Significance
Chemical and biomedical sciences depend heavily on interaction assays, particularly those providing structural insights. Here, we show interferometric, free-solution, label-free studies report conformation and hydration changes, and present a new way for interpreting these methods. Intrinsic property changes are the mechanism allowing for unprecedented sensitivities (picomolar to femtomolar) in complex milieu, even when individual binding partners are undetectable. We establish that the existing theory for label-free assay methods such as surface plasmon resonance (SPR) is not applicable and propose a model for the free-solution response function (FreeSRF), validated and highly predictive when combined with quality structural data and reliable calculations of solvent-addressable surface area. The model allows for interpretation of solution-phase, label-free interactions and could facilitate obtaining structural information from a simple mix-and-read assay.
Keywords: backscattering interferometry, assay methodology, molecular interactions, conformation change, hydration change
Abstract
Interaction/reaction assays have led to significant scientific discoveries in the biochemical, medical, and chemical disciplines. Several fundamental driving forces form the basis of intermolecular and intramolecular interactions in chemical and biochemical systems (London dispersion, hydrogen bonding, hydrophobic, and electrostatic), and in the past three decades the sophistication and power of techniques to interrogate these processes has developed at an unprecedented rate. In particular, label-free methods have flourished, such as NMR, mass spectrometry (MS), surface plasmon resonance (SPR), biolayer interferometry (BLI), and backscattering interferometry (BSI), which can facilitate assays without altering the participating components. The shortcoming of most refractive index (RI)-based label-free methods such as BLI and SPR is the requirement to tether one of the interaction entities to a sensor surface. This is not the case for BSI. Here, our hypothesis is that the signal origin for free-solution, label-free determinations can be attributed to conformation and hydration-induced changes in the solution RI. We propose a model for the free-solution response function (FreeSRF) and show that, when quality bound and unbound structural data are available, FreeSRF correlates well with the experiment (R2 > 0.99, Spearman rank correlation coefficients >0.9) and the model is predictive within ∼15% of the experimental binding signal. It is also demonstrated that a simple mass-weighted dη/dC response function is the incorrect equation to determine that the change in RI is produced by binding or folding event in free solution.
Contemporary assays enabling single-molecule detection (1, 2) have accelerated the sequencing of the human genome (3) and facilitated imaging with extraordinary resolution without labels (4). To most closely approximate the natural state, an interaction assay methodology would interrogate the processes (reaction, molecular interaction, protein folding event, etc.) without perturbation. Label-free chemical and biochemical investigations (5, 6) transduce the desired signal without an exogenous label (fluorescent, radioactive, or otherwise) representing an essential step toward this goal. Many label-free methods require one of the interacting species to be either tethered or immobilized to the sensor surface, introducing a potential perturbation to the natural state of the species (7, 8). However, back-scattering interferometry (BSI) is a free-solution label-free technique with the added benefit of sensitivity that rivals fluorescence (9). There are other techniques performed in free solution, such as MS (10, 11) and NMR (12, 13) and the widely used isothermal titration calorimetry (ITC) (14, 15). As with NMR, ITC has many advantages, but exhibits modest sensitivity and often requires large sample quantities. Another increasingly popular free-solution approach is microscale thermophoresis (MST). However, for MST to operate label-free, one of the binding partners must have a significant absorption/fluorescence cross-section (16, 17). BSI represents an attractive alternative to these methods because of its high sensitivity, small sample volume requirement, optical simplicity, and broad applicability (18–21). Whereas ITC and MST are well known, the fundamental mechanistic basis for the signal observed in BSI is less well understood.
Herein, we attempt to address the fundamental basis for the signal observed in label-free, free-solution interaction studies performed with an interferometer. We present a hypothesis for the mechanism of signal generation in free-solution assays (assumed to be label-free from this point forward) and pose a preliminary model for interaction studies. Our model is a work in progress and as such has limitations. Here, it is our intent to stimulate additional investigations and to address two questions: (i) How can interactions be measured label-free and in free solution, in the absence of absorbance, a significant mass change, or a thermal signature? (ii) What intrinsic property allows unprecedented sensitivities (picomolar to femtomolar) in complex milieu, when neither of the individual binding partners is detectable at those levels?
In addressing these questions, we show that free-solution methods properly performed by interferometry have a unique, enabling signal transduction mechanism and that the signal magnitude correlates with changes in quantifiable intrinsic properties. We also demonstrate that the free-solution response function (from hereon, FreeSRF) can be quantified and within defined parameters can be predictive.
Results presented establish that the relative measurements performed in free solution allow the solution refractive index (RI) to provide a reproducible, robust, and quantifiable readout of chemical reaction progression (interaction), principally due to conformation and hydration changes upon binding. We illustrate that the changes leading to a FreeSRF cannot be considered simply as mass-weighted dη/dC responses, even though the interferometer exhibits a dη/dC response for a single analyte (salt, sugar, protein, antibody, or DNA strand) (22). We describe how to properly use BSI and configure the free-solution assay to ensure quantitation of binding affinities for a wide range of species [e.g., ion binding a protein (9), a sugar binding a lectin (8), hydrogen bonds forming in nonaqueous media (23), small molecules to membrane proteins embedded in cell-derived vesicles (18), merazoite proteins to intact human erythrocytes (21), and protein folding (24)]. Finally, we show how to estimate the magnitude of free-solution signal using protein database-derived information.
Background
Nearly two decades ago, we published observations indicating that our unique interferometer could be used to measure protein folding (24). The importance of these preliminary studies was not more fully realized until 2007, when our group showed that binding events, such as ion–protein, protein–protein, and small molecule–protein interactions, could be measured using a RI technique in free solution and without labels (9). Numerous examples have validated that free-solution measurements by interferometry can be used to quantity interactions of widely different affinities (micromolar to picomolar) and on interacting pairs with significant mass differences (>10,000-fold) (18–20). Although we, and others, have postulated the origin of the free-solution signal, no explicit explanation for the physical phenomenon has emerged. Here, we capitalize upon the pioneering observations by Sota and others including Pitner and Koch (25–28), using techniques typically thought to be insensitive to bulk RI changes (25) that couple energy into an immobilized sensor surface layer, which suggest that a theory based purely on dη/dC considerations does not adequately describe the response for optical methods performed in free solution. The background necessary to support this supposition is provided below and in SI Appendix.
A wide range of surface techniques have dη/dC signal dependence, where the performance is bounded by the relative change in mass–concentration at the surface (SI Appendix, Eqs. S1–S3). These observations have led to a reasonable, but mistaken assumption that signal transduction in free solution is the same as deflection, refraction, or wavelength shift, techniques in which performance is bounded by the relative change in volume at the surface (mass or concentration). The dη/dC formalism was established in 1988 in one of the first papers describing surface plasmon resonance (SPR) (29). Other descriptions on how to relate these changes to adsorbed films appeared thereafter (30–32). In one of these reports, researchers showed the SPR response was linear with surface concentration of protein (in nanograms per square millimeter) for adsorbed species, and introduced the refractive index increment (RII), which was defined as dη/dC in milliliters per gram (32). They illustrated that the RI of the surface layer was the sum of the concentration weighted RII (SI Appendix, Eq. S1). Using a Matthews report (33) indicating the fractional solvent content of a globular protein crystals ranges from 30% to 78%, they then estimate the probed surface thickness to range from 60 and 200 nm. However, the closely packed protein crystal representation omitted values for solvent content and specific volume.
Yee and coworkers (30) recast the Lorenz–Lorenz equation (SI Appendix, Eq. S2), further establishing the paradigm by showing (SI Appendix) that ηprotein = 1.57 RIU for the water-free (unreacted) protein was close to that of crystalline proteins of 1.60 RIU as confirmed by Schuck and coworkers (34). Importantly, these values are greater than those estimated for “adsorbed protein films” using ellipsometric approaches assuming a single optical thickness, because the film volume does indeed include a great deal of water (30, 35, 36). Yee and coworkers referred to that part of the film that “are made of protein material itself, not water (italics added).” They also noted, “we believe this approach, which neglects the intermixed solvent (italics added) in the adlayer, is more direct and general for quantitative analysis of adsorbate coverages for proteins and adsorbates in general” (30). A report by Marsh and coworker (37) also suggests that hydration/conformation are important predictors of binding-induced structural changes.
In 2000, Davis and Wilson (38) reported on an approach to determine the RII of small molecules for correction of SPR data. They too used the formalism of a concentration weighted RII (SI Appendix, Eq. S3) and predicted the maximum (BIACORE) SPR instrument response for binding of a single ligand (Eq. 1):
| [1] |
where (RUpred)max is the predicted maximum instrument response in resonance units for binding at a single site, RUM is the experimental amount of macromolecule immobilized on the chip in resonance units, MWL is the molecular weight of the ligand, MWM is the molecular weight of the immobilized macromolecule, and (dη/dC)L/(dη/dC)M is the RII of the macromolecule. The relative mass was also shown to be an important parameter in predicting the maximal signal flow injection gradient SPR systems (39).
The model described above and in SI Appendix used to define the response for RI (bio)sensing methods, does not take into account the possibility that the signal may be impacted by conformation and hydration changes upon chemical or biochemical transformation (binding, folding, shedding, or gaining waters of hydration). To our knowledge, Sota et al. (25) were the first to question this supposition by reporting the detection of conformational changes in an immobilized protein using an SPR biosensor. They observed that the SPR signal of the tethered protein and the molar ellipticity of dihydrofolate reductase in solution responded similarly to pH changes. Combined with tethered protein sensor surface measurements in the pH range of 0.12–7.80, they postulated that their observations were “consistent with the interpretation that changes in the SPR signal reflect conformational changes occurring during acid denaturation” (25).
Numerous others have questioned the paradigm of RI sensing exhibiting simply a mass-weighted response. In 2000, Boussaad, Pean, and Tao (40) used multiwavelength SPR to show that altering the reduction potential of a solution caused a change in the conformation of cytochrome C and a corresponding change in signal. Salamon et al. (41) reported the use of an SPR-related technique, coupled plasmon-waveguide resonance spectroscopy, to study ligand-induced conformational changes in a G-protein–coupled receptor embedded in a lipid bilayer. Gestwicki et al. (27) exploited the observation that the SPR response is not strictly dictated by the RII to enhance small-molecule detection. In this work and a subsequent patent (42), they demonstrated that ligand-induced conformational changes can be used to report small-molecule binding to immobilized maltose-binding protein and tissue transglutaminase by SPR, without the need for a high-MW competitor. For ligands binding to a receptor, they showed (i) there is a net negative ΔRI with a decrease in hydrodynamic radius; and (ii) when the interaction increases hydrodynamic radius, a net positive ΔRI results. Using response reversibility and similarities between reported and SPR-determined equilibrium dissociation constants, they confirmed that these signals could not be attributed to the addition of analyte molecular mass to the surface as would be predicted by SI Appendix, Eqs. S1–S3, and Eq. 1. They showed, with a high level of confidence, the observations were a consequence of specific receptor–ligand interactions.
The details of the experiments by Gestwicki et al. are important here. X-ray crystal structures show that maltose binding to maltose-binding protein (MBP) induces a conformational change. The hinge–twist structure change between the two domains of MBP causes a net decrease in hydrodynamic radius (27). As predicted for a negative net change in hydrodynamic radius, the event produced a negative ΔRI. Also consistent with their hypothesis was the observation that a positive ΔRI resulted when calcium binds to tissue transglutaminase (tTG), which is allosterically regulated by Ca2+. The positive ΔRI of roughly +1,000 resonance units (RU) reported for Ca2+ binding to tTG, from a change in hydrodynamic radius resulting from a ∼15° rotation, was much greater than the expected +28 RU from mass alone. Thus, the conformationally active form of tTG increased the SPR signal intensity by 36-fold upon calcium binding. Unlike calcium, maltose is of sufficient molecular mass (360 Da) to be detected directly by SPR. However, when maltose binds to MBP, a net negative change in RI is recorded in the SPR sensorgram, apparently because the binding event-induced conformational change overwhelms the positive response due to accumulation of mass from maltose (MBP–maltose interaction gave a net 5- to 30-RU signal).
After a period of silence, several SPR papers appeared on the topic of conformation-dependent sensing despite many considering the reports to be simply anomalous behavior (43–47), attributed to the following: (i) buffer mismatch, (ii) volume exclusion due to ligand density differences (43–45), (iii) nonspecific matrix interaction (46), and (iv) nonspecific reference interactions (47). Regardless, a recent paper reported that the RI sensing figures of merit were dependent on shape and the size of the Au nanoparticles (48) with sensitivities generally increasing as the nanoparticles became elongated and their apexes become sharper. When nanobipyramids’ aspect ratio was increased from 1.5 to 4.7, signal increased from 1.7 to 4.5. Then in 2010, SPR was used to quantify E. coli DNA ligase using a hairpin DNA to probe self-structure change during the ligation process (49). Clearly, no change in MW was necessary to provide signal for this assay.
Because of the significant difference in the mass of the binding pair, ion–protein interactions are an important class of interactions to consider. Christopeit, Gossas, and Danielson (50) showed that SPR detects these interactions, provided that a large conformational change is induced upon binding. Here, a Ca2+-induced conformational change of C-reactive protein (CRP) made the protein more compact, decreasing the hydrodynamic radius, leading to an “unexpectedly” negative ΔRI change.
Recently, the Koch group contributed two studies (28, 51) further bolstering the argument that free-solution signals correlate to binding-induced conformational changes. In 2010, they reported that SPR sensorgram amplitudes for saturated Ca2+-binding of protein-coated surfaces greatly exceeded the theoretical values (51), concluding the SPR signal was a consequence of the concerted Ca2+-binding–induced protein conformational change in the vicinity of the protein–dielectric, rather than being due to a mere mass effect. Using NMR structures of Ca2+-free (apo) and Ca2+-bound myristoylated recoverin (mRec), reported as average structures, they illustrated Ca2+-free recoverin has a tense (T), compact conformation in which the myristoyl group is sequestered in a hydrophobic pocket (SI Appendix, Fig. S1A). However, in the Ca2+-loaded form, it undergoes a transition to a released (R), more extended conformation, where the myristoyl is solvent exposed (SI Appendix, Fig. S1B). The Ca2+-induced transition is characterized by both an increase in the radius of gyration (Rgyr) and total solvent-accessible surface area (SASA). They postulated that the solvation shell of the R conformation differs significantly from that of the T conformation, which affects the surrounding water structure and the overall change in RI signal measured. Recently, this group also showed a correlation between SPR, dynamic light scattering (DLS), and size-exclusion chromatography (28), confirming that conformational changes under conditions of molecular crowding yield fingerprint profiles reflecting different hydrodynamic properties for each Ca2+-sensor protein under changing Ca2+ conditions. These properties were extremely sensitive to even small alterations of structure/conformation induced by point mutations. We have made similar observations in free solution for binding of folate to histone demethylase, LSD1 (52).
The work reported by the Koch group indicates that the site-specific homogeneous immobilization of the proteins enhances the intensity of the phenomenon, but the RI changes induced by concerted Ca2+-binding/conformational transitions are essentially isotropic. Using DLS, circular dichroism (CD), and ellipsometry as confirmatory and complementary methods, they concluded, “conformational changes can be detected even via the p-polarized resonance excited by the commercial SPR systems (Biacore, GE)…likely to be a direct result of the heterogeneous orientation…a different case (than) the anisotropic immobilization of membrane receptors in lipid bilayers that require both s- and p-polarizations to be fully characterized as in plasmon waveguide resonance spectroscopy [PWRS] (53)” (51). Thus, Koch predicts that conformational changes are less likely to be observed in the absence of both polarizations with highly organized systems.
Like PWRS, dual polarization interferometry (DPI) (54), employs both s- and p-polarizations of light. DPI is based on the observation that a laser-illuminated waveguide stack produces an interference fringe pattern that undergoes a phase change when an immobilized layer on the top stack changes. By introducing alternating polarization states [transverse electric (TE) and transverse magnetic (TM)], at right angles to each other into the stack, the difference in response allows two independent surface measurements. Using classical optical theory, these two outputs give a measure of thickness and density for the (protein) layer. Comparison of the TM and TE output has enabled shape, orientation, binding, and molecular conformation changes to be studied (55).
Overall, the collective literature described above is consistent with our hypothesis that systems with significant conformation and hydration changes do not conform to the theory that predicts RI changes are equal the sum of the mass-weighted RI values (SI Appendix, Eqs. S1–S3, and Eq. 1). Too much evidence exists, from a diverse set of chemical/biochemical processes, for these observations to be anomalous or spurious in origin, particularly because the signal was observed in the presence of a large background arising from a mass adsorbed at the surface. Hence, configuring an assay to reduce or eliminate competing RI signals should result in a measurement that reports conformation/hydration changes. In 2007, we tested this hypothesis, showing that, under the proper conditions, a sensitive RI sensor can transduce solution-phase binding events (9). Since this original report, BSI has been used widely (18–21, 23, 24, 56), and benchmarked extensively, with comparisons to ITC, SPR, and other established assay platforms.
Results and Discussion
A unique aspect of our free-solution methodology is that often the sensitivity of the assay far exceeds that for detecting the individual participating species. In other words, the ligand alone has no quantifiable dη/dC response under the conditions of the assay, but when comparing the bound sample to the receptor/target (reference) there is robust and reproducible signal (SI Appendix, Figs. S2 and S3). This observation has raised the two important questions: (i) How it is physically possible to perform these label-free studies in free solution? (ii) What is the signal source? Here, we aim to definitively answer these questions, putting forth an explanation and a preliminary model for the free-solution signal and detailed transduction procedures by interferometry.
Conformation and Hydration Changes Are the Origin of Free-Solution Signals.
To quantify interactions in free solution, the experiment must be designed in a manner that places the chemical and optical focus on changes in conformation, hydrodynamic volume, hydration state, and to a lesser extent, the electronic state. Proper handling of index-matched sample and reference enable chemical focusing, while correct instrument alignment and operation maximize signal transduction by the interferometer. Our methodology should also apply to systems where there is no mass change, as in protein folding or where the difference in mass for the binding pair is large. We describe here how the relative mass of the binding partners plays a minor role in determining the FreeSRF.
Many years ago, we demonstrated protein folding on very small sample quantities with an early-generation capillary interferometer, showing that a readout for ubiquitin folding could be obtained (24). In retrospect, we now realize the importance of this observation, which illustrated that, in the absence of any mass change, we could use an RI sensor to follow conformation changes in free solution. Recent efforts to construct an assay for respiratory syncytial virus provides additional evidence for our hypothesis that free-solution assays are reporting changes in conformation and hydration (56). We found that the BSI sensitivity was not only related to the number of unpaired nucleotides, but also to the structure of the targeted regions of the RNA sequence. For example, locked nucleic acid probes showed a fourfold sensitivity improvement compared with DNA probes of the same sequence. To explain this non-dη/dC behavior, we investigated how the free-solution signal was impacted by changes in the duplex structure. Using titration and incubation of DNA:DNA duplexes with trifluroethanol, an established method for converting the duplex structure (57, 58), we induced the transition from the B-form to A-form (SI Appendix, Fig. S4). We then monitored these structural transformations with CD and ellipticity at 270 nm, showing that the BSI free-solution readout reports structural transformations in the DNA duplex. Other experiments performed by us and others (59) involving positional DNA mismatch binding experiments further validate our hypothesis, showing free-solution signal enhancement emanates from induced alterations to the helical geometry of the nucleic acid hybrid and not a dη/dC change.
The Interferometer.
The technology used to perform free-solution studies represents a unique interferometry configuration (9). The optical train depicted in Fig. 1 is quite simple for a highly sensitive, small-volume interferometer, consisting of a coherent source, an object (channel in a chip or capillary), and a transducer. Probing the object with an unfocused He–Ne beam at nearly 90° (±7° to allow fringes to be viewed), results in a high-contrast interference fringe pattern (Fig. 1) in the backscattered direction. Depending on configuration, tracking the position of the fringes enables RI changes to be quantified in the range from 10−4 to 10−9 (60, 61), within picoliter to nanoliter probe volumes. A long effective path length results from multiple reflections at the fluid–channel interface and leads to the unprecedented sensitivity in constrained volumes (62). We, and others, still use capillaries, yet the most common interferometer configuration is based on a microfluidic chip containing a nearly semicircular isotropically etched channel that is 100-µm deep and 210-µm wide. Based on empirical observations, we have found that fringe selection is best accomplished by filling the channel with the analysis solution (buffer, serum, etc.) and counting approximately five fringes from the centroid, then windowing or selecting five to seven fringes in this region that exhibit a nearly single spatial frequency (SI Appendix, Alignment and Fringe Selection). Although the fringes closest to the centroid appear to exhibit a greater shift (63), a binding signal has yet to elude us in the region described above. With proper alignment (SI Appendix), the fringe contrast ratio approaches 99%, and this metric, combined with response to a change in RI (detection limits with glycerol solutions), serves to consistently produce the desired outcome. Good thermal stabilization and environmental isolation is also necessary and allows the device to produce a detection limit of ΔRI < 5 × 10−7. Typically, the sample/chip is probed with both planes of polarization as a result of coupling a linearly polarized laser into a nonpolarization maintaining single-mode fiber coupler. Misalignment will lead to slanted fringes and/or fringes with poor contrast. All of the configurations of BSI we have investigated exhibit a classical dη/dC and dη/dT response expected of an RI detector.
Fig. 1.
BSI block diagram showing orientation of the beam relative to the chip, a photograph of the fringe pattern, the line profile of the region of interest (ROI) for a good fringe pattern, and the FFT spectrum for that ROI.
It is likely that the multipass optical configuration (SI Appendix) of BSI contributes to success in performing free-solution and label-free measurements as does the ultrasmall (constrained) volume of BSI, but it is unlikely these alone are enabling characteristics. Although additional research is needed, we can state that there are a combination of factors that enable our free-solution measurements. These include the following: (i) the use of the proper assay methodology involving informed choice of reference and control and RI matching, (ii) careful sample handling, (iii) prudent instrument design with respect to temperature and pressure control, and (iv) informed fringe selection as described below.
If the conformation/hydration hypothesis described here has a physical basis, free-solution assays should be detectable by a device with comparable ΔRI sensitivity to those used in the SPR reports noted above. Detection limits vary for SPR, but consistently reach ΔRI = 10−6. In our hands, the BSI detection limit is ΔRI = 10−6 or 10-fold below this level (19, 20, 62). Therefore, using proper methodology the signal to noise ratio (S/N) of our interferometer should enable molecular interactions to be measured. As shown in SI Appendix, Table S1 and Fig. S2, (i) the actual ΔRI measured by BSI for a binding event is well within the instrument detection limit, and (ii) as recently suggested (64), the predicted ΔRI using dη/dC considerations (SI Appendix, Eqs. S13–S17) would be undetectable.
The BSI signal is not calorimetric, as such reactions and binding events can add or remove energy from a solution, changing the solution temperature. This property has long been used to study interactions by calorimetry. Heat can also perturb the optical properties of a solution, in great part because of the relatively high dη/dT response exhibited by most fluids (1 × 10−4 °C for water). To rule out dη/dT perturbations as free-solution signals, we have shown that the quantities of sample interrogated in the interferometer volume cannot generate a temperature change large enough to produce a detectable ΔRI signal (9) (SI Appendix). Also, the absence of signal decay by conduction to a heat sink in stop-flow kinetic binding studies further confirms that the heat of reaction is not the source of the free-solution signal (9). Virtually all assays performed since 2008 have been run as end-point determinations, with samples prepared, mixed, allowed to equilibrate (sometimes for hours), and then read by the interferometer. The end-point scenario excludes reaction-based calorimetric contributions to the signal.
The Free-Solution RI Signal for Interactions/Reactions.
Consider the reaction between the two species A and B. As a chemist, it is tempting to write the equation for this reaction as follows: A + B → A–B, but this disregards the complexity of the interaction. When A and B react they undergo electronic transitions, lose or gain hydration, and experience significant changes in the atomic geometry. So the product is just that, an entirely new species allowing the reaction to written as: A + B → C. If this is the case, then the product formed from the interaction of A and B must have a unique and different dielectric constant or molecular dipole moment. The new species therefore responds differently to the probing electromagnetic radiation, in a manner analogous to the pH-change induced “structural” transformations in a dye molecule that lead to a significant change in absorption (color). For example, even the subtle change produced by ionizing phenol to phenolate results in quantifiably different absorbance spectra.
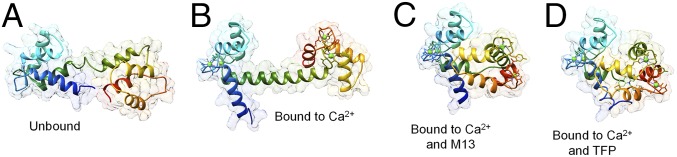
To aid in visualizing the free-solution transduction phenomena, we use the structural diversity found in calmodulin (CaM) (Fig. 2). Binding of Ca2+ to CaM (Fig. 2A) leads to a significant conformation and hydration change (65), resulting in a new complex, Ca2+–CaM (Fig. 2B), which has a considerable and quantifiably different electromagnetic cross-section (dielectric constant). Then, if the Ca2+–CaM complex reacts with the M13 protein kinase (Fig. 2C), the subsequent complex is unique and has a quantifiably different RI due to induced structural and hydration changes (66). Binding the small-molecule inhibitor, trifluoperazine (TFP), induces changes in atomic arrangement and hydration that leads to yet another unique chemical entity (Fig. 2D) (67). These graphical representations, generated from X-ray structure found in the Protein Data Bank [Research Collaboratory for Structural Bioinformatics (RCSB)/Protein Data Bank (PDB)], for bound and unbound species pictorially illustrate that the potential magnitude of free-solution signal can be large (approximately >10−4 RIU) under the proper conditions. Calculations of ΔRI (SI Appendix, Table S1 and Figs. S2 and S3) further illustrate this point.
Fig. 2.
Comparison of ribbon drawings for calmodulin unbound and bound with various ligands: (A) unbound calmodulin (PDB ID code 1CFD), (B) calmodulin bound to calcium (PDB ID code 1OSA), (C) calmodulin bound to M13 (PDB ID code 1CDL), and (D) calmodulin bound to TFP (PDB ID code 1CTR).
Although FreeSRF is not proportional to the sum of the mass-weighted change in RI for the reactants (SI Appendix, Eqs. S13–S17, and Figs. S2 and S3), this property does not preclude nonreacting or noninteracting analytes from exhibiting an RI response. Only the proper preparation of the sample and reference, typically from the same matrix, enables the extraction of the free-solution signal by canceling out (often) very large bulk RI background signals. In other words, the determination is not made by comparing samples with huge RI differences, but nearly identical ΔRI values. For example, we do not compare η1 = 1.33131 to η2 = 1.39131, but samples with RI values of 1.391312 and 1.391318 (ΔRI = 6 × 10−6). The use of relative measurements ensure that the interaction is the predominant signal. Furthermore, to minimize the influence of nonspecific binding at the surface, we establish a baseline with the receptor present in the buffer or matrix under investigation and then to the best of our ability using rinse solutions, reestablish this level before introduction of every new concentration for the assay.
The free-solution response function and an expression predicting performance.
Our first attempt to formulate a model for label-free, free-solution assays was heuristic and based on the assumption that binding-induced change in hydrodynamic radius dominated the signal for CaM interactions. Preliminary calculations used the RCSB/PDB structural information to estimate the radius of gyration (Rgyr) and SASA of the bound and unbound species (ΔRgyr and ΔSASA). A simple multivariable linear equation was obtained that relates the interferometry signal in phase, to change in Rgyr and SASA for the CaM system (ΔBSI = 1.0 + 2.6 × 10−4 ΔSASA + 0.054 ΔRgyr). SI Appendix, Fig. S5A, shows the correlation between the predicted and experimental values for free-solution interaction studies of CaM binding Ca2+, Ca2+-CaM–TFP, Ca2+-CaM–calcineurin, Ca2+-CaM–M13 peptide. The surprisingly good correlation (R2 = 0.88) between the actual and predicted signal magnitude for these binding events encouraged us to further our investigation.
Next, we expanded our formalism and applied it to a training set of binding pairs. Our expression mirrors Beer’s law in its simplest form, which equates the absorbance of a species to the experimental parameters of the determination (path length and concentration) and the intrinsic property of the species (molar absorptivity). Here, we propose defining the response for free-solution sensing to be expressed as follows:
| [2] |
where ρ is the FreeSRF measured in radians, χ is the molar refractometry in RI units per moles per liter, β is the instrument response function in radians per RI unit, and C is the concentration in moles per liter. This simple equation demonstrates that the fringe shift (in radians) quantified by an interferometer when measuring a folding, binding, or hybridization event in free solution (no labels) is directly proportional to (i) the magnitude of structural change (predominantly conformation and hydration) of the sample; (ii) the dθ/dn sensitivity of the interferometer (which incorporates the optical path length); and (iii) the concentration of the analyte. Below, we show that ρ is not a function of the RII or the relative mass of the interacting species and that it can be estimated for a binding pair with reasonable confidence.
Free-solution assays are predicated on the assumption that the solution is interrogated and not the surface (see below). The equation for FreeSRF states that the signal magnitude, ρ, is proportional to the number and type of transformations (see, for example, ref. 56), not just the number of bonds formed or broken. Measurement of ρ is obtained as a change in RI reported by a fringe shift or estimated from χ, the species concentration and instrument performance. As such, the most significant contributors to error in ρ are instrument drift, run-to-run reproducibility, and the uncertainty in χ.
In defining χ as the molar refractometry, we recognize that the structural changes observed are a consequence of processes (folding, interactions, chemical reactions, biochemical transformations, etc.) that lead to changes in the analyte intrinsic properties, such as the dielectric constant, the molecular dipole moment, or other third-order parameters. This premise is supported by evidence from complementary techniques including CD, ellipsometry (SI Appendix, Fig. S4), ITC, DLS (28), and NMR. Accuracy in χ is dominated by the quality of the initial training set data and the correctness of the structure prediction method and data derived from it. Several combined resources can provide quality structural data: (i) the PDB; (ii) PYMOL/MOLMOL (molecular analysis and display programs) and (iii) M-FOLD for structure prediction; and (iv) Chimera for structural analysis.
Note that β appears in the equation to account for path length variations, interferometer sensitivity (S/N) differences from device-to-device, laboratory-to-laboratory, or even operator-to-operator. Currently dθ/dη is expressed as milliradians per RI unit, but other sensible units that accurately express the instrument figures of merit can be used for β. In consideration of β, it should be recognized that signal extraction from an interferometric fringe shift is enabled by proper optical alignment, as well as careful selection and handling of references and controls. As a cautionary note, it is our observation that BSI fringes do not exhibit uniform behavior with respect to free-solution sensitivity (SI Appendix). What we have found is that optimized optical alignment for fringes 6–13 (counted from the centroid), yields a single spatial frequency when using fast Fourier transform (FFT) (60) (Fig. 1) that has always reported the free-solution signal.
The magnitude of FreeSRF scales with concentration; therefore, the addition of more protein always increases ρ, but it must be recognized that C is the product concentration, the quantity of the new shape or complex. So circumstances can be imagined where increasing the amount of receptor does not produce a directly proportional change in ρ. For KD determinations, this can be dealt with by avoiding a scenario where a high product concentration is reached in the assay. At this juncture for KD determinations, we perform FreeSRF most often with target concentrations near the assumed affinity or at KD/10. Additionally, we are mindful that error in C impacts FreeSRF, contributing uncertainty to the training set used to define χ and then used to predict ρ for a new system.
As with Beer’s law, which exhibits nonlinearity for three major reasons (68), we do expect refined versions of our theory to take on higher order terms that could effect ρ similarly. Although nonlinearities may be identified, our preliminary observations conform well to the simple expression proposed. We do acknowledge that our model can be improved and support that it will benefit from further investigation.
Testing the validity of FreeSRF.
As with other models (37, 69–72), it was necessary to use a learning set to establish the appropriate relationships and weighting parameters for FreeSRF. In our case, we determined χ from ρexp for a training set of well-characterized binding systems (Table 1). Multiple users performed the assays on several different interferometers (of similar configuration) to ensure confidence in the result and minimize operator or device biases.
Table 1.
Table comparing χmodel to χexp
| Large model | Small model | ||||||||
| Receptor | Ligand | Experimental χ (RIU/M) | Model χ (RIU/M) | Percent error, % | Receptor | Ligand | Experimental χ (RIU/M) | Model χ (RIU/M) | Percent error, % |
| IL-2 antibody | Interleukin-2 | 827,964 | 823,965 | 0.5 | Calmodulin | TFP | 75.2 | 73.6 | 2.2 |
| β2AR | Alprenolol | 591,423 | 604,924 | 2.3 | Calmodulin | TFP | 75.2 | 78.7 | 4.6 |
| β2AR | Isoproterenol | 290,953 | 278,649 | 4.2 | Calmodulin | TFP | 75.2 | 73.2 | 2.7 |
| Basigin | Rh5 | 215,777 | 212,174 | 1.7 | Carbonic anhydrase II | Sulpiride | 62.0 | 60.2 | 2.9 |
| Carbonic anhydrase II | Acetazolamide | −57,291 | −42,419 | 26.0 | Calmodulin | Calmodulin-Ca2+ | 56.1 | 56.5 | 0.6 |
| Carbonic anhydrase II | Acetazolamide | −57,291 | −37,288 | 34.9 | HIV PR | Pepstatin 1F1N | 13.7 | 13.7 | 0.5 |
| Calmodulin | Calcineurin | 46,087 | 37,389 | 18.9 | HIV PR | Pepstatin 1F1 | 10.2 | 7.2 | 29.9 |
| Calmodulin | Calcineurin | 46,087 | 51,594 | 11.9 | Con A | Mannose | 7.8 | 14.4 | 85.4 |
| Calmodulin | M13 | 16,458 | 15,393 | 6.5 | Con A | Mannose | 7.8 | 7.7 | 1.2 |
| Thrombin | Bock | 9,409 | 16,261 | 72.8 | Con A | Glucose | 2.6 | −1.3 | 149.2 |
| Thrombin | Tasset | 7,109 | 12,462 | 75.3 | Con A | Glucose | 2.6 | 4.6 | 78.1 |
| Carbonic anhydrase II | Benzene sulfonamide | −1,379 | −16,771 | 1,116.4 | Recoverin | Ca2+ | 78.0 | 78.1 | 0.1 |
| Carbonic anhydrase II | Benzene sulfonamide | −1,379 | −4,607 | 234.2 | |||||
| Carbonic anhydrase II | Sulfanilamide | 782 | −17,018 | 2,276.2 | |||||
| Carbonic anhydrase II | Dansylamide | −34,377 | −40,557 | 18.0 | |||||
For each of the training systems, the reference-corrected phase shift (ρexp in milliradians) was experimentally quantified at known concentrations of ligand. These values were used to determine the FreeSRF values for the experimental conditions: ρexpBmax, βexp, and CBmax, which in turn facilitates the calculation of values for χexp for the training set at the final concentration of product:
| [3] |
Running a dη/dC calibration experiment allows β to be determined in radians per RI unit for the specific instrument used in the binding assay. This experiment consists of measuring the phase shift as a function of glycerol concentration in millimolar concentration (or another suitable analyte). From this linear relationship, we obtain the slope, expressed in radians per millimolar concentration. For example, the response of BSI4 (instrument 4 of 9) for a glycerol calibration curve was found to be 0.011 radians per millimolar concentration, a typical value for our chip-based device. Then we express β in RI units per millimolar glycerol using a conversion factor from the CRC for dη/dC; in the case of glycerol, this parameter is 1.04863 × 10−5 RIU/mM (73). Thus, for BSI4:
| [4] |
To obtain the desired values for χBmax, we must know the concentration of the product, [Complex], detected upon physical transformation. Several approaches can be used to find this value. Here, we used an equilibrium solver written in Excel to determine the [Complex] at each concentration of ligand. The solver uses the mass balance equation, the receptor concentration, ligand concentration, and KD to calculate product concentration (SI Appendix). Then the maximal concentration of product is determined by plotting the product vs. ligand concentrations and fitting the curve using a single-site binding isotherm. Bmax is equal to the maximal concentration of product that is formed under the experimental conditions with high accuracy and has less bias than results produced at lower concentrations with a reduced S/N. To check the validity of using Bmax for the [Complex] and our solver, we used the quadratic equation to solve the equilibrium mass balance equation for the concentration of the complex at each point on the saturation isotherm produced from the end-point binding assay. Results shown in SI Appendix, Table S2, illustrate that using the solver for Bmax produces comparable values to the more computational intensive approach based on the quadratic expression.
With ρexp/CBmax and β in hand, we have the experimentally determined value χexp for each of the training set species and can turn to the task of determining χ. From our experience with CaM and observations by others (see above), the hypothesis that the free-solution signal has its origin in the physical transformations upon binding or folding emerges. Therefore, χ should be principally proportional to reaction/binding-induced conformation and hydration changes. Thus, allowing us to propose the expression for χ to be the following:
| [5] |
where ΔSASA is the difference in solvent-addressable surface area for bound complex and the unbound species in square angstroms, the aveSASA is the sum of SASA values divided by the number of values (PDB structures), ΔRGyr is difference for the radius of gyration for the unbound species and that of the complex (bound species) in angstroms, aveRGyr is sum of radius of gyration values divided by the number of values (PDB structures), and A, B, C, D, and E are fitting coefficients. The inclusion of the average quantities for Rgyr and SASA was motivated by a report by Marsh and Teichmann (37) where they demonstrate that that the absolute SASA value of a protein taken from a complex is an indicator for the amount of conformational change expected upon binding and is thus expected to affect χmodel. The absolute Rgyr value is required to normalize the effects of the absolute SASA value with protein size as described by Marsh and coworker in their equation 2 (37).
It is noteworthy that the quality and accuracy of the database structures used to determine the hydrodynamic properties directly impacts our predicted outcome. Here, we used the RSCB/PDB (SI Appendix, Table S3) to calculate of Rgyr and SASA with methods described below. In some cases, the PDB files were only available for corresponding ligand/receptor pairs in varying multiples of subunits (for example, unbound calmodulin was found as a monomer, but calmodulin bound to calcineurin was found as a homodimer). In cases where appropriate, these multimers were split into monomers using Chimera (74).
Numerous approaches exist to quantify Rgyr (75–77). Here, we used a Chimera script obtained from plato.cgl.ucsf.edu/trac/chimera/wiki/Scripts, enabling the calculation of Rgyr using the following expression:
| [6] |
where r is the position and m is the mass of each atom in the molecule. Hydrogens were removed for this calculation for consistency across species, because the Chimera program automatically adds these when displaying a new PDB file. Noninteracting species, such as ions, solvents, and accessory ligands were also removed before determining the Rgyr. The results for these calculations are compiled in SI Appendix, Table S4. The values obtained from Chimera correlated well with a self-written MatLab script using the same coordinates obtained from the PDB files.
Chimera was also used to aid in calculation of the SASA values. As recommended, solvent-excluded molecular surfaces were created with the help of the MSMS package: mgltools.scripps.edu/packages/MSMS/. Typically the SASA of only the main protein chain is used, with the surface area obtained using the “Surface” command in the MSMS program, which uses four different algorithms to determine surface area. Nonprotein molecules were discarded (including solvents, ions, and ligand) before calculations, except when the ligand was also a protein undergoing its own “significant” structural change. To calculate the SASA of the molecule, a “probe” (sphere of radius 1.4 Å) is “rolled” across the surface of the molecule. To begin, the first atom is selected, and the probe is placed at a distance of the radius of the atom, and then moved around the atom in the tangential direction until the probe comes into contact with the nearest neighbor atom. Then, the probe is moved along a path of equal radial distance between the two atoms until it encounters a third atom. This process is repeated to find the junctions between all atoms and their neighbors until the probe has been moved across the entire structure and the whole surface of the structure has been constructed. SI Appendix, Table S4, presents the values for SASA for each of the learning set structures.
We now have ΔRgyr, ΔSASA, and their average values (from PDB structure) allowing us determine the theoretical value for χmodel for each interaction. Using the experimentally determined value for χexp, obtained from ρexp/Cβ and theoretical χ values for our entire training set, we determined the coefficients A, B, C, D, and E for Eq. 5 (SI Appendix, Table S5) by performing a linear regression in Matlab. Using a wide range of χ values, this simple model produced a “good” fit with a high correlation coefficient (SI Appendix, Fig. S5B), but with a modest Spearman correlation coefficient of ρs = 0.853 (a nonparametric measure of statistical dependence between variables that indicates the relationship is not random and that the correlation between the variables can be described using a monotonic function). However, a relatively large residual error (20,249) (SI Appendix, Table S5) and percent difference between χexp and χmodel enhances the possibility of poor prediction accuracy, particularly for systems with a relatively small FreeSRF (ρ).
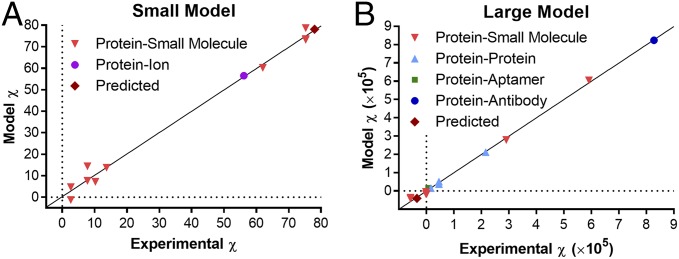
A better fit was found by separating the binding systems into two sets, “large” and “small” responders, based on the size of FreeSRF (ρexp). Because the interferometer reports the magnitude of structural changes (not the binding species MW), some proteins will populate both sets upon interaction with different ligands. For example, CaM can be found in both training sets. Using the signal-size segregation approach produces the plots shown in Fig. 3. These plots clearly illustrate that the relation between the χexp vs. χmodel predicted a priori produces two excellent results, with linear correlation coefficients of R2 = 0.991 and 0.998 and P values of 2.76 × 10−6 and 3.13 × 10−12 for the small and large FreeSRF models, respectively. Further evaluation of the relationship yields Spearman rank correlation coefficients of ρs = 0.936 and 0.979, respectively (SI Appendix, Table S5). It is important to note that our results do not necessarily split into two best-fit models, and we acknowledge that there are likely some scaling factors we have not identified which could impact the quality of the fit. The model can really be split in any number of ways (two subsets or three, four, five subsets) and provide similar results, yet a division into just large and small sets results in a relatively simple and easy-to-use model that produces a reasonably high quality result. It is also possible that, as the training set expands, a group of intermediate species will emerge. To the best of knowledge, there is not really a way to ascribe a physical property to E, which is the error term, disturbance term, or noise. This variable captures all other factors which influence the dependent variable yi other than the regressors xi and is dissimilar for the large/small sets because of the significant difference in error between the two models.
Fig. 3.
Plots showing correlation of χexp and χmodel when the learning sets are split into small (A) and large (B) χ values.
It is noteworthy that the training set used has a significant level of diversity, including ion–protein, protein–protein, small molecule–protein, protein–aptamer, membrane protein targets analyzed as cell-derived vesicles, an antibody–antigen pair, and unaltered human erythrocytes (21). Furthermore, the highly correlated results were obtained over a period spanning more than 3 years, by numerous BSI operators and on six different BSI instruments. Overall, the model provides values for the molar refractometry, χ, which correlate well with those derived from the binding experiment (Table 1), suggesting it can be used to estimate the FreeSRF for systems where binding-induced conformation and hydration changes can be obtained with reasonable accuracy.
Predicting/estimating FreeSRF.
There are two levels of prediction applicable to FreeSRF: one is to determine whether the small or large model should be used, and the other is to estimate the free-solution signal for a molecular interaction not currently part of the training set. Fig. 4A illustrates the work flow used to estimate the applicability of using the small vs. large model. First, the structural information (PDB) and the large and small model-fitting parameters (A, B, C, D, and E) are used to calculate χmodel. Then, because most assays are run under these conditions, we estimate the final complex concentration by setting it equal to KD/10 (SI Appendix, Table S1). Upon performing these two calculations and using the FreeSFR relationship, either a nonsense or sensible answer for the predicted change in ΔRI (e.g., detectable or not by BSI) emerges. Following the flowchart in Fig. 4A and using a conservative estimate for the operating range for the interferometer (approximate ΔRI of 3 × 10−3 to 3 × 10−6) allows successful ranking of the binding pair with respect to large or small model. Fig. 4B illustrates that in most cases (23 of 27) or 85% of the time the prediction properly classifies the binding pair. An additional calculation (SI Appendix) using the instrument response function β enables the determination of the actual ΔRI produced for a binding pair (SI Appendix, Table S1). Two important observations can be gleaned from this table. The first is that the experimentally measured value ρ correlates well with the predicted signal. Second, the table and SI Appendix, Figs. S2 and S3, illustrates that the magnitude of ΔRI for a binding event is relatively large.
Fig. 4.
(A) Flow diagram for predicting the suitable model (small or large) for a binding pair. (B) Results for predicting the model for the entire learning set.
Armed with the small vs. large selection method, we tested the capability of the model to estimate the free-solution signal for two molecular interaction pairs not used in the training set. These are Ca2+–recoverin protein–ion interaction and the dansylamide–carbonic anhydrase (CAII) enzyme–inhibitor system. Using the PDB and Eq. 5, we first calculated χmodel for each of the two test systems. Based on ΔSASA, aveSASA, ΔRgyr, aveRgyr, we obtain χmodel of 78.1 RIU/M and −40,557 RIU/M for Ca2+–recoverin and dansylamide–CAII, respectively (Table 1).
To estimate FreeSRF (ρpred) we combine χmodel with β for the instrument to be used and the values of C determined from the KD using our solver and the concentrations to be used to generate a binding isotherm. We commonly use a receptor concentration of ∼KD/10 and a ligand concentration of 4- to 10-fold larger than KD to reach saturation (Bmax). For example, for Ca2+–recoverin, use of the mass balance equation, a receptor concentration of 5.40 × 10−7 M, and a KD = 0.27 × 10−6 M (78), allows that the BSI equilibrium concentrations can be predicted (SI Appendix, Table S2). Substitution into the FreeSRF relationship, ρpred = χmodelβexpC, yields Eq. 7:
| [7] |
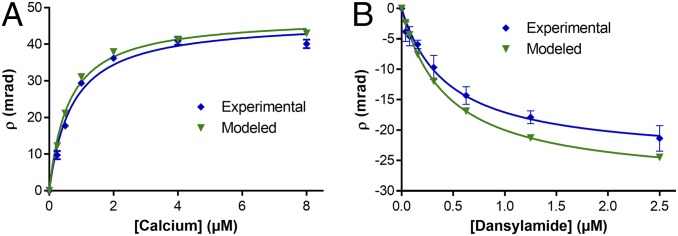
which allows the ρc at each ligand concentration to be calculated. Plotting ρc-model vs. the ligand concentration gives the predicted free-solution binding assay (green curve, Fig. 5A). The same procedure was performed for the dansylamide–CAII binding pair, producing values for C and the modeled FreeSRF (ρc-model) (SI Appendix, Table S2). Plotting these results gives the green binding curve displayed in Fig. 5B.
Fig. 5.
Experimental (blue) and modeled (green) FreeSRF binding curves for (A) recoverin–Ca2+ and (B) carbonic anhydrase II–dansylamide.
Independent of our prediction, we performed free-solution measurements with BSI to determine the KD for both the recoverin and CAII systems. The saturation isotherm binding curves for these experiments are presented as blue lines in Fig. 5. To further illustrate the correlation between measured and estimated FreeSRF, we plotted the χexp values on Fig. 3 showing where they lie on the training-set line. The percent difference from χmodel was 0.13% for Ca2+–recoverin and 18.0% for dansylamide–CAII.
Overall, there is a very good correlation between the empirical and theoretical results. The relative difference between the ρexp and ρmodel was found to be less than ∼29%, except for one value for one dansylamide–CAII concentration reaching 37.8% (SI Appendix, Table S2). It is not surprising that the largest difference in ρ values occurs at the lowest concentrations on the binding curve, a region of lowest instrumental S/N, which typically reports the smallest phase change. As expected for the cluster of systems that have relatively small experimental FreeSRF signals (Fig. 3), a larger difference in χmodel will lead to a comparable error in the prediction. However, the ability to confidently estimate the signal for a binding event within a factor of 2, given only a KD value and the structure, should enable rapid assay optimization, advancing the study of intermolecular interactions.
Also of note, our model accurately predicted a negative FreeSRF (ρ) value (relative to glycerol) for the CAII system, which was subsequently reported in the binding curve by the interferometer. This phenomenon, having been ascribed to a reduction in hydrodynamic diameter, was also observed by SPR for numerous binding systems. Directionality of the signal is a poorly studied parameter at this stage for free-solution measurements, requiring considerably more investigation to provide meaningful mechanistic insights. It is under intense investigation and does appear to inform about the binding mechanism as suggested by others (27, 42).
Even though the molecular shape and hydration changes predict the free-solution signal, it may be necessary to use additional parameters to more accurately describe the molecular dipole, dielectric constant, or electronic structure. For example, systems that undergo oxidation/reduction may produce an electronic structure redistribution that would require use of a third-order term in the equation for χ. We opted for a linear model for simplicity and because it produces an excellent correlation. However, we do recognize that the dependence of RI from changes in structural and dynamical parameters could be more complex than the linear model we fit to, in part because the fitting was done with a somewhat limited number of experimental data points. Interestingly, Marsh and Teichmann (37) make a compelling argument that the absolute value of SASA correlates with the amplitude of conformational change, so ascribing the dependence of RI changes upon binding to alterations in SASA and Rgyr is quite reasonable. We acknowledge that, with additional experimental data and a better understanding of the physical basis of these transformations, a more complex model could emerge that more accurately predicts changes in RI. Going forward, we will be continually evaluating and adding systems to expand our training set, which should improve model performance. To enable broader use by the community, we will be establishing a website to make our model available and for other investigators to contribute to the learning set.
The presentation of these results provides adequate evidence to address a recent assertion made by Sørensen and coworkers (64) that “so far no explicit explanation into how binding could physically generate a BSI signal has been provided.”
Experimental parameters for effective free-solution measurements.
Free-solution investigations are comparative analyses, and it is only under the proper conditions that these relative RI signals become quantifiable. Therefore, we clearly define in SI Appendix the experimental conditions needed to ensure others can successfully perform free-solution assays.
Concluding Remarks
We have presented a hypothesis: “the signal origin for label-free, free-solution studies can be principally ascribed to alterations in structure upon reaction, interaction, or folding. These changes in conformation and hydration produce a quantifiable RI signal.” Numerous examples from the SPR literature describe unanticipated RI signals that could only be attributed to binding-induced changes of this type, findings that further support our supposition. An expression for the FreeSRF is proposed, ρ = χβC, which relates structural changes (χ) to the fringe shift ρ. A training set of a diverse set of binding systems was tested on multiple instruments over a several-year period and used to calculate the coefficients for χ derived from a linear relationship between the experimental and predicted value for FreeSRF (ρ). Using this relationship, we were able to predict, a priori, with reasonable accuracy the FreeSRF for two binding systems. To enable others to use this approach and further refine the model, we have described in full detail the conditions and methodologies needed to perform free-solution assays. Using a carefully designed sample-reference assay that constrains background dη/dC changes and working within the S/N of our interferometer, it is possible to study processes in the absence of a mass change as in protein folding or molecular interactions when the two interacting species are at undetectable starting concentrations.
Based on the strong correlation between FreeSRF and the structural changes detected by the system, our model and BSI enables the use ρexp to determine or predict χ. This capability could provide insights into mechanism of action, allow expedited medicinal chemistry activities, and potentially predict the impact on structure and/or affinity with environment. Additionally, quantitative free-solution assays can be rapidly optimized using our FreeSRF model.
Although an excellent correlation between our theory and experiment has been obtained, we acknowledge more refinement of the model could be advantageous. As with Beer’s law, higher-order terms may be required for some processes (particularly for electronic redistribution) to fully describe a label-free, free-solution experiment.
We predict that the availability of a user-friendly interferometric instrument could usher in a new era for label-free, free-solution chemical, biochemical, and medical analyses.
Supplementary Material
Acknowledgments
We acknowledge Jonathan Sheehan for useful discussion and training of M.N.K. in the use of computational structural biology methodologies. Molecular graphics and analyses were performed with the University of California, San Francisco Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institute of General Medicine Sciences Grant P41-GM103311). Advanced Systems Analysis Program (ASAP) Optical Engineering Software was provided by the Breault Research Organization. D.J.B. and R.A.F. acknowledge support from National Science Foundation (NSF) Grant CHE 1307899. Work in the laboratory of J.M. was supported through NIH Grants R01 GM080403, R01 GM099842, R01 DK097376, R01 HL122010, and R01 GM073151, and NSF Grant CHE 1305874.
Footnotes
Conflict of interest statement: D.J.B. and A.K. have a financial interest in Molecular Sensing, Inc., a company that is commercializing BSI. The other authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.R.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515706113/-/DCSupplemental.
References
- 1.Betzig E, Chichester RJ. Single molecules observed by near-field scanning optical microscopy. Science. 1993;262(5138):1422–1425. doi: 10.1126/science.262.5138.1422. [DOI] [PubMed] [Google Scholar]
- 2.Levene MJ, et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299(5607):682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous The human genome. Unsung heroes. Science. 2001;291(5507):1207. [PubMed] [Google Scholar]
- 4.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19(11):780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 5.Liedberg B, Nylander C, Lundström I. Biosensing with surface plasmon resonance—how it all started. Biosens Bioelectron. 1995;10(8):i–ix. doi: 10.1016/0956-5663(95)96965-2. [DOI] [PubMed] [Google Scholar]
- 6.Yu Y, Ramachandran PV, Wang MC. Shedding new light on lipid functions with CARS and SRS microscopy. Biochim Biophys Acta. 2014;1841(8):1120–1129. doi: 10.1016/j.bbalip.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreira BG, You Y, Behlke MA, Owczarzy R. Effects of fluorescent dyes, quenchers, and dangling ends on DNA duplex stability. Biochem Biophys Res Commun. 2005;327(2):473–484. doi: 10.1016/j.bbrc.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Olmsted IR, Kussrow A, Bornhop DJ. Comparison of free-solution and surface-immobilized molecular interactions using a single platform. Anal Chem. 2012;84(24):10817–10822. doi: 10.1021/ac302933h. [DOI] [PubMed] [Google Scholar]
- 9.Bornhop DJ, et al. Free-solution, label-free molecular interactions studied by back-scattering interferometry. Science. 2007;317(5845):1732–1736. doi: 10.1126/science.1146559. [DOI] [PubMed] [Google Scholar]
- 10.Cubrilovic D, et al. Quantifying protein-ligand binding constants using electrospray ionization mass spectrometry: A systematic binding affinity study of a series of hydrophobically modified trypsin inhibitors. J Am Soc Mass Spectrom. 2012;23(10):1768–1777. doi: 10.1007/s13361-012-0451-6. [DOI] [PubMed] [Google Scholar]
- 11.Kaltashov IA, et al. Advances and challenges in analytical characterization of biotechnology products: Mass spectrometry-based approaches to study properties and behavior of protein therapeutics. Biotechnol Adv. 2012;30(1):210–222. doi: 10.1016/j.biotechadv.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu H, Sheehan JH, Chazin WJ. The mode of action of centrin. Binding of Ca2+ and a peptide fragment of Kar1p to the C-terminal domain. J Biol Chem. 2004;279(49):50895–50903. doi: 10.1074/jbc.M404233200. [DOI] [PubMed] [Google Scholar]
- 13.Tzeng SR, Kalodimos CG. Protein dynamics and allostery: An NMR view. Curr Opin Struct Biol. 2011;21(1):62–67. doi: 10.1016/j.sbi.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Ababou A, Ladbury JE. Survey of the year 2005: Literature on applications of isothermal titration calorimetry. J Mol Recognit. 2007;20(1):4–14. doi: 10.1002/jmr.803. [DOI] [PubMed] [Google Scholar]
- 15.Liang Y. Applications of isothermal titration calorimetry in protein folding and molecular recognition. J Iran Chem Soc. 2006;3(3):209–219. [Google Scholar]
- 16.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Duhr S, Baaske P, Laue E. Microscale thermophoresis for the assessment of nuclear protein-binding affinities. Methods Mol Biol. 2014;1094:269–276. doi: 10.1007/978-1-62703-706-8_21. [DOI] [PubMed] [Google Scholar]
- 18.Baksh MM, Kussrow AK, Mileni M, Finn MG, Bornhop DJ. Label-free quantification of membrane-ligand interactions using backscattering interferometry. Nat Biotechnol. 2011;29(4):357–360. doi: 10.1038/nbt.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kussrow A, Enders CS, Bornhop DJ. Interferometric methods for label-free molecular interaction studies. Anal Chem. 2012;84(2):779–792. doi: 10.1021/ac202812h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olmsted IR, et al. Toward rapid, high-sensitivity, volume-constrained biomarker quantification and validation using backscattering interferometry. Anal Chem. 2014;86(15):7566–7574. doi: 10.1021/ac501355q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saetear P, et al. Quantification of Plasmodium-host protein interactions on intact, unmodified erythrocytes by back-scattering interferometry. Malar J. 2015;14:88. doi: 10.1186/s12936-015-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Swinney K, Bornhop DJ. Attomole sensitivity for unlabeled proteins and polypeptides with on-chip capillary electrophoresis and universal detection by interferometric backscatter. Electrophoresis. 2003;24(5):865–873. doi: 10.1002/elps.200390109. [DOI] [PubMed] [Google Scholar]
- 23.Pesciotta EN, Bornhop DJ, Flowers RA., 2nd Backscattering interferometry: An alternative approach for the study of hydrogen bonding interactions in organic solvents. Org Lett. 2011;13(10):2654–2657. doi: 10.1021/ol200757a. [DOI] [PubMed] [Google Scholar]
- 24.Houlne MP, Hubbard DS, Makhatadze GI, Bornhop DJ. Refractive index-based calorimetric studies of RNase T1 unfolding in small volumes using interferometric backscatter. Proc Soc Photo Opt Instrum Eng. 1996;2982:159–167. [Google Scholar]
- 25.Sota H, Hasegawa Y, Iwakura M. Detection of conformational changes in an immobilized protein using surface plasmon resonance. Anal Chem. 1998;70(10):2019–2024. doi: 10.1021/ac9713666. [DOI] [PubMed] [Google Scholar]
- 26.Crauste C, et al. Unconventional surface plasmon resonance signals reveal quantitative inhibition of transcriptional repressor EthR by synthetic ligands. Anal Biochem. 2014;452:54–66. doi: 10.1016/j.ab.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Gestwicki JE, Hsieh HV, Pitner JB. Using receptor conformational change to detect low molecular weight analytes by surface plasmon resonance. Anal Chem. 2001;73(23):5732–5737. doi: 10.1021/ac0105888. [DOI] [PubMed] [Google Scholar]
- 28.Sulmann S, Dell’Orco D, Marino V, Behnen P, Koch KW. Conformational changes in calcium-sensor proteins under molecular crowding conditions. Chemistry. 2014;20(22):6756–6762. doi: 10.1002/chem.201402146. [DOI] [PubMed] [Google Scholar]
- 29.Daniels PB, Deacon JK, Eddowes MJ, Pedley DG. Surface-plasmon resonance applied to immunosensing. Sens Actuators. 1988;15(1):11–18. [Google Scholar]
- 30.Jung LS, Campbell CT, Chinowsky TM, Mar MN, Yee SS. Quantitative interpretation of the response of surface plasmon resonance sensors to adsorbed films. Langmuir. 1998;14(19):5636–5648. [Google Scholar]
- 31.Sjölander S, Urbaniczky C. Integrated fluid handling system for biomolecular interaction analysis. Anal Chem. 1991;63(20):2338–2345. doi: 10.1021/ac00020a025. [DOI] [PubMed] [Google Scholar]
- 32.Stenberg E, Persson B, Roos H, Urbaniczky C. Quantitative-determination of surface concentration of protein with surface-plasmon resonance using radiolabeled proteins. J Colloid Interface Sci. 1991;143(2):513–526. [Google Scholar]
- 33.Matthews BW. The Proteins III. Academic; New York: 1977. pp. 403–590. [Google Scholar]
- 34.Zhao H, Brown PH, Schuck P. On the distribution of protein refractive index increments. Biophys J. 2011;100(9):2309–2317. doi: 10.1016/j.bpj.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golander CG, Kiss E. Protein adsorption on functionalized and ESCA-characterized polymer-films studied by ellipsometry. J Colloid Interface Sci. 1988;121(1):240–253. [Google Scholar]
- 36.Haussling L, Ringsdorf H, Schmitt FJ, Knoll W. Biotin-functionalized self-assembled monolayers on gold - surface-plasmon optical studies of specific recognition reactions. Langmuir. 1991;7(9):1837–1840. [Google Scholar]
- 37.Marsh JA, Teichmann SA. Relative solvent accessible surface area predicts protein conformational changes upon binding. Structure. 2011;19(6):859–867. doi: 10.1016/j.str.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis TM, Wilson WD. Determination of the refractive index increments of small molecules for correction of surface plasmon resonance data. Anal Biochem. 2000;284(2):348–353. doi: 10.1006/abio.2000.4726. [DOI] [PubMed] [Google Scholar]
- 39.Tumolo T, Angnes L, Baptista MS. Determination of the refractive index increment (dn/dc) of molecule and macromolecule solutions by surface plasmon resonance. Anal Biochem. 2004;333(2):273–279. doi: 10.1016/j.ab.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 40.Boussaad S, Pean J, Tao NJ. High-resolution multiwavelength surface plasmon resonance spectroscopy for probing conformational and electronic changes in redox proteins. Anal Chem. 2000;72(1):222–226. doi: 10.1021/ac990947n. [DOI] [PubMed] [Google Scholar]
- 41.Salamon Z, et al. Plasmon resonance studies of agonist/antagonist binding to the human delta-opioid receptor: New structural insights into receptor-ligand interactions. Biophys J. 2000;79(5):2463–2474. doi: 10.1016/S0006-3495(00)76489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsieh HV, Pitner JB, Gestwicki JE. 2003. US Patent 6,576,430 B1. [DOI] [PubMed]
- 43.Frostell-Karlsson A, et al. Biosensor analysis of the interaction between immobilized human serum albumin and drug compounds for prediction of human serum albumin binding levels. J Med Chem. 2000;43(10):1986–1992. doi: 10.1021/jm991174y. [DOI] [PubMed] [Google Scholar]
- 44.Karlsson R. Real-time competitive kinetic analysis of interactions between low-molecular-weight ligands in solution and surface-immobilized receptors. Anal Biochem. 1994;221(1):142–151. doi: 10.1006/abio.1994.1390. [DOI] [PubMed] [Google Scholar]
- 45.Karlsson R, et al. Biosensor analysis of drug-target interactions: Direct and competitive binding assays for investigation of interactions between thrombin and thrombin inhibitors. Anal Biochem. 2000;278(1):1–13. doi: 10.1006/abio.1999.4406. [DOI] [PubMed] [Google Scholar]
- 46.Metzger J, et al. Biosensor analysis of beta2-glycoprotein I-reactive autoantibodies: Evidence for isotype-specific binding and differentiation of pathogenic from infection-induced antibodies. Clin Chem. 2007;53(6):1137–1143. doi: 10.1373/clinchem.2006.079632. [DOI] [PubMed] [Google Scholar]
- 47.Rich RL, Myszka DG. Survey of the year 2001 commercial optical biosensor literature. J Mol Recognit. 2002;15(6):352–376. doi: 10.1002/jmr.598. [DOI] [PubMed] [Google Scholar]
- 48.Chen H, Kou X, Yang Z, Ni W, Wang J. Shape- and size-dependent refractive index sensitivity of gold nanoparticles. Langmuir. 2008;24(10):5233–5237. doi: 10.1021/la800305j. [DOI] [PubMed] [Google Scholar]
- 49.Luan Q, Xue Y, Yao X, Lu W. Hairpin DNA probe based surface plasmon resonance biosensor used for the activity assay of E. coli DNA ligase. Analyst (Lond) 2010;135(2):414–418. doi: 10.1039/b920228e. [DOI] [PubMed] [Google Scholar]
- 50.Christopeit T, Gossas T, Danielson UH. Characterization of Ca2+ and phosphocholine interactions with C-reactive protein using a surface plasmon resonance biosensor. Anal Biochem. 2009;391(1):39–44. doi: 10.1016/j.ab.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 51.Dell’Orco D, Müller M, Koch KW. Quantitative detection of conformational transitions in a calcium sensor protein by surface plasmon resonance. Chem Commun (Camb) 2010;46(39):7316–7318. doi: 10.1039/c0cc02086a. [DOI] [PubMed] [Google Scholar]
- 52.Luka Z, Moss F, Loukachevitch LV, Bornhop DJ, Wagner C. Histone demethylase LSD1 is a folate-binding protein. Biochemistry. 2011;50(21):4750–4756. doi: 10.1021/bi200247b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hruby VJ, Tollin G. Plasmon-waveguide resonance (PWR) spectroscopy for directly viewing rates of GPCR/G-protein interactions and quantifying affinities. Curr Opin Pharmacol. 2007;7(5):507–514. doi: 10.1016/j.coph.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swann MJ, Peel LL, Carrington S, Freeman NJ. Dual-polarization interferometry: An analytical technique to measure changes in protein structure in real time, to determine the stoichiometry of binding events, and to differentiate between specific and nonspecific interactions. Anal Biochem. 2004;329(2):190–198. doi: 10.1016/j.ab.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 55.Cross GH, et al. A new quantitative optical biosensor for protein characterisation. Biosens Bioelectron. 2003;19(4):383–390. doi: 10.1016/s0956-5663(03)00203-3. [DOI] [PubMed] [Google Scholar]
- 56.Adams NM, Olmsted IR, Haselton FR, Bornhop DJ, Wright DW. The effect of hybridization-induced secondary structure alterations on RNA detection using backscattering interferometry. Nucleic Acids Res. 2013;41(9):e103. doi: 10.1093/nar/gkt165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivanov VI, Minchenkova LE, Minyat EE, Frank-Kamenetskii MD, Schyolkina AK. The B to A transition of DNA in solution. J Mol Biol. 1974;87(4):817–833. doi: 10.1016/0022-2836(74)90086-2. [DOI] [PubMed] [Google Scholar]
- 58.Kypr J, Kejnovská I, Renciuk D, Vorlícková M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37(6):1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pesciotta EN, Bornhop DJ, Flowers RA., 2nd Back-scattering interferometry: A versatile platform for the study of free-solution versus surface-immobilized hybridization. Chem Asian J. 2011;6(1):70–73. doi: 10.1002/asia.201000614. [DOI] [PubMed] [Google Scholar]
- 60.Markov D, Begari D, Bornhop DJ. Breaking the 10(-7) barrier for RI measurements in nanoliter volumes. Anal Chem. 2002;74(20):5438–5441. doi: 10.1021/ac020403c. [DOI] [PubMed] [Google Scholar]
- 61.Wang Z, Bornhop DJ. Dual-capillary backscatter interferometry for high-sensitivity nanoliter-volume refractive index detection with density gradient compensation. Anal Chem. 2005;77(24):7872–7877. doi: 10.1021/ac050752h. [DOI] [PubMed] [Google Scholar]
- 62.Swinney K, Markov D, Bornhop DJ. Chip-scale universal detection based on backscatter interferometry. Anal Chem. 2000;72(13):2690–2695. doi: 10.1021/ac000261r. [DOI] [PubMed] [Google Scholar]
- 63.Swinney K, Markov D, Bornhop DJ. Ultrasmall volume refractive index detection using microinterferometry. Rev Sci Instrum. 2000;71(7):2684–2692. [Google Scholar]
- 64.Jepsen ST, et al. Evaluation of back scatter interferometry, a method for detecting protein binding in solution. Analyst (Lond) 2015;140(3):895–901. doi: 10.1039/c4an01129e. [DOI] [PubMed] [Google Scholar]
- 65.Project E, Friedman R, Nachliel E, Gutman M. A molecular dynamics study of the effect of Ca2+ removal on calmodulin structure. Biophys J. 2006;90(11):3842–3850. doi: 10.1529/biophysj.105.077792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Török K. Calmodulin conformational changes in the activation of protein kinases. Biochem Soc Trans. 2002;30(2):55–61. doi: 10.1042/bst0300055. [DOI] [PubMed] [Google Scholar]
- 67.Vandonselaar M, Hickie RA, Quail JW, Delbaere LT. Trifluoperazine-induced conformational change in Ca2+-calmodulin. Nat Struct Biol. 1994;1(11):795–801. doi: 10.1038/nsb1194-795. [DOI] [PubMed] [Google Scholar]
- 68.Skoog DA, West DM, Holler FJ, Crouch SR. Fundamentals of Analytical Chemistry. 9th Ed Brooks/Cole; Belmont, CA: 2014. [Google Scholar]
- 69.Dickinson TA, White J, Kauer JS, Walt DR. A chemical-detecting system based on a cross-reactive optical sensor array. Nature. 1996;382(6593):697–700. doi: 10.1038/382697a0. [DOI] [PubMed] [Google Scholar]
- 70.Gharagheizi F, Ilani-Kashkouli P, Kamari A, Mohammadi AH, Ramjugernath D. Group contribution model for the prediction of refractive indices of organic compounds. J Chem Eng Data. 2014;59(6):1930–1943. [Google Scholar]
- 71.Katritzky AR, Sild S, Karelson M. Correlation and prediction of the refractive indices of polymers by QSPR. J Chem Inf Comput Sci. 1998;38(6):1171–1176. [Google Scholar]
- 72.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph. 1996;14(1):51–55, 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 73. Lide DR, ed (1996) CRC Handbook of Chemistry and Physics (Chemical Rubber Publishing Company, Boca Raton, FL), 77th Ed.
- 74.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 75.Grosberg AY, Khokhlov AR. Statistical Physics of Macromolecules. American Institute of Physics; New York: 1994. [Google Scholar]
- 76.Fixman M. Radius of gyration of polymer chains. II. Segment density and excluded volume effects. J Chem Phys. 1962;36(12):3123–3129. [Google Scholar]
- 77.Sun ST, Nishio I, Swislow G, Tanaka T. The coil-globule transition—radius of gyration of polystyrene in cyclohexane. J Chem Phys. 1980;73(12):5971–5975. [Google Scholar]
- 78.Permyakov SE, et al. Effects of mutations in the calcium-binding sites of recoverin on its calcium affinity: Evidence for successive filling of the calcium binding sites. Protein Eng. 2000;13(11):783–790. doi: 10.1093/protein/13.11.783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.