Abstract
The peach fruit moth Carposina sasakii is an economically important pest on dozens of fruits from Rosaceae and Rhamnaceae in Northeast Asia. We developed novel microsatellite markers for C. sasakii from randomly sequenced regions of the genome using next-generation sequencing. In total, 95,153 microsatellite markers were isolated from 4.70 GB genomic sequences. Thirty-five polymorphic markers were developed by assessing in 63 individuals from two geographical populations. The allele numbers ranged from 2 to 9 with an average value of 4.60 per locus, while the polymorphism information content ranged from 0.075 to 0.696 with an average value of 0.407. Furthermore, the observed and expected heterozygosity varied from 0.000 to 0.677 and 0.062 to 0.771, respectively. The microsatellites developed provide abundant molecular markers for investigating genetic structure, genetic diversity, and existence of host-plant associated biotypes of C. sasakii.
Keywords: peach fruit moth, Carposina sasakii, simple sequence repeat, next-generation sequencing
1. Introduction
The peach fruit moth Carposina sasakii Matsumura (Lepidoptera: Carposinidae), is an important orchard pest in Northeast Asia [1,2]. In China, this pest was distributed throughout the country, except for Tibet. Its larvae can inflict direct damage on dozens of fruits, including peach, apple, pear, jujube, wild jujube, apricot, hawthorn, and pomegranate [3,4,5,6,7] by boring into fruitage. Differences in number of generations, emergence time of overwintering and diapause generation were found among populations on different host species [8,9], likely leading to low gene flow among host-plant populations. Thus, several studies attempted to reveal the differentiation of those moths occurring on different host plants [10,11].
To date, three types of molecular marker have been used to examine the existence of host biotypes in C. sasakii. Using esterase isozyme, Hua et al. (1995) [11] reported that there is nearly no differentiation in isozyme-spectra between C. sasakii collected from jujube and wild jujube; however, populations collected from above two hosts were obviously different from those collected from apple orchard. Using RAPD (random amplified polymorphic DNA) to compare the populations collected from six kinds of host plants, including apple, hawthorn, peach, apricot, jujube, and wild jujube, Xu and Hua revealed that there were remarkable genetic differentiation between populations from apricot and those from other hosts [12]. Although a recent study using one region of mitochondrial DNA (mtDNA) sequences of cytochrome coxidase subunit I (COI) found that there was no evidence for associations between the variation of populations and host plants, the genetic differentiation showed significant correlation with the geographical distance [13]. Varied genetic markers used in the studies obviously lead to different results. To address this issue, more polymorphic and stable molecular markers are required.
Microsatellite is a kind of special sequence comprised by tandem repeats of one to six nucleotides. It always has high polymorphism, widely dispersed in both coding and noncoding regions of all prokaryotic and eukaryotic genomes [14]. Due to their codominant inheritance, high polymorphism, easy detection by polymerase chain reaction (PCR), and broad distribution in the genome, microsatellites are widely used for population genetic studies [15,16,17].
The traditional approach of microsatellite development, such as an enriched library followed by gene cloning, is time-consuming and labor-intensive. New approaches based on next-generation sequencing can be a good alternative. With the advantage of this technology, it is possible to develop a huge number of microsatellites, which are capable of generating tens of millions of short DNA sequence reads at a relatively low cost [18,19,20,21].
In the present study, we aimed to isolate microsatellites for C. sasakii from randomly obtained genomic sequences. This is the first report of novel microsatellites for C. sasakii. The markers developed will be helpful in investigating genetic structure, genetic diversity, and existence of host-plant associated biotypes of C. sasakii.
2. Results and Discussion
2.1. Microsatellite Marker Development
We generated 4.70 GB paired-end (PE) sequences with read length of 300 base pairs (bp), including 15,725,132 reads from a 500 bp insert DNA library constructed by Illumina MiSeq system. Raw data sequences were submitted to the National Center for Biotechnology Information (NCBI) Short Read Archive under accession number SRP068817. After removing low quality reads using SolexaQA software [22], the remaining high-quality reads were assembled into 1,902,994 contigs by SOAPdenovo2 [23]. They were with mean size of 252 bp and N50 of 286 bp, which are much shorter compared to a similar study in Dorcus hopei (Coleoptera) (N50 = 1218) [24]. This might be due to the method of assembly and the coverage of sequencing reads. However, the number of primer pairs designed in our study is reasonable (totally 8074 primer pairs / 479 Mb), as in other studies [19,24,25,26].
A total of 95,153 microsatellite loci were discovered using MSDB version 2.4.3 software (http://msdb.biosv.com/) from the assembled contigs, which will be provided upon request. The detected microsatellites included 54,559 (57.34%) dinucleotide, 34,957 (36.74%) trinucleotide, 5591 (5.88%) tetranucleotide, and 46 (0.05%) pentanucleotide repeats (Table 1). There are no hexanucleotide repeats found under our searching conditions of microsatellite loci (a minimum of 25, 5, 5, 5, 5 and 5 repeats were used to identify the mono-, di-, tri-, tetra-, penta-, and hexanucleotide motifs, respectively). Dinucleotide repeats are more than the higher order motif, which is in agreement with the previous report of Arthropoda in insects like Aphis glycines (Hemiptera) [27] and Coccinella septempunctata (Coleoptera) [28], and in species of Arachnida [29]. According to the distribution of microsatellite (Table 1), it seems that the quantity of loci decreases followed with the increase of corresponding motif repeats.
Table 1.
Distribution of microsatellites with different motifs and repeat numbers in Carposina sasakii.
| Repeat Motif | Number of Repeats | Total Frequency (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| AC/GT | 7521 | 2765 | 1747 | 1543 | 1433 | 720 | 76 | 2 | 16.612 |
| AG/CT | 2080 | 862 | 512 | 387 | 415 | 340 | 72 | 2 | 4.908 |
| AT/AT | 14,053 | 5758 | 4165 | 4491 | 3380 | 732 | 42 | 2 | 34.285 |
| CG/CG | 1325 | 109 | 12 | 4 | 3 | 3 | 1 | 2 | 1.533 |
| AAC/GTT | 92 | 34 | 18 | 3 | 0.154 | ||||
| AAG/CTT | 2074 | 747 | 142 | 3 | 3.117 | ||||
| AAT/ATT | 8333 | 4004 | 438 | 3 | 13.429 | ||||
| ACC/GGT | 45 | 12 | 6 | 1 | 0.067 | ||||
| ACG/CGT | 457 | 121 | 38 | 3 | 0.651 | ||||
| ACT/AGT | 218 | 71 | 50 | 3 | 0.359 | ||||
| AGC/CTG | 77 | 27 | 14 | 3 | 0.127 | ||||
| AGG/CCT | 49 | 7 | 7 | 1 | 0.067 | ||||
| ATC/ATG | 11,707 | 5017 | 435 | 3 | 18.036 | ||||
| CCG/CGG | 436 | 162 | 93 | 3 | 0.729 | ||||
| AAAC/GTTT | 257 | 4 | 0.274 | ||||||
| AAAG/CTTT | 355 | 4 | 0.377 | ||||||
| AAAT/ATTT | 1798 | 4 | 1.894 | ||||||
| AACC/GGTT | 14 | 4 | 0.019 | ||||||
| AACG/ | 1 | 1 | 0.002 | ||||||
| AACT/AGTT | 42 | 4 | 0.048 | ||||||
| AAGG/CTTC | 8 | 2 | 0.011 | ||||||
| AAGT/ACTT | 62 | 4 | 0.069 | ||||||
| AATC/GATT | 146 | 4 | 0.158 | ||||||
| AATG/CATT | 283 | 4 | 0.302 | ||||||
| AATT/AATT | 68 | 3 | 0.075 | ||||||
| ACAG/CTGT | 112 | 4 | 0.122 | ||||||
| ACAT/ATGT | 1448 | 4 | 1.526 | ||||||
| ACCT/AGGT | 114 | 3 | 0.123 | ||||||
| ACGC/ACGC | 20 | 4 | 0.025 | ||||||
| ACGG/CGTC | 51 | 4 | 0.058 | ||||||
| ACGT/ACGT | 3 | 0.003 | |||||||
| ACTC/GTGA | 20 | 3 | 0.024 | ||||||
| AGAT/ATCT | 473 | 4 | 0.501 | ||||||
| ATCC/ATGG | 216 | 4 | 0.231 | ||||||
| ATGC/ATGC | 6 | 1 | 0.007 | ||||||
| AGGC/CCTG | 10 | 2 | 0.013 | ||||||
| CGAG/CTCG | 2 | 0.002 | |||||||
| CGGC/CGGC | 3 | 0.003 | |||||||
| CTAG/CTAG | 3 | 0.003 | |||||||
| GACT/TCAG | 2 | 1 | 0.003 | ||||||
| GCAA/GCAA | 2 | 0.002 | |||||||
| OTHERS | 46 | 0.048 | |||||||
| DNR | 24,979 | 9494 | 6436 | 6425 | 5231 | 1795 | 191 | 8 | 57.338 |
| TNR | 23,488 | 10,202 | 1241 | 26 | 36.738 | ||||
| TTNR | 5519 | 72 | 5.876 | ||||||
| PNR | 46 | 0.048 | |||||||
DNR: dinucleotide repeats; TNR: trinucleotide repeats; TTNR: tetranucleotide repeats; PNR: pentanucleotide repeats.
Sixty-four primer pairs designed according to the sequences that are flanking trinucleotide repeats were selected for initial validation in eight individuals. Of them, 35 loci have polymorphic amplifications, 16 loci were monomorphic and 13 primer pairs did not produce any visible amplicon. These polymorphic loci can serve as candidate markers for future research, such as genetic diversity and relatedness analysis of different populations.
2.2. Characteristics of Validated Microsatellite Loci
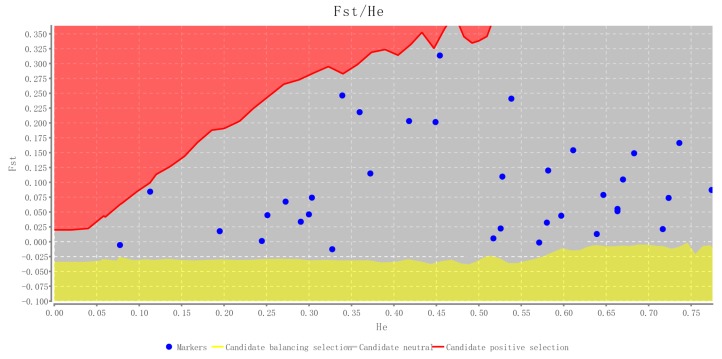
The polymorphic loci obtained were assessed with two C. sasakii natural populations, including 31 individuals from Beijing and 32 individuals from Hubei province, China (Table 2 and Table S1). The 35 microsatellite markers had allele numbers ranging from 2 to 9 with an average value of 4.60 per locus. The polymorphism information content (PIC) revealed a range from 0.075 to 0.696 with an average value of 0.407. The observed (HO) and expected (HE) heterozygosity ranged from 0.000 to 0.677 and 0.062 to 0.771, respectively. The inbreeding coefficient (FIS) ranged from −0.240 to 1.00. The significantly high FIS in locus CS21, CS38 and CS82 might be caused by the low HO, rather than sampling bias since most loci showed low FIS in the two populations. The loci CS31 and CS33 showed significant linkage disequilibrium only across Beijing population (corrected by Holm’s correction, p < 0.05). It is speculated that the linkage disequilibrium observed at certain loci in some populations may be due to substructure of population or bottleneck [30]. Eight loci in Beijing population and 13 loci in Hubei population significantly deviated from Hardy-Weinberg equilibrium (HWE), while 5 loci (CS05, CS17, CS21, CS29 and CS82) showed significant value in the both tested populations. The loci deviated from HWE might be resulted by heterozygote deficiency, because HO is much lower than HE in these loci (Table 2). Heterozygote deficiency can be caused by the Wahlund Effect [31] or the presence of null alleles, for which Lepidoptera species are notorious [32,33,34,35,36]. It was considered that the present of null alleles is very common in this order due to the flanking region with repetitive sequences and multiple copies of loci [37,38,39]. Random sequences of C. sasakii genome obtained by the Illumina MiSeq system may cover coding regions. Thus, they are probably linked to sites under selection, which cannot reflect facticity of population diversity and structure. A neutrality test was done with all of the 35 loci. Interestingly, all of the loci were under neutral expectations (Figure 1). Therefore, deviating from HWE is not necessarily due to the characteristics of loci. It may imply the distinct population structure, biological property of the species, or just sampling error, e.g., examined individuals from the same egg brood can also lead to deviation from HWE [40].
Table 2.
Characteristics of 35 microsatellite loci validated in 63 individuals of Carposina sasakii.
| Locus | Dye | Repeat Motif | Primer Sequence (5′–3′) | Allele No. | Size Range (bp) | HWE | r | HO | HE | FIS | PIC | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beijing | Hubei | Beijing | Hubei | Beijing | Hubei | Beijing | Hubei | Beijing | Hubei | |||||||
| CS03 | ROX | (AGT)6 | F: TAAAAGCGATTCGTTGGGAC | 5 | 209–218 | 0.608 | 1.000 | 0.000 | 0.000 | 0.419 | 0.125 | 0.387 | 0.122 | −0.086 | −0.029 | 0.250 |
| R: ATGGCGTCATATCTTCGACC | ||||||||||||||||
| CS04 | FAM | (ACT)6 | F: TTCCGTGCATGTCGTAAGAG | 6 | 120–139 | 0.012 | 0.016 | 0.100 | 0.011 | 0.484 | 0.406 | 0.655 | 0.468 | 0.265 | 0.133 | 0.531 |
| R: CGCGTTTAGCATCAATCTCA | ||||||||||||||||
| CS05 | HEX | (ACG)6 | F: ACACTAGTTGAGTGATTTCAACCG | 5 | 101–113 | 0.000 | 0.000 | 0.323 | 0.269 | 0.097 | 0.188 | 0.622 | 0.631 | 0.847 | 0.706 | 0.572 |
| R: GCATCTGGCTAGATTCTGATGA | ||||||||||||||||
| CS06 | HEX | (CCG)6 | F: ACCGACCAGTCCATTCGAT | 4 | 106–123 | 0.460 | 0.856 | 0.000 | 0.005 | 0.613 | 0.469 | 0.539 | 0.489 | −0.139 | 0.041 | 0.412 |
| R: CTCCTTAGGTCTCTGCGTCG | ||||||||||||||||
| CS07 | HEX | (AAT)6 | F: AGCAGCCTGCATCCAACC | 9 | 99–122 | 0.738 | 0.000 | 0.000 | 0.106 | 0.581 | 0.581 | 0.643 | 0.771 | 0.098 | 0.250 | 0.696 |
| R: ACACACTCCCAATTCGCTTC | ||||||||||||||||
| CS101 | HEX | (AAC)6 | F: TTGGTTCATGGATCTAGGAGG | 4 | 104–115 | 0.007 | 0.006 | 0.145 | 0.132 | 0.161 | 0.219 | 0.309 | 0.354 | 0.483 | 0.385 | 0.304 |
| R: TCCTAAGTCTACCTAACTTTATGTGTT | ||||||||||||||||
| CS102 | FAM | (AGT)6 | F: CCGTAATAATTCGACACAAGCA | 5 | 131–147 | 1.000 | 0.004 | 0.000 | 0.159 | 0.226 | 0.219 | 0.211 | 0.448 | −0.071 | 0.516 | 0.325 |
| R: CCTATACTCGTATACTTAAACAACTGA | ||||||||||||||||
| CS103 | HEX | (AAC)6 | F: AGTATCAAAAGAAACCCCTAA | 4 | 111–120 | 1.000 | 0.700 | 0.011 | 0.036 | 0.355 | 0.594 | 0.373 | 0.661 | 0.049 | 0.104 | 0.506 |
| R: ATCGGCATTATTTGTAAGGT | ||||||||||||||||
| CS11 | HEX | (AAG)6 | F: CCTCGTATTAGATTAGGCGGAA | 4 | 95–112 | 1.000 | 0.000 | 0.000 | 0.200 | 0.065 | 0.250 | 0.063 | 0.560 | −0.017 | 0.558 | 0.343 |
| R: CCCAAGTTGAATGGGAACAG | ||||||||||||||||
| CS14 | HEX | (AGT)6 | F: TGCGACAAAATGCCAGAATA | 6 | 106–136 | 0.020 | 0.952 | 0.129 | 0.000 | 0.355 | 0.594 | 0.590 | 0.554 | 0.403 | −0.074 | 0.489 |
| R: GCCGATGTATTCTAATGAAGCC | ||||||||||||||||
| CS17 | HEX | (AAG)6 | F: CTCAAGAGTTCTATATACGGGG | 5 | 102–117 | 0.000 | 0.001 | 0.294 | 0.170 | 0.233 | 0.219 | 0.751 | 0.448 | 0.693 | 0.516 | 0.592 |
| R: GGCGATGGGATAGCTGTTAC | ||||||||||||||||
| CS18 | HEX | (AAT)6 | F: AGATAGCTCGTTGACAAAGTT | 3 | 111–117 | 0.402 | 0.000 | 0.041 | 0.183 | 0.194 | 0.125 | 0.228 | 0.344 | 0.155 | 0.640 | 0.272 |
| R: TGTTTTGGAAGCAACAAACG | ||||||||||||||||
| CS19 | HEX | (AGT)6 | F: CCAATGTGTCGTACAACGTG | 7 | 113–134 | 0.291 | 0.015 | 0.062 | 0.089 | 0.516 | 0.438 | 0.631 | 0.561 | 0.184 | 0.222 | 0.568 |
| R: CCTCAAGTAAATATAATCAGGGCG | ||||||||||||||||
| CS20 | FAM | (ACT)6 | F: CAAATCCTTGGCAATGTGAA | 4 | 109–126 | 0.030 | 0.000 | 0.076 | 0.224 | 0.462 | 0.156 | 0.646 | 0.496 | 0.290 | 0.688 | 0.478 |
| R: AGAAAAGATTCACCTGCGCT | ||||||||||||||||
| CS21 | FAM | (ACT)6 | F: CGCATTTGCTACTCACCTGT | 4 | 105–120 | 0.000 | 0.000 | 0.201 | 0.248 | 0.000 | 0.063 | 0.178 | 0.383 | 1.000 | 0.839 | 0.256 |
| R: ACTTACATTCACGTTGCCCA | ||||||||||||||||
| CS22 | FAM | (CCG)6 | F: GTAACGAGCGCAATTGATGA | 3 | 122–128 | 0.050 | 1.000 | 0.108 | 0.000 | 0.032 | 0.063 | 0.094 | 0.062 | 0.659 | −0.008 | 0.075 |
| R: CGCGCTAATCTGGTTAATACG | ||||||||||||||||
| CS24 | ROX | (CCG)6 | F: TCTAAGGAGTGTCCGAAGGC | 2 | 247–248 | 1.000 | 1.000 | 0.000 | 0.013 | 0.452 | 0.469 | 0.444 | 0.496 | −0.017 | 0.055 | 0.373 |
| R: TCAAGTACCGTGTGCGGATA | ||||||||||||||||
| CS26 | FAM | (CCG)6 | F: ACCCGAGTAAAGACCCGACT | 4 | 123–135 | 0.000 | 0.105 | 0.272 | 0.097 | 0.129 | 0.065 | 0.535 | 0.182 | 0.762 | 0.649 | 0.360 |
| R: TGTTAACCCTAGAAGGCCCG | ||||||||||||||||
| CS28 | FAM | (ACT)6 | F: GCTGGTGTGGATGGCATAGT | 7 | 126–147 | 0.023 | 0.061 | 0.082 | 0.099 | 0.484 | 0.438 | 0.637 | 0.591 | 0.243 | 0.263 | 0.615 |
| R: AACTTCGAATTTCCATTGCG | ||||||||||||||||
| CS29 | FAM | (ACC)6 | F: TCGGTCACGTTATTTTAGCAA | 9 | 89–147 | 0.000 | 0.000 | 0.173 | 0.266 | 0.290 | 0.290 | 0.504 | 0.525 | 0.428 | 0.451 | 0.494 |
| R: CATGGTCAGTGCTAGGCAGA | ||||||||||||||||
| CS31 | FAM | (ACT)6 | F: CGGACTTCTGAAACCGTGAT | 6 | 129–148 | 0.086 | 0.000 | 0.028 | 0.137 | 0.484 | 0.484 | 0.563 | 0.698 | 0.143 | 0.310 | 0.601 |
| R: GCCAATTCAGTTATGAGGGC | ||||||||||||||||
| CS32 | FAM | (AGG)6 | F: CTAGGTACACCAATCGGCCA | 2 | 134–137 | 0.054 | 0.495 | 0.111 | 0.037 | 0.194 | 0.438 | 0.317 | 0.500 | 0.394 | 0.127 | 0.360 |
| R: GCTGCCATTTCACCAGTCTT | ||||||||||||||||
| CS33 | FAM | (ACT)6 | F: AATAGGGCTCCTCCACACCT | 8 | 130–156 | 0.392 | 0.706 | 0.030 | 0.003 | 0.677 | 0.531 | 0.769 | 0.571 | 0.121 | 0.071 | 0.643 |
| R: GATCTGCAAATCTGCCTGTG | ||||||||||||||||
| CS34 | FAM | (AGT)6 | F: CGCCCTAGACGAACCTACAC | 4 | 130–143 | 0.587 | 1.000 | 0.026 | 0.000 | 0.258 | 0.219 | 0.283 | 0.205 | 0.091 | −0.069 | 0.227 |
| R: GCCTATGTTCAGCAGAAGACG | ||||||||||||||||
| CS35 | ROX | (AAG)6 | F: CAAAGATAATGTACAAAGACGTG | 5 | 113–142 | 0.001 | 0.040 | 0.215 | 0.121 | 0.269 | 0.531 | 0.652 | 0.750 | 0.592 | 0.296 | 0.655 |
| R: CAACTGTCTGCAACACAGCA | ||||||||||||||||
| CS36 | ROX | (CCG)6 | F: CACCGATTTGTTTTATCGCA | 7 | 138–159 | 0.284 | 1.000 | 0.025 | 0.000 | 0.581 | 0.063 | 0.604 | 0.062 | 0.039 | −0.008 | 0.351 |
| R: GGCGCTAATGTCTACCCTCA | ||||||||||||||||
| CS37 | ROX | (ACC)6 | F: TAAGAAGATCCTCGCCCAGA | 2 | 145–148 | 0.159 | 0.300 | 0.081 | 0.000 | 0.097 | 0.406 | 0.151 | 0.329 | 0.362 | −0.240 | 0.215 |
| R: TACATCGTTGTAGGACCGCC | ||||||||||||||||
| CS38 | ROX | (AGC)6 | F: CAAACAAATTATCCGCGTCC | 3 | 147–153 | 0.022 | 0.001 | 0.140 | 0.176 | 0.000 | 0.000 | 0.148 | 0.235 | 1.000 | 1.000 | 0.181 |
| R: GACAGAAACAATAACAACGACGA | ||||||||||||||||
| CS41 | ROX | (AAC)6 | F: CCACTGGGCTATCACTGCTAT | 6 | 140–168 | 0.118 | 0.132 | 0.040 | 0.052 | 0.581 | 0.281 | 0.664 | 0.360 | 0.128 | 0.221 | 0.509 |
| R: TGCAACAGTGACATCACAAGA | ||||||||||||||||
| CS44 | ROX | (AGT)6 | F: AGTGGGCGCCACCTGCAT | 3 | 149–155 | 1.000 | NA | 0.000 | 0.001 | 0.226 | 0.000 | 0.207 | 0.000 | −0.094 | NA | 0.102 |
| R: CCATCTTTGGCTCAGAAAGC | ||||||||||||||||
| CS45 | ROX | (ACT)6 | F: TGGCCGTTATATCATCCACA | 2 | 155–158 | 1.000 | 1.000 | 0.000 | 0.000 | 0.065 | 0.469 | 0.063 | 0.448 | −0.017 | −0.047 | 0.254 |
| R: GGTAGTCCTGGTCAGAGGCA | ||||||||||||||||
| CS47 | ROX | (AGT)7 | F: ACCGGTATTGCTGTATTTGT | 5 | 151–163 | 0.001 | 0.764 | 0.163 | 0.000 | 0.400 | 0.625 | 0.666 | 0.592 | 0.404 | −0.056 | 0.573 |
| R: CAATTTGTGATTAGGTATTTGTTTCAA | ||||||||||||||||
| CS48 | ROX | (AAT)6 | F: TGTAGCAGTCAAGGTCACGG | 3 | 156–162 | 0.048 | 0.000 | 0.071 | 0.219 | 0.484 | 0.194 | 0.666 | 0.497 | 0.277 | 0.614 | 0.556 |
| R: CGCTATAAAAGTGAACGGCG | ||||||||||||||||
| CS53 | ROX | (AAG)6 | F: TCACGTAACCGTCTGGTTCA | 3 | 137–176 | 1.000 | 0.802 | 0.000 | 0.015 | 0.097 | 0.438 | 0.094 | 0.469 | −0.035 | 0.068 | 0.274 |
| R: TCGTCTTTTCTTTCCATCGG | ||||||||||||||||
| CS82 | HEX | (AGT)6 | F: AAAGGCAGATTAACCGACTAGTGT | 2 | 89–106 | 0.000 | 0.000 | 0.293 | 0.198 | 0.000 | 0.000 | 0.389 | 0.173 | 1.000 | 1.000 | 0.247 |
| R: AAATATTTTCGCGTTCATTTCG | ||||||||||||||||
F: forward primer; R: reverse primer; r: frequency of null allele; HO: observed heterozygosity; HE: expected heterozygosity; PIC: polymorphism information content; FIS: inbreeding coefficient; HWE: exact p-value of Hardy-Weinberg Equilibrium; BLASTx/BLASTn: results of BLASTx/BLASTn. NA: not available.
Figure 1.
Potential candidates for selection. Loci located in the red region are candidates for positive selection, grey region for neutral, and yellow region for balancing selection. All of the 35 loci are under neutral expectations.
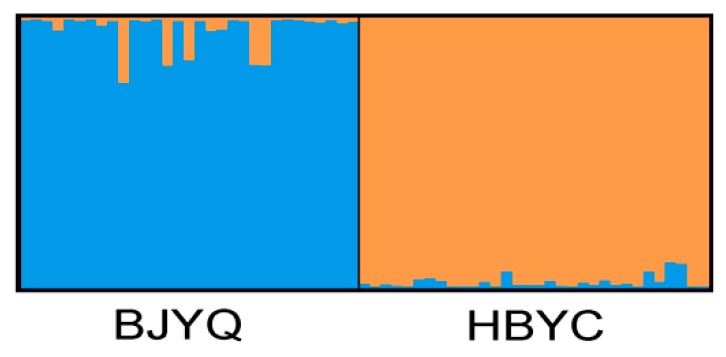
The population structure of C. sasakii was inferred with the dataset of 35 microsatellite markers. The 63 individuals from two geographic populations were divided into two clusters. As can be clearly seen in Figure 2, there are genetic differences between two populations, indicating that the microsatellite markers validated could be used to discriminate geographic populations and other genetic study of C. sasakii.
Figure 2.
Population structure of K = 2 inferred by Bayesian clustering approaches based on 35 microsatellite markers. BJYQ: population of Yanqing from Beijing; HBYC: population of Yichang from Hubei province.
3. Materials and Methods
3.1. Sample Collection and DNA Extraction
A total of 63 larvae were collected from two geographic regions in China, of which 31 samples were from Yanqing of the Beijing (N 40°27′20.05″, E 115°58′8.14″), named BJYQ, and 32 specimens came from Yichang of Hubei province (N 30°41′39.43″, E 111°16′50.77″), named HBYC. Additionally, eight individuals from eight sampled sites were used for the initial test. Samples were stored in ethanol absolute and frozen at −80 °C prior to use. Genomic DNA were extracted from half of an individual larva using DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions.
3.2. Sequencing, Microsatellites Searching and Primer Design
One larva of C. sasakii from Beijing was used to prepare the library with the Illumina TruSeq DNA PCR-Free HT Library Prep Kit (Illumina, San Diego, CA, USA), and then sequenced on a Illumina MiSeq Sequencer using the MiSeq Reagent Kit v3 (Illumina, San Diego, CA, USA). Generated genomic sequences were assembled by SOAPdenovo program [23].
The microsatellite isolation from the genomic sequences and primer design for loci was conducted in the software QDD [41]. The searching criteria were as follows: at least six motif repeats for target microsatellites, and PCR product lengths ranged between 90 and 350 bp. For primer design, the annealing temperature ranged from 52 to 68 °C, and the difference in annealing temperature in one pairwise primer was <5 °C. The remaining parameters were at default settings.
3.3. Primer Testing and Polymorphism Detection
Firstly, in order to improve efficiency and lower cost, we added a PC tail (Primer tail C) (5′ CAGGACCAGGCTACCGTG 3′) to the 5′ end of the candidate forward primer [42]. Eight larvae of C. sasakii from eight different populations were used for the initial test. Amplification was carried out in a final volume of 10 μL, containing 0.5 μL (12.5 ng) of template DNA, 5 μL of Master Mix (Promega, Madison, WI, USA), 0.25 μL of forward primer (modified by the PC tail) at a final concentration 0.25 µM, 0.25 μL (10 µM) of reverse primer at a final concentration 0.25 µM, and 4 μL of ddH2O. The amplification program was as follows: 4 min at 94 °C; 35 cycles of 30 s at 94 °C, 30 s at 56 °C, and 45 s at 72 °C, with a final 10-min extension at 72 °C. PCR products were visualized on agarose gel (1.5%) electrophoresis. This step was taken to screen primers that can amplify PCR fragment.
Secondly, primers selected in previous steps were tested using a capillary sequencer. Amplification was performed in a final volume of 10 μL, containing 0.5 μL (12.5 ng) of template DNA, 5 μL of Master Mix (Promega, Madison, WI, USA), 0.08 μL of forward primer (modified by the PC tail) at a final concentration 0.08 µM, 0.16 μL of reverse primer at a final concentration 0.16 µM, 0.32 μL of PC tails modified by fluorescence (FAM (blue), HEX (green), and ROX (red)) including different color at a final concentration 0.32 µM, and 3.94 μL of ddH2O. The amplification program was the same as above. The ABI 3730xl DNA Analyzer (Applied Biosystems, Foster, CA, USA) was used to analyze the amplified PCR fragments with the GeneScan 500 LIZ size standard (Applied Biosystems).
Finally, marker primers screened out by the first two steps were validated in 63 samples from two regions. Amplification mixture, amplification program, and analysis of PCR fragments were the same as the second step.
3.4. Statistical Analysis
Genotyping data was identified, and errors were corrected by MICRO-CHECKER [43]. Diversity statistics including allele frequencies, Ho, He and PIC were estimated by the macros Microsatellite Tools [44]. Tests for linkage disequilibrium among loci within each population and deviation from HWEat each locus/population pair, and estimation of FIS for each population, were performed in GENEPOP v4.0 (Applied Biosystems). Additionally, the null allele test was conducted with FREENA [34]. The program LOSTAN [45] was used to detect putative loci potentially under selection with two options: neutral mean FST’ and force mean FST’. Corresponding sequences of polymorphic loci were screened using BLASTx and BLASTn in the NCBI database (http://www.ncbi.nlm.nih.gov/). Population differentiation was investigated using the Bayesian clustering approach implemented in the program STRUCTURE, version 2.3.3 [46]. Simulations were run for 200,000 Markov chain Monte Carlo with a burn-in of 100,000 iterations under admixture ancestry and correlated allele frequency models. We performed 15 independent runs for each K (from 1 to 6) to confirm consistency across runs. The most accurate number of groups (K) was visually examined when plotting K against delta-K and using the Evanno method in the online program STRUCTURE HARVESTER [47].
4. Conclusions
We characterized and developed microsatellite markers for C. sasakii from random regions of the genome generated by using next-generation sequencing. The loci assessed in our study could reveal the genetic structure in two geographical populations. This method provides fast way for high throughput development of microsatellite markers from non-model species without reference genome.
Acknowledgments
We thank Zong-Jiang Kang and Jiang-Wen Qin for collection of specimens. This research was supported by the National Basic Research Program (2013CB127600), the National Natural Science Foundation (31472025), and the Special Fund for Agro-scientific Research in the Public Interest (201103024), China.
Supplementary Materials
The following are available online at www.mdpi.com/1422-0067/17/3/362/s1.
Author Contributions
Shu-Jun Wei and Jia-Ying Zhu conceived and designed the experiments; You-Zhu Wang and Li-Jun Cao performed the experiments and analyzed the data; You-Zhu Wang and Shu-Jun Wei wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Kim D.S., Lee J.H., Yiem M.S. Spring emergence pattern of Carposina sasakii (Lepidoptera: Carposinidae) in apple orchards in Korea and its forecasting models based on degree-days. Environ. Entomol. 2000;29:1188–1198. doi: 10.1603/0046-225X-29.6.1188. [DOI] [Google Scholar]
- 2.Liu Y.S., Cheng J.A., Mou J.Y. Review of the advances of the peach fruit-borer (Carposina sasakii Matsmura) J. Shandong Agric. Univ. 1997;28:113–120. [Google Scholar]
- 3.Han X.J. The damage and prevention of peach friut moth on pomegranate. China Rurac Sci. Technol. 2002;5:17. [Google Scholar]
- 4.Hua B.Z., Zhang A.J., Lu X.Z., Hua L. Seasonal history of Carposina sasakii Matsumura on apricot orchard in Qinling mountain region, Shaanxi. Acta Agric. Boreali-Occident. Sin. 1998;7:32–35. [Google Scholar]
- 5.Jiao R.L. The occurrence and prevention of peach fruit moth. Plant Dr. 2006;19:19. [Google Scholar]
- 6.Wang X.M., Liu M. A detailed study of the living history, medicament prevention, cure and its relations with meteorology. J. Shenyang Univ. 1999:63–71. (In Chinese) [Google Scholar]
- 7.Cai P., Ding W.Z. Briefing of peach fruit moth damage on pomegranate. China Fruits. 1990:35–37. (In Chinese) [Google Scholar]
- 8.Hua L., Shen B.C. Preliminary study on the peach fruit borer of wild jujube. Plant Prot. Sci. 1992:15–17. (In Chinese) [Google Scholar]
- 9.Hua L., Hua B.Z., Huang W.L. The bionomics of peach fruit borer damaged on apricot tree. Acta Phytophylacica Sin. 1998;25:141–144. [Google Scholar]
- 10.Hua L., Hua B.Z. Preliminary study on the host-biotypes of peach fruit borer. Acta Phytophylacica Sin. 1995;22:165–170. [Google Scholar]
- 11.Hua L., Wang J.F., Hui X.X. The esterase isozymes of the Carposina sasakii from different hosts. Acta Agric. Boreali-Occident. Sin. 1995;4:95–96. [Google Scholar]
- 12.Xu Q.G., Hua B.Z. RAPD analysis on the speciation in host races of Carposina sasakii Matsumura (Lepidoptera: Carposinidae) Acta Entomol. Sin. 2004;47:379–383. [Google Scholar]
- 13.Wang J., Yu Y., Li L.L., Guo D., Tao Y.L., Chu D. Carposina sasakii (Lepidoptera: Carposinidae) in its native range consists of two sympatric cryptic lineages as revealed by mitochondrial COI gene sequences. J. Insect Sci. 2015;15:1–6. doi: 10.1093/jisesa/iev063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishitani G., Nagai S., Lian C.L., Yamaguchi H., Sakamoto S., Yoshimatsu S., Oyama K., Itakura S., Yamaguchi M. Development of compound microsatellite markers in the harmful red tide species Chattonella ovata (Raphidophyceae) Mol. Ecol. Notes. 2007;7:1251–1253. doi: 10.1111/j.1471-8286.2007.01847.x. [DOI] [Google Scholar]
- 15.Bruford M.W. Microsatellite and their application to population genetic studies. Curr. Opin. Genet. Dev. 1993;3:939–943. doi: 10.1016/0959-437X(93)90017-J. [DOI] [PubMed] [Google Scholar]
- 16.Wei S.J., Cao L.J., Gong Y.J., Shi B.C., Wang S., Zhang F., Guo X.J., Wang Y.M., Chen X.X. Population genetic structure and approximate Bayesian computation analyses reveal the southern origin and northward dispersal of the oriental fruit moth Grapholita molesta (Lepidoptera: Tortricidae) in its native range. Mol. Ecol. 2015;24:4094–4111. doi: 10.1111/mec.13300. [DOI] [PubMed] [Google Scholar]
- 17.Wei S.J., Shi B.C., Gong Y.J., Jin G.H., Chen X.X., Meng X.F. Genetic structure and demographic history reveal migration of the diamondback moth Plutella xylostella (Lepidoptera: Plutellidae) from the southern to northern regions of China. PLoS ONE. 2013;8:362. doi: 10.1371/journal.pone.0059654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zalapa J.E., Cuevas H., Zhu H., Steffan S., Senalik D., Zeldin E., McCown B., Harbut R., Simon P. Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am. J. Bot. 2012;99:193–208. doi: 10.3732/ajb.1100394. [DOI] [PubMed] [Google Scholar]
- 19.Yang T., Fang L., Zhang X., Hu J., Bao S., Hao J., Li L., He Y., Jiang J., Wang F., Tian S., Zong X. High-throughput development of SSR markers from pea (Pisum sativum L.) based on next generation sequencing of a purified chinese commercial variety. PLoS ONE. 2015;10:e0139775. doi: 10.1371/journal.pone.0139775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X.M., Sun J.T., Xue X.F., Zhu W.C., Hong X.Y. Development and characterization of 18 novel EST-SSRs from the western flower thrips, Frankliniella occidentalis (Pergande) Int. J. Mol. Sci. 2012;13:2863–2876. doi: 10.3390/ijms13032863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malausa T., Gilles A., MeglÉCz E., Blanquart H., Duthoy S., Costedoat C., Dubut V., Pech N., Castagnone-Sereno P., DÉLye C., et al. High-throughput microsatellite isolation through 454 GS-FLX Titanium pyrosequencing of enriched DNA libraries. Mol. Ecol. Resour. 2011;11:638–644. doi: 10.1111/j.1755-0998.2011.02992.x. [DOI] [PubMed] [Google Scholar]
- 22.Cox M.P., Peterson D.A., Biggs P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:1–6. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang T.H., Han S.H., Park S.J. Development of seven microsatellite markers using next generation sequencing for the conservation on the Korean population of Dorcus hopei (E. Saunders, 1854) (Coleoptera, Lucanidae) Int. J. Mol. Sci. 2015;16:21330–21341. doi: 10.3390/ijms160921330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berman M., Austin C.M., Miller A.D. Characterisation of the complete mitochondrial genome and 13 microsatellite loci through next-generation sequencing for the New Caledonian spider-ant Leptomyrmex pallens. Mol. Biol. Rep. 2014;41:1179–1187. doi: 10.1007/s11033-013-2657-5. [DOI] [PubMed] [Google Scholar]
- 26.Nam H.Y., Coates B., Kim K.S., Park M., Lee J.-H. Characterization of 12 novel microsatellite markers of Sogatella furcifera (Hemiptera: Delphacidae) identified from next-generation sequence data. J. Insect Sci. 2015;15 doi: 10.1093/jisesa/iev069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai X., Zhang W., Orantes L., Jun T.H., Mittapalli O., Mian M.A., Michel A.P. Combining next-generation sequencing strategies for rapid molecular resource development from an invasive aphid species, Aphis glycines. PLoS ONE. 2010;5:362. doi: 10.1371/journal.pone.0011370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouanani M.A., Magne F., Lecompte E., Crouau-Roy B. Development of 18 novel polymorphic microsatellites from Coccinella septempunctata and cross-species amplification in Coccinellidae species. Conserv. Genet. Resour. 2015;7:445–449. doi: 10.1007/s12686-014-0392-4. [DOI] [Google Scholar]
- 29.Meglécz E., Nève G., Biffin E., Gardner M.G. Breakdown of phylogenetic signal: A survey of microsatellite densities in 454 shotgun sequences from 154 non model Eukaryote species. PLoS ONE. 2012;7:362. doi: 10.1371/journal.pone.0040861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arunkumar K.P., Kifayathullah L., Nagaraju J. Microsatellite markers for the Indian golden silkmoth, Antheraea assama (Saturniidae: Lepidoptera) Mol. Ecol. Resour. 2009;9:268–270. doi: 10.1111/j.1755-0998.2008.02414.x. [DOI] [PubMed] [Google Scholar]
- 31.Dharmarajan G., Beatty W.S., Rhodes O.E. Heterozygote deficiencies caused by a Wahlund effect: Dispelling unfounded expectations. J. Wildl. Manag. 2013;77:226–234. doi: 10.1002/jwmg.458. [DOI] [Google Scholar]
- 32.Amsellem L., Risterucci A.M., Benrey B. Isolation and characterization of polymorphic microsatellite loci in Lobesia botrana Den. & Schiff. (Lepidoptera: Tortricidae) Mol. Ecol. Notes. 2003;3:117–119. [Google Scholar]
- 33.An B., Deng X., Shi H., Ding M., Lan J., Yang J., Li Y. Development and characterization of microsatellite markers for rice leaffolder, Cnaphalocrocis medinalis (Guenee) and cross-species amplification in other Pyralididae. Mol. Biol. Rep. 2014;41:1151–1156. doi: 10.1007/s11033-013-2961-0. [DOI] [PubMed] [Google Scholar]
- 34.Chapuis M.P., Estoup A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007;24:621–631. doi: 10.1093/molbev/msl191. [DOI] [PubMed] [Google Scholar]
- 35.Coates B.S., Sumerford D.V., Hellmich R.L., Lewis L.C. Mining an Ostrinia nubilalis midgut expressed sequence tag (EST) library for candidate genes and single nucleotide polymorphisms (SNPs) Insect Mol. Biol. 2008;17:607–620. doi: 10.1111/j.1365-2583.2008.00833.x. [DOI] [PubMed] [Google Scholar]
- 36.Groenewald C.W., Born C., Chown S.L., van Vuuren B.J. Development of a microsatellite library for the flightless moth Pringleophaga marioni Viette (Lepidoptera: Tineidae) Conserv. Genet. Resour. 2011;3:291–294. doi: 10.1007/s12686-010-9344-9. [DOI] [Google Scholar]
- 37.Meglecz E., Petenian F., Danchin E., D’Acier A.C., Rasplus J.Y., Faure E. High similarity between flanking regions of different microsatellites detected within each of two species of Lepidoptera: Parnassius apollo and Euphydryas aurinia. Mol. Ecol. 2004;13:1693–1700. doi: 10.1111/j.1365-294X.2004.02163.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhang D.X. Lepidopteran microsatellite DNA: Redundant but promising. Trends Ecol. Evol. 2004;19:507–509. doi: 10.1016/j.tree.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 39.Cao L.J., Wen J.B., Wei S.J., Liu J., Yang F., Chen M. Characterization of novel microsatellite markers for Hyphantria cunea and implications for other Lepidoptera. Bull. Entomol. Res. 2015;105:273–284. doi: 10.1017/S0007485315000061. [DOI] [PubMed] [Google Scholar]
- 40.A’Hara S., Cottrell J. Development and characterisation of ten polymorphic microsatellite markers for the pine-tree lappet moth Dendrolimus pini (Lepidoptera: Lasiocampidae) Conserv. Genet. Resour. 2013;5:1135–1137. doi: 10.1007/s12686-013-9977-6. [DOI] [Google Scholar]
- 41.Meglécz E., Costedoat C., Dubut V., Gilles A., Malausa T., Pech N., Martin J.F. QDD: A user-friendly program to select microsatellite markers and design primers from large sequencing projects. Bioinform. 2010;26:403–404. doi: 10.1093/bioinformatics/btp670. [DOI] [PubMed] [Google Scholar]
- 42.Blacket M.J., Robin C., Good R.T., Lee S.F., Miller A.D. Universal primers for fluorescent labelling of PCR fragments—an efficient and cost—effective approach to genotyping by fluorescence. Mol. Ecol. Resour. 2012;12:456–463. doi: 10.1111/j.1755-0998.2011.03104.x. [DOI] [PubMed] [Google Scholar]
- 43.Van Oosterhout C., Hutchinson W.F., Wills D.P.M., Shipley P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. doi: 10.1111/j.1471-8286.2004.00684.x. [DOI] [Google Scholar]
- 44.Park S.D.E. Ph.D. Thesis. University of Dublin; Ireland, UK: 2001. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection. [Google Scholar]
- 45.Antao T., Lopes A., Lopes R.J., Beja-Pereira A., Luikart G. LOSITAN: A workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinform. 2008;9:323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;7:574–578. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Earl D.A., vonHoldt B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2011;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.