Abstract
The CMG helicase is composed of Cdc45, Mcm2-7 and GINS. Here we report the structure of the S. cerevisiae CMG determined by cryo-EM at a resolution of 3.7–4.8 Å. The structure reveals that GINS and Cdc45 scaffold the N-tier of the helicase while enabling motion of the AAA+ C-tier. CMG exists in two alternating conformations, compact and extended, suggesting that the helicase functions like an inchworm. The N-terminal regions of the Mcm2-7, braced by Cdc45-GINS, form a rigid platform upon which the AAA+ C-domains make longitudinal motions, nodding up and down like an oil rig pumpjack makes nodding motions attached to a stable platform. The Mcm ring is remodeled in CMG relative to the inactive Mcm2-7 double-hexamer. The Mcm5 winged helix domain is inserted into the central channel, blocking entry of dsDNA, and supporting a steric exclusion DNA unwinding model.
Keywords: CMG, helicase, cryo-electron microscopy, replication, DNA translocation, strand exclusion
Introduction
In eukaryotes, two Mcm2-7 rings are loaded onto DNA at origins of replication in G1 phase1,2. The double hexamer is inactive as a helicase, but is activated upon entry into S phase by Dbf4-dependent Cdc7 kinase, cyclin-dependent kinase, and several transient factors that activate origins, by assembling Cdc45 and GINS onto each Mcm2-7 complex to form two active CMG helicases that surround each strand of parental DNA3,4. CMG then translocates along single-strand (ss) DNA in the 3’ to 5’ direction for bidirectional replication5–7. Several archaeal MCM crystal structures are known8–12, along with the atomic model of the yeast double hexamer of Mcm2-7 by cryo-EM13. These structures reveal that the Mcm2-7 core forms a two-tiered ring structure: a N-terminal domain (NTD) tier composed of a helical sub-domain, a Zn binding motif, and an OB motif, while the C-terminal domain (CTD) tier contains the AAA+ motors. The structure of the 11-protein CMG complex is known only at low resolution: single particle EM of fly and yeast CMG show that GINS and Cdc45 bind to one side of the Mcm2-7 hexamer, forming a double-ring-like structure14–16.
Most studies of replicative helicases focus on homohexamers such as papilloma virus E1, SV40 T-antigen, archaeal Mcm, E. coli DnaB and bacteriophage T7 gene 4 protein17. In comparison, studies of the asymmetric 11-protein CMG are scarce and many fundamental questions remain. Why is CMG an asymmetric ring? How does it translocate along DNA? Does it function differently from the rotary homohexameric helicases? Why are some ATP sites dispensable for CMG helicase activity, while others are essential? Where is the location in CMG where dsDNA is separated? Two general models for the unwinding point are: the steric exclusion model in which the dsDNA split point is on the C-surface, with leading strand ssDNA passing through the central channel while lagging ssDNA is excluded to the outside of the ring; or the duplex is split inside the hexamer channel and one strand is extruded out a side channel between the CTD and NTD tiers, referred to as the ploughshare, or side channel extrusion model18.
Translocation of a helicase along DNA is presumed to require at least two DNA binding sites that differ in relative distance to one another during the ATP cycle, promoting movement along DNA17. Structural studies of papilloma virus E1 helicase propose six equivalent sites, the DNA binding loops in the motor domains that undergo relative distance changes in a rotary fashion, escorting ssDNA through the ring in steps of one nucleotide per ATP10. The structure of bacterial DnaB also reveal that the motor domain of each subunit binds ssDNA, and it is proposed that whole protomers translocate along ssDNA as a rotary “staircase” in steps of two nucleotides per ATP19. CMG lacks the six-fold symmetry of homohexamers, and whether CMG acts in a rotary fashion is unknown.
To address the conformation changes during CMG translocation, and its mode of unwinding (steric exclusion or side channel extrusion), we have used cryo-EM to derive an atomic model of yeast CMG. We find that the Cdc45-GINS-Mcm interfaces are sculpted to rigidify the Mcm2-7 NTD ring while still allowing movements within the Mcm2-7 CTD motor ring. Thus the CTD ring adopts two different conformations, tilted and untilted, relative to a fixed NTD ring which gives rise to compact and extended forms of CMG with a maximum difference of about 20 Å. Five of the Mcm2-7 subunits contain C-terminal winged helix domains (WHD) that extend outward from the CTD and appear flexible and ill defined in the double hexamer13. But in CMG the WHD of Mcm5 and Mcm6 are localized, and the WHD of Mcm5 is pushed into the interior axial channel of the CTD motor ring, where it restricts the large diameter pore to a size too small for dsDNA. This supports a steric exclusion mechanism for CMG in which leading ssDNA threads through the axial channel, excluding the lagging strand as indicated by biochemical studies in the Xenopus system20. The high resolution structures of the two CMG conformers observed here indicates that the CTD motor ring tilts up and down relative to a rigid NTD ring-Cdc45-GINS platform, much like the “nodding head” of an oil rig pumpjack. Alternating between these structures suggests that CMG might move on DNA in a linear fashion, ratcheting along ssDNA during unwinding without need for rotary action.
Results
3D reconstruction of CMG helicase
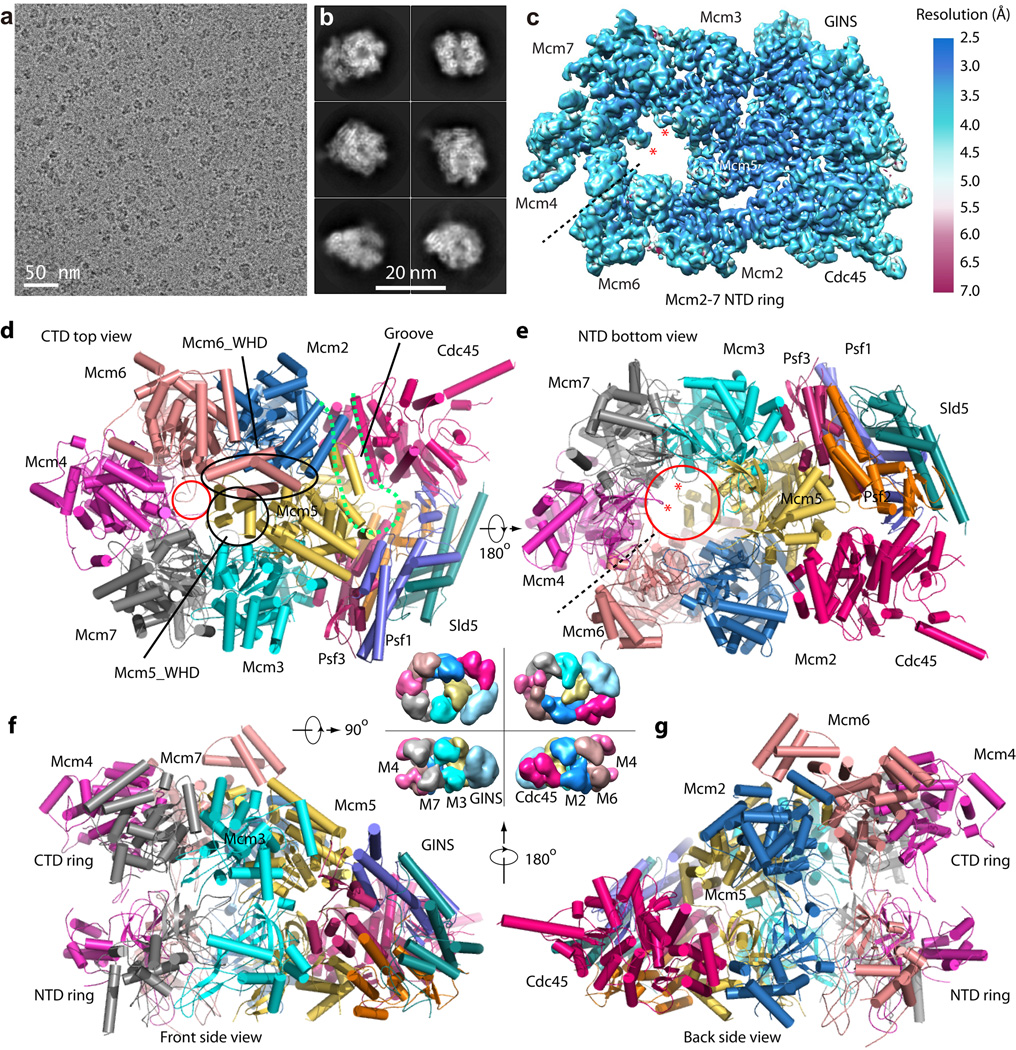
Recombinant Saccharomyces cerevisiae CMG was expressed and purified from yeast, and cryo-EM images were recorded on a Titan Krios microscope with a K2 summit direct electron detector (Fig. 1a–b). After 2D and 3D classification, we initially reconstructed from ~470,000 particle images a 3D density map of the full CMG complex at 3.8 Å resolution (Supplementary Table I and Supplementary Figs. 1–2). Interestingly, the Cdc45-GINS-Mcm2-7 NTD ring had solid density, but the density of the Mcm2-7 CTD ring was much weaker, indicating partial flexibility. Excluding the flexible CTD motor region, the 3D density map has an estimated resolution of 3.7 Å (Fig. 1c, Supplementary Fig. 3). We subsequently carried out focused 3D classification in the CTD motor region, and obtained two conformers for the CMG helicase, at 4.8 Å and 4.7 Å, respectively (Supplementary Fig. 4–6, discussed below). The lower resolution was probably a result of a decreased number of particles belonging to each conformer, but could also be an indication that the helicase exists in a continuum of conformations21. This possibility was underscored by the gold-standard Fourier shell correlation curve that started to drop as early as 6.5 Å. We first built the Cdc45, GINS and the Mcm2-7 NTD ring based on the 3.7 Å resolution density map as well as the previously reported inactive yeast Mcm2-7 structure and homolog GINS structure13,22–24. We then used the 3.7 Å structure and the 3.8 Å structure of the CTD ring of the inactive Mcm2-7 to model CMG conformers I and II.
Figure 1. Cryo-EM and overall structure of the S. cerevisiae CMG complex.
(a) A typical motion-corrected raw image of frozen CMG particles recorded on a direct detector. (b) Selected six 2D averages representing the particles in different views. (c) 3D cryo-EM map of CMG color coded by local resolution. Overall resolution is 3.7 Å. This density map includes Cdc45-GINS-Mcm2-7 NTD ring. The two red asterisks mark the hairpin loop in the NTD OB-subdomain of Mcm4 and Mcm7 that protrude into the axial channel. (d–g), Cartoon view of the full-length atomic model of CMG conformer I as viewed from the CTD motor ring (d), NTD bottom (e), Front side (f) and back side (g). The dashed black line in (c, e) marks the smaller interface between NTDs of Mcm4 and Mcm6. The dashed magenta curve in (d) marks the groove between Cdc45 and Mcm2-Mcm5. The smaller red circle in (d) marks the constricted opening (10 Å) at the CT motor ring. The large red circle in (e) marks the 20 Å channel opening at the bottom. The black circle and the oval in (d) mark the WHD of Mcm5 and Mcm6, respectively. The central four surface representations of CMG between panels (d)–(g) are an aid to understanding the high resolution structures. They are lower resolution EM map of CMG (EMDB 6463)16.
Overall structure of CMG
CMG has a relatively large ~20 Å diameter opening at the bottom NTD ring where the leading strand presumably exits the Mcm2-7 axial channel (Fig. 1c–g). The axial channel in the NTD ring is partially obstructed by two protruding loops that connect the first two β-strands of the OB sub-domains in Mcm4 and Mcm7 (Fig. 1c, 1e). These loops bind ssDNA in the archaeal MCM hexamer, and two conserved DNA binding arginine residues in the yeast Mcm4 and Mcm7 OB domains are essential for function25. In CMG, these OB domains are positioned directly below the ssDNA pore of the CTD-ring where DNA enters (Fig. 1d). Therefore the leading strand coming from the CTD ring will likely encounter and interact with the two channel-lining OB folds in the NTD ring.
The Mcm2-7 NTD-ring is highly stabilized by an extensive network of bracing interactions with Cdc45 and GINS. Interestingly, the only contacts of Cdc45 with Mcm2 and Mcm5 are with the NTD, and Cdc45 makes no contact with the CTDs, resulting in a remarkably wide groove between the Mcm2-7 CTD and Cdc45 (Fig. 1d–g). This architectural facet enables major changes in the CTD ring between the two CMG conformers while still stabilizing the NTD ring and holding it rigid. Specifically, in conformer I the CTD ring is tilted relative to the NTD ring through upward displacement of the motor domains of the Mcm 2,6,4 subunits, and the interface between the CTDs of Mcm2 and Mcm5 springs open. The fact that ATP sites are located at the CTD subunit interfaces suggest that ATP hydrolysis at the Mcm2-5 interface leads to conformer I in which this interface is cracked. The other function that the Cdc45-GINS-Mcm NTD architecture may serve is to hold onto DNA when the Mcm2-5 interface opens, as DNA is known to pass through this interface during origin activation14,26,27. If DNA were to escape the CTD ring via the dynamic Mcm2-5 interface, the Cdc45-GINS subunits that span the Mcm2-5 interface would prevent the helicase from falling off DNA as indicated by CMG-DNA cross-linking studies28. Within the Mcm2-7 NTD ring, the interface with the least buried surface area is the Mcm4-6 interface, shown as a dashed line (Fig. 1c, 1e). If CMG needed to open for bypass of a leading strand block, the Mcm4-6 interface may be the “pressure release valve” for NTD ring opening.
Compared to the inactive double hexamer structure, Mcm2-7 is substantially remodeled in the active helicase. We observed defined WHD densities for Mcm5 and Mcm6, unlike the Mcm2-7 double hexamer13 in which the WHDs are flexible and ill defined on the CTD surface outside the central channel (Fig. 1d–g, Supplementary Figs. 5–6). Surprisingly, the WHD of Mcm5 is stuffed inside the central pore and the Mcm6 WHD stacks on top of it. The interior location of the Mcm5 WHD is consistent with our earlier mass spectrometry analysis demonstrating that the only WHD that crosslinks to residues lining the channel is the Mcm5 WHD, even though all other Mcm subunits, except Mcm2, also contain a WHD (Supplementary Fig. 7a–c)16. The location of the Mcm5 WHD constricts the diameter of the central channel to ~ 10 Å, too small for dsDNA that is 20 Å in diameter (Fig. 1d). Therefore, the Mcm5 WHD may provide a structural basis for the steric exclusion mechanism of DNA unwinding. The most dramatic alteration between CMG and the double hexamer is the existence of at least two distinct conformers in the CTD AAA+ motor ring of CMG (described below).
The surface of CMG is mostly of mixed charge (Supplementary Fig. 7d–g), while the orifice of the CTD pore is positively charged and suitable for entry of ssDNA. The axial channel is positively charged in the NTD ring, largely due to the channel-lining loops of OB domains previously shown to interact with ssDNA25. Furthermore, there is a nearly continuous and positively charged path on the outer surface of Mcm3 that extends from the top CTD to the bottom of the CMG structure. Because the lagging strand interacts with the outer surface of an archaeal Mcm helicase29, it is possible that the lagging strand follows the positively charged route to reach the Pol α polymerase-primase, which our earlier structural study showed is below the NTD ring of CMG16.
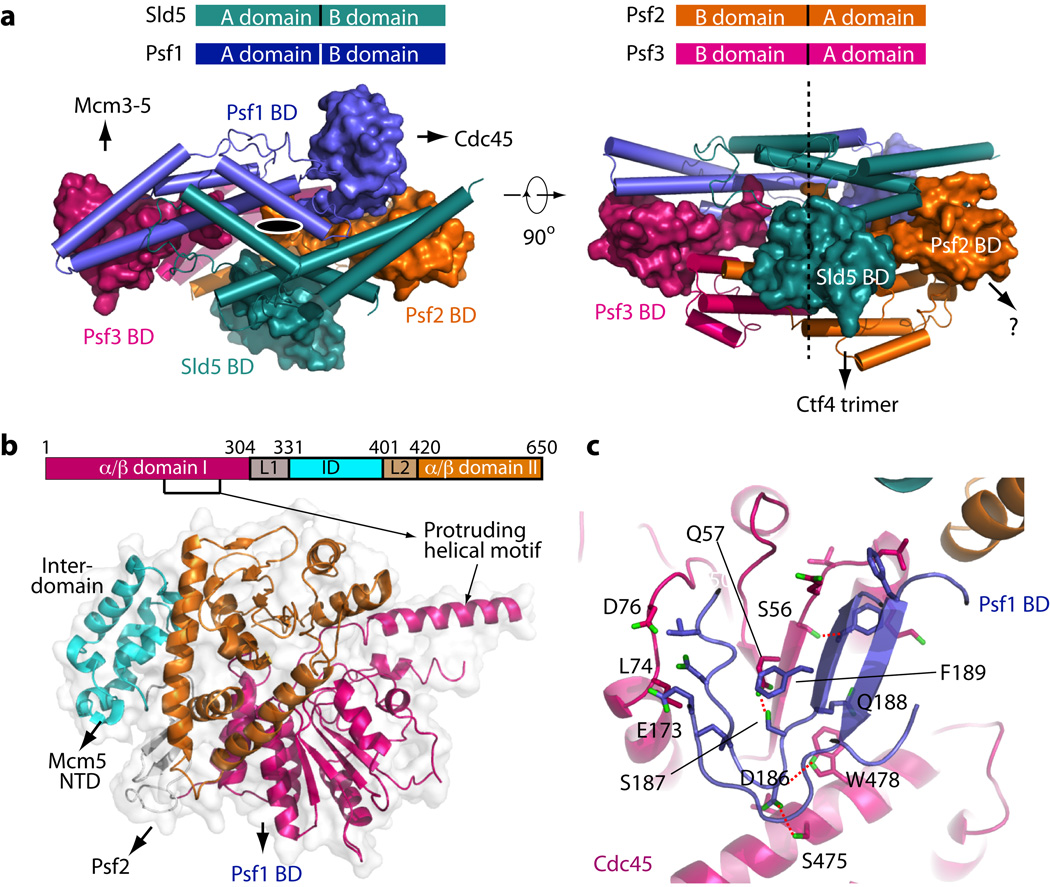
In the context of CMG complex, the function of individual GINS subunits as scaffolding points for different proteins became clear. Each GINS subunit has a two-domain architecture: a 4-helix bundle region (A-domain) and a α/β region (B-domain (BD)). As with the human GINS22–24, the A-domains of the 4 subunits stack to form a pseudo 2-fold symmetric flat core onto which the 4 BDs are attached at the periphery (Fig. 2). The Psf1 BD is not visible in the crystal lattice of human GINS22–24. The Psf1 BD is not needed for GINS complex formation, but it is essential for CMG formation and is proposed to bind Cdc4524. In previous low-resolution EM maps of CMG, a density between GINS and Cdc45 was tentatively assigned to the Psf1 BD14,16. The CMG structure clearly showed this density is the Psf1 BD, which folds into an α/β motif similar to the corresponding domain in Sld5 (Fig. 2). The Psf1 BD interacts with Cdc45 by forming a β-sheet core crossing the two proteins. Interaction between the Psf1 BD and Cdc45 is of a mixed nature and extensive, burying an interface of 820 Å2. The Psf3 BD clearly functions to interact with Mcm3-5, strengthening the Mcm3-5 interface (Figs 1e,f). The Sld5 BD is located at the bottom, and together with a short N-terminal helix that was not resolved in the current structure, binds the Ctf4 trimer16,30. By analogy to the other B-domains, the Psf2 BD may bind a protein that is not yet identified.
Figure 2. Structure and interactions of the yeast GINS and Cdc45.
(a) The full-length GINS structure in top and side views. Domain A is shown in cartoon, and domain B in surface. The top sketch shows that all four subunits have a similar two-domain architecture, but domain A and B in Psf2 and Psf3 are inverted compared to Sld5 and Psf1. The black oval and dashed line indicate the pseudo two fold symmetric arrangement of the GINS complex. The question mark indicates the Psf2 B domain may have an unknown interacting partner in a functional replisome. (b) The full-length structure of Cdc45 in cartoon representation superimposed on the transparent surface view. The two α/β-domains are shown in magenta and brown, respectively. The middle all-helix inter-domain (ID) is connected to the NT and CT α/β-domains via two linkers (L1 and L2). Part of L1 and L2 further forms a two stranded β-sheet. The ID mainly interacts with NTDs of Mcm2 and Mcm5. The L1 loop region interacts with Psf2. (c) Extensive interactions between GINS Psf1 C-terminal BD (blue) and Cdc45 (magenta). The two β-strands of Psf1 BD adjoin with the 4 β-strands in the α/β-domain I of Cdc45, forming a 6-strand β-sheet core across the two proteins. Four H-bonds are drawn as dashed red lines.
Cdc45 displayed a prominent helical motif that extends far beyond the previously determined low resolution profile of CMG (Fig. 1d,e and Fig. 2b)16. This helical protrusion is proximal to the catalytically active domain of Pol ε, as a lysine at the tip of the helix cross-links to the N-terminal catalytic domain of Pol216. Cdc45 is predicted to have a RecJ-like fold31–33. The structure showed that yeast Cdc45 contains two RecJ-like α/β-domains interspersed by a small helical inter-domain (ID) (Fig. 2b). The helical ID contacts and stabilizes the NTDs of Mcm2 and Mcm5. The double α/β-domain fold is also found in GT-B type glycosyltransferases34. Full-length RecJ forms an “O”-like structure with a pore in the middle surrounded by its four domains, and there is a ssDNA binding groove between the N-terminal two α/β-domains35,36. However, yeast Cdc45 did not form an analogous central pore or DNA binding groove, and supported the α helical protrusion instead.
CMG fluctuates between planar and tilted forms
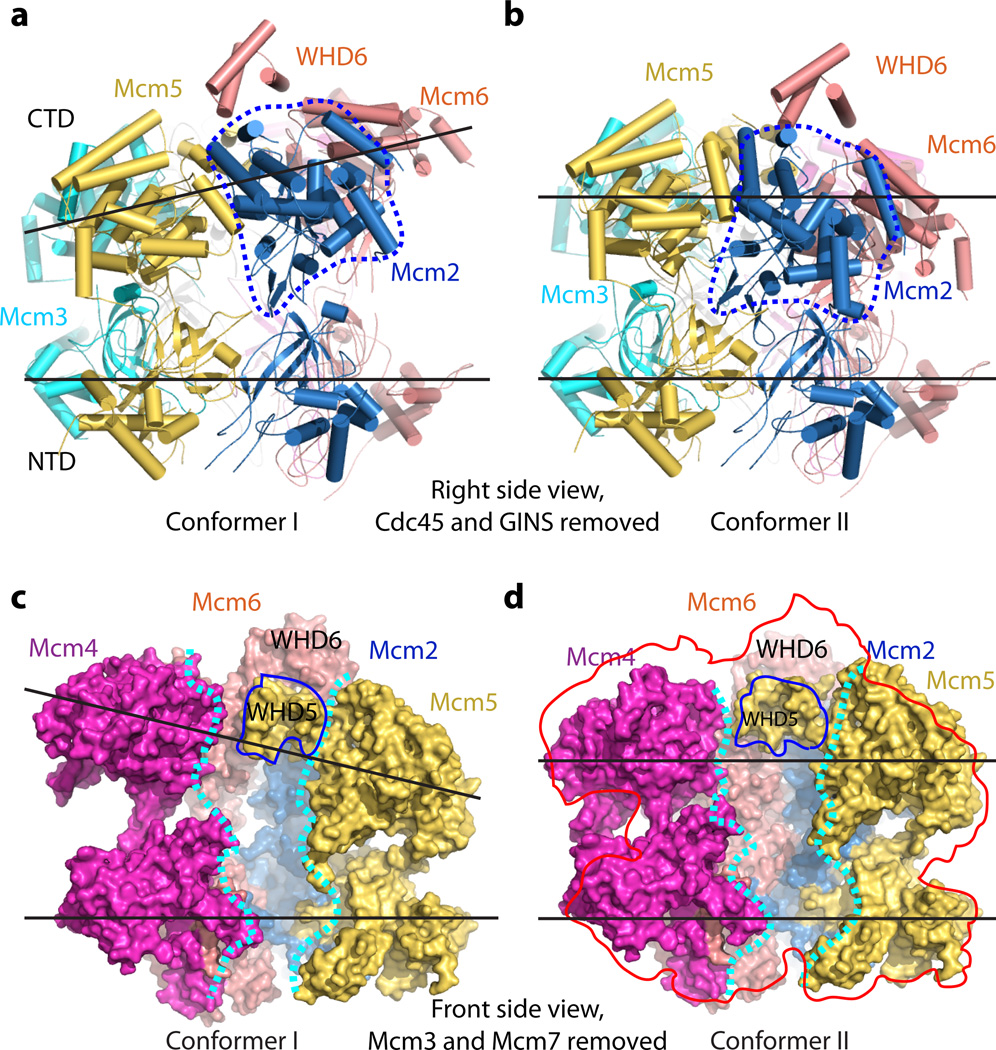
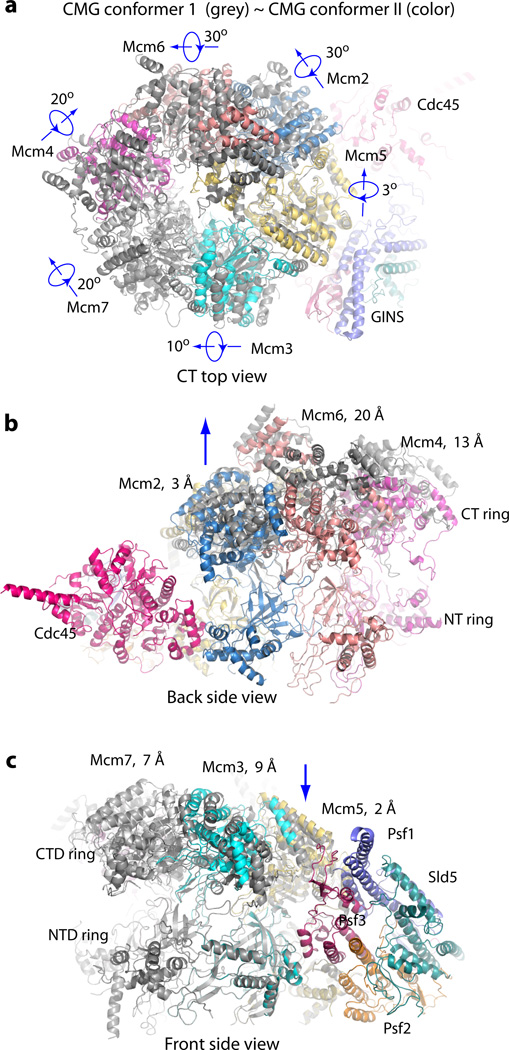
The cryo-EM images were recorded using pure CMG with no added nucleotides or DNA, so the structure is expected to be the apo-form. Indeed, no nucleotide or DNA density was observed in any of our 3D reconstructions. Among the ~470,000 CMG particles used for 3D reconstruction, ~40% of the particles existed in the extended and tilted form termed conformer I, and ~20% of the particles were in the untilted planar form termed conformer II (Fig. 3, Supplementary Figs 4–5). In conformer I, when viewed from the Cdc45-GINS side of the structure, the CTD ring containing the AAA+ motors of Mcm2-7 is tilted by ~10° with respect to the NTD ring, leading to an approximate spiral arrangement of domains comprising the CTD ring (Fig. 3a). In conformer II, the CTD motor ring is approximately parallel to the NTD ring and CMG is more compact than conformer I (Fig. 3b). The Mcm5 WHD was inside the axial channel in both conformers (Fig. 3c–d). By superimposing the two conformers, each of the six Mcm2-7 CTD AAA+ motor domains undergo both rotation and translation, to varying degrees (Fig. 4a–c and Supplementary Videos 1 and 2). Compared to conformer II, the Mcm6 AAA+ motor domain in conformer I was translated upward 20 Å, while the AAA+ motor domains of the neighboring Mcm2 and Mcm4 were shifted up by 3 Å and 13 Å, respectively (Fig. 4b). When viewed from the right side (Cdc45 and GINS), the Mcm2 AAA+ motor domain in conformer I rotates 30° and moves away from the Mcm5 AAA+ domain, compared to conformer II, leaving a small gap between Mcm2 and Mcm5 (Fig. 3a–b). In overview, conformer I is an expanded form of CMG relative to the more compact conformer II. It is possible that the CTD motor ring may have additional conformations that were not captured in the current focused 3D classification, as the two conformers accounted for ~60% of the CMG particles. Earlier low resolution EM studies of CMG noted two forms, an apo form having an opening in the AAA+ tier between Mcm2-5, and an ATP form with both tiers closed14,15. These forms likely correspond to conformers I and II, respectively.
Figure 3. Side-by-side comparison of conformer I and conformer II in the Mcm2-7 region of CMG helicase.
(a–b) Comparison of the two conformations shown in cartoon representation and viewed from the right side, from Cdc45 and GINS (which are both removed for clarity) with the CTD motor ring on top and NTD ring at the bottom. The two black lines show that the CTD motor ring is tilted by ~ 10° with respect to the NTD ring in (a) but nearly parallel in (b). The dashed blue line encircles the Mcm2 CTD. This domain is rotated by 30° clockwise in (b) compared to (a). (c–d) Front side surface view of conformer I (c) and conformer II (d), with the front Mcm3 and Mcm7 removed to show Mcm2-7 axial channel, which is approximately demarcated by two dashed white curves. Cdc45 and GINS at the right side are also removed for clarity. In both conformers, the Mcm5 WHD is located inside the axial channel. The Mcm6 WHD sits on top of the Mcm5 WHD. The red shape in (d) is a contour of (c), superimposed on conformer II to show the changes in the CTD motor ring between the two conformers.
Figure 4. Superposition of CMG conformers I and II.
(a) Top view, (b) back side view showing the movement of the Mcm2-6-4 half motor ring, and (c) Front side view showing the smaller movements of the Mcm 5-3-7 half ring. Conformer I is shown in dark gray and conformer II in color. The distances shown in b–c are measured for the AAA+ domains between the two conformers.
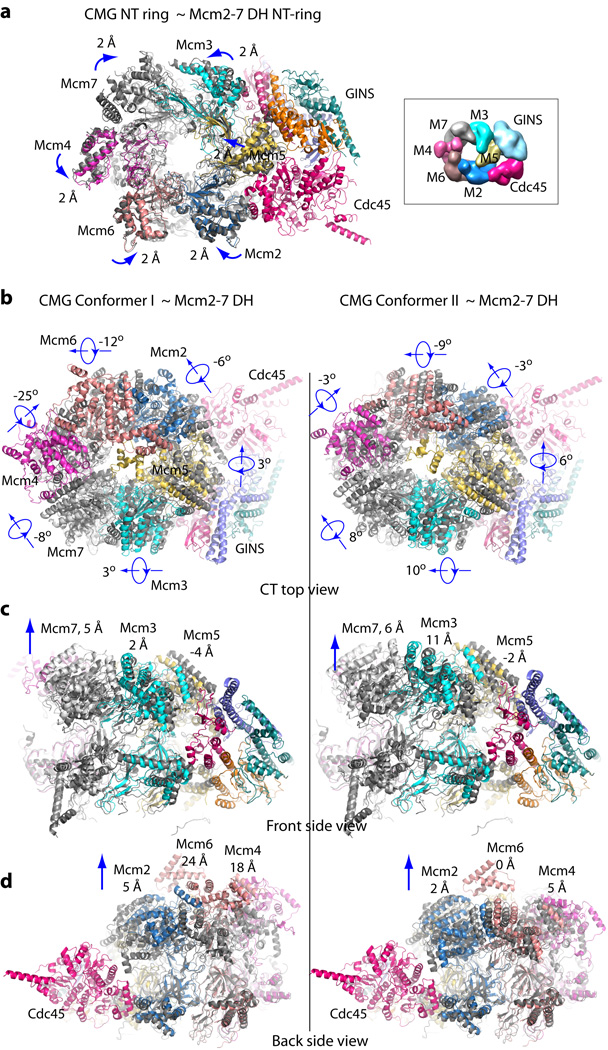
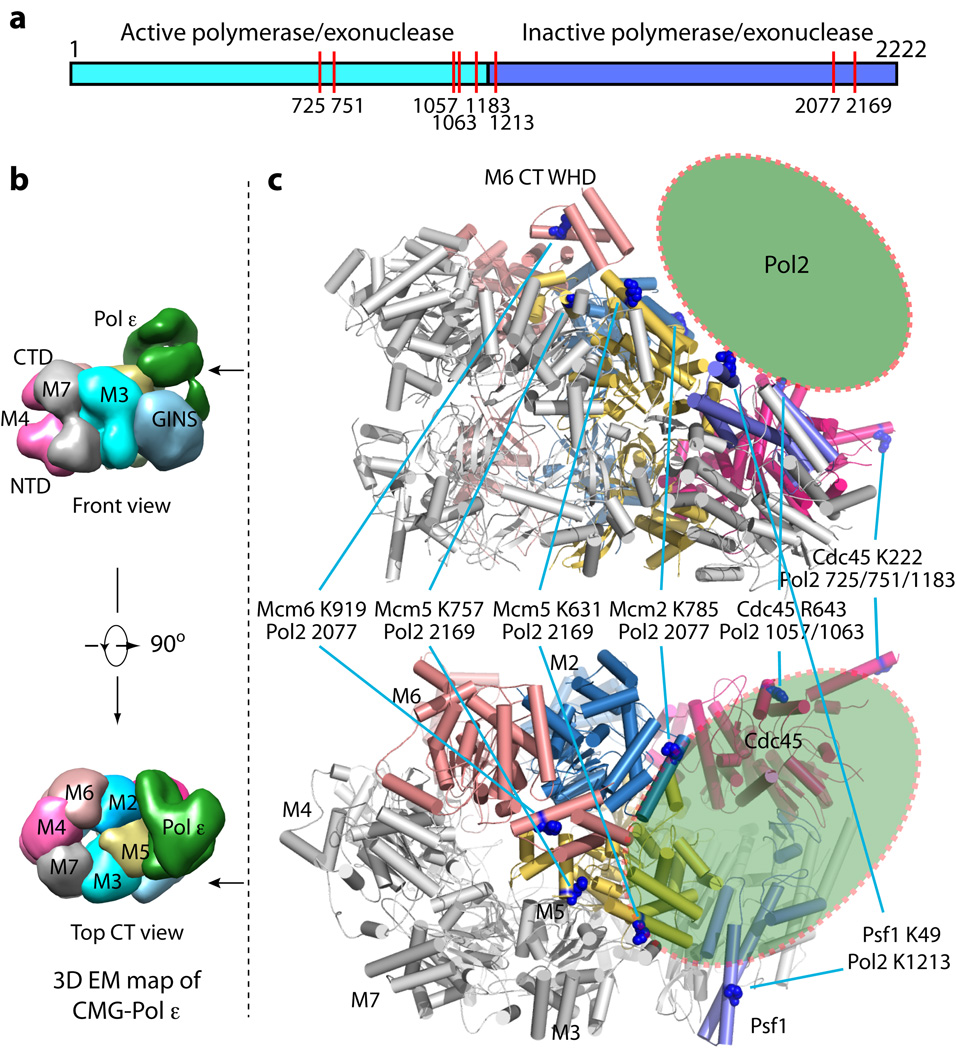
The two distinct conformers of CMG mark a major difference to the single conformation of the inactive Mcm2-7 double-hexamer. Remodeling of the Mcm2-7 ring transiting from the double-hexamer to the two CMG conformers requires numerous domain translations of up to 20 Å and rotations of up to 30° (Fig. 5). The previous study of the CMG-Pol ε structure determined numerous intra-peptide cross-links using a mass spectrometry approach16. The precise structures of the GINS-Cdc45 within CMG now permit mapping crosslinks between CMG and the Pol2 subunit of Pol ε, and revealed a Pol ε footprint on the helicase that further supports the conclusion that Pol ε rides ahead rather than trailing behind the helicase (Fig. 6)16. Two crosslinks between the active N-half of Pol2 and Cdc45 locate to the tip of the protruding helix of Cdc45, suggesting a possible role of Cdc45 in communication between helicase and polymerase activity (Fig. 6).
Figure 5. Remodeling changes between the Mcm2-7 in the double-hexamer (DH) and in the active CMG conformers.
(a) Superimposition of the Mcm2-7 NTD-tier ring of CMG (color cartoon representation) with that of the inactive double-hexamer (gray cartoon representation). The double-hexamer Mcm2-7 structure is (EMDB 6338)13. The direction and distance of rigid body domain movements are noted by each subunit. At the right (boxed) is a surface representation of the NTD view of the previously reported 3D density map of CMG (EMDB 6463)16. (b) Comparison of the top view of the CTD-tier of Mcm2-7 of the double-hexamer with the Mcm2-7 CTD-tier of CMG conformer I (left column) and CMG conformer II (right column), and in: (c) Front side view, and in (d) Back side view. Each CTD AAA+ domain undergoes a combination of rotation and translation, with the degree of rotation shown in (b) and the vertical translation in (c) for Mcm3, Mcm5, Mcm7, and (d) for Mcm2, Mcm4, and Mcm6. Another significant change is the insertion of the Mcm5 WHD into the interior channel in the active helicase, but this is not illustrated here because the deposited PDB coordinates of the inactive Mcm2-7 hexamer do not contain the Mcm5 WHD. The Mcm5 WHD in DH is documented and explained in Extended Data Figure 3d–e13.
Figure 6. Pol2 footprint on the atomic model of CMG helicase.
(a) The two-domain architecture of Pol2, the catalytic subunit of the Pol ε complex. The N-terminal half contains the polymerase and exonuclease activities. The C-terminal half is homologous to a B-family polymerase but without enzymatic activity. The red vertical bars indicate the locations of the residues in Pol2 that crosslink to subunits of CMG16. (b) Surface-rendered front side and top views of the previously reported 3D density map of the CMG-Pol ε complex (EMDB 6465)16. (c) The top and bottom panels are corresponding front side and top views of the atomic model of CMG helicase. The helicase residues that cross-link to Pol2 are shown in blue spheres. The green oval represents the approximate Pol2 position on CMG as predicted from the crosslink footprint.
Nodding pumpjack unwinding model
Constriction of the ~25 Å axial channel in Mcm2-7 to ~10 Å by the Mcm5 WHD restricts the CTD channel to entry of ssDNA (Fig. 3c–d, Fig. 4a), consistent with a steric exclusion mechanism of unwinding and studies in the Xenopus system20. Translocation of homohexameric helicases is proposed to occur in rotary fashion in which an opening between motor domains circularly permutes around the ring with ATP hydrolysis10,17,19. The heterohexamic Mcm2-7 within CMG may function in rotary fashion, but there are compelling data that may suggest otherwise. First, in a rotational model, the opening and vertical displacement observed between CMG conformers I and II upon opening the Mcm2-5 interface would need to occur in a sequentially permuted fashion at each Mcm subunit interface around the ring. However, the GINS interacts with the AAA+ domain of Mcm5 and possibly Mcm3, and this may block rotary propagation of large AAA+ motions. Second, only two ATP sites are necessary for CMG helicase activity (Mcm5-2 and Mcm3-5), while two ATP sites (Mcm 4-7 and Mcm 6-4) can be mutated without observable effect; mutations in the remaining two sites only reduce helicase activity by half6. The two required ATP sites are at the hinge point between the two conformers, and the non-essential sites are the most distant from the hinge. Rotary models generally require all, or most of the ATP sites. Finally, the compact and extended shapes of the two CMG conformers immediately suggest another type of mechanism. We propose a provocative and speculative new “pumpjack” model that is linear instead of rotational. We freely concede that there is little data to back this proposal, and further structural advances (e.g. CMG-DNA structures), biochemical experiments and single-molecule studies are needed to test its applicability to CMG and generality to other helicases. Our intent is that the model helps foster new ideas and future experimentation to test the hypothesis.
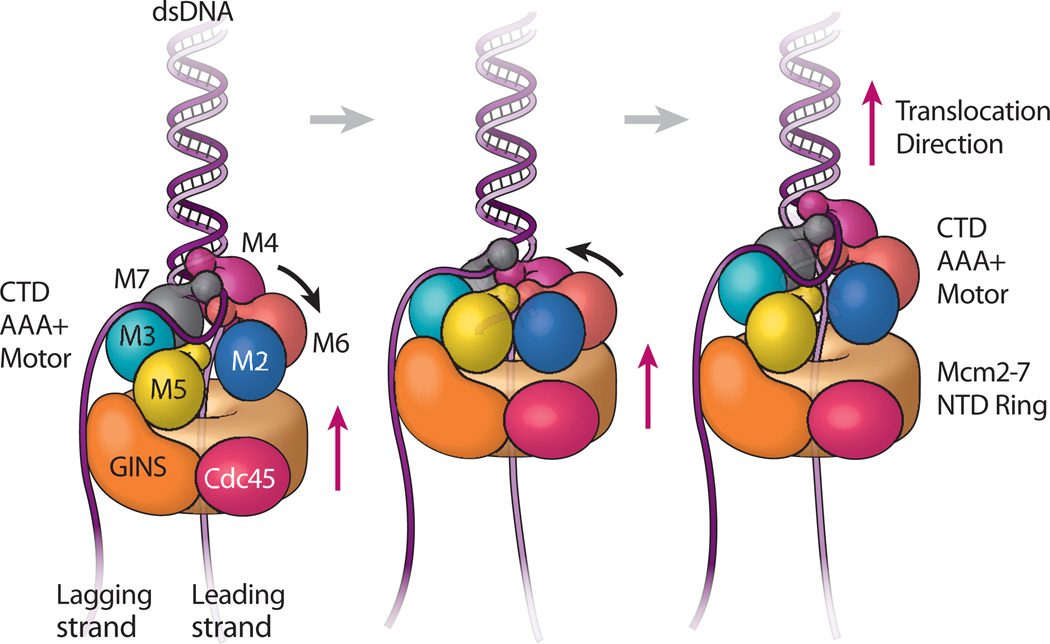
If one alternates between the two conformers, we see a ratcheting, or inchworm type of expansion/contraction process (Supplementary Videos 1 and 2). A more accurate analogy of the movements is the old-fashioned oil rig pumpjack, which looks like a horse head that nods up and down, and sits on top of an immobile platform. In the case of CMG, the Mcm2-7 NTD ring, along with their Cdc45-GINS side brace, appear rigid, like a platform, and the Mcm2-Mcm6-Mcm4 act like the nodding horse head. Because the Mcm5 WHD stays inside the channel in both conformers, it may function as a pin to facilitate strand separation as ssDNA is pulled through the motor domains. Based on these structural features, we propose a nodding pumpjack mechanism of DNA unwinding that proceeds in simple linear fashion along DNA (Supplemental Video 3). In the nodding pumpjack model the binding of ATP brings the interface between Mcm2 and Mcm5 closed (conformer II), and this causes the Mcm2-6-4 horse head to nod, forming a more compact form of CMG (Fig. 7a–to-b). ATP hydrolysis enables CMG to spring open at the Mcm2-Mcm5 interfacial ATP site (conformer I), causing Mcm2 to rotate by 30°, and the subsequent upward movement of the Mcm2-6-4 horse head (Fig. 7b–to-c), thereby readying the helicase for the next nodding cycle. In this scheme, the Cdc45-GINS-Mcm2-7 NTD ring acts as a platform upon which the CTD motor ring strokes up and down, powered by ATP binding and hydrolysis. The pumpjack model is essentially an inchworm-ratchet that has extensive precedent in non-replicative monomeric helicases37–43.
Figure 7. Nodding pumpjack model of CMG translocation.
The leading strand (light purple) goes through the Mcm2-7 ring while the lagging strand (dark purple) is excluded to the outside. The Mcm2-7 is composed of the CTD AAA+ motor ring (subunits labeled and colored), and the NTD ring (drawn as a solid tan cylinder). The Mcm NTD ring, Cdc45 and GINS form a stable platform upon which the Mcm2-7 CTD AAA+ motor ring moves up and down. The small knobs attached to Mcm AAA+ domains are the WHDs. The Mcm5 WHD plugs the central channel, preventing dsDNA entry and reinforcing the steric exclusion mode. Left illustration: CMG conformer I is the expanded form. Middle illustration: ATP binding results in nodding of the CTD ring, forming the compact conformer II, which results in bringing the NTD, GINS, Cdc45 platform upward. Right illustration: ATP hydrolysis leads back to the expanded conformer I which translocates the CTD motor domain ring upward. Although all six AAA+ domains move during the DNA translocation cycle, the major movement is by Mcm2-6-4, as revealed by the two conformers captured by cryo-EM. Repeated cycles of ATP hydrolysis result in continued CMG translocation along the leading strand.
Discussion
An enzyme is currently viewed as a population of conformations, with individual proteins rapidly sampling many conformations, each with a role in catalysis44–46. Therefore, we expect the two conformers of apo CMG are relevant to function, and that ATP binding and hydrolysis act to direct them, as shown previously14,47. The two CMG conformers display a large change in distance between the CTD and NTD, and thus the axial channel for DNA becomes shorter or longer by alternating between the two forms. The simplest interpretation is that ATP drives successive cycles between the compact and extended forms, resulting in linear translocation (Supplemental Video 3).
Directional translocation along DNA can be produced by a protein that binds DNA at two sites and change distance in two conformers that alternate with ATP hydrolysis. This would enable a hand-over-hand action like a person climbing a rope. The DNA binding sites in papillomavirus E1 are six equivalent interior loops in the motor domains that are spirally arranged, and the two conformers in the E1 crystal suggest that they move in rotary fashion to escort ssDNA through the central channel10,11. Bacterial DnaB helicase is shaped as a spiral, and each subunit binds DNA via interior loops in the six motor domains; it is proposed that whole protomers move along DNA in a rotary staircase process19. During rotary motion in E1 and DnaB, there is a gap between motor domains at the top and bottom of the spiral, and with each ATP hydrolysis event the DNA binding element at the bottom of the spiral leapfrogs the sites in the other subunits to assume a new position at the top of the spiral10,19. This leapfrog motion permutes the gap sequentially around the ring with ATP hydrolysis. The Rho, T7gp4, SV40 large T-antigen, and the 19S proteasomal cap are hexameric ATPases that have also been discussed in terms of rotational processes17.
It is reasonable to propose that a gap between two motor domains of a circular homohexamer rotationally permute around the ring. But CMG is asymmetric, and while it may still function by a rotational model, there are compelling reasons to propose a linear model instead (discussed earlier). Furthermore, both E1 and DnaB have a gap between two motor domains that is proposed to remain open as it permutes around the ring without closing. In contrast one CMG conformer is a closed ring with no gap, and the other has a Mcm2-5 gap. It is possible that the open conformer is only used for origin activation, but since the Mcm2-5 gap is sealed in CMG by the GINS-Cdc45-NTD ring, strand passage cannot occur and suggests the conformer is an intermediate in helicase action. It also seems reasonable that other interfaces of CMG may undergo open-closed transitions.
Translocation by a linear pumpjack ratchet only needs two DNA binding sites. Therefore we propose only two DNA sites in CMG, although additional sites could be involved. One DNA site in CMG is probably the same as established in homohexameric helicases, formed by the six DNA binding loops in the AAA+ regions of the CTD motor domains8–10. DNA binding loops in AAA+ domains were previously established in AAA+ clamp loader heteropentamers and we propose that they function as an amalgam of one DNA binding site in CMG, as in the clamp loaders48–50. We propose the second DNA binding site is in the NTD, formed by the OB folds that are well established DNA binding elements in an archaeal Mcm NTD, and are required for yeast Mcm2-7 function in vivo25. Bacterial Rho also binds ssRNA using six OB folds located in the NTD51. Despite this intelligence, other elements may comprise the DNA binding sites in CMG, for example the WHDs of Mcms 3,4,6,7. Clearly, additional structural information of CMG bound to DNA is needed to identify possible DNA sites for structure directed mutagenesis.
Translocation requires that DNA binding sites not only change distance, but also switch affinity for DNA during an ATP hydrolysis cycle. The CTD DNA binding site in the ATPase motor domains seems the most likely site to change affinity in response to the ATP state. In its most simple form, the CTD-DNA binding affinities would alternate between tight and loose relative to an intermediate fixed affinity NTD-DNA binding site. For example, in the ATP state, tight CTD binding relative to the NTD site would release DNA from the NTD faster than the CTD. In the ADP or apo state, weak CTD binding relative to the NTD site would release DNA from the CTD faster than the NTD. This would result in translocation along DNA with cycles of ATP hydrolysis. Reverse polarity of tracking could result from binding ssDNA in the opposite orientation, or by reversing the relative affinity of ATP vs ADP states in the CTD.
Why is CMG circular if it doesn’t act in rotary fashion? In fact, many helicases (non replicative) are monomers or dimers, and thus helicase action per se does not require circularity37–42. Perhaps replicative helicases are ring shaped for processivity, as proposed earlier10. It should be noted that results to date do not specifically exclude a linear pumpjack model for the homohexameric helicases. In fact, hexameric archaeal helicases can function when they are “doped” with up to 3 mutated subunits52, suggesting a non-sequential, and possibly a non-rotational mechanism. In summary, the CMG structure makes unexpected and specific predictions about its mechanism, possibly extending to other replicative helicases and provides a foundation for further dissection of the eukaryotic replisome.
ONLINE METHODS
Sample preparation and electron microscopy
Saccharomyces cerevisiae CMG was purified as previously described16. To prepare EM grids, we first diluted each sample with 20 mM Tris-Acetate (pH 7.5), 40 mM K-Glutamate, 2 mM DTT and 0.1 mM EDTA. Before EM grid preparation of CMG, we checked the sample homogeneity by negative-stain electron microscopy. Then we applied 3 µl of CMG sample at a final concentration of 0.6 mg/ml to glow-discharged C-flat 1.2/1/3 holey carbon grids, incubated for 10 s at 6 °C and 90% humidity, blotted for 3 s then plunged into liquid ethane using an FEI Vitrobot IV. We loaded the grids into an FEI Titian Krios electron microscope at 300 keV and collected images automatically using low-dose mode at a magnification of ×29,000 and a pixel size of 1.01 Å per pixel. A Gatan K2 summit direct electron detector was used for image recording with a defocus range from 1.5 to 3.5 µ under super resolution mode. The dose rate was 10 electrons per Å2 per second and total exposure time was 5 seconds. The total dose was divided into a 25-frame movie and each frame was exposed for 0.2 s.
Image processing and 3D reconstruction
Approximately 8000 raw movie micrographs were collected. The movie frames were first aligned and superimposed by the program motioncorr53. Contrast transfer function parameters of each aligned micrograph were calculated using the program CTFFIND454. All the remaining steps, including particle autoselection, 2D classification, 3D classification, 3D refinement, and density map post-processing were performed using Relion-1.3 or Relion-1.455,56. We manually picked ~10,000 particles from different views to generate 2D averages, which were used as templates for subsequent automatic particle selection. Automatic particle selection was then performed for the entire data set. About one million particles were initially selected. Particles were then sorted by similarity to the 2D references; the 10% of particles with the lowest z-scores were deleted from the particle pool. 2D classification of all remaining particles was performed and particles in unrecognizable classes by visual inspection were removed. A total of 687,794 particles were used for 3D classification. We derived six 3D models from the dataset, and found three models were similar to each other and their associated particles were combined for further refinement; the other three models were distorted and those particles were discarded, leading to a dataset size of 469,818 particles. This final dataset was used for further 3D refinement, resulting in a 3.8 Å 3D density map. The resolution of the map was estimated by the so-called gold-standard Fourier shell correlation, at the correlation cutoff value of 0.143. The 3D density maps were corrected for the detector modulation transfer function and sharpened by applying a negative B-factor of −144 Å2. The particles had some preference for end-on views but because of the large number of particles used virtually all of the angular space was well sampled. In order to improve the density of the flexible C-terminal AAA+ ring of Mcm 2-7, we performed a “focused” 3D classification procedure. We first generated two soft-edge masks, one from the Mcm2-7 CTD ring region and the other from the entire CMG complex. Subtraction of the Mcm2-7 CTD ring mask from the entire CMG complex mask was performed using relion_image_handler from which we obtained a mask that only covered the Mcm2-7 NTD ring, GINS, and Cdc45. This mask was then applied to the 3.8 Å density map. The resulting masked map without the Mcm 2-7 CTD ring was used to subtract that region from the original particles, and 3D classification was performed on the Mcm 2-7 CTD ring only particles without further image alignment. Two distinct conformations were found: the CTD ring of conformer I was “tilted” compared to the NTD ring, while conformer II had roughly “parallel” CTD and NTD rings. These two maps were further refined resulting in two 3D maps with an estimated resolution of 4.7 Å and 4.8 Å, respectively. Local resolution estimation was calculated using ResMap.
Model building, refinement, and validation
The initial model of S. cerevisiae Cdc45 was built by Rosetta based on the extracted density map, using homolog RecJ as a reference35,57. The fold of this model was essentially the same as the crystal structure of human Cdc45 (personal communication, Luca Pellegrini, University of Cambridge). The initial models of the S. cerevisiae GINS subunits were generated from the crystal structure of the human GINS complex (PDB ID: 2Q9Q)22 using the SWISS-MODEL server58. Six NTDs of the yeast Mcm2-7 were directly extracted from the cryo-EM structure of the yeast Mcm2-7 double hexamer (PDB code 3JA8)13. These models were docked into the 3.7-Å resolution 3D density map of Cdc45-GINS-Mcm2-7 NTD-tier ring in COOT59, and fitted into the density using Chimera60. After assignment of the models, remaining density corresponded to the C-terminal B-domain of Psf1, which was absent from the crystal structure. We manually built this domain de novo from a poly-Ala model. Then the entire atomic model was manually adjusted and built in COOT. Clearly resolved bulky residues such as Phe, Tyr, Trp, and Arg were crucial for sequence registration.
The manually built structure was then refined in real space by phenix.real_space_refine61 and manually adjusted in COOT. The reciprocal space refinement was also performed by Phenix62, with secondary structure and stereochemical constraints applied. The structure factors (including phases) were obtained from the Fourier transform of the experimental density map by phenix.map_to_structure_factors. The atomic models were validated by MolProbity with a percentile score of 95-97%.
The final model was cross validated using a method described previously63. We randomly added 0.1 Å noise to the coordinates of the final model using the PDB tools in Phenix, then refined the noise-added against the first half map (Half1) that was produced from one half of the particle dataset during refinement by RELION. We performed one round of coordinate refinement, followed by a B-factor refinement. The refined model was then correlated with the 3D maps of the two half maps (Half1, Half2) in Fourier space to produce two FSC curves: FSCwork (model versus Half1 map) and FSCfree (model versus Half2 map), respectively. A third FSC curve was calculated between the refined model and the final 3.7-Å resolution density map produced from all particles. The general agreement of these curves was taken as an indication that the model was not over-fitted.
To build the complete models for CMG conformers I (4.8 Å) and II (4.7 Å), we first used the SWISS-MODEL server to derive two WHD models of MCM5 and MCM6, based on the crystal structure of a GntR family transcriptional regulator (PDB code 3C7J) and NMR structure of WHD of the human MCM6 (PDB code 2KLQ)64, respectively. We then fitted them along with the above-built model of Cdc45-GINS-Mcm2-7 N-tier ring, and the six CT AAA+ domain structure extracted from the cryo-EM structure of yeast Mcm2-7 (PDB code 3JA8), in COOT and Chimera. The structures were then refined in Phenix and manually adjusted in COOT. All modeled structures were validated using MolProbity65. Structural figures were prepared in Chimera and Pymol (https://www.pymol.org).
Supplementary Material
Acknowledgments
Cryo-EM data was collected on Titan Krios I at HHMI Janelia Farm. We also collected a cryo-EM dataset on an FEI Polara with a K2 detector at the University of Texas Health Science Center. We thank the staff at these facilities for help with data collection. We also thank Luca Pellegrini (University of Cambridge) for sharing the human Cdc45 structure before publication. This work was funded by the US National Institutes of Health (GM111472 and OD12272 to H.L. and GM115809 to M.O.D.) and Howard Hughes Medical Institute (M.O.D.).
Footnotes
Accession Codes The 3D cryo-EM maps of CMG at 3.7 Å, 4.8 Å, and 4.7 Å resolution have been deposited at the EMDB database with accession codes EMD-6534, EMD-6536, EMD-6535 respectively. The corresponding atomic models were deposited at RCSB PDB bank with accession codes 3JC6, 3JC7, 3JC5, respectively.
Author Contributions Y.Z, L.B., J.S., R.G., M.O.D. and H.L. designed experiments. Z.Y., L.B., J.S., R.G., and J. L. performed experiments. Z.Y., L.B., J.S., H.L. analyzed the data. M.O.D. and H.L. wrote the manuscript.
REFERENCES
- 1.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botchan M, Berger J. DNA replication: making two forks from one prereplication complex. Molecular Cell. 2010;40:860–861. doi: 10.1016/j.molcel.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015 doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarova KS, Koonin EV, Kelman Z. The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biol Direct. 2012;7:7. doi: 10.1186/1745-6150-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slaymaker IM, Chen XS. MCM structure and mechanics: what we have learned from archaeal MCM. Subcell Biochem. 2012;62:89–111. doi: 10.1007/978-94-007-4572-8_6. [DOI] [PubMed] [Google Scholar]
- 9.Brewster AS, et al. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci U S A. 2008;105:20191–20196. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 11.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 12.Miller JM, Arachea BT, Epling LB, Enemark EJ. Analysis of the crystal structure of an active MCM hexamer. Elife. 2014;3:e03433. doi: 10.7554/eLife.03433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li N, et al. Structure of the eukaryotic MCM complex at 3.8 A. Nature. 2015;524:186–191. doi: 10.1038/nature14685. [DOI] [PubMed] [Google Scholar]
- 14.Costa A, et al. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. Elife. 2014:e03273. doi: 10.7554/eLife.03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa A, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J, et al. The architecture of a eukaryotic replisome. Nat Struct Mol Biol. 2015 doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Struct Biol. 2011;21:240–248. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bochman ML, Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell. 2012;151:267–277. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu YV, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen B, Frank J. Two promising future developments of cryo-EM: capturing short-lived states and mapping a continuum of states of a macromolecule. Microscopy (Oxf) 2015 doi: 10.1093/jmicro/dfv344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang YP, Wang G, Bermudez V, Hurwitz J, Chen XS. Crystal structure of the GINS complex and functional insights into its role in DNA replication. Proc Natl Acad Sci U S A. 2007;104:12685–12690. doi: 10.1073/pnas.0705558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi JM, Lim HS, Kim JJ, Song OK, Cho Y. Crystal structure of the human GINS complex. Genes Dev. 2007;21:1316–1321. doi: 10.1101/gad.1548107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamada K, Kubota Y, Arata T, Shindo Y, Hanaoka F. Structure of the human GINS complex and its assembly and functional interface in replication initiation. Nat Struct Mol Biol. 2007;14:388–396. doi: 10.1038/nsmb1231. [DOI] [PubMed] [Google Scholar]
- 25.Froelich CA, Kang S, Epling LB, Bell SP, Enemark EJ. A conserved MCM single-stranded DNA binding element is essential for replication initiation. Elife. 2014;3:e01993. doi: 10.7554/eLife.01993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun J, et al. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol. 2013;20:944–951. doi: 10.1038/nsmb.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samel SA, et al. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev. 2014;28:1653–1666. doi: 10.1101/gad.242404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petojevic T, et al. Cdc45 (cell division cycle protein 45) guards the gate of the Eukaryote Replisome helicase stabilizing leading strand engagement. Proc Natl Acad Sci U S A. 2015;112:E249–E258. doi: 10.1073/pnas.1422003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothenberg E, Trakselis MA, Bell SD, Ha T. MCM forked substrate specificity involves dynamic interaction with the 5'-tail. J Biol Chem. 2007;282:34229–34234. doi: 10.1074/jbc.M706300200. [DOI] [PubMed] [Google Scholar]
- 30.Simon AC, et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature. 2014;510:293–297. doi: 10.1038/nature13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onesti S, Macneill SA. Structure and evolutionary origins of the CMG complex. Chromosoma. 2013 doi: 10.1007/s00412-013-0397-x. [DOI] [PubMed] [Google Scholar]
- 32.Krastanova I, et al. Structural and functional insights into the DNA replication factor Cdc45 reveal an evolutionary relationship to the DHH family of phosphoesterases. J Biol Chem. 2012;287:4121–4128. doi: 10.1074/jbc.M111.285395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez-Pulido L, Ponting CP. Cdc45: the missing RecJ ortholog in eukaryotes? Bioinformatics. 2011;27:1885–1888. doi: 10.1093/bioinformatics/btr332. [DOI] [PubMed] [Google Scholar]
- 34.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 35.Wakamatsu T, et al. Structure of RecJ exonuclease defines its specificity for single-stranded DNA. J Biol Chem. 2010;285:9762–9769. doi: 10.1074/jbc.M109.096487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamagata A, Kakuta Y, Masui R, Fukuyama K. The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity. Proc Natl Acad Sci U S A. 2002;99:5908–5912. doi: 10.1073/pnas.092547099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J Biol Chem. 2007;282:27076–27085. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- 38.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 39.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rad B, Kowalczykowski SC. Efficient coupling of ATP hydrolysis to translocation by RecQ helicase. Proc Natl Acad Sci U S A. 2012;109:1443–1448. doi: 10.1073/pnas.1119952109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bjornson KP, Wong I, Lohman TM. ATP hydrolysis stimulates binding and release of single stranded DNA from alternating subunits of the dimeric E. coli Rep helicase: implications for ATP-driven helicase translocation. J Mol Biol. 1996;263:411–422. doi: 10.1006/jmbi.1996.0585. [DOI] [PubMed] [Google Scholar]
- 43.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 44.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 45.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 46.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 47.Lyubimov AY, Costa A, Bleichert F, Botchan MR, Berger JM. ATP-dependent conformational dynamics underlie the functional asymmetry of the replicative helicase from a minimalist eukaryote. Proc Natl Acad Sci U S A. 2012;109:11999–12004. doi: 10.1073/pnas.1209406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowman GD, O'Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 49.Simonetta KR, et al. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell. 2009;137:659–771. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelch BA, Makino DL, O'Donnell M, Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011;334:1675–1680. doi: 10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skordalakes E, Berger JM. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 52.Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
References for Online Methods
- 53.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheres SH. Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol. 2015;189:114–122. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang RY, et al. De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nat Methods. 2015;12:335–338. doi: 10.1038/nmeth.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 60.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 61.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei Z, et al. Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6. J Biol Chem. 2010;285:12469–12473. doi: 10.1074/jbc.C109.094599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.