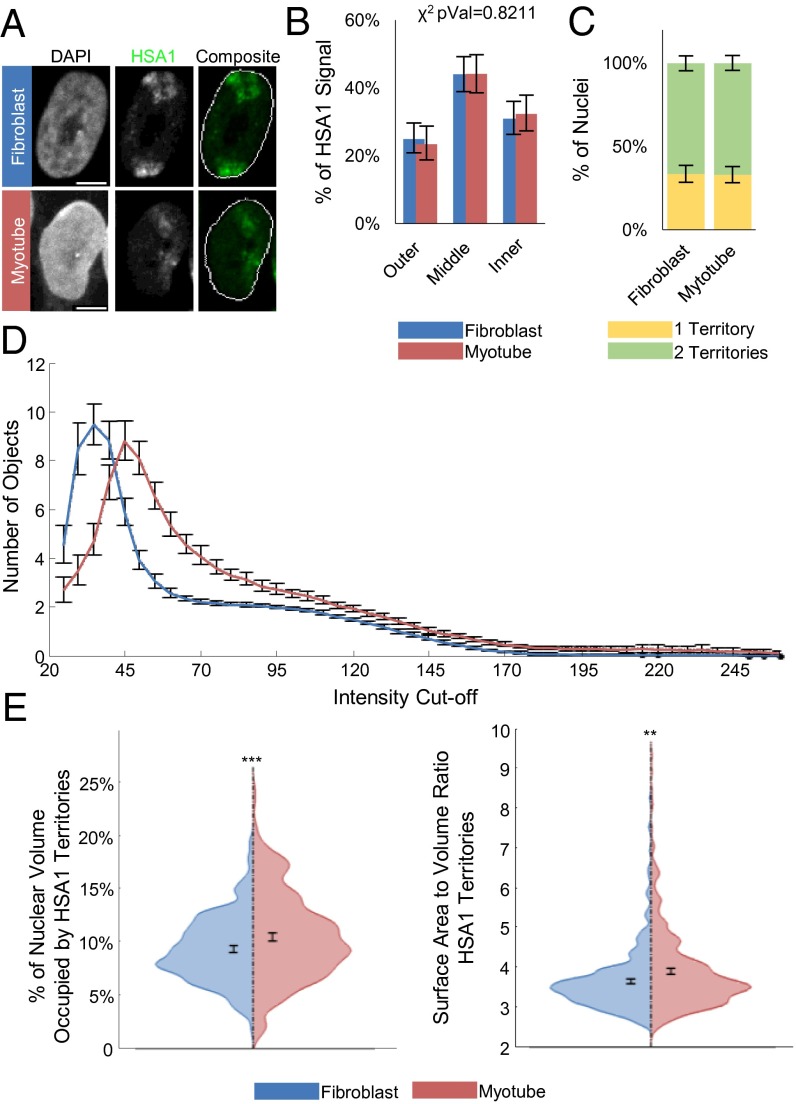
For this study, we used a well-characterized system of viral transduction-mediated myogenic differentiation (46). In this approach, overexpression of MyoD in primary human fibroblasts (IMR-90s) induces myogenesis in a manner analogous to the process occurring in biopsy-derived myoblasts, such that myogenic genes that are not expressed in fibroblasts become strongly induced (SI Appendix, Fig. S3). Importantly, it has been shown that this process results in increased MyoD binding at physiological sites, but not the formation of novel binding sites (47). We first assayed the nuclear position of HSA1 with whole chromosome paint (WCP) 3D FISH by creating three concentric shells of equal area and measuring the percentage of HSA1 signal per shell (Fig. 2A). This analysis revealed that HSA1 does not significantly reposition relative to the nuclear periphery during myogenesis by a χ2 test of independence (P = 0.8211), although there is a trend toward internalization (Fig. 2B). As opposed to what we have reported previously (15), the degree of homologous HSA1 coalescence during myogenic differentiation is also invariant (Fig. 2C). Therefore, myogenic gene expression is not correlated with changes in the nuclear positioning of HSA1.
Fig. 2.
Behavior of the myogenic gene-enriched HSA1 during MyoD-induced myogenesis. (A) Example FISH images of cells transduced with either GFP (fibroblast) or MyoD (myotube), showing the HSA1 CT (green). (B) Percentage of total signal from HSA1 paint staining residing in three zones of equal area in maximum intensity projections. Reported χ2 values represent the probabilities that the myotube and fibroblast values are from the same underlying distribution. (C) Percentage of nuclei with a single coalesced HSA1 territory versus those with two distinct regions. (D) Number of discrete objects larger than 10 μm3 in volume as a function of the intensity cutoff used to generate binary masks for analysis. (E) Violin plots of individual territory properties. (Left) Percentage of the DAPI nuclear mask occupied by the HSA1 mask. (Right) Area of the sum of the faces of the perimeter voxels divided by the volume of the convex hulls of each territory. Significance was assayed by the two-sample t test with unequal variance (*P < 0.05; **P < 0.005; ***P < 0.0005). Error bars are 95% CIs of the mean. n >250 nuclei and n >430 territories. Results represent a pooling of the raw data from a combination of three biological replicates. (Scale bar: 5 μm.)
Interestingly, despite the lack of HSA1 reorganization, we did note morphological differences in the CTs during differentiation. Thus, we examined how the structure of HSA1, as represented by the WCP, is altered during myogenesis. To analyze the topological changes in the CTs during differentiation, we measured the number of discrete domains present at increasing intensity thresholds. This process allows for a description of the fluorescence signal in terms of its uniformity or lack thereof (SI Appendix, Materials and Methods) (17). Our analysis indicates that the HSA1 CT becomes fragmented into a larger number of objects (or domains) specifically in myotube nuclei, consistent with a more dissolute chromosome conformation during myogenesis (Fig. 2 A and D), as opposed to the morphological changes observed in three other HSAs (SI Appendix, Fig. S4). These changes in HSA1 topology are corroborated by an increase in the total percentage of nuclear volume occupied by homologous CTs and an increase in the surface area-to-volume ratio of individual CTs (Fig. 2E). Therefore, the HSA1 CT reveals lineage-specific changes to its internal structure as opposed its overall nuclear positioning.
LE-TADs Are Nonrandomly Organized in Myotube Nuclei.
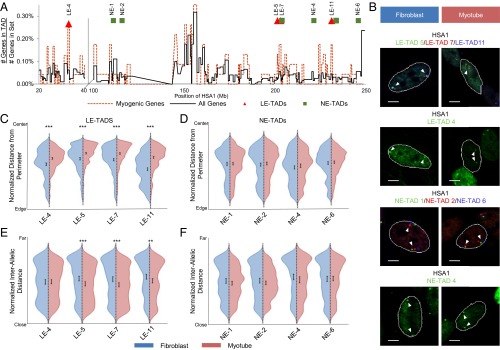
As described above, genes are positioned nonrandomly within the interphase nucleus, including both their relationship to the nuclear periphery as well as to themselves, i.e., allelic association/proximity (38). We tested whether domains within HSA1 CT were repositioned during myogenesis according to the defined radial and proximity metrics of interphase nuclear organization. The Euclidian (or “straight line”) distance between two chromosomal regions separated by an equal number of base pairs is smaller when the domains are found within a single TAD (18); thus, for this analysis we chose four LE-TADs (4, 5, 7, 11) and four nonenriched TADs (NE-TADs) (1, 2, 4, 6) based on their linear distribution and myogenic-specific gene density (Fig. 3A and SI Appendix, Table S2). Importantly, the NE-TADs are interspersed among the LE-TADs, which permits a direct test of domain organization versus general chromatin changes. TAD boundaries are reportedly relatively invariant (18, 19); however, to ensure the conformity of the TADs that we have analyzed, we compared their boundaries from IMR-90s with those identified in hESCs. We found the TAD boundaries to be well preserved between the two developmentally distinct cell types, suggesting that the IMR-90 boundaries should be represented in myotubes (SI Appendix, Table S3). To control for levels of gene expression, NE-TAD 2 serves a double purpose as it encodes the most highly induced gene (CASQ2) found on HSA1 during myogenesis, but no other differentially expressed genes. In comparison, LE-TAD 7 contains both the MYOG (Myogenin) gene locus, which was previously implicated in repositioning during myogenesis (10, 11), and a significant number of myogenic-restricted genes (SI Appendix, Table S2).
Fig. 3.
Behavior of myogenic gene-enriched domains of HSA1 during MyoD-induced myogenesis. (A) Relative gene density map for the regions of interest on HSA1 denoting the positioning of the LE-TADs and NE-TADs assayed. (B) Example FISH images of cells transduced with either GFP (fibroblast) or MyoD (myotube), showing the positioning of the LE-TADs or NE-TADs. (C) Violin plots of the normalized distances from the nuclear perimeters of the LE-TADs. (D) Violin plots of the normalized distances from the nuclear perimeters of the NE-TADs. (E) Violin plots of the normalized distances between homologous alleles of the LE-TADs. (F) Violin plots of the normalized distances between homologous alleles of the LE-TADs. Significance was assayed by the two-sample t test with unequal variance for distance from the nuclear perimeter and the Wilcoxon rank-sum test for interallelic distance (*P < 0.05; **P < 0.005; ***P < 0.0005). Error bars are 95% CIs of the mean. n >400 individual alleles and n >200 pairs. Results represent a pooling of the raw data from a combination of at least two biological replicates. (Scale bar: 5 μm.)
On our 3D FISH analysis, all four LE-TADs demonstrated significant repositioning toward the nuclear interior of both alleles during myogenesis (Fig. 3 B and C and SI Appendix, Fig. S5), whereas the four NE-TADs remain unchanged (Fig. 3 B and D and SI Appendix, Fig. S5). Compared with a uniform population, a χ2 goodness-of-fit test indicated that all of these TADs were positioned nonrandomly relative to the nuclear periphery (P < 1 × 10−11). Although all of the TAD positions were nonrandom, only LE-TADs showed differential positioning as a function of cellular identity. Our assay of interallelic distances between homologous TAD pairs showed no change in the NE-TADs, but significantly reduced distances among the three proximal LE-TADs (5, 7, 11) (Fig. 3 E and F), which was unexpected because the HSA1 CTs did not exhibit increased association during differentiation (Fig. 2C). Interestingly, the distal LE-TAD 4 (which is ∼167 Mb from the beginning of the clustered LE-TADs) did not demonstrate a reduction in interallelic distance, indicating that proximity is not a direct consequence of allele internalization and suggesting regional expression-dependent CT confirmations.
To further validate our findings, we performed multicolor FISH with LE-TAD 7 and NE-TAD 4 within the same nuclei, given that any bias introduced during image processing will affect objects in the same image equally. This approach confirmed that LE-TADs and NE-TADs behave differently, and that our previous findings were not an effect of image processing (SI Appendix, Fig. S6). Because only LE-TADs exhibit differentiation-dependent localization, these data support the hypothesis that myogenesis induces a change in the internal structure of HSA1, but not in its nuclear repositioning. Moreover, because NE-TADs are not relocalized despite being interspersed among LE-TADs, we suggest that LE-TAD organization informs HSA1 CT structure during myogenesis.
The Relative Positioning of LE-TADs Defines CT Structure During Myogenesis.
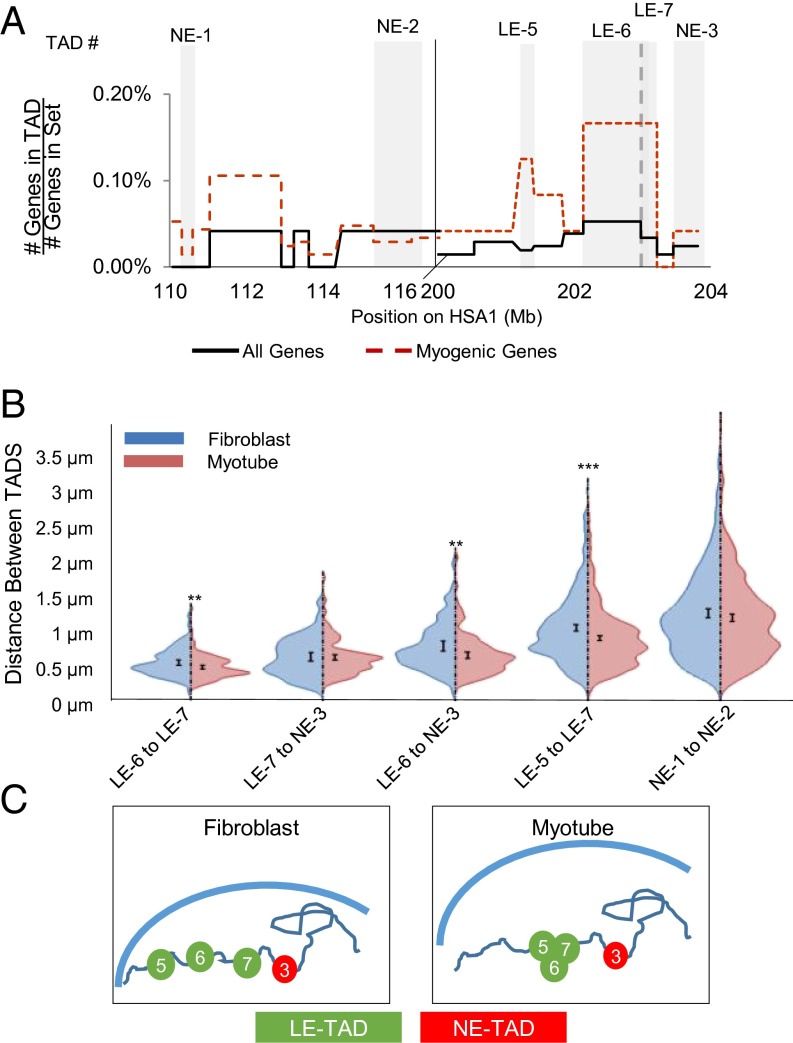
To determine whether LE-TAD reorganization influences CT morphology, we examined the positioning of LE-TADs and NE-TADs relative to one another within single CTs. Using multicolor FISH with various combinations of probes, we measured a series of pairwise Euclidian distances between TADs in fibroblasts and derived myotubes (Fig. 4A). Of note, we were interested specifically in the change in distance between two TADs that are not subject to binary cutoffs implicit in colocalization, given our belief that assessing a continuous variable, such as distance, better captures the probabilistic nature of genome organization. This analysis revealed that LE-TADs were significantly closer to one another in myotubes, whereas the distances among NE-TADs and LE-TADs were unchanged between cell types (Fig. 4B). Surprisingly, LE-TAD 5 was significantly more proximal to LE-TAD 7, even though the linearly adjacent NE-TAD 3 was not (Fig. 4 B and C). Thus, we reasoned that the inherent affinity between LE-TADs supersedes linear proximity.
Fig. 4.
Behavior of LE-TADs on a single HSA1 during myogenesis. (A) Gene density map of the TADs interrogated for pairwise interactions. Gray backgrounds highlight the TADs assayed. (B) Violin plots of the pairwise distances, in microns, between TADs of interest in GFP (fibroblast)-expressing or MyoD (myotube)-expressing cells. n >90 individual alleles. (C) Model of local domain positioning changes in response to differentiation, showing all of the TADs between LE-TAD 5 and LE-TAD 7 clustering in myotubes while the distance between LE-TAD 7 and NE-TAD 3 remained static. Significance was assayed by the two-sample t test with unequal variance (*P < 0.05; **P < 0.005; ***P < 0.0005). Error bars represent 95% CIs of the mean.
To further examine how the preferential association of LE-TADs may affect neighboring chromatin, we assayed the localization of NE-TADs 3 and 5, which are 0.5 Mb and 1 Mb telomere-proximal to LE-TADs 7 and 11, respectively. This analysis revealed that LE-TAD reorganization affected the positioning of adjacent regions that span multiple TADs (SI Appendix, Fig. S7) in the context of a CT in which not all NE-TADs showed differential positioning (Fig. 2). Therefore, the movement of LE-TADs affects regions larger than single TADs but not as large as an entire chromosome, suggesting that a CT is composed of multiple independently acting topological units.
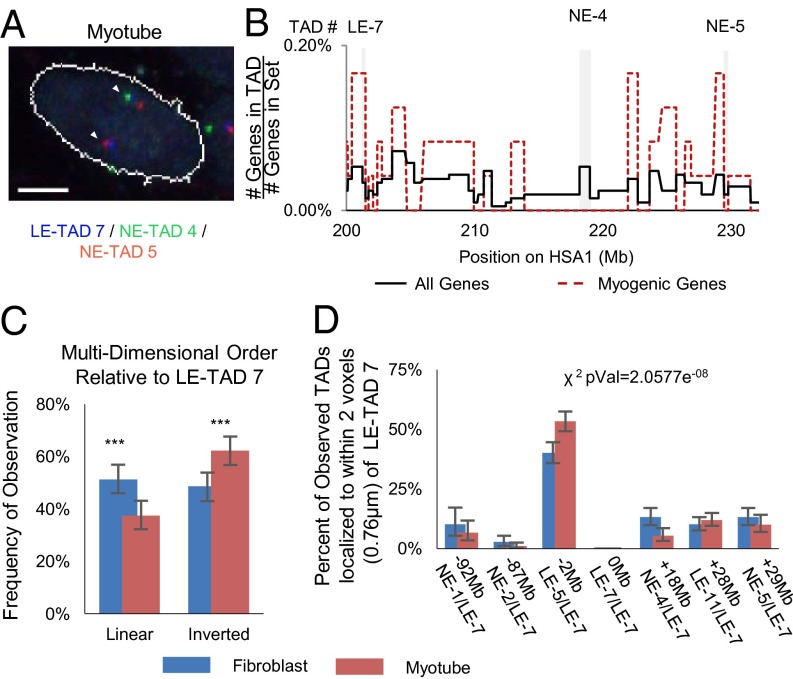
To gain insight into the nature of these independent movements, we focused on a short (∼2-Mb) region containing multiple LE-TADs and NE-TADs (SI Appendix, Fig. S8A). Multicolor FISH and multiplane optical sectioning confocal microscopy allowed us to identify and analyze three unique highly overlapping loci per chromosome (SI Appendix, Fig. S8B). This 2-Mb region of HSA1 centromere proximal to LE-TAD 7 becomes more compact and moves into closer proximity to LE-TAD 7 during myogenesis (Fig. 4B). We generated triangles that represent the pairwise relationships among LE-TAD 6, LE-TAD 7, and NE-TAD 3. The 3D coordinates of LE-TAD 7 were placed at the origin, and a standard distance between LE-TAD 7 and NE-TAD 3 was applied, thereby creating a defined localization for LE-TAD 6 (SI Appendix, Fig. S8C). The resulting map revealed that in myotubes the domains exhibit a more compact structure, indicating an overall decrease in the heterogeneity of local organization (Fig. 4C). Taken together, these findings lead us to conclude that LE-TAD 7 may be nucleating a cluster of coregulated genes.
Having established increased physical proximity among LE-TADs over a 2-Mb domain, we next wanted to determine the relative positions of LE-TADs over a longer linear distance. We performed multicolor FISH with probes to LE-TAD 7, the LE-TAD proximal NE-TAD 5 (∼30 Mb distal), and intervening NE-TAD 4 (Fig. 5 A and B). The expected relationship between linear and 3D proximity is that because domains are a greater number of base pairs apart, there would be on average a concomitantly greater Euclidian distance between them within the context of an interphase nucleus, but if there is no relationship between linear and 3D distance, then an equal proximity should be observed between any two sets of domains. Our analysis demonstrated that in myotubes there was a clear bias for the linearly distal NE-TAD 5 to be physically more proximal to LE-TAD 7 than NE-TAD 4, suggesting that NE-TAD 4 is “looped out” in 3D space, conferring an inverted conformation (Fig. 5C). Therefore, distal LE-TADs appeared to act independently of local chromatin and had a propensity to cluster, giving rise to a specific CT topology.
Fig. 5.
Intrachromosomal LE-TADs distances are not a function of linear distance. (A) Example FISH images of a cell expressing MyoD, showing three domains. The linearly distal domains, NE-TAD 5 (red) and LE-TAD 7 (blue), are proximal, and NE-TAD 3 (green) is looped out. (B) Gene density map of the chosen TADs highlighted with gray backgrounds. (C) Quantification of the frequency at which the three TADs (LE-7, NE-3, and NE-4) are found in 3D distances predicted by linear sequence. Significance was assayed by the two-sample t test with unequal variance (*P < 0.05; **P < 0.005; ***P < 0.0005). Error bars represent 95% CIs of the mean. n >300 clusters. (Scale bar: 5 μm.) (D) Percentage of observations in which the TAD denoted on the x-axis is found within two voxels (0.7898 μm) of LE-TAD 7. The reported χ2 value represents the probability that the myotube and fibroblast values are from the same underlying distribution.
Three-dimensional clustering also can be assayed by determining the percentage of pairwise observations between LE-TAD 7 and regions of interest within 780 nM (two voxels) of each other. This method once again led to an observed increase in association between LE-TAD 7 and other LE-TADs within myotube nuclei, regardless of the length of the intervening linear sequence (Fig. 5D). We suggest that the combined effects of these individual alterations in LE-TAD localization resulted in an emergent CT topology. This model of emergent order, although here confined to a single CT, is mechanistically analogous to what has been proposed to occur on the level of the whole genome (39).
The Subnuclear Localization of LE-TADs Impacts Allelic Transcriptional Variance.
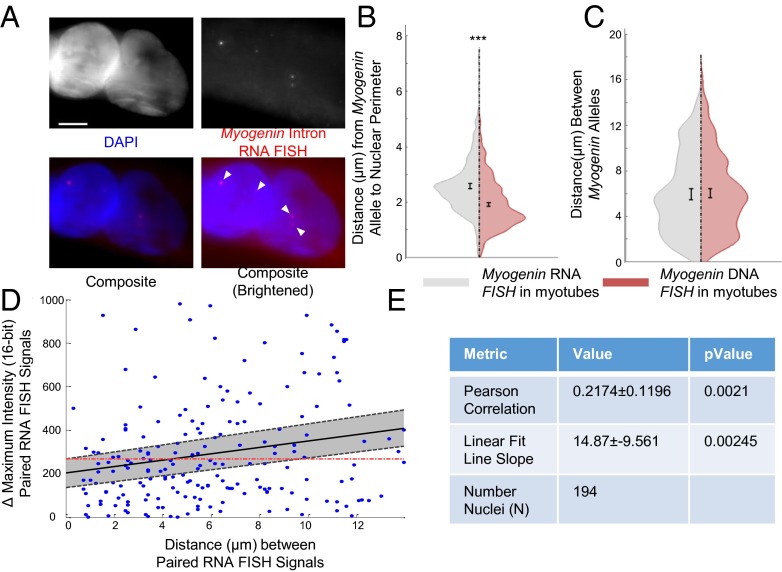
Having observed nonrandom positioning of LE-TADs within myotube nuclei, we performed nascent RNA FISH with a probe to introns of Myogenin, which is contained within LE-TAD 7, to test the functional outcome of these lineage-specific topologies. Using intronic sequences as a probe allows for the detection of unprocessed transcripts, capturing the real-time expression state of the gene at the time of fixation (Fig. 6A). By comparing the subnuclear position of LE-TAD 7 as defined by nascent RNA FISH to that of DNA FISH localization, topologies that are more likely to be active can be determined. We first analyzed two common features of activity-based genome organization, radial positioning and allele proximity. As expected, the Myogenin RNA foci observed in myotube nuclei paralleled the increased proximity of the gene loci observed during differentiation (Fig. 3E). In support of the nuclear interior being permissive to transcription (15), the RNA foci were further from the periphery than the Myogenin gene loci (Fig. 6B).
Fig. 6.
Subnuclear positioning of the Myogenin alleles attenuates stochastic differences in biallelic expression. (A) Example nascent Myogenin intron RNA FISH images of cells expressing MyoD; nuclear counterstaining with DAPI (blue) or Myogenin intron RNA FISH (red). Arrows and image brightening highlight nascent RNA FISH signals. (Scale bar: 5 μm.) (B) Violin plot of the distances from the perimeter in maximum intensity projections of the nascent RNA FISH foci versus DNA loci. (C) Violin plot of the distances between alleles in maximum intensity projections of the nascent RNA FISH foci versus DNA loci. (D) Scatterplot comparing the distances among RNA FISH foci to the changes in maximal peak intensity of the foci. The dashed line and gray shading represent the 95% CI of the best-fitting line. The red dashes indicate the expected position of the fitted line with a slope of 0. (E) Summary statistics for the measurement of Pearson correlation and the slope of the best-fitting line. ***P < 0.0005. Error bars represent 95% CIs of the mean. n >350 individual alleles and n >175 pairs. The results represent a pooling of the raw data from a combination of three biological replicates.
We next compared the interallelic distances between RNA and DNA signals in myotubes and, unexpectedly, found very similar distributions (Fig. 6C). Thus, although both RNA foci and gene loci were significantly closer in myotube nuclei, biallelic transcriptional activity occurred over a range of distances. Therefore, we hypothesized that reducing interallelic distance might affect other aspects of transcription; specifically, allelic proximity may stabilize the differences in transcriptional activity between the two alleles. Mechanistically, this could occur by having the two alleles experience the same subnuclear milieu, thereby minimizing the difference in stochastic binding events during gene activation and reducing intrinsic noise (26).
To test this hypothesis, we quantified the difference between maximal peak intensity of nascent Myogenin RNA FISH signals in nuclei with two signals relative to the distance between them. We observed a positive Pearson correlation (r = 0.217) between the two values that was significantly different from an uncorrelated (r = 0) population (P = 0.001) (Fig. 6D). Although there was variability around the best-fitting line, the slope of this line was significantly unlikely to be zero (P = 0.0021) (Fig. 6D). Thus, a positive relationship existed between transcriptional output and reduced interallelic distance, indicating that as homologous alleles move into closer proximity, their expression variance is reduced. Therefore, although reducing interallelic distance in not a strict requirement for gene activation, it attenuates variability, or intrinsic noise. Our results imply disparate but overlapping roles for two commonly studied features of nuclear organization, interallelic and radial distance, and establish the potential for a functional role for lineage-specific nuclear topologies.
Cell Division Is Required for Establishing Changes in Nuclear Organization During Myogenesis.
How lineage-specific genome topologies emerge is largely not understood. Although it has been observed that chromatin organization in cycling cells maintains a defined organization from interphase through mitosis and to daughter nuclei, this pattern has not been reproducibly demonstrated (35, 36, 48). Moreover, analyzing the role of the cell cycle in establishing a new nuclear topology during cellular development has remained an intractable problem owing to the tight coupling of proliferation and differentiation. Therefore, we developed a strategy to uncouple cell division and differentiation using the cell surface protein myomaker, which can mediate the fusion of fibroblasts into fully differentiated myotubes (49). Specifically, cultures of IMR-90 hTert cells were either infected with GFP-MyoD or infected with myomaker and then pulsed with BrdU. After infection and selection, the two populations of cells were mixed in a coculture. BrdU staining was used to trace the nuclei derived from the myomaker infections, and GFP staining indicated MyoD+ nuclei (Fig. 7A). The BrdU molecules were stably integrated into the genomic DNA of myomaker-infected cells and thus could not move into other nuclei in the myotube; however, the same was not true for cells expressing MyoD. Importantly, the fusion event placed a nonmyogenic nucleus from the myomaker-infected cells into a myogenic context but in the postmitotic environment of terminal myotubes (50), unlike the MyoD-infected cells, which were also in a myogenic context but were able to initially undergo cell division in the presence of MyoD before the formation of myotubes (Fig. 7B). This dichotomy of cellular environment at the time of MyoD introduction is the key factor leading to the decoupling of cell division and differentiation. Importantly, myomaker ectopic expression alone did not induce myogenesis or any myogenic topological changes (SI Appendix, Fig. S9).
Fig. 7.
Cell division is required for repositioning of the Myogenin-containing LE-TAD 7. (A) Experimental method used to generate and differentially label nuclei within a chimeric syncytium in which nuclei expressing myomaker are labeled with BrdU. (B) Immunofluorescence images of chimeric myotubes generated through coculture or the cognate monocultures. (C) Schematic showing the relationship between cellular identity and differentiation time for either MyoD- expressing cells or myomaker-expressing cells that fuse into a myotube. (D) Example images of how nuclear identity was scored. (Left) LE-TAD 7 DNA FISH (red) and BrdU (green). (Right) Myosin heavy chain (red) and BrdU (green). The DNA counterstain outline is in white. (E) Normalized distance of LE-TAD 7 to the nuclear perimeter. (F) Normalized LE-TAD 7 interallelic distances. (G) Volume of the convex hull of the DAPI stain in BrdU+ versus BrdU− nuclei; n = 35. Significance was assayed by the two-sample t test with unequal variance, except for interallelic distance, in which a one-tailed Wilcoxon rank-sum test was used (*P < 0.05; ***P < 0.0005; ^P < 0.05, one-tailed test). Error bars represent 95% CIs of the mean. n >300 individual alleles and n >150 pairs. Results represent pooling of the raw data from a combination of at least four biological replicates. (Scale bar: 10 μm.)
Previous investigators have suggested that mitosis is necessary for the repositioning of genes during differentiation (7, 30, 31, 51). Our chimeric myotube strategy allowed us to directly test whether changes in lineage-specific nuclear topology are dependent on cell division. Using BrdU as a nuclear marker for actively dividing myomaker-transduced cells, we performed DNA immuno-FISH on chimeric myotubes. Each nucleus was then assigned a BrdU status (positive or negative), and the positioning of LE-TAD 7 was analyzed in terms of both the nuclear periphery and the interallelic distance (Fig. 7C). In coculture experiments, the Myogenin alleles were found further from the periphery in nuclei derived from BrdU− (MyoD-transduced) cells than in nuclei derived from BrdU+ (myomaker-transduced) cells (Fig. 7D). Compared with MyoD-expressing nuclei, myomaker-expressing nuclei also had longer interallelic distances for Myogenin loci and were larger, with a morphology similar to that of GFP-infected cells (Fig. 7 E and F), suggesting that global changes in nuclear organization also depend on cell division. Taken together, our data demonstrate that differentiation-dependent (i.e., MyoD expression) cell division is required for the emergence of lineage-specific nuclear topologies.
Discussion
We have examined the relationship between linear gene order and 3D nuclear organization during cellular differentiation using myogenesis as a model. Our initial bioinformatics analysis revealed that genes coregulated during myogenesis have a significant tendency to be colinear in the human genome (TGAs). This result bolsters our hypothesis that the organization of developmentally regulated gene expression is nonrandom. By interrogating the linear distribution of coregulated myogenic genes in the context of TAD boundaries, we observed a class of TADs significantly enriched for lineage-restricted genes (Fig. 1B). Although the set of chromosomes found to be enriched for lineage-enriched TADs (LE-TADs) did not entirely overlap with those enriched for TGAs, this was to be expected, given that the latter are defined solely as having no interceding genes that are coregulated, whereas LE-TADs require only a general density of genes, not a strict sequential relationship (Fig. 1A and SI Appendix, Fig. S1).
Our assay of these LE-TADs in the context of their spatial localization revealed their involvement in expression-dependent changes in genome organization. Individual genes expressed during differentiation and/or for a particular cellular activity do not necessarily demonstrate differentiation-dependent changes in subnuclear positioning (52). Therefore, our results expand on the important role of gene colinearity in nuclear organization (4). Specifically, we provide evidence that TADs, the perceived unchanging building blocks of CT organization, can in fact influence lineage-specific genome organization. Moreover, LE-TADs represent a unique way of identifying candidate genes/regions that display differentiation-dependent repositioning with a high degree of accuracy (Fig. 3). Our results demonstrate that LE-TADs contribute to emergent genomic order and reveal a new means of testing the general functional relevancy of TAD organization.
Myogenic LE-TADs on HSA1 were repositioned even though NE-TADs and the CT on which they reside did not demonstrate marked changes as a function of differentiation (Figs. 2 and 3). These changes in LE-TAD subnuclear and intrachromosomal localization created preferential local (Fig. 4) and long-range (Fig. 5) associations between genes located on HSA1 during cellular differentiation. Thus, TADs have the capacity to intermingle within a single CT, which has not been reported previously. The pronounced invariance in TAD boundaries reported across various cells types (18) makes changes in TAD identity an unlikely explanation for the changes in CT organization that we observed. In light of this model of chromosome organization, the remodeling in HSA1 may represent a repositioning of TADs relative to one another. A slightly larger (∼5 Mb) interaction domain, known as a compartment, also can be observed on the heat maps generated by the Hi-C experiments that originally defined TADs (18, 53, 54). Compartments are composed of multiple associated TADs and, unlike TAD boundaries, are highly cell type-dependent and cannot be predicted from sequence identity alone (55, 56). Our observation of cell-type specific LE-TAD association leads us to the conclusion that lineage-specific compartments of chromosomal order may be composed of coregulated genes that are themselves within LE-TADs. The organization of these LE-TAD–mediated intrachromosomal compartments relative to one another would then serve as the basis for an emergent lineage-specific CT morphology.
Changes in genome organization as a function of linear gene density are critical to understanding how lineage-specific nuclear topologies emerge; however, the significance import of nonrandom organization on transcriptional regulation remains incompletely understood. To ascribe a functional outcome to the decreased distances observed between Myogenin alleles, we performed nascent RNA FISH to concurrently detect their level of activity and subnuclear position (Fig. 6). Intrinsic transcriptional noise occurs due to the inherently stochastic nature of general transcription machinery and the regulatory factors associated with their target sequences (21, 26). Importantly, our RNA FISH analysis indicates that the physical proximity of the Myogenin loci in myotube nuclei attenuates differences in their biallelic transcriptional output, reducing intrinsic noise. Increases in transcriptional noise have been identified as detrimental to organismal fitness (57, 58); therefore, features of nuclear organization that are able to modulate it may provide an evolutionary advantage. Our data suggest that allelic proximity acts as one such feature. We propose that the attenuation of variance is due to a reduction in the stochastic contribution of transcription initiation by creating a shared microenvironment. It was recently demonstrated that chromatin remodeling is sufficient to induce gene relocalization (9). One possible explanation for movement preceding expression is to allow for the modulation of noise during all stages of transcriptional activation.
Changes in genome organization during cellular differentiation require that genes and chromosomes reorganize in a dense nuclear environment. During mitosis, the physical constraints of chromosome movement are mitigated as chromosomes condense and the nuclear envelope breaks down. Therefore, it has been hypothesized that mitosis is an ideal time for lineage-specific changes in nuclear organization to occur (48). Unfortunately, the tight coupling of cell division and differentiation made this an untestable hypothesis before the present study. Using ectopic myomaker expression in coculture, as reported previously (49), we overcame this limitation and decoupled mitosis and differentiation. Our approach revealed that repositioning of endogenous gene loci is dependent on cell division events in the context of differentiation (Fig. 7). This observation is in contrast to the dynamic changes in nuclear organization observed under stress response conditions (13, 30, 31), in which loci move in less time than is required for a cell cycle. The nuclear topology did not return to its prestress configuration until after release from a mitotic block, however (30). Therefore, not all changes in genome organization require mitosis, as we have observed in our study. However, unlike the stress response, cell differentiation establishes a new stable state in which the immediacy of transcription is less important than the fidelity of expression.
We suggest that modes of nuclear reorganization may depend on the competing requirements for speed and stability in gene expression. In the context of lineage commitment, the tight relationship between differentiation and cell proliferation across various biological systems suggests an evolutionary advantage for the emergence of topologies derived from coordinated gene expression.
Materials and Methods
Identification of Myogenic Genes.
The raw data from a whole-genome microarray expression profile using the HumanHT-12 v4 Expression BeadChip platform (Illumina) from MacQuarrie et al. (44) was downloaded from the National Center for Biotechnology Gene Expression Omnibus (NCBI-GEO; accession no. GSE50411). To determine gene expression changes between myoblast and myotubes, we used the R limma package for analysis of Expression BeadChips to compare only the data from myoblasts and myotubes. This method reports the degree of differential expression across the replicates for the two different experimental conditions for all genes on the microarray. Among these genes, we removed all nonautosomal genes and genes with unannotated or ambiguously annotated chromosomal locations. Multiple annotated start positions across more than one megabase of sequence were considered ambiguous. Finally, we extracted genes with a >1.75-fold change in expression from the list and treated them as differently regulated genes during myogenesis. In doing this, we identified 2,275 genes in the myogenic gene set.
Identification/Enrichment of Linear Gene Clusters.
TAD domain boundaries were used as described previously in IMR-90 cells (18) and transformed to GRCh37/hg19 positions using the liftOver tool (UCSC Genome Browser). In the rare cases when a position from hg18 did not exist in hg19, the location of the boundary was estimated using TAD size and the location of linearly proximal TAD boundaries. All DNA positions reported are in terms of GRCh37/hg19 positions. The total gene set and myogenic gene set were mapped onto chromosomes within defined TADs by their annotated starting positions. Using the entire gene list, a probability distribution for gene per TAD was established. These distributions allow for the assessment of an expected number of genes to be observed within the TAD when a smaller subset of the whole gene set is mapped into TADs. When the probability of observing the deviation that was empirically determined was <5% (P < 0.05), the TAD was classified as a significant domain, or an LE-TAD.
For the assessment of TGAs, the annotated start positions of the myogenic genes were considered in conjunction with the classification of the genes whose annotated start positions immediately precede and follow. If either of the linearly proximal genes is found to be myogenic, then the two genes are said to be in a tandem gene array. This process is continued until the flanking genes are both nonmyogenic, at which point the scope of the TGA has been identified.
To calculate the enrichment of TGAs and LE-TADs, a simulated gene set was created by randomly sampling the entire gene set for populations of 2,275 genes, the number of genes in the myogenic set. This process was repeated 1,000 times to create a large collection of mock gene sets. For each of these mock gene sets, the numbers of TGAs and LE-TADs were identified and combined to generate a histogram representing the likelihood that a random collection of 2,275 genes taken from our defined total gene set would contain a defined number of TGAs or LE-TADs. Thus, by comparing the empirically derived number of TGAs and LE-TADs to these histograms, we were able to determine the probability that a random collection of sampled genes would contain more TGAs or LE-TADs. Performing 1,000 simulations limits the P value to a minimum of 0.001. For enrichment of a per-chromosome basis, the same analysis was performed, but the number of TGAs and LE-TADs were considered per chromosome as opposed to the whole genome.
DNA Constructs.
The MyoD and GFP viral expression constructs were created by cloning the PCR-amplified cDNAs into the Retro-X Q vector (pQXCIP) expression plasmid (Clontech), using restriction enzymes. The MyoD cDNA (GenBank accession no. BC064493.1) was PCR-amplified to add NotI and BamHI sites for cloning into the MCS of pQXCIP. The GFP cDNA was PCR-amplified from a pEGFP vector (Clontech) to add NotI and EcoRI sites for cloning into the MCS of pQXCIP. The hTert retroviral expression vector pQXCIH-hTert was kindly provided by Drs. Robert Goldman and Takeshi Shimi (Northwestern University). A cDNA library was generated from MyoD-transduced IMR-90–derived myotubes by performing a reverse-transcriptase reaction with oligoDT primers. From this library, the myomaker (Tmem8C) cDNA was PCR-amplified with the following primers: 5′-CCGGATCCTCAGAGCACCTGGTATATC-3′ and 5′-GCGAATTCTC AGACACAAGCACAGCACA-3′. The resulting product was cloned into the pCR-Blunt-II Topo plasmid through the topoisomerase reaction (Invitrogen; 450245). The product was then cut out using the EcoRI restriction enzyme and cloned into a pBabe-Puro retroviral expression vector. The correct orientation and copy number for insertion were assessed through sequencing with the common pBabe 5′ and 3′ primers. The resulting myomaker cDNA was identified through sequencing as NM_001080483.2.
Cell Culture.
IMR-90 fibroblast cells (American Type Culture Collection) were cultured in MEM-α, supplemented with 15% FBS and 2 mM L-glutamine, 100 U mL−1 penicillin, and 100 μg mL−1 streptomycin. Fibroblasts were kept at 37 °C in 5% CO2 and passaged by splitting 1:3 once they reached confluence, approximately every 3–4 d. Primary human skeletal-derived myoblasts were acquired from Cook Myosite and passaged or differentiated with the provided media (Cook Myosite).
Retroviral Transduction of Fibroblasts.
GP-293 packaging cells (Clontech) cultured in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 100 U mL−1 penicillin, and 100 μg mL−1 streptomycin were transfected with polyJET reagent (SignaGen Laboratories). In brief, 1 mL of fresh medium was added to cells in a single well of a six-well plate. Then 3 μL of polyJET was diluted in 50 μL of serum-free DMEM, and 0.5 μg of expression vector and 0.5 μg of VSV-G vector were diluted in 50 μL of serum-free DMEM. PolyJET/DNA solution was added dropwise to GP-293 cells and the cells were incubated for approximately 16 h, at which point the medium was changed. After another 24 h of incubation, the supernatant was collected, filtered through a 0.45-μm filter, diluted 1:6 in MEM-α with 4 μg mL−1 polybrene (Sigma-Aldrich; h9268), and applied to cells. A second infection was performed after an additional 24 h in the same manner. Selection was carried out on infected cells in 2 μg mL−1 puromycin (Invitrogen; A11138-02) or 200 μg mL−1 hygromycin (Invitrogen; 0687-010) in accordance with specific experimental requirements.
IMR-90 hTert passage-elongated fibroblasts were generated by infecting IMR-90 cells as described above with a pQCXIH-hTert expression vector and selected with 200 μg mL−1 hygromycin (Invitrogen; 0687-010) for 72 h. The cells were then expanded in culture and frozen for future use.
Generation of Myotubes by Transduction.
At 72 h before the initial infection, IMR-90 hTert cells were seeded at ∼3.5 E4 cells per cm−2 of growth area. The cells were then infected with virus derived from either pQCXIP-GFP or pQCXIP-MyoD expression plasmids as described previously. At 24 h after the secondary infection, the cells were selected in 2 μg mL−1 puromycin (Invitrogen; A11138-02) for ∼16 h. The cells that survived drug selection were then trypsinized and reseeded at a density of 1.5–2.5 E5 cell per cm−2 in MEM-α, 15% FCS, and 2 μg mL−1 puromycin (Invitrogen; A11138-02) onto glass coverslips pretreated with a 0.1% (wt/vol) solution of porcine gelatin (Sigma-Aldrich; G1936) in double-distilled H20 at 37 °C for at least 1 h. The myotubes were then allowed to develop for 72 h before being fixed in 4% formaldehyde for immunofluorescence-based assays or in TRIzol (Invitrogen) for RNA analysis.
Chimeric myotubes were generated through coculture experiments. One plate of IMR-90 hTert cells was infected with pQXCIP-MyoD as described above. A second plate was infected with a 1:6 dilution of virus made from the pBabe-Puro-myomaker expression clone. The myomaker-infected population of cells was treated with 50 μM BrdU (Life Technologies; 00-0103) for 30 min before each infection and before the initial drug selection. At 16 h after drug selection, the cells infected with either MyoD or myomaker were trypsinized, mixed at a 1:1 ratio, and reseeded at a final density of 3 E5 cells/cm−2 per cell type in MEM-α, 15% FCS, and 2 μg mL−1 puromycin (Invitrogen; A11138-02) onto glass coverslips coated with 0.1% porcine gelatin. Cells were then allowed to develop for 72 h and then fixed in 4% formaldehyde.
FISH.
Paraformaldehyde-fixed cells (4%) were permeabilized for 30 min in 0.5% Triton X-100, and 3D-FISH was carried out as described previously (59). Glycerol treatment and flash-freezing in liquid nitrogen were omitted to preserve the structural integrity of the large syncytium of the myotube. In addition, the denaturation step before overnight hybridization was held at 75 °C for but elongated to a total time of 6 min. All probes were labeled by nick translation with DIG-11-dUTP (Roche; 11558706910), BIO-16-dUTP (Roche; 11093070910), or DNP-11-dUTP (PerkinElmer; NEL551001EA) with Nick Translation Mix (Roche; 11745808910) according to the manufacturer’s protocol. Chromosome painting was performed with the same protocol using the XCP XCyting Chromosome Paints (Metasystems).
Immuno-FISH was performed as described previously (59), with a biotinylated secondary antibody. BrdU detection was performed during detection of the nick-translated probes. Primary antibodies used for detection were 1:100 sheep α-BrdU (Abcam; ab1893), 1:100 mouse α-MHC (DSHB; A4.1025), 1:250 rabbit α-myogenin (Santa Cruz Biotechnology; M-225), 1:500 rabbit α-dinitrophenol (Life Technologies; A-6430). The following secondary antibodies were used for detection: 1:250 sheep α-DIG fluorescein (Roche; 11207741910), 1:250 mouse α-Bio Alexa Fluor 647 (Jackson ImmunoResearch; 200-602-211), 1:500 goat α-rabbit Alexa Fluor 594 (Molecular Probes; A-11012), 1:250 donkey α-sheep FITC (Life Technologies; A10642), 1:250 donkey α-sheep FITC (Life Technologies; A10642), and 1:250 donkey α-rabbit Alexa Fluor 594 (Life Technologies; A-21207).
BACs.
All of the BACs used in this study where acquired from the BACPAC Resources Center at the Children’s Hospital Oakland Research Institute and are listed in SI Appendix, Table 2. All BACs used were obtained from the RP11 library, which is constructed on the pBACe3.6 vector backbone (GenBank accession no. U80929), allowing conformation of BAC identity by sequencing with either SP6 or T7 sequencing primers. Most BACs were also validated by FISH, ensuring that BACs on the same chromosomes appeared close in interphase nuclei and on the same molecule in metaphase spreads.
RNA FISH.
To detect nascent transcripts, custom Stellaris FISH probes were designed against the introns of Myogenin (MYOG; NM_002479.5) using the Stellaris FISH probe designer (Biosearch Technologies). The exact probe sequences are listed in SI Appendix, Table S4. Cells were hybridized with the MYOG intronic Stellaris FISH probe set labeled with Quasar 570 (Biosearch Technologies) following the manufacturer’s instructions for adherent cells. Slides were imaged using a Leica DMI6000 B microscope equipped with a Lumen 200 illumination system (Prior Scientific), a Plan Apo 63× 1.4 NA oil immersion objective lens, and a CoolSNAP HQ2 CCD camera (Photometrics). Leica A4 and TX2 filter cubes were used to image DAPI and Quasar 570, respectively. Images were acquired with 0.2-µm z-steps in each channel using a motorized stage controlled by the Leica Application Suite Advanced Fluorescence software.
Image Acquisition and Analysis.
Unless noted otherwise, all images were acquired on a Nikon A1R+ line scanning confocal microscope equipped with photomultiplier tubes. Imaging was done with a Plan Apo VC 60× 1.4 NA oil objective as a multidimensional z-stack with a consistent step size of 0.25 um using the Nikon Elements software. The acquired 3D image stacks were then fed through an image processing pipeline that uses a series of scripts written in-house within the MATLAB environment. In brief, this pipeline performs the functions of image segmentation, distance normalization, and measurements in 3D space while accounting for gross changes in nuclear shape (SI Appendix, Fig. S10). Detailed explanations of the components of the image processing pipeline are provided in SI Appendix, Materials and Methods.
Statistics.
For the enrichment of LE-TADs and TGAs within the myogenic gene set, the number of observed LE-TADs or TGAs was compared with a distribution of LE-TADs or TGAs generated by finding the number of LE-TADs or TGAs in each of the 1,000 simulated gene sets. The P value was then reported as the percentage of observed events in the distribution equal to or greater than the number of LE-TADs or TGAs in the myogenic gene set. For chromosome-by-chromosome analysis, the same method was used, but LE-TADs and TGAs were first separated by chromosome before the comparison was made. For statistical comparisons, two-sample t tests with unequal variance, nonparametric Wilcoxon rank-sum tests, and bootstrap-based methods were used to determine significance. Unless specifically noted, all of these methods yielded the same levels of significance.
When determining the significance of the correlation observed between two variables multiple tests were used. The probability of observing an uncorrelated or negatively correlated R value was calculated by performing a Pearson correlation of 10,000 random subsamplings with replacement of the observed values and the reporting the percent of simulation that resulted in an R value ≤ 0. The slope and 95% CIs of the slope of the linear best-fitting line were also calculated in MATLAB using a linear regression model and estimating the 95% CIs of the coefficients. If the range of the 95% CI of the slope did not contain 0, then we concluded that the best-fitting line had a nonzero slope, suggesting a nonrandom relationship between the two variables being modeled.
Supplementary Material
Acknowledgments
We thank Zizhen Yao and Stephen Tapscott for the whole genome expression data, and Takeshi Shimi and Stephen A. Adam for reagents and technical assistance. Imaging work was performed at Northwestern University’s Center for Advanced Microscopy, generously supported by National Cancer Institute Grant CCSG P30 CA060553, awarded to the Robert H. Lurie Comprehensive Cancer Center. This work was funded by National Institutes of Health New Innovator Award DP2 OD008717-01 (to S.T.K.). D.S.N. is supported by National Institute of General Medical Sciences Cellular and Molecular Basis of Disease Training Grant T32 GM08061. A.G.G.-G. is supported by National Cancer Institute Oncogenesis and Developmental Biology Training Grant T32 CA080621.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521826113/-/DCSupplemental.
References
- 1.Kmita M, Duboule D. Organizing axes in time and space: 25 years of colinear tinkering. Science. 2003;301(5631):331–333. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- 2.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18(10):1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mischke D, Korge BP, Marenholz I, Volz A, Ziegler A. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (“epidermal differentiation complex”) on human chromosome 1q21. J Invest Dermatol. 1996;106(5):989–992. doi: 10.1111/1523-1747.ep12338501. [DOI] [PubMed] [Google Scholar]
- 4.Kosak ST, Groudine M. Gene order and dynamic domains. Science. 2004;306(5696):644–647. doi: 10.1126/science.1103864. [DOI] [PubMed] [Google Scholar]
- 5.Roy PJ, Stuart JM, Lund J, Kim SK. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418(6901):975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- 6.Kosak ST, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296(5565):158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 7.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452(7184):243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 8.Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394(6693):592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- 9.Therizols P, et al. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346(6214):1238–1242. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moen PT, Jr, et al. Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol Biol Cell. 2004;15(1):197–206. doi: 10.1091/mbc.E03-06-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao J, Fetter RD, Hu P, Betzig E, Tjian R. Subnuclear segregation of genes and core promoter factors in myogenesis. Genes Dev. 2011;25(6):569–580. doi: 10.1101/gad.2021411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulbricht T, et al. PML promotes MHC class II gene expression by stabilizing the class II transactivator. J Cell Biol. 2012;199(1):49–63. doi: 10.1083/jcb.201112015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khanna N, Hu Y, Belmont AS. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol. 2014;24(10):1138–1144. doi: 10.1016/j.cub.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42(1):53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosak ST, et al. Coordinate gene regulation during hematopoiesis is related to genomic organization. PLoS Biol. 2007;5(11):e309. doi: 10.1371/journal.pbio.0050309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2(4):292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 17.Stadler S, et al. The architecture of chicken chromosome territories changes during differentiation. BMC Cell Biol. 2004;5(1):44. doi: 10.1186/1471-2121-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485(7398):381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gómez-Díaz E, Corces VG. Architectural proteins: Regulators of 3D genome organization in cell fate. Trends Cell Biol. 2014;24(11):703–711. doi: 10.1016/j.tcb.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 22.McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci USA. 1997;94(3):814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sánchez A, Kondev J. Transcriptional control of noise in gene expression. Proc Natl Acad Sci USA. 2008;105(13):5081–5086. doi: 10.1073/pnas.0707904105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453(7194):544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teles J, et al. Transcriptional regulation of lineage commitment: A stochastic model of cell fate decisions. PLOS Comput Biol. 2013;9(8):e1003197. doi: 10.1371/journal.pcbi.1003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raser JM, O’Shea EK. Noise in gene expression: Origins, consequences, and control. Science. 2005;309(5743):2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neems D, Kosak ST. Turning down the volume on transcriptional noise. Nat Cell Biol. 2010;12(10):929–931. doi: 10.1038/ncb1010-929. [DOI] [PubMed] [Google Scholar]
- 28.Kosak ST, Groudine M. Form follows function: The genomic organization of cellular differentiation. Genes Dev. 2004;18(12):1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- 29.Pombo A, Dillon N. Three-dimensional genome architecture: Players and mechanisms. Nat Rev Mol Cell Biol. 2015;16(4):245–257. doi: 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- 30.Mehta IS, Kulashreshtha M, Chakraborty S, Kolthur-Seetharam U, Rao BJ. Chromosome territories reposition during DNA damage-repair response. Genome Biol. 2013;14(12):R135. doi: 10.1186/gb-2013-14-12-r135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mehta IS, Amira M, Harvey AJ, Bridger JM. Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol. 2010;11(1):R5. doi: 10.1186/gb-2010-11-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol. 2002;12(6):439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- 33.Marshall WF, et al. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol. 1997;7(12):930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- 34.Naumova N, et al. Organization of the mitotic chromosome. Science. 2013;342(6161):948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerlich D, et al. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112(6):751–764. doi: 10.1016/s0092-8674(03)00189-2. [DOI] [PubMed] [Google Scholar]
- 36.Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol. 2003;160(5):685–697. doi: 10.1083/jcb.200211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parada LA, Roix JJ, Misteli T. An uncertainty principle in chromosome positioning. Trends Cell Biol. 2003;13(8):393–396. doi: 10.1016/s0962-8924(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 38.Wood AM, Garza-Gongora AG, Kosak ST. A Crowdsourced nucleus: Understanding nuclear organization in terms of dynamically networked protein function. Biochim Biophys Acta. 2014;1839(3):178–190. doi: 10.1016/j.bbagrm.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajapakse I, et al. The emergence of lineage-specific chromosomal topologies from coordinate gene regulation. Proc Natl Acad Sci USA. 2009;106(16):6679–6684. doi: 10.1073/pnas.0900986106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown G, Hughes PJ, Michell RH. Cell differentiation and proliferation: Simultaneous but independent? Exp Cell Res. 2003;291(2):282–288. doi: 10.1016/s0014-4827(03)00393-8. [DOI] [PubMed] [Google Scholar]
- 41.Zhu L, Skoultchi AI. Coordinating cell proliferation and differentiation. Curr Opin Genet Dev. 2001;11(1):91–97. doi: 10.1016/s0959-437x(00)00162-3. [DOI] [PubMed] [Google Scholar]
- 42.Hardwick LJ, Ali FR, Azzarelli R, Philpott A. Cell cycle regulation of proliferation versus differentiation in the central nervous system. Cell Tissue Res. 2015;359(1):187–200. doi: 10.1007/s00441-014-1895-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrés V, Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J Cell Biol. 1996;132(4):657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacQuarrie KL, et al. Comparison of genome-wide binding of MyoD in normal human myogenic cells and rhabdomyosarcomas identifies regional and local suppression of promyogenic transcription factors. Mol Cell Biol. 2013;33(4):773–784. doi: 10.1128/MCB.00916-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volpi EV, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci. 2000;113(Pt 9):1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- 46.Choi J, et al. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc Natl Acad Sci USA. 1990;87(20):7988–7992. doi: 10.1073/pnas.87.20.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao Z, et al. Comparison of endogenous and overexpressed MyoD shows enhanced binding of physiologically bound sites. Skelet Muscle. 2013;3(1):8. doi: 10.1186/2044-5040-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strickfaden H, Zunhammer A, van Koningsbruggen S, Köhler D, Cremer T. 4D chromatin dynamics in cycling cells: Theodor Boveri’s hypotheses revisited. Nucleus. 2010;1(3):284–297. doi: 10.4161/nucl.1.3.11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Millay DP, et al. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature. 2013;499(7458):301–305. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Endo T, Nadal-Ginard B. Reversal of myogenic terminal differentiation by SV40 large T antigen results in mitosis and apoptosis. J Cell Sci. 1998;111(Pt 8):1081–1093. doi: 10.1242/jcs.111.8.1081. [DOI] [PubMed] [Google Scholar]
- 51.Bridger JM, Boyle S, Kill IR, Bickmore WA. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr Biol. 2000;10(3):149–152. doi: 10.1016/s0960-9822(00)00312-2. [DOI] [PubMed] [Google Scholar]
- 52.Wiblin AE, Cui W, Clark AJ, Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J Cell Sci. 2005;118(Pt 17):3861–3868. doi: 10.1242/jcs.02500. [DOI] [PubMed] [Google Scholar]
- 53.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: Interpreting chromatin interaction data. Nat Rev Genet. 2013;14(6):390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49(5):773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148(5):908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCullagh E, Seshan A, El-Samad H, Madhani HD. Coordinate control of gene expression noise and interchromosomal interactions in a MAP kinase pathway. Nat Cell Biol. 2010;12(10):954–962. doi: 10.1038/ncb2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bahar R, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441(7096):1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 59.Cremer M, et al. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol Biol. 2008;463:205–239. doi: 10.1007/978-1-59745-406-3_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.