Abstract
The Sox transcription factor family is characterized with the presence of a Sry-related high-mobility group (HMG) box and plays important roles in various biological processes in animals, including sex determination and differentiation, and the development of multiple organs. In this study, 27 Sox genes were identified in the genome of the Nile tilapia (Oreochromis niloticus), and were classified into seven groups. The members of each group of the tilapia Sox genes exhibited a relatively conserved exon-intron structure. Comparative analysis showed that the Sox gene family has undergone an expansion in tilapia and other teleost fishes following their whole genome duplication, and group K only exists in teleosts. Transcriptome-based analysis demonstrated that most of the tilapia Sox genes presented stage-specific and/or sex-dimorphic expressions during gonadal development, and six of the group B Sox genes were specifically expressed in the adult brain. Our results provide a better understanding of gene structure and spatio-temporal expression of the Sox gene family in tilapia, and will be useful for further deciphering the roles of the Sox genes during sex determination and gonadal development in teleosts.
Keywords: Nile tilapia, Sox gene, genomic structure, transcriptome, gene expression
1. Introduction
Sox transcriptional factors are characterized as Sry-related high-mobility group (HMG) box proteins in metazoans. With the availability of whole genome sequence, genome-wide characterization of Sox genes has been performed in several animals [1,2,3,4,5], and in total more than 40 members of the Sox family have been identified. Based on the sequences of both DNA and proteins, Sox gene family is currently divided into 11 groups from A to K [2,5,6]. To date, Sox genes have been reported to be involved in not only sex determination and differentiation [7,8,9,10,11], but also the formation of multiple organs, including neuronal system [12,13,14,15], gonad [16,17,18], eye [19,20], pancreas [21,22,23], and cartilage [14,24,25].
Previous reports revealed that the numbers of Sox genes greatly varied in animals, namely five in the nematode (Caenorhabditis elegans) [26], seven in the calcareous sponge (Sycon ciliatum) [1], seven in the sea lamprey (Petromyzon marinus) [4], seven in the sea squirt (Ciona intestinalis) [27,28], eight in the fruit fly (Drosophila melanogaster) [3], nine in the silkworm (Bombyx mori) [29], 11 in the sea urchin (Strongylocentrotus purpuratus) [30], 14 in the cnidarians (Nematostella vectensis) [31], and 20 in the mouse (Mus musculus) and human (Homo sapiens) [32]. In teleosts, it has been reported that there are 19 in the medaka (Oryzias latipe) [2], and 24 Sox genes in the pufferfish (Fugu rubripes) [5]. Recently, new versions of genome sequences of the Nile tilapia (Oreochromis niloticus), zebrafish (Danio rerio), and common carp (Cyprinus carpio) have been published [33,34,35]. A genome-wide comparative analysis of the Sox gene family between the tilapia and other animals including other teleost fishes will be helpful for deciphering the evolutionary process of this gene family.
Previous studies have investigated the potential roles of Sox genes in the growth and development of the teleost fishes. For example, several members of the medaka Sox family exhibit differential expressions during embryonic development and may play a variety of roles in embryogenesis [2]. Importantly, the medaka Sox9b has been shown to be indispensible for the proper proliferation and survival of germ cells in gonads [36]. In addition, evidence from the zebrafish suggests that Sox7 and Sox18 play redundant roles in both arteriovenous specification and vascular development [37,38], and Sox21a functions as a transcriptional repressor in dorso-ventral patterning during embryonic development [39]. Moreover, only three Sox genes, namely, Sox2, Sox14, and Sox30, have been studied in the tilapia [40,41], and Sox30 has been confirmed to be specifically expressed in gonads [41]. Recently, the transcripomes of multiple adult tissues and different stages of gonadal development in the tilapia have been examined via RNA-Seq method [33,42]. This enables us to carry out transcriptome-based expression profiling of the tilapia Sox genes and to obtain more functional evidence for the Sox genes in teleosts.
In this study, based on the genome sequence and transcriptome data of the tilapia and other animals, we performed a genome-wide identification and evolutionary analysis of the tilapia Sox gene family, and further profiled their spatio-temporal expressions. Our goal is to provide new insight into the evolution and functions of the Sox genes in teleosts.
2. Results
2.1. Identification of the Sox Genes in the Tilapia Genome
We used the amino acids sequence of conserved HMG-box domain of Sox transcription factors as query to search against the tilapia genome by a basic local alignment search tool (BLAST). As a result, a total of 27 Sox genes, including three previously identified Sox genes, namely Sox2, Sox14, and Sox30, were identified in the tilapia genome (Table 1). All the tilapia Sox genes could be classified into seven subfamilies, namely, eight members in group B (including five in B1 subgroup and three in B2 subgroup), four in group C, four in group D, six in group E, three in group F, one in group H, and one in group K (Table 1). Interestingly, each of the eight members of the ancestral vertebrate Sox genes, namely, Sox1, Sox4, Sox6, Sox8, Sox9, Sox10, Sox11, and Sox14, has two copies in the tilapia genome, indicating that these Sox genes experienced a duplication during the evolution of the tilapia.
Table 1.
Inventory of Sox genes in the tilapia genome.
| Group | Gene | NCBI ID | Linkage Group | Position | Intron Number | Length (aa) | HMG-Box Position |
|---|---|---|---|---|---|---|---|
| B1 | Sox1a | XP_005457843.1 | 16–21 | 12,271,122–12,353,293 | 0 | 344 | 35–109 |
| Sox1b | XP_005450142.1 | unknown | 7,601,713–7,604,245 | 0 | 354 | 41–115 | |
| Sox2 | XP_003457401.1 | 17 | 280,838–283,149 | 0 | 322 | 38–112 | |
| Sox3 | XP_005467513.1 | 2 | 13,569,507–13,587,072 | 0 | 300 | 33–107 | |
| Sox19 | XP_003458966.1 | 3 | 282,331–286,532 | 1 | 307 | 57–131 | |
| B2 | Sox14a | XP_005451233.1 | 23 | 7,181,057–7,183,304 | 0 | 238 | 6–80 |
| Sox14b | XP_003438076.1 | 18 | 5,299,795–5,300,529 | 0 | 241 | 6–80 | |
| Sox21 | XP_003447333.1 | 16–21 | 14,804,010–14,806,286 | 0 | 248 | 6–80 | |
| C | Sox4a | XP_005450837.1 | 22 | 19,810,503–19,814,320 | 0 | 371 | 55–129 |
| Sox4b | XP_005450601.1 | 11 | 10,635,379–10,640,355 | 0 | 414 | 65–139 | |
| Sox11a | AAR01937.1 | 19 | 1,462,641–1,463,732 | 0 | 363 | 44–118 | |
| Sox11b | XP_005452537.1 | 15 | 7,943,145–7,945,227 | 0 | 433 | 48–122 | |
| D | Sox5 | XP_005451840.1 | 17 | 29,053,787–29,297,905 | 15 | 773 | 566–640 |
| Sox6a | XP_003442343.2 | 1 | 665,105–766,767 | 14 | 778 | 568–646 | |
| Sox6b | XP_005460016.1 | 7 | 667,787–732,752 | 17 | 838 | 613–691 | |
| Sox13 | XP_005450158.1 | 5 | 6,796,676–6,846,010 | 15 | 664 | 461–535 | |
| E | Sox8a | XP_003438722.1 | 8–24 | 6,051,795–6,054,972 | 2 | 479 | 96–170 |
| Sox8b | XP_003450163.1 | unknown | 1,216,205–1,219,881 | 2 | 464 | 98–172 | |
| Sox9a | XP_005448042.1 | 8–24 | 5,686,065–5,688,370 | 2 | 500 | 105–179 | |
| Sox9b | XP_003450167.1 | unknown | 1,462,602–1,466,515 | 2 | 484 | 102–176 | |
| Sox10a | XP_003447925.1 | 6 | 19,531,528–19,536,616 | 2 | 500 | 107–181 | |
| Sox10b | XP_005468484.1 | 4 | 7,506,891–7,515,424 | 2 | 503 | 105–179 | |
| F | Sox7 | XP_005460371.1 | 15 | 17,570,516–17,573,991 | 1 | 407 | 41–115 |
| Sox17 | XP_003439290.1 | 9 | 14,405,231–14,407,376 | 1 | 397 | 63–137 | |
| Sox18 | XP_003459774.1 | unknown | 18,060–22,872 | 2 | 565 | 90–164 | |
| H | Sox30 | XP_003447014.1 | unknown | 3,119,307–3,123,369 | 4 | 353 | 122–196 |
| K | Sox32 | XP_003439409.1 | 9 | 3,127,363–3,128,440 | 1 | 310 | 35–109 |
2.2. Genomic Structure of the Tilapia Sox Genes
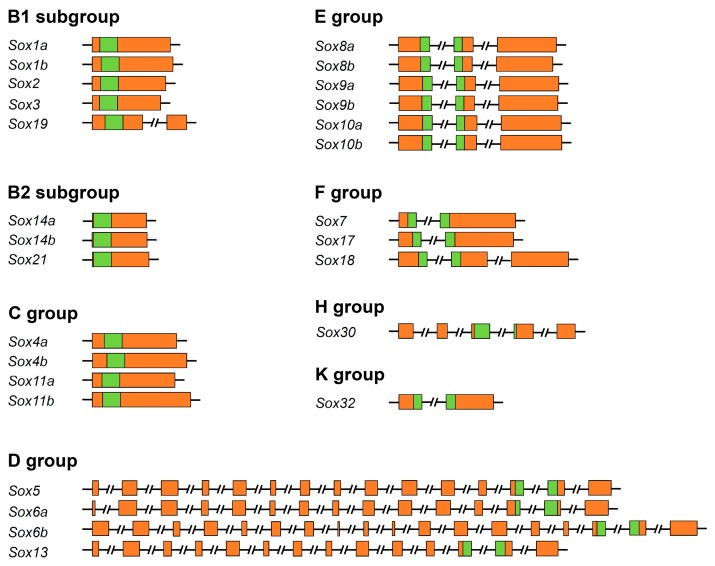
The exon–intron structure of the tilapia Sox genes was further characterized. The results showed that the numbers of intron in each Sox gene varied from zero to 17 (Figure 1 and Table 1). No intron was found in 11 of the tilapia Sox genes, namely, Sox1a, Sox1b, Sox2, Sox3, Sox4a, Sox4b, Sox11a, Sox11b, Sox14a, Sox14b, and Sox21. Interestingly, we noted that the Sox genes from the same subfamily generally contained similar, even same intron number (Figure 1). For example, all Sox genes in group B (including subgroups B1 and B2) had no intron, except for Sox19. Two introns were found in Sox genes of the group E. More than 14 introns were present in all Sox genes that belong to group D. Notably, the HMG boxes in the Sox genes from groups D, E, F, H, and K contained only one intron.
Figure 1.
Exon–intron structure of the tilapia Sox genes. Rectangle and line with double slash indicate exon and intron, respectively. The HMG-box domain regions and the rest regions of the exons are highlighted with green and brown, respectively.
The amino acid sequences of the HMG boxes of the tilapia Sox proteins were aligned. As shown in Figure 2, the core motif of RPMNAFMVW (in the position of 5–13) in the HMG boxes of the tilapia Sox proteins, which is responsible for recognizing and binding cis-regulatory elements in the promoter of their target genes, is highly conserved. Especially, these motifs are the same among the tilapia Sox proteins, except for Sox30 and Sox32.
Figure 2.
Multiple alignment of HMG-box domain sequences of the tilapia Sox proteins. The HMG-box domain of each Sox protein was predicted online using SMART program (http://smart.embl-heidelberg.de). ClustalX program was used to carry out a multiple alignment of amino acid sequences of the HMG-box domain of all the tilapia Sox proteins.
2.3. Comparison of the Sox Genes Among the Tilapia and Other Animals
Given that the tilapia has undergone three rounds of whole genome duplication [43], and the whole genome duplication (WGD) can drive the expansion of gene families [44], we surveyed the number changes of the Sox gene members among the tilapia and other analyzed animals with different rounds of genome duplication, from first round (1R) to fourth round (4R). For a comprehensive comparison, we newly identified 49 Sox genes in the common carp genome, and updated the number of the Sox genes as 27 in the zebrafish (including four newly identified Sox genes, namely Sox12, Sox13, Sox14a, and Sox14b), 25 in the pufferfish (Sox32 was newly identified in this study), and 10 in the Florida lancelet (Branchiostoma floridae) (Table 2 and Table S1). Group K and group G only existed in teleost fishes and human, respectively. These results, together with the previous reports on the genome-wide identification of the Sox genes in other analyzed animals (Table 2 and Table S1), revealed that the number of the Sox genes have undergone an expansion following genome duplication in the teleost fishes, and this expansion of the Sox gene family mainly occurred in the groups of B, C, E, and K.
Table 2.
Number variation of Sox genes in the Nile tilapia and the other surveyed animals.
| Group | Common Carp (4R) | Nile Tilapia (3R) | Zebrafish (3R) | Pufferfish (3R) | Medaka (3R) | Human (2R) | Western Clawed (2R) | Chicken (2R) | Florida Lancelet (1R) | Fruit Fly |
|---|---|---|---|---|---|---|---|---|---|---|
| A | - | - | - | - | - | 1 | - | - | - | |
| B1 | 12 | 5 | 6 | 5 | 3 | 3 | 3 | 3 | 3 | 1 |
| B2 | 8 | 3 | 4 | 3 | 2 | 2 | 2 | 2 | 2 | 3 |
| C | 8 | 4 | 5 | 3 | 2 | 3 | 2 | 3 | 1 | 1 |
| D | 3 | 4 | 3 | 4 | 4 | 3 | 3 | 3 | 1 | 1 |
| E | 10 | 6 | 5 | 6 | 4 | 3 | 3 | 3 | 1 | 1 |
| F | 6 | 3 | 3 | 3 | 3 | 3 | 4 | 3 | 1 | 1 |
| G | - | - | - | - | - | 1 | - | - | - | - |
| H | - | 1 | - | - | - | 1 | - | 1 | 1 | - |
| I | - | - | - | - | - | - | 1 | - | - | - |
| J | - | - | - | - | - | - | - | - | - | - |
| K | 2 | 1 | 1 | 1 | 1 | - | - | - | - | - |
| Total | 49 | 27 | 27 | 25 | 19 | 20 | 18 | 18 | 10 | 8 |
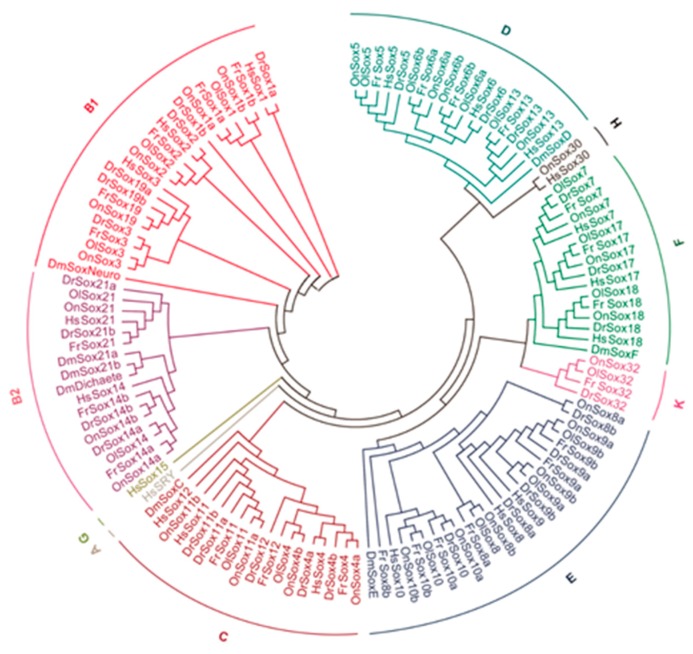
We further used amino acid sequences of the HMG-boxes of the Sox proteins to build phylogenetic tree of the Sox genes among the tilapia and other five selected animals, including zebrafish, pufferfish, medaka, human, and fruit fly. The result showed that all Sox genes were grouped into nine groups, including A, B (B1 and B2), C, D, E, F, G, H, and K (Figure 3 and Supplementary Figure S1). Notably, although group K is very close to group F, given that the Sox32 gene from group K only existed in teleosts and previous report has assigned the Sox32 gene of the teleost medaka to group K [2], we thus considered all Sox32 genes from teleosts as group K. Intriguingly, the phylogenetic tree, together with the number variation, revealed that several ancestral Sox genes have undergone duplication to form two copies in teleost fishes, including eight members in the tilapia (i.e., Sox1, Sox4, Sox6, Sox8, Sox9, Sox10, Sox11, and Sox14), six in the zebrafish (i.e., Sox1, Sox4, Sox9, Sox11, Sox19, and Sox21), six in the pufferfish (including Sox1, Sox6, Sox8, Sox9, Sox10, and Sox14), and two in the medaka (i.e., Sox6 and Sox9). The duplication of Sox9 gene occurred in all teleost fishes. In addition, one duplicate of each ancestral Sox gene in the tilapia firstly grouped well together with its orthologs in other fishes, then with the groups containing another duplicate. This indicates that the duplication of these Sox genes may have occurred prior to the radiation of teleosts but after the separation of the teleosts from other vertebrates.
Figure 3.
Phylogenetic tree of the Sox proteins from the tilapia and the other animals. The amino acid sequences of the HMG-box domain of the Sox proteins were used to build a neighbor-joining phylogenetic tree of the Sox genes by using MEGA 6.0 program. The sources of the sequences were described in the Materials and Methods Section. On: Oreochromis niloticus; Ol: Oryzias latipes; Dr: Danio rerio; Fr: Fugu rubripes; Hs: Homo sapiens; Dm: Drosophila melanogaster.
2.4. Spatial Expression of the Tilapia Sox Genes
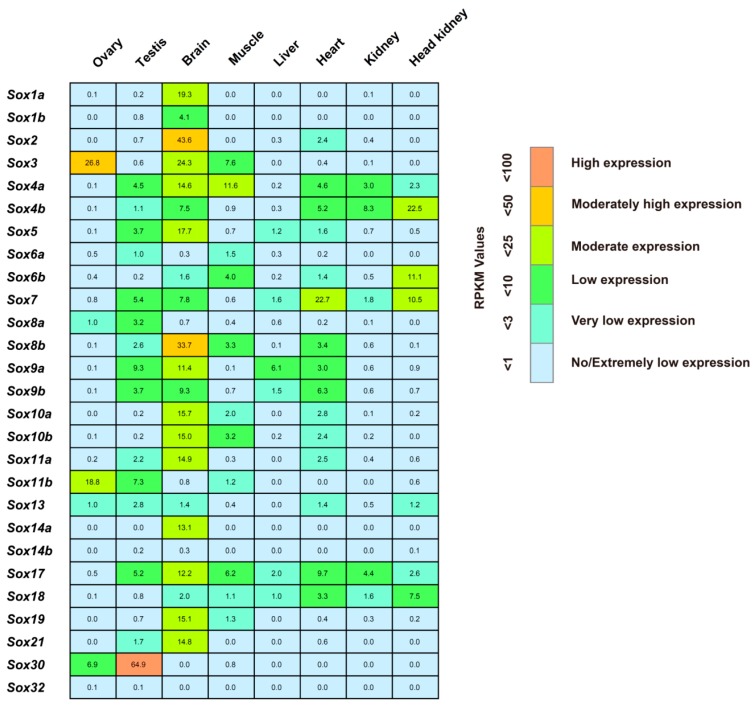
We next profiled the spatial expression of the tilapia Sox genes by using transcriptome data for eight adult tissues of the tilapia, including ovary, testis, brain, muscle, liver, heart, kidney, and head kidney. According to the criteria that a gene is regarded to be expressed if it exhibits an expression level with RPKM (reads per kb per million) value ≥ 1, we found that except for Sox14b and Sox32, the other 25 Sox genes were expressed in at least one of the adult tilapia tissues (Figure 4). The number of the Sox genes that were expressed in brain is the largest, reaching to 21. In addition, we observed the expressions of 15 Sox genes in testis, 15 in heart, 11 in muscle, seven in head kidney, six in liver, five in ovary, and five in kidney.
Figure 4.
Spatial expression profiles of the tilapia Sox genes. The transcriptomes (NCBI accession number: PRJNA78915 and SRR1916191) of multiple tissues of the adult tilapia were used to profile spatial expression of the tilapia Sox genes. Numbers within the box indicate the RPKM values.
Notably, four Sox genes showed high expression with RPKM value ≥25, including Sox3 in ovary, and Sox30 in testis, and Sox2 and Sox8b in brain (Figure 4). Moreover, 16 Sox genes were moderately expressed in different adult tissues, showing RPKM value that were greater than 10 and less than 25, including Sox11b in ovary, Sox1a, Sox3, Sox4a, Sox5, Sox9a, Sox10a, Sox10b, Sox11a, Sox14a, Sox17, Sox19, and Sox21 in brain, Sox4a in muscle, Sox7 in heart, and Sox4b, Sox6b, and Sox7 in head kidney. Interestingly, several Sox genes showed a tissue-specific expression, including Sox8a in ovary, and Sox1a, Sox1b, Sox2, Sox14a, Sox19, and Sox21 in brain.
2.5. Temporal Expression of the Sox Genes in the Tilapia Gonads
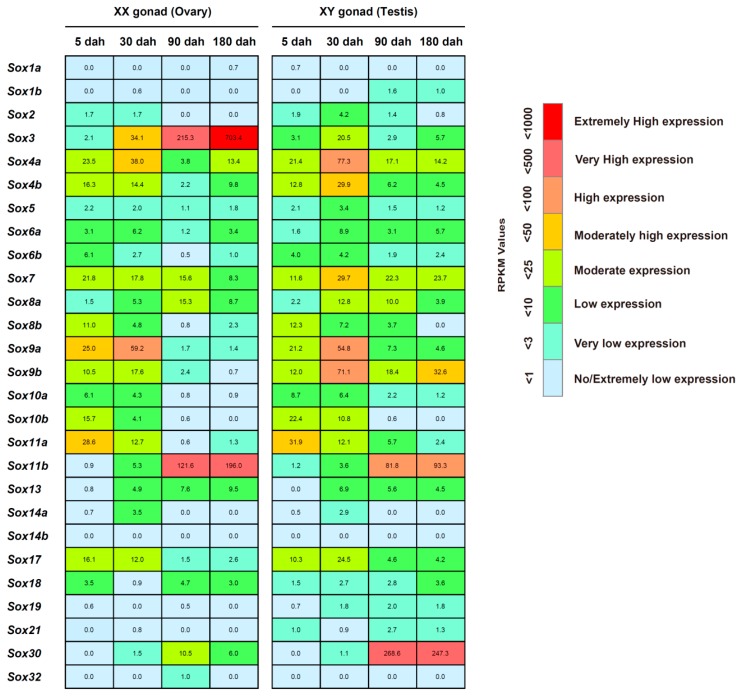
We used transcriptome data of the tilapia XX (ovary) and XY (testis) gonads at four developmental stages, namely 5, 30, 90 and 180 days after hatching (dah), to profile the temporal expression of the tilapia Sox genes. The results revealed that except for Sox1a, the other 26 Sox genes were expressed in XX (ovary) and/or XY (testis) gonads in at least one development stage (Figure 5). Among these expressed Sox genes, nine members (i.e., Sox3, Sox4a, Sox4b, Sox7, Sox9a, Sox9b, Sox11a, Sox11b, and Sox30) and four members (i.e., Sox8a, Sox8b, Sox10b, and Sox17) presented high expression and a moderate expression in XX (ovary) and/or XY (testis) gonads in at least one developmental stage, respectively.
Figure 5.
Temporal expression profiles of the Sox genes in the XX and XY gonads of the tilapia. The transcriptomes (NCBI accession number: SRA055700) of the tilapia gonads in four developmental stages comprising of 5, 30, 90 and 180 dah were used to analyze the expression profiles of the Sox genes during gonadal development. Numbers within the box indicate the RPKM values.
We further characterized the expression change of each Sox gene during gonadal development of the tilapia. As shown in Figure 5, the expressions of several Sox genes were gradually elevated during gonadal development, such as Sox3 and Sox11b in XX gonads (ovary) as well as Sox11b and Sox30 in XY gonads (testis), which showed great elevation in the late two stages of 90 dah and 180 dah. Moreover, in XX (ovary) and/or XY (testis) gonads, nine Sox genes, namely Sox4a, Sox4b, Sox8b, Sox9a, Sox9b, Sox10a, Sox10b, Sox11a, and Sox17, exhibited high expression in the early two stages of 5 dah and 30 dah, and of which the expressions of two Sox genes (Sox10b and Sox11a) and four Sox genes (i.e., Sox4a, Sox4b, Sox9a, and Sox9b) were very highly enriched at 5 dah and 30 dah, respectively.
2.6. Sexually Dimorphic Expression of the Sox Genes in the Tilapia Gonads
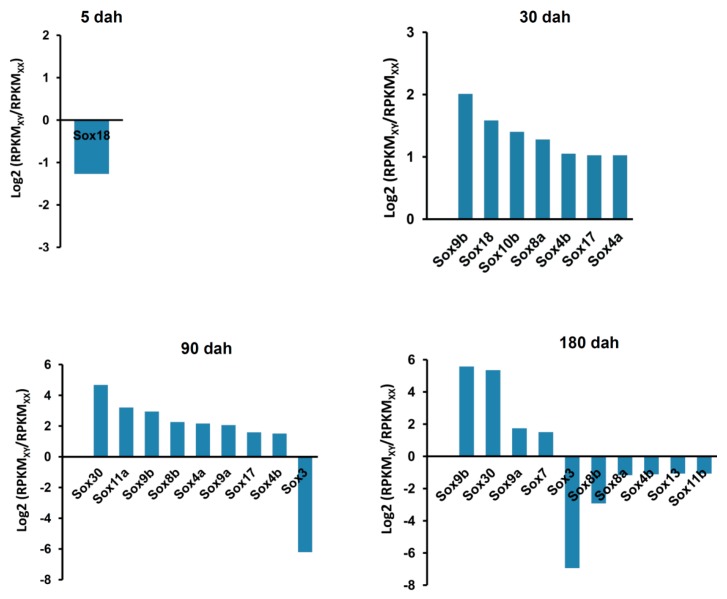
We examined whether the tilapia Sox genes exhibited sexually dimorphic expressions in gonads. First, a paired comparative analysis demonstrated that several Sox genes were specifically expressed at a time point during the development of the tilapia gonads (Figure 5), such as Sox1b and Sox2 in XY gonads (testis) at both 90 dah and 180 dah, and Sox8b at 180 dah and Sox32 at 90 dah in XX gonads (ovary). But, the expression level of these tilapia Sox genes was very low.
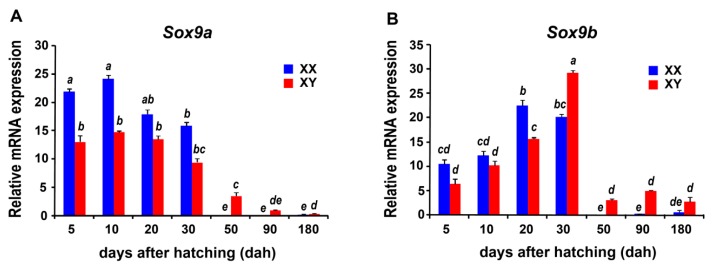
Second, we observed there were 1, 7, 10, and 10 Sox genes to display sexually dimorphic expression at 5 dah, 30 dah, 90 dah, and 180 dah, respectively (Figure 5 and Figure 6). In detail, only Sox18 was up-regulated in XX gonads (ovary) at 5 dah. At 30 dah, seven Sox genes, containing Sox4a, Sox4b, Sox8a, Sox9b, Sox10b, Sox17, and Sox18, were up-regulated in XY gonads (testis). In addition, we noted that eight Sox genes (i.e., Sox4a, Sox4b, Sox8b, Sox9a, Sox9b, Sox11a, Sox17, and Sox30) and four Sox genes (i.e., Sox7, Sox9a, Sox9b, and Sox30) were up-regulated in XY gonads (testis) at 90 and 180 dah, respectively. Conversely, several Sox genes showed up-regulation in XX gonads (ovary), including Sox3 at 90 dah and 6 Sox genes (i.e., Sox3, Sox4b, Sox8a, Sox8b, Sox11b, and Sox13) at 180 dah. Intriguingly, although Sox9a was highly expressed in both XX (ovary) and XY (testis) gonads at 5 and 30 dah (Figure 5 and Figure 6), it began to be differentially expressed in gonads, showing an up-regulation in XY gonads (testis) at 90 and 180 dah. Besides, Sox9b expression exhibited a significant up-regulation in XY gonads (testis) at 30, 90 and 180 dah. Consistently, quantitative RT-PCR examination revealed similar developmental and sexually dimorphic expressions of Sox9a and Sox9b in the tilapia gonads, and further confirmed that compared to XY gonads (testis), the expression of the Sox9a gene displayed a significant up-regulation in XX gonads (ovary) at 5 and 10 dah.
Figure 6.
Differential expressions of the Sox genes between XX and XY gonads of the tilapia at different developmental stages. The ratio of RPKM value for the expression of each Sox gene in XX (ovary) and XY (testis) gonads in each developmental stage was calculated. If the log2 of the ratio is ≥1 or ≤−1, this Sox gene was considered as sexual dimorphically expressed Sox genes in a developmental stage.
3. Discussion
Sox transcription factor family is exclusively discovered in animals to date and contributes to modulate various biological processes, like sex determination and gonadal development [45]. Recently, genome-wide characterization of the Sox gene family has been extensively performed in the metazoan, such as the nematode, insect, mammal and teleost. In this study, based on the recently published genome sequences for two lineages of teleost fishes, the tilapia and zebrafish [33,34], we identified 27 Sox genes in the tilapia and 27 Sox genes in the zebrafish.
Comparative analysis revealed several evolutionary perspectives of the Sox gene family during the separation of fish species from other animals. First, the Sox genes have undergone a continuous expansion in the teleost fishes following their whole genome duplication, which is supported by our finding shown in Table 2 and previous finding [46]. Intriguingly, although the orthologs of several mammalian Sox genes in the teleost fishes with 3R whole genome duplication have undergone duplication to generate two copies, different duplicates from an ancestral Sox gene have been demonstrated to exhibit splitting roles, like Sox9, Sox11, and Sox21 in the zebrafish [25,47,48]. Second, several Sox genes are exclusively present in vertebrates. For instance, the group K member Sox32 gene was specifically identified in teleosts. Previous study demonstrated that Sox32 is essential for the endodermal differentiation in the zebrafish [49], suggesting that it may be evolved to control the formation of specific organs in teleosts. In addition, the homolog of the human and chicken Sox30 gene was also discovered in the tilapia genome, consistent with our previous observation [41], but absent in other four teleosts, namely the zebrafish, pufferfish, medaka, and common carp. Curiously, Sox30 could be found in the teleosts guppy and channel catfish [41]. Undoubtedly, the evolution and functions of Sox30 in fishes and other vertebrates are somewhat complex and are worthy to be further investigated.
Sox transcription factors are involved in diverse physiological processes in animals through transcriptional activation and/or repression of their target genes in tissue- or development-specific manners [45,50]. It is very interesting that six Sox genes that belong to group B of the Sox family, including Sox1a, Sox1b, Sox2, Sox14a, Sox19, and Sox21, exhibited a specific expression in brain of the adult tilapia (Figure 5). Notably, four members of the SoxB1 subfamily, Sox1a, Sox1b, Sox2, and Sox19, have been characterized as the markers of the neural progenitor and stem cells throughout the vertebrate central nervous system (CNS) including brain, and contribute to not only regulating pluripotency but also mediate self-renewal and differentiation of neural progenitor and stem cells [51]. Sox21 from the SoxB2 subfamily functions as a counteracting partner of the SoxB1 genes to regulate neuron differentiation and promotes neurogenesis in vertebrate [52,53]. Accordingly, we proposed that the specific expression of the group B Sox genes in the adult brain may be necessary for the neurogenesis or the maintenance of specific biological processes in the tilapia brain.
Generally, several key biological events occur at these different time points during gonadal development of the tilapia, like sex determination and differentiation around 5–10 dah, the initiation of germ cell meiosis and oogenesis in the XX gonads (ovary) at 30 dah, the initiation of spermatogenesis in the XY gonads (testis) at 90 dah, and sperm maturation in the XY gonads (testis) and vitellogenesis in the XX gonads (ovary) at 180 dah [42,54]. A striking finding of our study is that several Sox genes exhibited a stage-specific and/or sexually dimorphic expression in the tilapia gonads, which provides new insights into their potential roles in gonadal development of the tilapia.
Our results revealed that Sox3 and Sox30 were very highly expressed in XX (ovary) and XY (testis) gonads during 90–180 dah, respectively (Figure 5 and Figure 6), indicating that these two genes may be required for oogenesis and spermatogenesis but not for sex determination. In fact, previous reports have demonstrated that the homolog of the tilapia Sox3 gene is required for gonadal function in the mouse and for oogenesis in the protogynous hermaphrodite fish (Halichoeres poecilopterus) [55,56], and the mice Sox30 gene is highly expressed in testis and regulates spermatogonial differentiation and spermatogenesis during testis development [57].
We also noted, as shown in Figure 5, Figure 6 and Figure 7, both Sox9a and Sox9b were highly expressed in gonads before 30 dah, a period that the completion of sex determination and the initiation of sex differentiation occur [54,58], indicating they may be involved in these processes during gonadal development of the tilapia. However, Sox9a expression was significantly higher in XX gonads (ovary) than that in XY gonads (testis) at 5 and 10 dah, whereas Sox9b expression was significantly higher in XY gonads (testis) than that in XX gonads (ovary) at 30 dah, indicating Sox9a may be mainly involved in the regulation of sex determination and ovarian differentiation, and Sox9b may regulate testicular differentiation in the tilapia.
Figure 7.
Quantitative RT-PCR examination of the Sox9a (A) and Sox9b (B) expressions in the XX and XY gonads of the tilapia. The quantitative RT-PCR experiment was performed in triplicates using pooled monosex fish cDNAs in each of the selected developmental stages. Values represent the relative mRNA expression in relation to the internal control (β-actin gene). Data were expressed as the mean ± SD of the triplicates. Column error bars with the same letter are not significantly different at p < 0.05 as determined using Least Significant Difference (LSD) test.
In addition, Sox17 was confirmed to be highly expressed in both XX (ovary) and XY (testis) gonads of the tilapia at 5 dah (sex determination and differentiation) and 30 dah (initiation of germ cell meiosis and oogenesis in the XX gonads (ovary). Given that previous observation in mouse that Sox17 mediates the specification of parietal endoderm cells during embryogenesis [59,60], the early expression of the tilapia Sox17 suggests that it might also be involved in the differentiation and specification of the tilapia gonads. Interestingly, Sox11a and Sox11b were highly expressed at 5–30 and 90–180 dah (the initiation of spermatogenesis in the XY gonads (testis) at 90 dah, and sperm maturation in the XY gonads (testis) as well as vitellogenesis in the XX gonads (ovary) at 180 dah), respectively. This suggests that Sox11a may regulate gonadal differentiation while Sox11b may be involved in spermatogenesis and vitellogenesis in the tilapia. Undoubtedly, the real roles of these Sox genes in the development of the tilapia gonads need to be clarified in the future studies.
4. Materials and Methods
4.1. Animal Rearing
The Nile tilapia fishes used in this study were reared in large tanks with recirculating freshwater at ambient temperature (26 °C) and under natural photoperiod. All females (XX) and males (XY) progenies were obtained by crossing the normal female (XX) with the sex-reversed XX pseudomale and YY supermale, respectively [61]. All animal experiments were performed following the regulations of the Guide for Care and Use of Laboratory Animals at Southwest University, Chongqing, China.
4.2. Genome-Wide Identification of the Sox Genes
The genome sequences and predicted protein-coding gene sets of the tilapia, zebrafish and common carp were downloaded from the online databases (http://asia.ensembl.org/Oreochromis_niloticus/Info/Index; http://asia.ensembl.org/Danio_rerio/Info/Index; http://www.carpbase.org/download_home.php). To identify candidate Sox genes in these three fish species, we first used the protein sequence of conserved HMG box domain (InterPro ID: IPR009071) for Sox protein to search against their predicted protein-coding gene sets by using local BLASTP program, with an E value threshold of 10−5. Secondly, given that the annotation of the zebrafish genes should be more precise, we used the amino acid sequences of each Sox gene from the zebrafish to search against the genome assemblies of tilapia and common carp via TBLASTN program with an E value threshold of 10−5, and the results from this search could be used to check the accuracy of the predicted Sox genes from the tilapia and common carp. The identified Sox genes were named according to the principle described in the previous report [6]. In addition, the genomic distribution of the Sox genes from these three fish species were characterized by mapping the amino acid sequences of each Sox gene on the genome assembly by using TBLASTN program.
To perform a comparative analysis, we collected the previously identified Sox genes of seven animals in the databases of NCBI (http://www.ncbi.nlm.nih.gov) and Ensembl (http://www.ensembl.org/), including the fruit fly, pufferfish, medaka, human, western clawed frog (Xenopus tropicalis), and chicken (Gallus gallus), according to the previous reports [2,3,5,32].
4.3. Phylogenetic Analysis
The amino acid sequence of the HMG box of all Sox proteins from six analyzed species, including the tilapia, zebrafish, fruit fly, pufferfish, medaka, and human were extracted base on the SMART analysis [62]. Multiple alignment of the HMG box of Sox proteins was performed using ClustalX program [63]. The neighbor-joining phylogenetic tree of the Sox genes were constructed by using MEGA 6.0 program [64], with a bootstrap of 1000 replicates.
4.4. Transcriptome-Based Analysis of Expression Profiling of the Sox Genes
The transcriptome data of the developing gonads and adult tissues were used to profile the temporal-spatial expressions of the tilapia Sox genes. Our previous study has sequenced the transcriptomes (NCBI accession number: SRA055700) of XX (ovary) and XY (testis) gonads at four different stages of the tilapia development, namely 5, 30, 90 and 180 days after hatching (dah) [42]. In addition, the transcriptomes (NCBI accession number: PRJNA78915 and SRR1916191) of the tilapia adult tissues were generated from brain, muscle, liver, heart, kidney, ovary, testis, and head kidney [33,65].
A normalized measure of RPKM value was used to characterize the expressions of the tilapia Sox genes. A threshold of RPKM value ≥ 1 was used to determine a reasonable expression for each Sox gene in a specific tissue at a specific time points [66,67]. The method described in our previous report was used to identify Sox genes sexually dimorphically expressed in gonads (XX or XY gonads) at each developmental stage [42]. Briefly, at each stage, Sox genes that were expressed specifically either in the XX or XY gonad only were classified as XX or XY-specific, whereas among the Sox genes expressed in both XX and XY gonads, those meeting the statistical criteria of both “FDR ≤ 10−2” and “|log2 (XX_RPKM/ XY_RPKM)| ≥ 1 or ≤ −1” were classified as differentially expressed candidates.
4.5. Gene Expression Profiling by Quantitative RT-PCR
Quantitative RT-PCR experiment was used to confirm the temporal-spatial expressions of the tilapia Sox genes. The gonads from monosex fishes (XX and XY) were dissected at 5, 10, 20, 30, 50, 90, and 180 dah. Different amount of gonads were collected from each sex at each developmental stage as a pooled sample, namely, approximate 50 gonads for each of two early stages (5 and 10 dah), 10 gonads for each of two stages (20 and 30 dah), 5 gonads for 50 dah, and 3 gonads for each of the two late stages (90 and 180 dah). Three samples were prepared for each stage to perform qRT-PCR experiments in triplicate. Total RNA was extracted from each sample, then treated by DNase, and immediately reverse-transcribed into cDNA using M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Quantitative RT-PCR examination was carried out according to the protocol of PlatinumSYBR Green qPCR SuperMix UDG kit (Invitrogen). The tilapia β-actin gene (NCBI accession number: EF206796) was used as an internal control. The primers used here were listed in Supplemental Table S2. The primer pair covers exons 1 and 2 and spanned intron 1 of the tilapia Sox9a and Sox9b. The raw data were analyzed by using one-way ANOVA and a Fisher's Least Significant Difference (LSD) test, and p < 0.05 was considered to be significant.
5. Conclusions
Sox transcription factors play important roles in animal development. In this study, a genome-wide analysis identified the varied numbers of the Sox genes in the tilapia (27), zebrafish (27), and common carp (49). Comparative analysis revealed that the Sox genes have undergone duplication in teleosts following their whole genome duplication after their separation from the other vertebrate species. Transcriptome-based expression profiling uncovered the tissue-, stage-, or sex-specific expressions of the tilapia Sox genes. The exact roles of these differentially expressed Sox genes during the tilapia development need to be precisely characterized in the future.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31302170 and 91331119), the Specialized Research Fund for the Doctoral Program of Higher Education of China (20130182120021 and 20130182130003), the Municipal Natural Science Foundation of Chongqing (cstc2013jcyjA80003), the Training Program of Scientific and Technological Talents of Chongqing (CSTC2013kjrc-tdjs80003), the Specialized Foundation for the Postdoctoral Research Program of Chongqing (Xm2014078), the National High Technology Research and Development Program of China (2011AA100404), and the Fundamental Research Funds for the Central Universities of China (XDJK2015C036).
Abbreviations
- HMG
high-mobility group
- 1R
first round of genome duplication
- 4R
fourth round of genome duplication
- dah
days after hatching
- RPKM
reads per kb per million read
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/17/3/270/s1.
Author Contributions
Deshou Wang and Ling Wei conceived and designed the project. Ling Wei and Wenjing Tao carried out computational analysis and transcriptome-based expression profiling. Ling Wei and Chao Yang performed the experiments. Ling Wei and Deshou Wang wrote the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Fortunato S., Adamski M., Bergum B., Guder C., Jordal S., Leininger S., Zwafink C., Rapp H.T., Adamska M. Genome-wide analysis of the sox family in the calcareous sponge Sycon ciliatum: Multiple genes with unique expression patterns. EvoDevo. 2012;3:270. doi: 10.1186/2041-9139-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui J., Shen X., Zhao H., Nagahama Y. Genome-wide analysis of Sox genes in Medaka (Oryzias latipes) and their expression pattern in embryonic development. Cytogenet. Genome Res. 2011;134:283–294. doi: 10.1159/000329480. [DOI] [PubMed] [Google Scholar]
- 3.Cremazy F., Berta P., Girard F. Genome-wide analysis of Sox genes in Drosophila melanogaster. Mech. Dev. 2001;109:371–375. doi: 10.1016/S0925-4773(01)00529-9. [DOI] [PubMed] [Google Scholar]
- 4.Uy B.R., Simoes-Costa M., Sauka-Spengler T., Bronner M.E. Expression of Sox family genes in early lamprey development. Int. J. Dev. Biol. 2012;56:377–383. doi: 10.1387/ijdb.113416bu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koopman P., Schepers G., Brenner S., Venkatesh B. Origin and diversity of the Sox transcription factor gene family: Genome-wide analysis in Fugu rubripes. Gene. 2004;328:177–186. doi: 10.1016/j.gene.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Bowles J., Schepers G., Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 7.Kashimada K., Koopman P. Sry: The master switch in mammalian sex determination. Development. 2010;137:3921–3930. doi: 10.1242/dev.048983. [DOI] [PubMed] [Google Scholar]
- 8.Sekido R., Lovell-Badge R. Sex determination and SRY: Down to a wink and a nudge? Trends Genet. 2009;25:19–29. doi: 10.1016/j.tig.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Kent J., Wheatley S.C., Andrews J.E., Sinclair A.H., Koopman P. A male-specific role for SOX9 in vertebrate sex determination. Development. 1996;122:2813–2822. doi: 10.1242/dev.122.9.2813. [DOI] [PubMed] [Google Scholar]
- 10.Jiang T., Hou C.C., She Z.Y., Yang W.X. The SOX gene family: Function and regulation in testis determination and male fertility maintenance. Mol. Biol. Rep. 2012;40:2187–2194. doi: 10.1007/s11033-012-2279-3. [DOI] [PubMed] [Google Scholar]
- 11.Kozhukhar V.G. SRY and SOX9: The main genetic factors of mammalian sex determination. Tsitologiia. 2012;54:390–404. [PubMed] [Google Scholar]
- 12.Jager M., Queinnec E., Houliston E., Manuel M. Expansion of the SOX gene family predated the emergence of the Bilateria. Mol. Phylogenet. Evol. 2006;39:468–477. doi: 10.1016/j.ympev.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Wegner M. SOX after SOX: SOXession regulates neurogenesis. Genes Dev. 2011;25:2423–2428. doi: 10.1101/gad.181487.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki T., Sakai D., Osumi N., Wada H., Wakamatsu Y. Sox genes regulate type 2 collagen expression in avian neural crest cells. Dev. Growth Differ. 2006;48:477–486. doi: 10.1111/j.1440-169X.2006.00886.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L.H., Zhu T.Y., Lin D., Zhang Y., Zhang W.M. A second form of Sox11 homologue identified in the orange-spotted grouper Epinephelus coioides: Analysis of sequence and mRNA expression patterns. Comp. Biochem. Phys. B. 2010;157:415–422. doi: 10.1016/j.cbpb.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Clarkson M.J., Harley V.R. Sex with two SOX on: SRY and SOX9 in testis development. Trends Endocrinol. Metab. 2002;13:106–111. doi: 10.1016/S1043-2760(01)00541-0. [DOI] [PubMed] [Google Scholar]
- 17.Barrionuevo F., Scherer G. SOX E genes: SOX9 and SOX8 in mammalian testis development. Int. J. Biochem. Cell Biol. 2010;42:433–436. doi: 10.1016/j.biocel.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Liu J.F., Liu S.J., Tao M., Li W., Liu Y. Isolation and expression analysis of testicular type Sox9b in allotetraploid fish. Mar. Biotechnol. 2007;9:329–334. doi: 10.1007/s10126-006-6123-4. [DOI] [PubMed] [Google Scholar]
- 19.Soriano N.S., Russell S. The Drosophila SOX-domain protein Dichaete is required for the development of the central nervous system midline. Development. 1998;125:3989–3996. doi: 10.1242/dev.125.20.3989. [DOI] [PubMed] [Google Scholar]
- 20.Mukherjee A., Shan X., Mutsuddi M., Ma Y., Nambu J.R. The Drosophila sox gene, fish-hook, is required for postembryonic development. Dev. Biol. 2000;217:91–106. doi: 10.1006/dbio.1999.9506. [DOI] [PubMed] [Google Scholar]
- 21.McDonald E., Krishnamurthy M., Goodyer C.G., Wang R. The emerging role of SOX transcription factors in pancreatic endocrine cell development and function. Stem Cells Dev. 2009;18:1379–1388. doi: 10.1089/scd.2009.0240. [DOI] [PubMed] [Google Scholar]
- 22.Gracz A.D., Magness S.T. Sry-box (Sox) transcription factors in gastrointestinal physiology and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;300:G503–G515. doi: 10.1152/ajpgi.00489.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lioubinski O., Muller M., Wegner M., Sander M. Expression of Sox transcription factors in the developing mouse pancreas. Dev. Dyn. 2003;227:402–408. doi: 10.1002/dvdy.10311. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda T., Kamekura S., Mabuchi A., Kou I., Seki S., Takato T., Nakamura K., Kawaguchi H., Ikegawa S., Chung U.I. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 2004;50:3561–3573. doi: 10.1002/art.20611. [DOI] [PubMed] [Google Scholar]
- 25.Yan Y.L., Willoughby J., Liu D., Crump J.G., Wilson C., Miller C.T., Singer A., Kimmel C., Westerfield M., Postlethwait J.H. A pair of Sox: Distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- 26.The C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 27.Dehal P., Satou Y., Campbell R.K., Chapman J., Degnan B., De Tomaso A., Davidson B., di Gregorio A., Gelpke M., Goodstein D.M., et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 28.Leveugle M., Prat K., Popovici C., Birnbaum D., Coulier F. Phylogenetic analysis of Ciona intestinalis gene superfamilies supports the hypothesis of successive gene expansions. J. Mol. Evol. 2004;58:168–181. doi: 10.1007/s00239-003-2538-y. [DOI] [PubMed] [Google Scholar]
- 29.Wei L., Cheng D., Li D., Meng M., Peng L., Tang L., Pan M., Xiang Z., Xia Q., Lu C. Identification and characterization of Sox genes in the silkworm, Bombyx mori. Mol. Biol. Rep. 2011;38:3573–3584. doi: 10.1007/s11033-010-0468-5. [DOI] [PubMed] [Google Scholar]
- 30.Howard-Ashby M., Materna S.C., Brown C.T., Chen L., Cameron R.A., Davidson E.H. Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus. Dev. Biol. 2006;300:90–107. doi: 10.1016/j.ydbio.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 31.Magie C.R., Pang K., Martindale M.Q. Genomic inventory and expression of Sox and Fox genes in the cnidarian Nematostella vectensis. Dev. Genes Evol. 2005;215:618–630. doi: 10.1007/s00427-005-0022-y. [DOI] [PubMed] [Google Scholar]
- 32.Schepers G.E., Teasdale R.D., Koopman P. Twenty pairs of Sox: Extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev. Cell. 2002;3:167–170. doi: 10.1016/S1534-5807(02)00223-X. [DOI] [PubMed] [Google Scholar]
- 33.Brawand D., Wagner C.E., Li Y.I., Malinsky M., Keller I., Fan S., Simakov O., Ng A.Y., Lim Z.W., Bezault E., et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513:375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C., Muffato M., Collins J.E., Humphray S., McLaren K., Matthews L., et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu P., Zhang X., Wang X., Li J., Liu G., Kuang Y., Xu J., Zheng X., Ren L., Wang G., et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 2014;46:1212–1219. doi: 10.1038/ng.3098. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura S., Watakabe I., Nishimura T., Toyoda A., Taniguchi Y., Tanaka M. Analysis of medaka sox9 orthologue reveals a conserved role in germ cell maintenance. PLoS ONE. 2012;7:270. doi: 10.1371/journal.pone.0029982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herpers R., van de Kamp E., Duckers H.J., Schulte-Merker S. Redundant roles for sox7 and sox18 in arteriovenous specification in zebrafish. Circ. Res. 2008;102:12–15. doi: 10.1161/CIRCRESAHA.107.166066. [DOI] [PubMed] [Google Scholar]
- 38.Cermenati S., Moleri S., Cimbro S., Corti P., Del Giacco L., Amodeo R., Dejana E., Koopman P., Cotelli F., Beltrame M. Sox18 and Sox7 play redundant roles in vascular development. Blood. 2008;111:2657–2666. doi: 10.1182/blood-2007-07-100412. [DOI] [PubMed] [Google Scholar]
- 39.Argenton F., Giudici S., Deflorian G., Cimbro S., Cotelli F., Beltrame M. Ectopic expression and knockdown of a zebrafish sox21 reveal its role as a transcriptional repressor in early development. Mech. Dev. 2004;121:131–142. doi: 10.1016/j.mod.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Cnaani A., Lee B.Y., Ozouf-Costaz C., Bonillo C., Baroiller J.F., D’Cotta H., Kocher T. Mapping of sox2 and sox14 in tilapia (Oreochromis spp.) Sex. Dev. 2007;1:207–210. doi: 10.1159/000102109. [DOI] [PubMed] [Google Scholar]
- 41.Han F., Wang Z., Wu F., Liu Z., Huang B., Wang D. Characterization, phylogeny, alternative splicing and expression of Sox30 gene. BMC Mol. Biol. 2010;11:98. doi: 10.1186/1471-2199-11-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tao W., Yuan J., Zhou L., Sun L., Sun Y., Yang S., Li M., Zeng S., Huang B., Wang D. Characterization of gonadal transcriptomes from Nile tilapia (Oreochromis niloticus) reveals differentially expressed genes. PLoS ONE. 2013;8:270. doi: 10.1371/journal.pone.0063604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glasauer S.M., Neuhauss S.C. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014;289:1045–1060. doi: 10.1007/s00438-014-0889-2. [DOI] [PubMed] [Google Scholar]
- 44.Kondrashov F.A. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. Biol. Sci. 2012;279:5048–5057. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wegner M. All purpose Sox: The many roles of Sox proteins in gene expression. Int. J. Biochem. Cell Biol. 2010;42:381–390. doi: 10.1016/j.biocel.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 46.Larroux C., Luke G.N., Koopman P., Rokhsar D.S., Shimeld S.M., Degnan B.M. Genesis and expansion of metazoan transcription factor gene classes. Mol. Biol. Evol. 2008;25:980–996. doi: 10.1093/molbev/msn047. [DOI] [PubMed] [Google Scholar]
- 47.Lan X., Wen L., Li K., Liu X., Luo B., Chen F., Xie D., Kung H.F. Comparative analysis of duplicated sox21 genes in zebrafish. Dev. Growth Differ. 2011;53:347–356. doi: 10.1111/j.1440-169X.2010.01239.x. [DOI] [PubMed] [Google Scholar]
- 48.De Martino S., Yan Y.L., Jowett T., Postlethwait J.H., Varga Z.M., Ashworth A., Austin C.A. Expression of sox11 gene duplicates in zebrafish suggests the reciprocal loss of ancestral gene expression patterns in development. Dev. Dyn. 2000;217:279–292. doi: 10.1002/(SICI)1097-0177(200003)217:3<279::AID-DVDY6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 49.Kikuchi Y., Agathon A., Alexander J., Thisse C., Waldron S., Yelon D., Thisse B., Stainier D.Y. Casanova encodes a novel Sox-related protein necessary and sufficient for early endoderm formation in zebrafish. Genes Dev. 2001;15:1493–1505. doi: 10.1101/gad.892301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lefebvre V., Dumitriu B., Penzo-Mendez A., Han Y., Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int. J. Biochem. Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abdelalim E.M., Emara M.M., Kolatkar P.R. The SOX transcription factors as key players in pluripotent stem cells. Stem Cells Dev. 2014;23:2687–2699. doi: 10.1089/scd.2014.0297. [DOI] [PubMed] [Google Scholar]
- 52.Sandberg M., Kallstrom M., Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat. Neurosci. 2005;8:995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- 53.Uchikawa M., Kamachi Y., Kondoh H. Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: Their expression during embryonic organogenesis of the chicken. Mech. Dev. 1999;84:103–120. doi: 10.1016/S0925-4773(99)00083-0. [DOI] [PubMed] [Google Scholar]
- 54.Ijiri S., Kaneko H., Kobayashi T., Wang D.S., Sakai F., Paul-Prasanth B., Nakamura M., Nagahama Y. Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus. Biol. Reprod. 2008;78:333–341. doi: 10.1095/biolreprod.107.064246. [DOI] [PubMed] [Google Scholar]
- 55.Yao B., Zhou L., Wang Y., Xia W., Gui J.F. Differential expression and dynamic changes of SOX3 during gametogenesis and sex reversal in protogynous hermaphroditic fish. J. Exp. Zool. Part A Ecol. Genet. Phys. 2007;307:207–219. doi: 10.1002/jez.361. [DOI] [PubMed] [Google Scholar]
- 56.Weiss J., Meeks J.J., Hurley L., Raverot G., Frassetto A., Jameson J.L. Sox3 is required for gonadal function, but not sex determination, in males and females. Mol. Cell. Biol. 2003;23:8084–8091. doi: 10.1128/MCB.23.22.8084-8091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osaki E., Nishina Y., Inazawa J., Copeland N.G., Gilbert D.J., Jenkins N.A., Ohsugi M., Tezuka T., Yoshida M., Semba K. Identification of a novel Sry-related gene and its germ cell-specific expression. Nucleic Acids Res. 1999;27:2503–2510. doi: 10.1093/nar/27.12.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakamura M., Kobayashi T., Chang X.T., Nagahama Y. Gonadal sex differentiation in teleost fish. J. Exp. Zool. 1998;281:362–372. doi: 10.1002/(SICI)1097-010X(19980801)281:5<362::AID-JEZ3>3.0.CO;2-M. [DOI] [Google Scholar]
- 59.Niimi T., Hayashi Y., Futaki S., Sekiguchi K. SOX7 and SOX17 regulate the parietal endoderm-specific enhancer activity of mouse laminin alpha 1 gene. J. Biol. Chem. 2004;279:38055–38061. doi: 10.1074/jbc.M403724200. [DOI] [PubMed] [Google Scholar]
- 60.Futaki S., Hayashi Y., Yamashita M., Yagi K., Bono H., Hayashizaki Y., Okazaki Y., Sekiguchi K. Molecular basis of constitutive production of basement membrane components—Gene expression profiles of Engelbreth-Holm-Swarm tumor and F9 embryonal carcinoma cells. J. Biol. Chem. 2003;278:50691–50701. doi: 10.1074/jbc.M304985200. [DOI] [PubMed] [Google Scholar]
- 61.Zhang X., Wang H., Li M., Cheng Y., Jiang D., Sun L., Tao W., Zhou L., Wang Z., Wang D. Isolation of doublesex- and mab-3-related transcription factor 6 and its involvement in spermatogenesis in tilapia. Biol. Reprod. 2014;91:270. doi: 10.1095/biolreprod.114.121418. [DOI] [PubMed] [Google Scholar]
- 62.Letunic I., Doerks T., Bork P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jeanmougin F., Thompson J.D., Gouy M., Higgins D.G., Gibson T.J. Multiple sequence alignment with Clustal x. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/S0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 64.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng Y.Y., Tao W.J., Chen J.L., Sun L.N., Zhou L.Y., Song Q., Wang D.S. Genome-wide identification, evolution and expression analysis of nuclear receptor superfamily in Nile tilapia, Oreochromis niloticus. Gene. 2015;569:141–152. doi: 10.1016/j.gene.2015.05.057. [DOI] [PubMed] [Google Scholar]
- 66.Hart T., Komori H.K., LaMere S., Podshivalova K., Salomon D.R. Finding the active genes in deep RNA-seq gene expression studies. BMC Genom. 2013;14:270. doi: 10.1186/1471-2164-14-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsagaratou A., Aijo T., Lio C.W., Yue X., Huang Y., Jacobsen S.E., Lahdesmaki H., Rao A. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proc. Natl. Acad. Sci. USA. 2014;111:E3306–E3315. doi: 10.1073/pnas.1412327111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.