Life on our planet evolved under predictable light:dark cycles and dependent rhythms of nutrient availability. Accordingly, a vast majority of living organisms, ranging from archaea to humans, have adopted molecular mechanisms to anticipate and respond to daily metabolic rhythms. Central to this timing mechanism in mammals is a cell-autonomous molecular circadian oscillator based on transcription–translation feedback loops. CLOCK, BMAL, and ROR classes of transcriptional activators, and CRY, PER, and REV-ERB classes of repressors act in concert to generate daily rhythms in their own protein levels as well as thousands of target genes (1). The circadian oscillator in the hypothalamic suprachiasmatic nucleus (SCN) functions as a master pacemaker by imposing a daily rhythm on activity–rest and feeding–fasting cycles. The SCN also responds to changes in ambient light in a time-of-day–specific manner (2) that ultimately leads to the activity–rest cycle adapting to seasonal changes in day length.
Time-series transcriptome studies in the SCN (2) and liver (3) of WT mice fed a standard diet demonstrated daily rhythms in thousands of transcripts in a tissue-specific manner. These rhythms in liver arising from synergistic action of both the cell-autonomous circadian clock and feeding–fasting driven molecular programs (4) are thought to temporally coordinate metabolism to the appropriate time of the day to sustain metabolic homeostasis. Clock-deficient mice lack both cell-autonomous circadian oscillations and feeding–fasting rhythms, compromising transcriptional rhythms in the liver. As a result, these mice exhibit disrupted metabolic homeostasis, giving rise to metabolic diseases (5). Furthermore, in WT mice chronic ad libitum feeding of a high-fat diet dampens the daily rhythm of both the molecular oscillator and feeding–fasting cycles, leading to metabolic diseases (6). An imposed feeding–fasting rhythm in these high-fat fed mice can prevent or reverse metabolic diseases (7, 8). Taken together, these observations have highlighted the paramount significance of circadian rhythms in metabolic homeostasis.
Despite this broad view of circadian rhythms in metabolic fitness, the underlying mechanisms are unclear. It is known that some transcriptional and metabolite rhythms can be restored or driven by an imposed feeding rhythm in clock-deficient mice (9). Furthermore, it is becoming increasingly apparent that the daily rhythms in protein accumulation and function can differ from that of transcriptional rhythms (10). At the subcellular level, mitochondria function as a central hub in metabolism by producing a large proportion of cellular energy, as well as several metabolites that are used as starting materials for anabolic synthesis of complex biomolecules in the cytoplasm. Therefore, identifying specific rhythmic nodes in mitochondrial metabolic processes is important for understanding diurnal regulation of metabolism. In PNAS, Neufeld-Cohen et al. (11) use a time series proteomics approach in purified liver mitochondria to identify mitochondrial proteins that exhibit daily oscillations, and test their relevance in mitochondrial function under specific genetic, diet, and eating pattern modulation.
Liver circadian transcriptome studies have identified cyclic mRNA levels of several mitochondrial proteins. However, circadian liver proteomics studies have failed to identify a sufficient number of mitochondrial components as rhythmic (10). By enriching liver mitochondria before proteomics analyses, Neufeld-Cohen et al. (11) identify nearly 38% of 590 mitochondrial proteins detected as cyclic. As opposed to poor correlation between rhythmic mRNA and protein levels identified in previous proteomics studies of whole liver extract, they found that nearly 86% of rhythmic mitochondrial proteins also had corresponding rhythmic mRNAs. The rhythmic mitochondrial proteins showed a median fold-change between peak and trough ∼1.3-fold, which is relatively modest compared with liver mRNA oscillations, which are often >1.5-fold (3). Therefore, subcellular fractionation, followed by proteomics, is likely a more sensitive approach to uncover daily oscillation in protein accumulation.
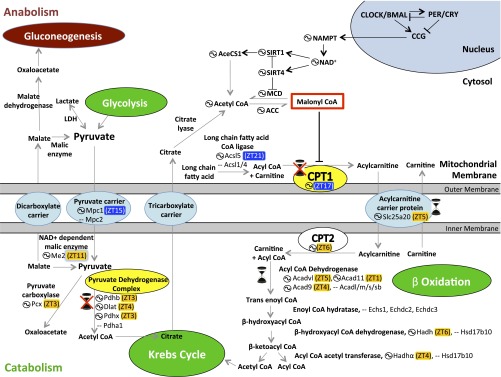
Functional annotation of the cycling proteome highlights the key catabolic and oxidative functions of mitochondria under diurnal fluctuation (Fig. 1). Specifically, the Neufeld-Cohen et al. study (11) unravels diurnal regulation of mitochondrial metabolism of pyruvate and fatty acids, which produce acetyl-CoA. In mitochondria, aceytyl-CoA is oxidized in the tricarboxylic acid (TCA) or Krebs cycle to produce GTP and NADH. NADH is used in numerous biochemical reactions, including its oxidation by the mitochondrial oxidative phosphorylation complex or electron transport chain (ETC) to produce ATP.
Fig. 1.
Circadian regulation of pyruvate metabolism and fatty acid uptake and oxidation. The circadian clock exerts transcriptional and translational control over many clock-controlled genes (CCG), including the NAMPT gene and other metabolic components. One mechanism in which the molecular clock regulates metabolism is through control of NAD+-dependent protein deacetylases (sirtuins, SIRT). Note, not all forms of circadian regulation of metabolism are depicted. Rate-limiting enzymes are indicated in yellow, metabolites are in gray, catabolic processes in green, and anabolic processes in brown. The hourglass ( ) signifies protein oscillations that were restored by nighttime feeding in Per1/2−/− mice (11). The circled wave (
) signifies protein oscillations that were restored by nighttime feeding in Per1/2−/− mice (11). The circled wave ( ) indicates oscillating proteins; peak protein accumulation is provided in zeitgeber time (ZT) (11). Protein levels peaking during the day are highlighted in orange, and during the evening in blue. ACC, acetyl-CoA carboxylase; AceCS1, acetyl-CoA synthetase; Hadh, hydroxyacyl-CoA dehydrogenase; LDH, lactate dehydrogenase; MCD, malonyl-CoA decarboxylase; NAMPT, nicotinamide phosphoribosyltransferase; PDH, pyruvate dehydrogenase. All acronyms that are not in bold are genes that code for the enzymes in bold above them.
) indicates oscillating proteins; peak protein accumulation is provided in zeitgeber time (ZT) (11). Protein levels peaking during the day are highlighted in orange, and during the evening in blue. ACC, acetyl-CoA carboxylase; AceCS1, acetyl-CoA synthetase; Hadh, hydroxyacyl-CoA dehydrogenase; LDH, lactate dehydrogenase; MCD, malonyl-CoA decarboxylase; NAMPT, nicotinamide phosphoribosyltransferase; PDH, pyruvate dehydrogenase. All acronyms that are not in bold are genes that code for the enzymes in bold above them.
Pyruvate is a key node in the metabolic pathways of the liver. It is produced in the cytoplasm from amino acids, or from glucose through glycolysis. Pyruvate can be used in several pathways to produce ethanol, amino acids, glucose, or to produce fatty acids or energy through acetyl-CoA. Cytoplasmic pyruvate enters the mitochondria through mitochondrial pyruvate carrier (MPC1/MPC2) or is produced de novo in mitochondria from malate by malic enzyme (ME2). Both MPC1 and ME2 proteins displayed circadian variation. Mitochondrial pyruvate is decarboxylated to acetyl-CoA by the pyruvate dehydrogenase complex or carboxylated to oxaloacetate by pyruvate carboxylase (PCX). PCX, as well as several components of the pyruvate dehydrogenase complex, were rhythmic with peak protein accumulation in the early morning (11).
Another source of mitochondrial acetyl-CoA is from fatty acid oxidation. Although short-chain fatty acids in the cytoplasm can diffuse into the mitochondria, long-chain fatty acids are transported as acylcarnitine through the mitochondria membrane. Rate-limiting enzyme carnitine palmitoyl transferase 1 (CPT1) and acylcarnitine carrier protein both exhibited diurnal oscillations. Inside the mitochondria, both long- and short-chain fatty acids are oxidized through acyl-CoA dehydrogenases (ACAD) to ultimately produce acetyl-CoA and acyl-CoA. Several ACADs and other enzymes for fatty acid oxidation showed daily oscillation, with peak protein levels in the daytime when mice typically eat less and are likely to use fat as an energy source. Mitochondrial acetyl-CoA is used in the TCA cycle, because it cannot cross the mitochondrial membrane. NADH produced by the TCA cycle is used in ATP production through the ETC complex. Finally, numerous components of the ETC complex, including some encoded by the mitochondrial genome, were also found to show diurnal oscillations.
In addition to oxidation of acetyl-CoA for NADH production, TCA cycle intermediates are also transported out of the mitochondria for synthesis of numerous biochemicals, including nonessential amino acids, nucleotides, fatty acids, cholesterol, bile acids, glucose, and porphyrin ring-containing compounds. Circadian oscillation of several key enzymes of the TCA cycle (malate dehydrogenase, isocitrate dehydrogenase, α-ketoglutarate dehydrogenase, and succinyl CoA synthase) likely help divert certain intermediates for anabolic metabolism and may contribute to the observed rhythms in these metabolites in mouse liver (12). Shuttling of TCA cycle intermediates between the mitochondria and the cytoplasm in hepatocytes also subserves the important task of carrying electrons from NADH across the mitochondrial membrane, as it is impervious to NADH. Previous studies have established a key role of the circadian clock in generation of NAD+ in the cytoplasm (13). Overall, the circadian system temporally coordinates the production and utilization of NAD/NADH in both the cytoplasm and the mitochondria.
To test whether these cyclic protein levels result in diurnal rhythms in mitochondria function, Neufeld-Cohen et al. (11) purified liver mitochondria throughout the 24-h day and tested their efficiency in oxidizing excess pyruvate or fatty acid (palmitate) as an energy source by measuring the oxygen consumption rate (OCR). More importantly, they systematically tested the contribution of the circadian oscillator, nutrient quality, and eating pattern on daily oscillations in mitochondrial function. CPT1 conjugates palmitoyl CoA with carnitine to produce palmitoyl carnitine, which can be translocated across the mitochondrial membrane and subsequently oxidized. Mitochondria from WT mice fed a standard diet displayed a daily oscillation in CPT1 protein levels and a parallel rhythm in utilization of palmitoyl CoA and carnitine. However, this rhythm is abolished and CPT1 level remains constitutively high in mice lacking the circadian oscillator repressors PER1/2. To test whether the predominantly night-eating behavior of WT mice, which is lost in Per1/2−/− mice, might be driving the CPT1 rhythm, Per1/2−/− mice were fed exclusively at night. This night-eating pattern did not restore CPT1 rhythms. Conversely, the high-fat diet, known to dampen circadian oscillation of several gene products, did not decrease CPT1 protein levels. These results suggest that the circadian clock regulates key enzymes that gate the entry of long-chain fatty acids into the mitochondria. When palmitoyl carnitine is supplied as a substrate bypassing CPT1, OCR still shows a diurnal rhythm, reflecting that mitochondrial β-oxidation is rhythmic. Although this rhythm is dampened in Per1/2−/− liver, night-feeding can partially rescue both the rhythm of ACAD11 protein and the parallel OCR rhythm, albeit with a slightly different phase of oscillation. Conversely, a high-fat diet can dampen the ACAD11 protein rhythm and OCR. Therefore, fatty acid oxidation in liver is sensitive to both eating pattern and diet composition.
Similar experiments with pyruvate and malate as substrates revealed that a daily rhythm in pyruvate dehydrogenase parallels with cyclic OCR. However, these rhythms are abolished in Per1/2−/− mice, cannot be rescued by night-feeding, and yet are dampened by a high-fat diet. Although night-feeding failed to restore rhythmic OCR in Per1/2−/− mitochondria when pyruvate + malate or palmitoyl CoA + carnitine were used as substrates, night-feeding did change the steady-state OCR.
These observations illustrate that diurnal rhythms in the mitochondrial proteome and their associated functions are heavily influenced by the molecular circadian clock, nutrient quality, and eating pattern. Each of these three factors differentially affects the overall level, rhythm, and phase of oscillation for several mitochondrial proteins. Such regulation likely extends to transcriptional and translational regulation in other organs and even in other organisms. In Drosophila, the pattern of global gene expression is influenced by eating pattern, nutrient quality, and the molecular clock, which can be leveraged to change the function of mitochondrial ETC in the heart and improve cardiac health (14). This finding opens up new questions and offers several practical implications for human health. For example, substituting a high-fat diet with a standard balanced diet can support diurnal rhythms in metabolism. As circadian rhythms dampen with age, time-restricted feeding (TRF) may be able to reinforce the weak genetic drive for metabolic oscillations. Such eating patterns restore robust daily rhythms in whole-body energy utilization in clock-deficient mice (4, 11). However, in Drosophila, TRF imparts cardiac benefits in aged flies only in the presence of an intact circadian clock (14). Hence, understanding the mechanisms by which TRF, clock, and nutrient quality affect the oscillation, phase, and overall levels of metabolic regulators has huge untapped potential to maintain metabolic homeostasis and extend a healthy lifespan.
Acknowledgments
Research in the S.P. laboratory is supported by National Institutes of Health Grants DK091618 and EY016807, Leona M. and Harry B. Helmsley Charitable Trust Grant 2012-PG-MED002, the Glenn Foundation, and American Federation of Aging Research Grant M14322.
Footnotes
The authors declare no conflict of interest.
See companion article on page E1673.
References
- 1.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 2.Hatori M, et al. Lhx1 maintains synchrony among circadian oscillator neurons of the SCN. eLife. 2014;3:e03357. doi: 10.7554/eLife.03357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106(50):21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zarrinpar A, Chaix A, Panda S. Daily eating patterns and their impact on health and disease. Trends Endocrinol Metab. 2016;27(2):69–83. doi: 10.1016/j.tem.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6(5):414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Chaix A, Zarrinpar A, Miu P, Panda S. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 2014;20(6):991–1005. doi: 10.1016/j.cmet.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatori M, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15(6):848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adamovich Y, et al. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014;19(2):319–330. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robles MS, Cox J, Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10(1):e1004047. doi: 10.1371/journal.pgen.1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neufeld-Cohen A, et al. Circadian control of oscillations in mitochondrial rate-limiting enzymes and nutrient utilization by PERIOD proteins. Proc Natl Acad Sci USA. 2016;113:E1673–E1682. doi: 10.1073/pnas.1519650113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asher G, Sassone-Corsi P. Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161(1):84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 13.Bass J. Circadian topology of metabolism. Nature. 2012;491(7424):348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 14.Gill S, Le HD, Melkani GC, Panda S. Time-restricted feeding attenuates age-related cardiac decline in Drosophila. Science. 2015;347(6227):1265–1269. doi: 10.1126/science.1256682. [DOI] [PMC free article] [PubMed] [Google Scholar]