ABSTRACT
Altered nuclear size is associated with many cancers, and determining whether cancer-associated changes in nuclear size contribute to carcinogenesis necessitates an understanding of mechanisms of nuclear size regulation. Although nuclear import rates generally positively correlate with nuclear size, NTF2 levels negatively affect nuclear size, despite the role of NTF2 (also known as NUTF2) in nuclear recycling of the import factor Ran. We show that binding of Ran to NTF2 is required for NTF2 to inhibit nuclear expansion and import of large cargo molecules in Xenopus laevis egg and embryo extracts, consistent with our observation that NTF2 reduces the diameter of the nuclear pore complex (NPC) in a Ran-binding-dependent manner. Furthermore, we demonstrate that ectopic NTF2 expression in Xenopus embryos and mammalian tissue culture cells alters nuclear size. Finally, we show that increases in nuclear size during melanoma progression correlate with reduced NTF2 expression, and increasing NTF2 levels in melanoma cells is sufficient to reduce nuclear size. These results show a conserved capacity for NTF2 to impact on nuclear size, and we propose that NTF2 might be a new cancer biomarker.
KEY WORDS: Nucleus, NTF2, Nuclear transport, Ran, Nuclear pore complex, Nuclear structure, Xenopus, Cancer, Melanoma, Development, Nuclear size, Nuclear scaling
Summary: NTF2 reduces the diameter of the NPC, nuclear import of large cargos and nuclear size in a Ran-binding-dependent manner, in both Xenopus and mammalian cells.
INTRODUCTION
Cell size varies when comparing different species and tissue types, and dramatic changes in cell size accompany early development and cell differentiation within the same organism. A fundamental question in cell biology is how the sizes of intracellular structures and organelles are regulated relative to cell size (Rafelski and Marshall, 2008; Chan and Marshall, 2012; Levy and Heald, 2012; Marshall, 2012). It has long been noted that the size of the nucleus tends to correlate with the size of the cell, a phenomenon referred to as nuclear scaling (Conklin, 1912; Wilson, 1925; Webster et al., 2009; Chan and Marshall, 2010; Levy and Heald, 2010; Walters et al., 2012; Hara et al., 2013; Jevtic and Levy, 2015). Altered nuclear scaling is associated with many cancers, and cytopathologists monitor graded increases in nuclear size for cancer diagnosis and prognosis (Zink et al., 2004; Chow et al., 2012; Jevtic and Levy, 2014). Importantly, nuclear size increases in cancer are frequently independent of gross ploidy changes (Lindberg, 1992; Miyamoto et al., 1992; Sørensen et al., 1992; Wang et al., 1992; Dangou et al., 1993; van Velthoven et al., 1995; Sørensen, 1996; Dey, 2010), implicating potential cytoplasmic contributions to altered nuclear size (Goehring and Hyman, 2012). It is unknown whether changes in nuclear size contribute to carcinogenesis, and answering this question necessitates an understanding of the mechanisms responsible for regulating nuclear size.
Previous studies have implicated nucleocytoplasmic transport in the regulation of nuclear size (Levy and Heald, 2012; Edens et al., 2013; Jevtic et al., 2014; Vukovic et al., 2016). Blocking nuclear export increases nuclear size in fission yeast (Neumann and Nurse, 2007), and altering nucleoporin (Nup) expression affects nuclear size in Tetrahymena, Arabidopsis, and Xenopus (Iwamoto et al., 2009; Tamura et al., 2010; Theerthagiri et al., 2010; Shaulov et al., 2011; Tamura and Hara-Nishimura, 2011). In addition, manipulating the expression of various nuclear transport factors causes nuclear size changes in Tetrahymena, worms and budding yeast (Malone et al., 2008; Gonzalez et al., 2009; Meyerzon et al., 2009). One specific nucleocytoplasmic transport factor implicated in nuclear size regulation is nuclear transport factor 2 (NTF2, also known as NUTF2). NTF2 serves as a recycling factor for the small GTPase Ran, binding to RanGDP in the cytoplasm and targeting it to the nuclear pore complex (NPC) through association with FG-repeat containing Nups, including Nup62 (Paschal and Gerace, 1995; Clarkson et al., 1996, 1997; Smith et al., 1998; Bayliss et al., 1999, 2002; Morrison et al., 2003). NTF2 translocates RanGDP into the nucleus where RCC1, the Ran guanine-nucleotide-exchange factor (GEF) associated with the chromatin, exchanges RanGDP for RanGTP (Ribbeck et al., 1998). In this way, NTF2 promotes efficient nuclear import by replenishing the nuclear RanGTP pool that is responsible for releasing imported cargo molecules bound to importin α or β proteins (Madrid and Weis, 2006; Stewart, 2007).
NTF2 concentrations contribute to nuclear size differences between two related frog species, Xenopus laevis and Xenopus tropicalis. Nuclei assembled in X. tropicalis egg extract, which contains relatively high levels of NTF2, expand more slowly than nuclei in X. laevis egg extract, which has nearly fourfold less NTF2. Supplementing X. laevis extract with increased amounts of NTF2 is sufficient to decrease nuclear size, whereas NTF2 immunodepletion or antibody inhibition in X. tropicalis extract leads to increased nuclear size (Levy and Heald, 2010). One mechanism contributing to these nuclear size effects was determined to be differential nuclear import of large cargo molecules. Addition of NTF2 to X. laevis extract results in increased levels of nuclear Ran and reduced nuclear import kinetics for large cargo molecules, including 20 MDa IBB-coated Qdots and lamin B3 (LB3) that exists as a >700 kDa complex as determined by gel filtration (Adam et al., 2008; Gambus et al., 2011), whereas import of smaller cargos is largely unaffected [e.g. a GST-tagged GFP containing a nuclear localization sequence (GST–GFP-NLS) and smaller Qdots]. Conversely, NTF2 inhibition in X. tropicalis is sufficient to increase the rate of large cargo import, without impacting on the import of small cargos (Levy and Heald, 2010). These data are consistent with studies in Xenopus oocytes showing that microinjection of NTF2 causes a reduction in the import of large nucleoplasmin-coated gold particles (Feldherr et al., 1998). From these studies, it was unclear how NTF2 might specifically regulate the import of large, but not small, cargo molecules. One possibility is that high levels of NTF2 bound to the NPC alter the effective diameter of the pore, potentially slowing the import of larger-sized cargos. Consistent with this idea, experimentally increasing Ran levels in Xenopus oocytes reduced the internal diameter of the pore of the NPC (Goldberg et al., 2000). Here, we test whether binding of Ran to NTF2 is required for NTF2 to reduce NPC size, large cargo import and nuclear size.
In this study, we use Xenopus egg extracts to investigate how NTF2 concentrations and mutants affect nuclear size. Egg extracts contain all the cytoplasmic proteins and membranes necessary for nuclear assembly, and addition of demembranated Xenopus sperm to interphasic extracts induces the formation and expansion of nuclei in vitro (Chan and Forbes, 2006; Edens and Levy, 2014a). Advantages of this open biochemical system over in vivo approaches include: (1) facile and precise manipulation of the cytoplasmic composition by immunodepletion or addition of recombinant proteins (e.g. NTF2 protein); (2) the ability to study individual steps of complex processes in isolation (e.g. nuclear growth and size); (3) the relative rapidity with which these experiments can be performed; and (4) the ability to study essential processes that might pose viability issues in vivo (Levy and Heald, 2015). We also study nuclear size using Xenopus embryo extracts that provide a powerful link between the in vitro and in vivo approaches. Embryo extracts offer many of the same in vitro advantages of egg extracts, while allowing us to study endogenous embryonic nuclei in their native cytoplasm (Edens and Levy, 2014a,b).
Using a mutant version of NTF2 that is defective for Ran binding, we find that the ability of NTF2 to inhibit nuclear growth and import of large cargos in Xenopus egg extract largely depends on its binding to Ran. We also show, by performing transmission electron microscopy (TEM), that wild-type NTF2, but not NTF2 defective for Ran binding, reduces the diameter of the NPC. Furthermore, we show that these in vitro findings are also relevant in vivo, as ectopic NTF2 expression in Xenopus embryos and mammalian tissue culture cells leads to altered nuclear size. Studying melanoma cell lines and tissue microarrays, we find that increases in nuclear size during melanoma progression correlate with reduced NTF2 expression, and that more normal nuclear sizes can be attained in melanoma cells by increasing NTF2 levels. These studies provide new insights into the mechanism of nuclear size regulation by NTF2. We propose that NTF2 bound to Ran at the NPC reduces the effective diameter of the NPC to inhibit import of large cargos and nuclear expansion, and that NTF2 might be a novel cancer biomarker.
RESULTS
Binding of Ran to NTF2 is required for NTF2 to affect nuclear size in Xenopus egg extract
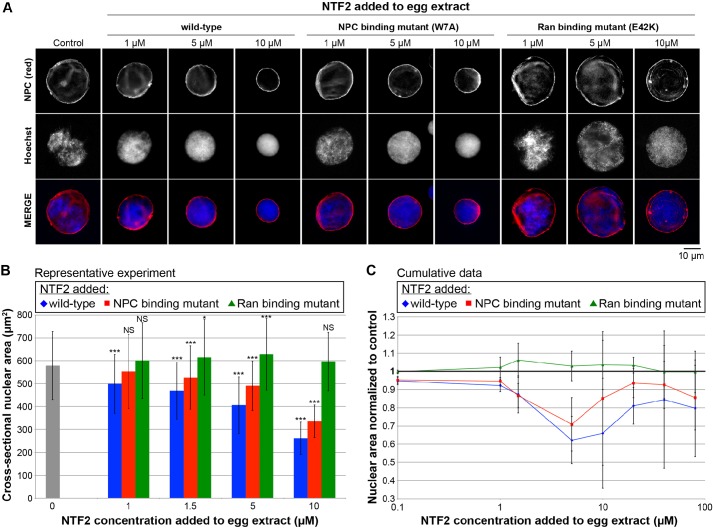
Distinct sites on NTF2 are responsible for Ran and NPC binding, and single point mutations in NTF2 have been previously identified that specifically reduced the affinity of NTF2 for RanGDP (E42K) or for the NPC (W7A). Importantly, structural studies have demonstrated that these mutations do not cause gross changes in protein folding or conformation (Clarkson et al., 1996, 1997; Kent et al., 1996; Bayliss et al., 1999; Morrison et al., 2003). It is worth noting that the W7A mutant retains some affinity for the NPC due to a secondary Nup-binding site that is not easily disrupted by mutation because it is composed of multiple amino acids forming an elongated hydrophobic stripe (Bayliss et al., 1999; Morrison et al., 2003). We purified recombinant versions of these mutant proteins and tested their ability to inhibit the growth of nuclei assembled in X. laevis egg extract. For most of these experiments, NTF2 protein was added to egg extract prior to nuclear assembly; however, we also performed experiments in which extract was supplemented with NTF2 protein after nuclear assembly, which resulted in similar effects on nuclear size. As previously reported, addition of wild-type NTF2 reduced the rate of nuclear expansion (Levy and Heald, 2010), and the effect was more pronounced with increasing NTF2 concentrations up to 5–10 µM. At this NTF2 concentration range, which represents an approximate tenfold increase over the endogenous NTF2 level, nuclear surface area was reduced by 40–50% compared to control-treated nuclei (Fig. 1). The E42K Ran-binding mutant completely failed to inhibit nuclear growth. Rather, in extract supplemented with this NTF2 mutant protein, nuclei were slightly enlarged relative to controls (Fig. 1). At 5–10 µM NTF2 concentrations, the W7A NPC-binding mutant exhibited an intermediate effect, resulting in nuclei that were smaller than controls but not as small as in extract supplemented with wild-type NTF2 protein (Fig. 1). At lower NTF2 concentrations, the effect of the W7A mutant on nuclear size was similar to that of wild-type NTF2. These data demonstrate that Ran binding to NTF2 is absolutely required for NTF2 to affect nuclear size in X. laevis egg extract.
Fig. 1.
Binding of Ran to NTF2 is required for NTF2 to affect nuclear size in X. laevis egg extract. (A) X. laevis egg extract was supplemented with recombinant wild-type and mutant NTF2 proteins at the indicated final concentrations, and the same total volume was added to each reaction. For the control reaction, an equal volume of NTF2 storage buffer was added. Nuclei were assembled and growth was allowed to proceed for 90 min at 20°C. Given that nuclei expand continuously in egg extract, this time point serves as a proxy for the nuclear expansion rate, and at this time nuclei have nearly reached their largest size before physical integrity of the nuclear envelope becomes compromised (Levy and Heald, 2010). Nuclei were fixed, spun onto coverslips, processed for immunofluorescence and stained with mAb414 to visualize the NPC (red) and Hoechst 33342 to visualize the DNA (blue). Representative images are shown. The NPC-binding mutant is NTF2 W7A, and the Ran-binding mutant is NTF2 E42K. (B) Nuclear size data are shown from one representative experiment as described in A. For each bar, the cross-sectional areas of 125–643 nuclei were measured from NPC-stained nuclei and averaged. The error bars are s.d. *P<0.05; ***P<0.001; NS, not significant (compared with the buffer addition control; two-tailed Student's t-test). (C) Cumulative data from 32 different X. laevis egg extracts treated as described in A. The average cross-sectional nuclear areas were measured as in B and normalized to the buffer addition control (bold horizontal line set at 1.0), due to variability between egg extracts. Normalized nuclear areas were averaged across multiple experiments. The error bars are s.d. A subset of experiments was performed in which NTF2 proteins were added to X. laevis egg extract containing nuclei pre-assembled for 35 min followed by an additional 75-min incubation period. Similar nuclear size effects were observed, so these data were averaged along with the data in which NTF2 protein was added to extract at the beginning of nuclear assembly. For the wild-type and NPC-binding mutant, all normalized nuclear area data points are statistically different from the buffer control (1.0) by at least P<0.05; for the Ran-binding mutant, the 10 µM data point is the only one that is statistically different from the buffer control, with P<0.05 (two-tailed Student's t-test).
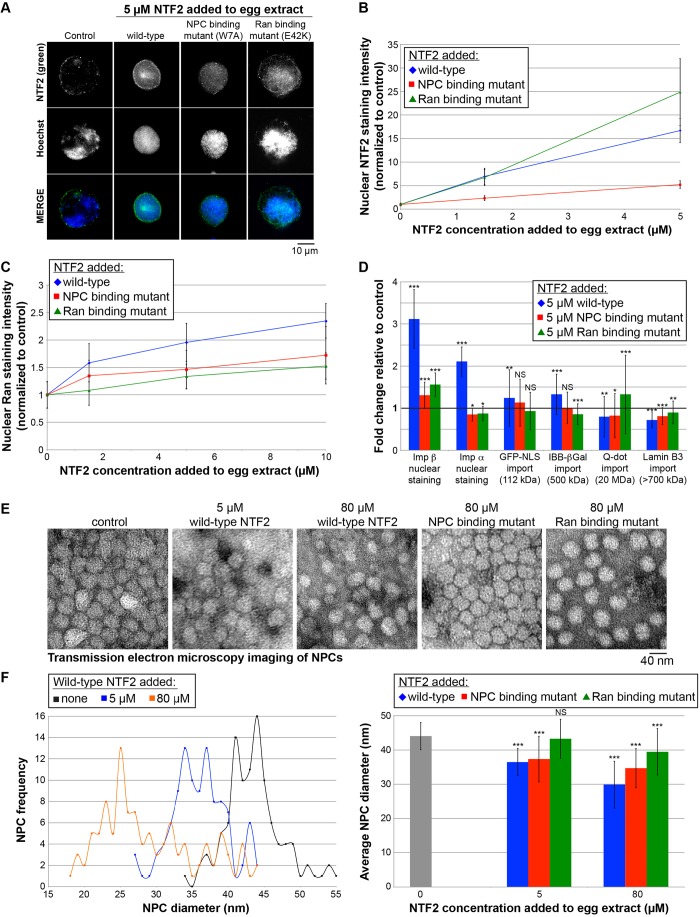
We wondered whether the ability of the W7A NTF2 mutant to partially inhibit nuclear growth might be due to residual affinity for the NPC, as previously shown by TEM (Bayliss et al., 1999). We performed immunofluorescent staining against NTF2 on nuclei assembled in egg extract supplemented with either wild-type or mutant NTF2 proteins. Increased nuclear NTF2 staining was observed with increasing concentrations of both wild-type NTF2 and the E42K Ran-binding mutant (Fig. 2A,B). This demonstrates that the E42K mutation does not interfere with targeting of that NTF2 mutant protein to the NPC. In extract supplemented with the NPC-binding mutant W7A, nuclear NTF2 staining was significantly reduced relative to extract supplemented with wild-type NTF2, showing that the mutant has reduced affinity for the NPC. However, the W7A mutation clearly does not abolish NPC binding, as nuclear NTF2 staining intensity still increased with increasing concentrations of the W7A mutant protein. For instance, addition of 5 µM wild-type protein increased nuclear NTF2 staining intensity greater than 15-fold relative to controls, and the W7A mutant still increased nuclear NTF2 localization by fivefold (Fig. 2A,B). This observation might explain why the W7A mutant still retains some ability to inhibit nuclear growth (Fig. 1).
Fig. 2.
Effects of wild-type and mutant NTF2 proteins on nuclear localization of transport factors, nuclear import, and NPC diameter. (A) X. laevis egg extract was supplemented with recombinant wild-type and mutant NTF2 proteins at the indicated final concentrations, and the same total volume was added to each reaction. For the control reaction, an equal volume of NTF2 storage buffer was added. Nuclei were assembled and growth was allowed to proceed for 90 min at 20°C. Nuclei were fixed, spun onto coverslips, processed for immunofluorescence, and stained with an antibody against NTF2 (green) and Hoechst 33342 to visualize the DNA (blue). Representative images acquired with the same exposure time are shown. (B) For each data point, the nuclear NTF2 fluorescence staining intensity per unit area was quantified for 102–117 nuclei, averaged and normalized to the buffer control (set at 1.0). The error bars are s.d. All data points are statistically different from the buffer control (1.0) by at least P<0.001 (two-tailed Student's t-test). One representative experiment out of three is shown. (C) Similar experiments were performed except that nuclei were stained with an antibody against Ran. For each data point, the nuclear Ran fluorescence intensity per unit area was quantified for 100–130 nuclei, averaged and normalized to the buffer control (set at 1.0). The error bars are s.d. All data points are statistically different from the buffer control (1.0) by at least P<0.05 (two-tailed Student's t-test). One representative experiment out of six is shown. (D) Similar experiments were performed except that only the 5 µM concentration of added NTF2 protein was tested. As indicated, nuclei were either stained with an antibody against importin β or importin α2, or import of the indicated cargos was quantified as previously described (Lyman et al., 2002; Levy and Heald, 2010). For each data point, the nuclear fluorescence intensity per unit area was quantified for 111–1108 nuclei, averaged and normalized to the buffer control (bold horizontal line set at 1.0). The error bars are s.d. One representative experiment out of three is shown. *P<0.05; **P<0.01; ***P<0.001; NS, not significant (two-tailed Student's t-test). The GST–GFP-NLS cargo is listed as 112 kD assuming it is dimeric. The lamin B3 cargo is listed as >700 kDa based on gel filtration experiments (Adam et al., 2008; Gambus et al., 2011). (E) Nuclei were assembled as described in A. Unfixed nuclei were negatively stained with uranyl acetate, and NPCs were visualized by TEM. Representative images are shown. Although this approach does not provide structural details of the NPC, we prefer this method for quantifying NPC diameters because the NPCs are not fixed and their structures are minimally perturbed during sample preparation. (F) For each sample, TEM images were acquired for 6–7 different nuclei, and the diameters of 103–123 NPCs were quantified using ImageJ. As negative staining outlines the entire NPC, here we are quantifying the diameter of the NPC and not the central channel. NPC frequency refers to the number of NPCs with the indicated NPC diameter. The error bars are s.d. ***P<0.001; NS, not significant (two-tailed Student's t-test).
To confirm that the E42K mutant was functionally defective in Ran binding, we performed immunofluorescent staining against Ran on nuclei assembled in egg extract supplemented with either wild-type or mutant NTF2 proteins. Although wild-type NTF2 increased nuclear Ran localization, this effect was significantly reduced with the E42K Ran-binding mutant, as expected (Fig. 2C). The W7A NPC-binding mutant also exhibited reduced nuclear Ran localization relative to the wild-type protein, showing that targeting of NTF2 to the NPC contributes to its ability to increase nuclear Ran localization (Fig. 2C).
Given that Ran binding to NTF2 is required to increase the level of nuclear-localized Ran and to reduce nuclear size, we hypothesized that these nuclear size reductions might be mediated by recruitment of additional Ran-binding proteins. To test whether increasing the levels of NTF2 led to concomitant increases in the recruitment of other Ran-binding proteins to the NPC or nucleus, we performed quantitative immunofluorescence. Although supplementing X. laevis egg extract nuclei with additional NTF2 increased nuclear Ran levels, there were no significant changes in the amounts of nuclear RanGAP1, RCC1, RanBP1, RanBP2, CAS (also known as CSE1L), TRN (also known as TNPO1) or CRM1 (also known as XPO1) (data not shown). These data suggest that these Ran-binding proteins are not mediators of the effect of NTF2 on nuclear size. Rather, we favor the model that increased amounts of NTF2 bound to Ran at the NPC alter the conformation of the pore in a way that reduces import of large cargo molecules, thereby impacting on nuclear import and size. To directly test this idea, we performed TEM on nuclei assembled in egg extract and quantified NPC diameters. Addition of 5 µM or 80 µM wild-type NTF2 reduced average NPC diameter by ∼20% or ∼30%, respectively (Fig. 2E,F). In contrast, the E42K Ran-binding mutant had no effect on NPC size at 5 µM, and only decreased average pore diameter by ∼10% at 80 µM, and the W7A NPC-binding mutant exhibited an intermediate effect, decreasing average pore diameter by ∼20% at 80 µM (Fig. 2E,F). For egg extract nuclei supplemented with NTF2, NPC clustering appeared reduced, as assessed by TEM imaging, whereas the overall NPC density was slightly increased as determined by quantitative NPC immunofluorescence (data not shown).
Previous studies have shown that increased NTF2 levels reduce import of 40-nm Qdot and 19–23-nm gold particles, whereas import of smaller Qdot and gold particles is not affected (Feldherr et al., 1998; Levy and Heald, 2010). Given that binding of Ran to NTF2 is required for NTF2 to reduce NPC size and nuclear size, a prediction is that binding of Ran to NTF2 would also be required to inhibit import of large cargos. To test this idea, we supplemented extract with 5 µM of wild-type or mutant NTF2 proteins and quantified the nuclear import of different sized cargos. Import of GST–GFP-NLS (112 kDa assuming a dimer) and IBB-βGal (importin-β-binding domain of human SRP1a fused to β-galactosidase that exists as a 500-kDa tetrameric complex) cargos was slightly increased by wild-type NTF2, whereas the Ran- and NPC-binding mutants had little effect on import of these smaller cargo molecules (Fig. 2D). As expected, import of the 20-MDa Qdot cargo was reduced by wild-type NTF2 and to a lesser degree by the NPC-binding mutant. Strikingly, import of this large cargo was actually increased by addition of the NTF2 Ran-binding mutant (Fig. 2D). A similar trend was observed for import of lamin B3 (LB3), in which LB3 import was most inhibited by wild-type NTF2 and least inhibited by the Ran-binding mutant (Fig. 2D). These data suggest that LB3 is imported as a large cargo, consistent with gel filtration studies showing that LB3 in X. laevis egg extract exists in a complex that is >700 kDa, indicating the complex is highly oligomeric and/or extended in conformation (Adam et al., 2008; Gambus et al., 2011). These results are consistent with the idea that NTF2 must bind Ran at the NPC in order to reduce pore diameter and import of large cargo molecules, including LB3, to inhibit nuclear expansion.
In the course of these experiments, we noted that the nuclear levels of importin β and importin α2 were increased when extract was supplemented with wild-type NTF2, and that this effect was much less pronounced with the W7A and E42K mutants (Fig. 2D). This led us to investigate whether even higher concentrations of NTF2 might increase nuclear expansion rates by further increasing the rate of bulk import and potentially overcoming the inhibitory effect of NTF2 on large cargo import. To test this idea, we increased the NTF2 concentration in egg extract to more than 100-fold above endogenous levels. Although nuclei were again smaller than controls, nuclear surface area was only reduced by around 20%, as opposed to 40–50% by lower levels of NTF2 (Fig. 1C). Import of Ran and GFP-NLS, as well as two native nuclear import substrates (150 kDa pentameric nucleoplasmin and 240 kDa NuMa), was increased at 80 µM NTF2, demonstrating that at high NTF2 levels there is an increase in bulk import and the NTF2 protein is not aggregating and becoming less effective (data not shown). As with lower NTF2 concentrations, the Ran-binding mutant had little effect on nuclear size at these higher NTF2 concentrations, and the NPC-binding mutant exhibited an intermediate effect, reducing nuclear surface area by ∼15% (Fig. 1C). These data suggest that the concentration-dependent effect of NTF2 on nuclear size might be dictated by a balance between its inhibitory effect on the import kinetics of large cargo molecules and its stimulatory effect on bulk import kinetics.
NTF2 levels affect nuclear size in vivo in Xenopus embryos
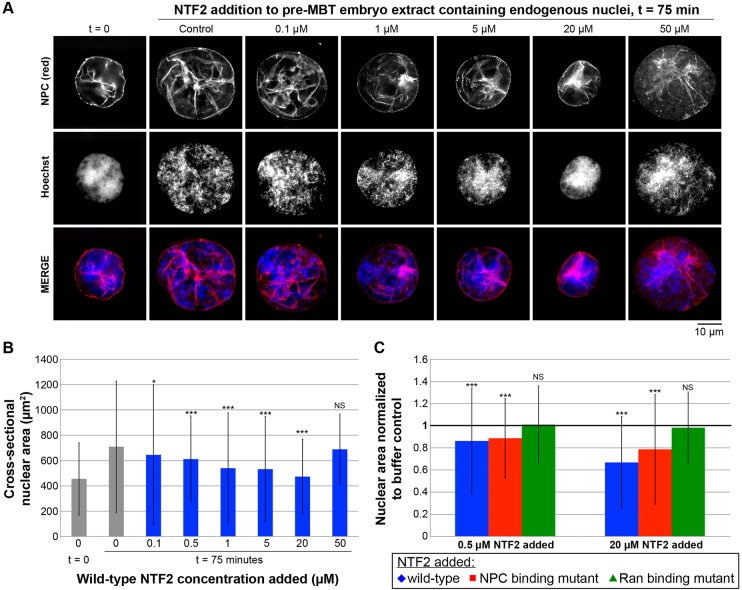
To determine whether our in vitro results in egg extract were relevant in an in vivo setting, we examined how NTF2 levels affect nuclear size in X. laevis embryos. As a transition between our in vitro and in vivo studies, we first tested the effect of increased NTF2 concentrations on nuclear size in X. laevis embryo extracts. We prepared embryo extract containing endogenous embryonic nuclei from early stage 8 embryos (Nieuwkoop and Faber, 1967), just prior to the activation of zygotic gene transcription associated with the midblastula transition (MBT). When these extracts were incubated at room temperature for 75 min, the surface area of nuclei in the extract increased by ∼50%, providing a system to test the effect of increased NTF2 levels on embryonic nuclear growth kinetics (Fig. 3A,B). When embryo extract was supplemented with increasing concentrations of recombinant wild-type NTF2 protein, nuclear expansion was inhibited, such that nuclei ultimately attained smaller sizes compared to the buffer-addition control. When 20 µM NTF2 was added, almost no nuclear growth was observed (Fig. 3A,B). Interestingly, NTF2 concentrations higher than 20 µM resulted in less inhibition of nuclear growth, consistent with these high NTF2 levels increasing bulk import (Fig. 3A,B). Effects of NTF2 mutant proteins on embryonic extract nuclear size were similar to those observed in egg extract. Nuclear growth was not significantly altered by the Ran-binding mutant, whereas the NPC-binding mutant reduced nuclear size but not to the same extent as wild-type NTF2, consistent with this mutant retaining some reduced affinity for the NPC (Fig. 3C).
Fig. 3.
NTF2 concentration affects nuclear size in X. laevis embryo extracts. (A) Embryo extracts containing endogenous embryonic nuclei were prepared from X. laevis embryos at early stage 8. Recombinant NTF2 protein was added to the indicated final concentrations, and the same total volume was added to each reaction. For the control reaction, an equal volume of NTF2 storage buffer was added. Nuclei were incubated for 75 min at 20°C, during which time some nuclear growth occurs. Nuclei were then fixed, spun onto coverslips, processed for immunofluorescence, and stained with mAb414 to visualize the NPC (red) and Hoechst 33342 to visualize the DNA (blue). Representative images are shown. (B) For each sample, the cross-sectional areas of 127–625 nuclei were measured from NPC-stained nuclei and averaged. Average nuclear size data from three different embryo extracts are shown. The error bars are s.d. For the statistical analysis, calculations were performed relative to the no-NTF2-added control at t=75 min. (C) Average cross-sectional nuclear areas were measured as in B and normalized to the buffer-addition control (bold horizontal line set at 1.0). Average nuclear size data from three different embryo extracts are shown. The error bars are s.d. For the statistical analysis, calculations were performed relative to the no-NTF2-added control at t=75 min. *P<0.05; ***P<0.001; NS, not significant (two-tailed Student's t-test).
To determine whether ectopic NTF2 expression within the embryo would alter nuclear size in later stages of development, we microinjected single-cell X. laevis embryos with four different amounts of mRNA encoding NTF2, allowed embryos to develop to late stage 8 or stage 9 (i.e. post-MBT), and then isolated and quantified endogenous nuclei from these embryos. Nuclei were smaller in embryos microinjected with low levels of NTF2 mRNA, exhibiting ∼20% smaller nuclear envelope surface areas compared to controls (Fig. 4). Higher amounts of NTF2 mRNA resulted in increased nuclear size relative to control-injected embryos, with anywhere from 10–50% increased nuclear envelope surface areas (Fig. 4). These data are consistent with our in vitro results, demonstrating that in vivo embryonic nuclear size is sensitive to NTF2 expression levels.
Fig. 4.
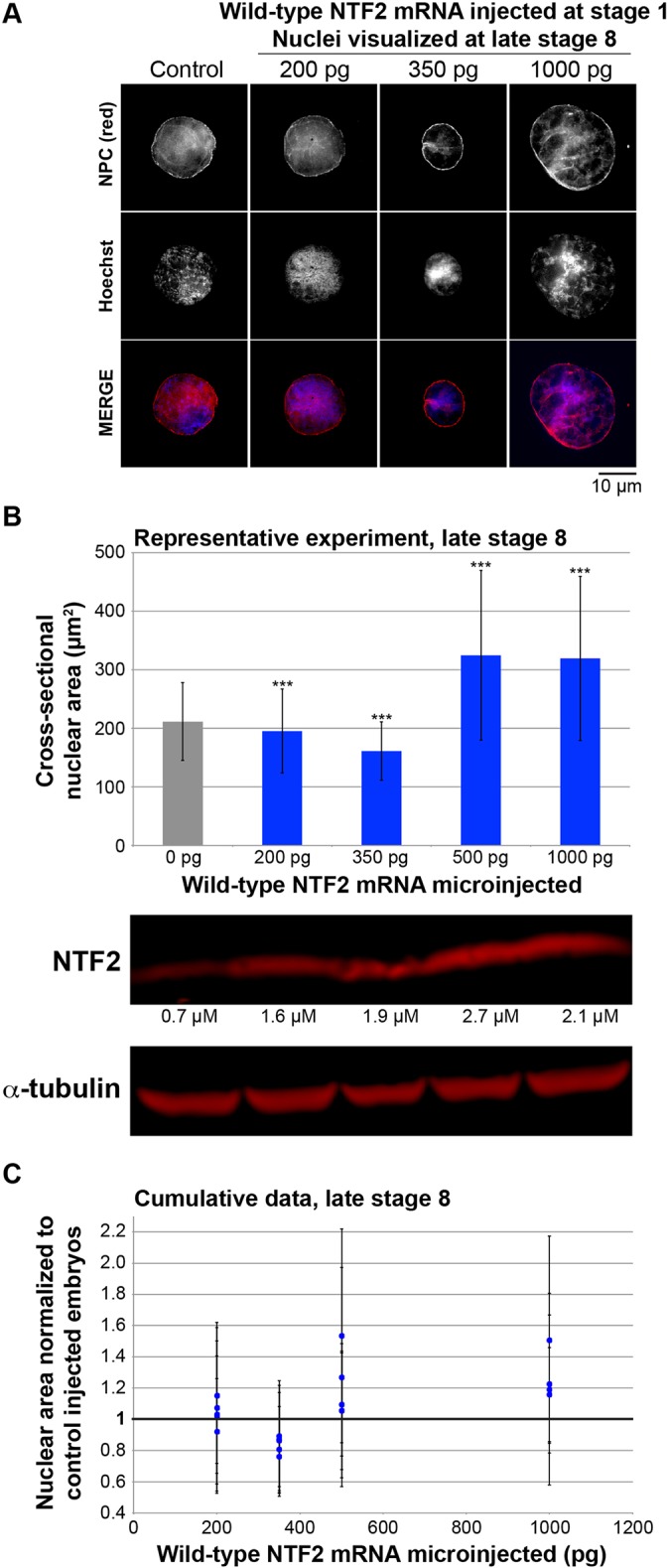
NTF2 concentration affects nuclear size in vivo in X. laevis embryos. (A) Single-cell X. laevis embryos were microinjected with the indicated amount of NTF2 mRNA or water as a control. Embryos were allowed to develop to late stage 8 and embryo extracts were prepared. Extracts containing endogenous embryonic nuclei were fixed, nuclei were spun onto coverslips, and nuclei were visualized and quantified as described in Fig. 1. Representative images are shown. (B) Nuclear size data are shown from one representative experiment. For each bar, 50–100 embryos were microinjected and the cross-sectional areas of 172–1605 NPC-stained nuclei were measured and averaged. The error bars are s.d. ***P<0.001 (two-tailed Student's t-test). To measure the level of ectopic NTF2 protein expression in embryos, western blots were performed on embryo extracts and quantified by infrared fluorescence as described in Materials and Methods. (C) Cumulative data from five different batches of microinjected X. laevis embryos are shown. Average cross-sectional nuclear areas were measured as in B and normalized to the control (bold horizontal line set at 1.0). The error bars are s.d. All normalized nuclear area data points greater than 1.05 and less than 1.0 are statistically different from the control (1.0) by at least P<0.05 (two-tailed Student's t-test).
NTF2 levels affect nuclear size in mammalian tissue culture cells
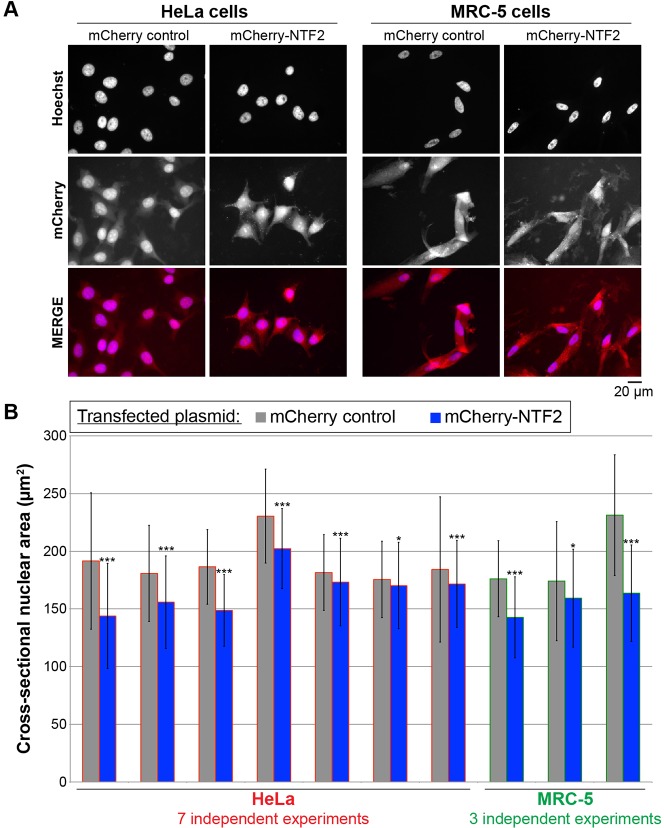
To extend our in vivo Xenopus results, we tested whether altering NTF2 expression levels also affected nuclear size in mammalian tissue culture cells. We transiently transfected HeLa and MRC-5 (normal human lung fibroblast) cells with an mCherry–NTF2 construct and verified ectopic expression of the fluorescently-labeled NTF2 by immunoblotting (data not shown) and by visualization of the fluorescently tagged protein (Fig. 5A). In multiple independent experiments, ectopic NTF2 expression always led to a reduction in nuclear size. The reduction in nuclear cross-sectional area induced by NTF2 expression was 3–25% in HeLa cells and 9–29% in MRC-5 cells (Fig. 5B). In HeLa cells, transfection with an NTF2 construct lacking mCherry similarly reduced nuclear size (data not shown). Nuclear size was similarly decreased by ectopic NTF2 expression in U2OS osteosarcoma cells (data not shown). Based on immunoblots of cell lysates, the level of ectopic NTF2 expression ranged from two- to three-fold over endogenous levels in all cell lines tested (data not shown). We also performed small interfering RNA (siRNA)-mediated knockdown of NTF2 in HeLa cells but found that cell viability was greatly reduced by this treatment, making it difficult to accurately assess nuclear size in these experiments (data not shown). It is worth noting that NTF2 expression is largely not cell cycle regulated (Ly et al., 2014) (data not shown). Taken together, these data demonstrate that nuclear size is also sensitive to NTF2 expression levels in mammalian cells.
Fig. 5.
NTF2 levels affect nuclear size in mammalian tissue culture cells. (A) HeLa and MRC-5 cells were transiently transfected with plasmids expressing mCherry or mCherry–NTF2. At 48 h after transfection, cells were fixed and stained with Hoechst 33342 to visualize the DNA (blue). Representative images are shown. (B) Nuclear cross-sectional areas were quantified from Hoechst-33342-stained cells that were identified as being transfected by mCherry expression. For each data point, the cross-sectional areas of 105–840 nuclei were measured and averaged. Error bars are s.d. *P<0.05, ***P<0.001 (two-tailed Student's t-test).
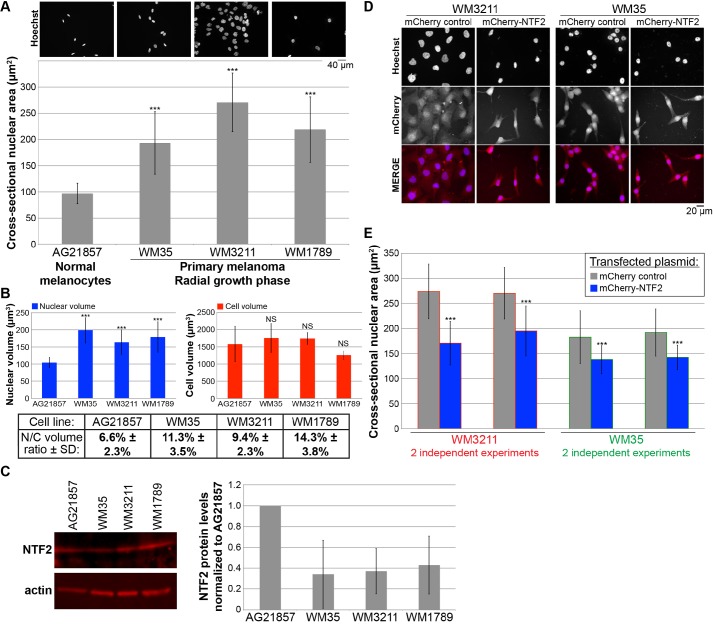
Nuclear size and NTF2 expression levels inversely correlate during melanoma progression
To begin to address whether changes in NTF2 expression might contribute to altered nuclear size in cancer cells, we examined normal melanocytes as a control and three primary radial growth phase melanoma cell lines. We first determined that nuclear cross-sectional area was greater in the melanoma cell lines by 2- to 2.8-fold compared to nuclei in normal melanocytes (Fig. 6A). To calculate nuclear-to-cell volume ratios for these cells, we quantified the volumes of trypsinized cells and of isolated nuclei. We determined that although normal and cancer cell volumes were similar, nuclear volume was greater in the primary melanoma cell lines compared to melanocytes, accounting for increased nuclear-to-cell volume ratios in the melanomas (Fig. 6B).
Fig. 6.
Increased nuclear size in melanoma cell lines can be reversed by ectopic NTF2 expression. (A) One normal melanocyte cell line (AG21857) and three different primary radial growth phase melanoma cell lines (WM35, WM3211 and WM1789) were grown on coverslips, fixed, and stained with Hoechst 33342. Representative images are shown. Nuclear cross-sectional areas were quantified for 226–703 nuclei and averaged. Error bars are s.d. ***P<0.001 (two-tailed Student's t-test). We validated, by performing confocal imaging and quantification of nuclear volumes, that cross-sectional nuclear area measurements could be used to accurately compare relative nuclear sizes in these cell lines. (B) Diameters of spherical trypsinized cells were quantified using a Countess II Automated Cell Counter (Invitrogen). Nuclei were isolated using an EZ Prep Nuclei Isolation kit (Sigma), and diameters of spherical nuclei were quantified using the Countess II. For each cell line, 158–457 cells were quantified, and 149–961 nuclei were quantified. Volumes for spherical cells and nuclei were estimated from diameter measurements and averaged. Nuclear-to-cell (N/C) volume ratios were calculated as average nuclear volume divided by average cell volume. Error bars are s.d. ***P<0.001; NS, not significant (two-tailed Student's t-test). To validate this approach, we performed confocal imaging of trypsinized cells to quantify nuclear-to-cell volume ratios on a per cell basis. This analysis yielded similar average nuclear-to-cell volume ratios: 9.3% versus 9.4% for WM3211, and 12.9% versus 11.3% for WM35 (data not shown). (C) Whole-cell lysates were prepared from the indicated tissue culture cells. Proteins were separated on 4–20% gradient SDS-PAGE gels, transferred onto PVDF membrane, and probed with antibodies against NTF2 and actin. Bands were detected and quantified by infrared fluorescence. One representative gel is shown. NTF2 band intensities were normalized to the actin signal, then normalized to the AG21857 normal melanocyte NTF2 protein level and averaged across four experiments. Error bars are s.d. (D) Primary radial growth phase melanoma cell lines (WM3211 and WM35) were transiently transfected with plasmids expressing mCherry alone as a control or mCherry–NTF2. At 48 h after transfection, cells were fixed and stained with Hoechst 33342. Representative images are shown. (E) Quantification was performed for the experiments described in D. Nuclear cross-sectional areas were quantified from Hoechst-33342-stained cells that were identified as being transfected by mCherry expression. For each bar, the cross-sectional areas of 88–223 nuclei were measured and averaged. Error bars are s.d. ***P<0.001 (two-tailed Student's t-test). For WM35, the average nuclear-to-cell volume ratio is reduced from 11.3% to 9.3% upon ectopic NTF2 expression (data not shown).
To determine whether NTF2 expression levels differed in these cell lines, we performed quantitative immunoblotting on cell lysates. Compared to normal melanocytes, the melanoma tissue culture cell lines exhibited an average 62% reduction in NTF2 expression (Fig. 6C). This inverse correlation between NTF2 expression and nuclear size suggested that NTF2 might serve as a novel biomarker for melanoma. To determine whether this was a robust trend that could be observed in patient tumor samples, we quantified nuclear size and NTF2 staining intensity for a number of biopsied tissue samples using a tissue microarray. Compared to nevi (i.e. benign melanocytic proliferations), melanomas (i.e. primary malignant melanocytic lesions) exhibited increased and more variable nuclear sizes that correlated with lower levels of NTF2 expression (Fig. 7). Based on data from tissue samples, average nuclear cross-sectional areas were increased in melanoma by ∼70% relative to nevi, and average NTF2 expression was reduced by ∼40% (Fig. 7). These data from actual patient tissue samples corroborate our results with cultured cells, alleviating concerns about artifacts with the tissue culture cell lines we analyzed. Progression into later stages of melanoma was not associated with further increases in nuclear size (data not shown).
Fig. 7.
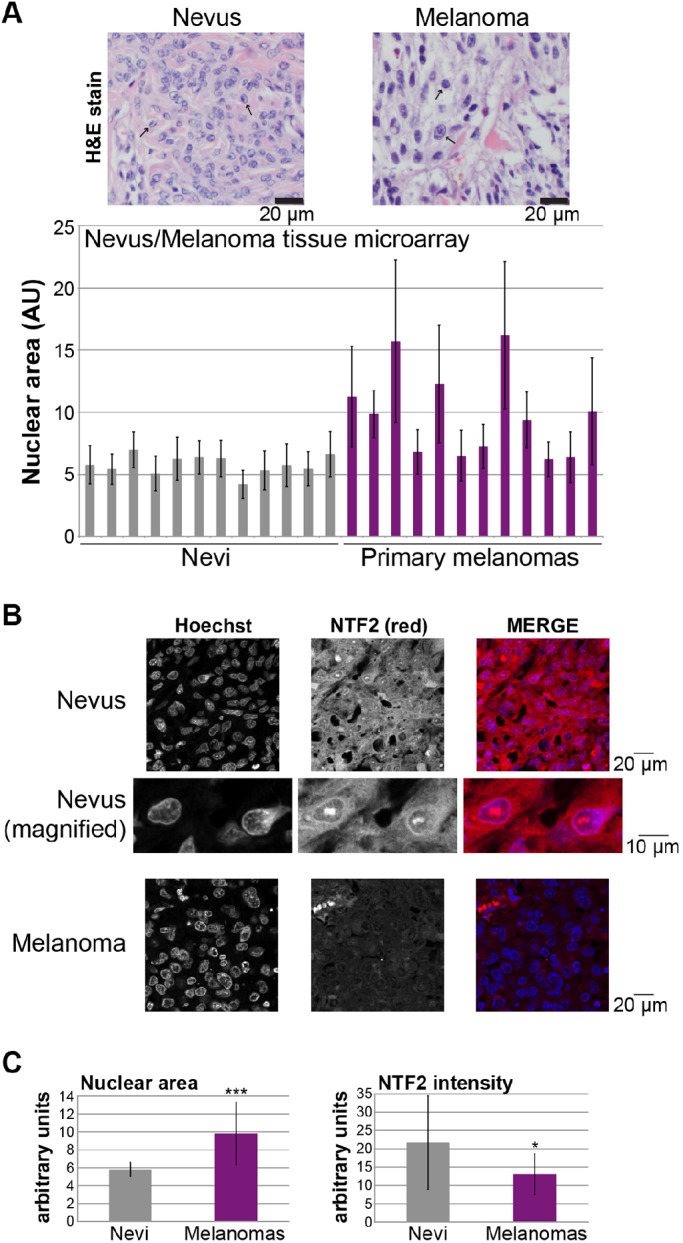
Nuclear size and NTF2 expression are inversely correlated during melanoma progression. (A) Nuclear cross-sectional areas were measured from an H&E-stained melanoma tissue microarray (ME1004c, US Biomax, Inc.). One representative image each of a nevus (i.e. benign melanocytic proliferation) and a melanoma (i.e. primary malignant melanocytic lesion) is shown, and representative blue-stained nuclei are indicated with arrows. For each tissue sample, the cross-sectional areas of 30 adjacent melanocyte nuclei within a single randomly selected field were measured and averaged. Because these are tissue sections and nuclei might not be sectioned through their widest plane, data are presented in arbitrary units (AU) to emphasize that only relative comparisons between samples can be accurately determined. Samples with extensive necrosis or pigment were avoided. Each bar in the graph represents a different tissue biopsy. Error bars are s.d. (B) An unstained ME1004c microarray was processed for immunofluorescence using an antibody against NTF2 (red), and nuclei were stained with Hoechst 33342 (blue). Images were acquired at the same exposure time. Representative images are shown from one nevus and one melanoma. The NTF2 is mostly localized to the cytoplasm and nuclear envelope rim. (C) The NTF2-stained microarray described in B was quantified. Images were acquired at the same exposure time. NTF2 staining intensity measurements were performed for more than 20 regions (each encompassing 1–3 cells) of each tissue sample, corrected for the background signal, normalized for the area of the measurement region and averaged. Brightly staining streaks and puncta suggestive of non-specific or necrotic staining were avoided. Instead, regions of viable tissue with similar cell densities were selected for quantification. Nuclear sizes and NTF2 staining intensities were measured for the same tissue samples. The left graph shows average nuclear areas for 12 nevi and 12 melanomas. The right graph shows the average NTF2 staining intensities for those same samples. Individual melanoma nuclear area measurements adopt a unimodal distribution (data not shown). Error bars are s.d. *P<0.05; ***P<0.001 (two-tailed Student's t-test).
Having observed an inverse correlation between nuclear size and NTF2 expression, we next tested the effects of increasing NTF2 levels in the melanoma cell lines by transient transfection. Ectopic NTF2 expression significantly reduced melanoma nuclear cross-sectional area by 25–40% in two different radial growth phase primary melanoma cell lines (Fig. 6D,E). When examined on a cell-by-cell basis, a weak inverse correlation was noted between the signal intensity of the mCherry-labeled NTF2 and nuclear size in both transfected HeLa and WM35 melanoma cells (data not shown). Taken together, these data show that increased nuclear size in melanoma correlates with reduced NTF2 expression, and that a more normal nuclear size can be restored in melanoma cells by increasing the level of NTF2 expression.
DISCUSSION
In this study, we have extended previous observations about how NTF2 affects nuclear size. We show that binding of Ran to NTF2 is absolutely required for NTF2 to inhibit nuclear expansion in X. laevis egg extract, and that the mechanism is through reducing the diameter of the NPC and import of large cargos. These effects are most pronounced at ∼5 µM NTF2, which corresponds to NTF2 levels in late stage Xenopus embryos (Peshkin et al., 2015) and adult Xenopus liver cells (data not shown). We have expanded our in vitro results into X. laevis embryos and mammalian tissue culture cells, demonstrating that NTF2 expression levels also affect nuclear size in vivo. Finally, we find that early stages of melanoma progression are associated with increased nuclear size and decreased NTF2 expression, and ectopic NTF2 expression in melanoma cell lines is sufficient to reduce nuclear size.
Our data clearly show that binding of Ran to NTF2 is necessary for NTF2 to affect nuclear size. The NTF2 E42K Ran-binding mutant failed to inhibit both nuclear growth and import of large cargos in X. laevis egg and embryo extracts. Based on our NTF2 immunofluorescence experiments, the E42K NTF2 mutant is recruited to the NPC as well as, or better than, wild-type NTF2. This shows that simple recruitment of NTF2 to the NPC is not sufficient to block the import of large cargos and associated nuclear expansion. Rather, NTF2 at the NPC must be bound to Ran to induce these effects. Strikingly, even though the Ran-binding mutant efficiently associated with the NPC, it had little effect on the size of the NPC, strongly arguing that the NTF2-induced conformational change in the pore is dependent on Ran binding. This result is in agreement with previous electron microscopy data indicating that Ran levels affect the conformation of the cytoplasmic filaments of the NPC, with high RanGTP levels reducing the mean diameter of the NPC pore (Goldberg et al., 2000).
The W7A NTF2 mutant with reduced NPC affinity partially inhibited nuclear growth, but not to the same extent as wild-type NTF2. We suspect the reason this mutant was not completely impaired in its ability to reduce nuclear size is because it still retains significant affinity for the NPC, as evidenced by our quantification of NTF2 immunofluorescence. In support of this idea, it has been shown that NTF2 possesses a secondary Nup-binding site that is not disrupted by the W7A mutation (Bayliss et al., 1999; Morrison et al., 2003). Furthermore, we observed an increase in nuclear levels of importin α and β and increased import of several cargos (GST–GFP-NLS, nucleoplasmin and NuMa) when Xenopus egg extract nuclei were supplemented with 80 µM W7A NTF2 (data not shown), suggesting that this mutant does retain sufficient residual NPC affinity to enhance Ran recycling and nuclear import when added at a sufficiently high concentration. The W7A mutant does not appear to be acting by sequestration of cytoplasmic Ran as we did not observe a dominant-negative effect on bulk import. Although the W7A mutant is clearly defective in its ability to modulate bulk import, it retains substantial activity to reduce NPC size and large cargo import (Fig. 2). We propose that these two activities are separable and partially uncoupled in this mutant, such that bulk import is more sensitive to the W7A mutation whereas the residual NPC affinity of this mutant is sufficient to modulate nuclear size. Taken together, we propose that increased NTF2 levels reduce nuclear size by binding Ran at the NPC, thereby reducing the effective diameter of the pore and slowing import of large cargo molecules, such as lamins in >700 kDa complexes (Adam et al., 2008; Gambus et al., 2011), that directly influence nuclear expansion (Jevtic et al., 2015).
Although low levels of ectopic NTF2 generally reduced nuclear size, we noted that further increases in NTF2 levels began to increase nuclear size. In egg extract, high NTF2 levels generated nuclei that approached the size of control-treated nuclei, and, in embryos, nuclear size was even increased relative to controls. We believe these increases in nuclear size can be explained by NTF2 increasing bulk import kinetics. We observed that supplementing egg extract with NTF2 resulted in increased nuclear Ran and importin levels. Consistent with these observations, computer simulations of nuclear import and cell microinjections have demonstrated that nuclear import rates increase with increasing NTF2 concentrations (Riddick and Macara, 2005). Conversely, NTF2 depletion in worms reduced import and nuclear size (Ladouceur et al., 2015). We also noted that addition of the E42K NTF2 Ran-binding mutant to egg extract led to increased import of large Q-dot cargos with a modest increase in nuclear size. Perhaps by occupying the NPC and excluding wild-type NTF2 molecules bound to Ran, this mutant can exhibit a slight dominant-negative effect. Interestingly, relatively low levels of NTF2 protein (∼2–3 µM) that decrease nuclear size in egg and pre-MBT embryo extracts were sufficient to increase nuclear size in post-MBT embryos. This effect might be explained by the different cellular context of late-stage embryos where zygotic transcription is active, as was recently noted for the effects of lamin expression on Xenopus nuclear size (Jevtic et al., 2015). As nuclear import kinetics are reduced in the post-MBT embryo, it seems reasonable that a small increase in NTF2 levels might be sufficient to increase bulk nuclear import and nuclear size. We propose that low ectopic levels of NTF2 predominantly act to inhibit large cargo import and reduce the rate of nuclear expansion, whereas higher levels of ectopic NTF2 begin to counteract this effect by increasing the kinetics of bulk import. Ultimately, it is the balance of these two activities that dictate nuclear size.
Our study of how NTF2 levels affect nuclear size has potential implications in disease. Nuclear size is usually increased in cancer cells, and altered nuclear morphology is a key diagnostic and prognostic feature used by pathologists (Zink et al., 2004; Chow et al., 2012; Jevtic and Levy, 2014). The changes in nuclear size that we report here in tissue culture cells fall within the clinically relevant range of nuclear size changes in cancer. For instance, it has been reported that nuclear area increases 29% from stage 1 to stage 4 breast cancer (Abdalla et al., 2009), 16% from primary to metastatic melanoma (Mossbacher et al., 1996), and 39% from benign hyperplasia to prostate carcinoma (Wang et al., 1992). Focusing on melanoma, we find that increased nuclear size in primary melanoma correlates with reduced NTF2 expression. In agreement with our data showing that NTF2 generally has a negative impact on nuclear size, we are able to reverse the nuclear enlargement in melanoma cell lines by ectopic NTF2 expression. These data suggest that NTF2 may be a novel cancer biomarker, and it will be important to examine if this trend holds true for other cancer types. Interestingly, another nuclear transport factor, importin α2, has been implicated as a biomarker for breast and non-small cell lung cancers (Dahl et al., 2006; Gluz et al., 2008; Wang et al., 2010). We note that ectopic NTF2 expression does not completely correct the nuclear size defect in melanoma cancer cells, suggesting that additional factors likely contribute to increased nuclear size in melanoma (Fig. 6). Aneuploidy is one potential contributor to increased nuclear size in melanoma, and we did find that the melanoma cell lines used in our study are roughly triploid relative to the normal diploid melanocytes (data not shown). The relationship between nuclear size and cancer cell phenotypes is still unclear. Our study represents one of the first to identify a specific protein whose expression levels correlate with nuclear size. Using the mechanistic information gained here, future studies will focus on investigating how nuclear size functionally impacts the formation and progression of cancer. These studies might ultimately suggest that altering nuclear size is a viable approach in cancer treatment, with NTF2 as a potential target for cancer therapeutics.
MATERIALS AND METHODS
Recombinant protein expression and purification
The wild-type, W7A, and E42K NTF2 mutants cloned into pET15b were obtained from Murray Stewart (MRC Laboratory of Molecular Biology, United Kingdom). The wild-type gene was subcloned into pET30a at NcoI/XhoI (pDL31). The W7A plasmid was used as is. The E42K NTF2 gene was subcloned into pET30a at NcoI/XhoI (pDL33). Bacterial expression and purification of these recombinant proteins were performed as previously described (Lane et al., 2000; Levy and Heald, 2010). Purified proteins were dialyzed into NTF2 storage buffer (100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 50 mM sucrose, 10 mM HEPES pH 7.8).
Xenopus laevis egg and embryo extracts
Egg and embryo extracts were prepared as previously described (Edens and Levy, 2014b). All Xenopus procedures and studies were conducted in compliance with the US Department of Health and Human Services Guide for the Care and Use of Laboratory Animals. Protocols were approved by the University of Wyoming Institutional Animal Care and Use Committee (Assurance #A-3216-01).
Nuclear assembly and NTF2 protein addition
For each nuclear assembly reaction, 2.5 µl of recombinant NTF2 protein diluted in NTF2 storage buffer were added to 25 µl of X. laevis egg extract. The concentration of the NTF2 stock solution was adjusted to vary the final concentration of NTF2 protein added to the reaction. For control reactions, 2.5 µl of NTF2 storage buffer was added. Nuclear assembly was initiated as previously described (Edens and Levy, 2014b), and the reaction was allowed to proceed for 90 min at 20°C prior to fixation. Stage 8 X. laevis embryo extracts were prepared as previously described (Edens and Levy, 2014b), and embryo extracts containing endogenous embryonic nuclei were supplemented with NTF2 proteins, in the same manner as for egg extracts, and incubated at room temperature for 60 min prior to fixation.
Embryo microinjections
NTF2 was cloned from a plasmid consisting of NTF2 in pRSET B obtained from Karsten Weis (Department of Biology, ETH Zurich, Switzerland) (Lane et al., 2000) into pCS2+ at EcoRI/XhoI (pDL18). The pDL18 plasmid was linearized with NotI, and mRNA was expressed from the SP6 promoter using the mMessage mMachine kit (Ambion). X. laevis embryos were prepared, microinjected with mRNA ranging from 200–1000 pg per embryo, cultured and used to prepare embryo extracts, as previously described (Jevtic and Levy, 2015).
Fixation and immunofluorescence
Nuclei in egg or embryo extract were fixed, centrifuged onto coverslips, and processed for immunofluorescence as previously described (Edens and Levy, 2014b). The primary antibody against the NPC was mAb414 (mouse, Covance, 1:1000). Antibodies used to detect NTF2, Ran, importin β, and importin α2 were as previously described (Levy and Heald, 2010). Secondary antibodies were Alexa-Fluor-568-conjugated anti-mouse-IgG and Alexa-Fluor-488-conjugated anti-rabbit-IgG antibodies (Molecular Probes, 1:500).
Nuclear import assays
Preparation of nuclear import cargos and quantification of nuclear import for GST–GFP-NLS, IBB-coated Q-dots 605, and lamin B3 (LB3) were performed as previously described (Levy and Heald, 2010). The IBB-βGal nuclear import cargo construct was obtained from Karsten Weis (pKW319), and the protein was prepared and its nuclear import detected as previously described (Weis et al., 1996; Lyman et al., 2002).
Microscopy and image quantification
Nuclei were visualized with an Olympus BX51 fluorescence microscope as previously described (Jevtic et al., 2015). Images for measuring fluorescence staining intensity were acquired using the same exposure times. Total fluorescence intensity and cross-sectional nuclear area were measured from the original thresholded images using MetaMorph software (Molecular Devices). Where noted, total nuclear fluorescence intensity was divided by nuclear cross-sectional area, to normalize for nuclear size. Where indicated, confocal imaging was performed on an Olympus IX71 spinning-disk confocal microscope. For Xenopus nuclei, measurements of total nuclear envelope surface area from confocal z-stacks agreed (within 4%) with estimates from the cross-sectional area (data now shown). We chose to quantify nuclear size from the cross-sectional area because it allowed for the acquisition of data from large numbers of nuclei (Levy and Heald, 2010). For electron microscopy of NPCs, nuclei assembled in egg extract (25 µl) were diluted in 1 ml of ELB (250 mM sucrose, 50 mM potassium chloride, 2.5 mM magnesium chloride, 10 mM HEPES pH 7.8), pelleted at 1600 g for 3 min and resuspended in 25 µl of ELB, as previously described (Edens and Levy, 2014b). 5 µl were applied onto a 200-mesh formvar carbon-coated copper grid (Electron Microscopy Sciences) for 2–3 min. This approach relies on the intact live nuclei breaking open and laying flat on the grid, exposing the cytoplasmic faces of the NPCs. The solution was then gently removed from the grid with filter paper. The grid was rinsed briefly with deionized water and negatively stained with 1% uranyl acetate for 1 min. Excess uranyl acetate was removed with filter paper. The grid was air-dried and observed with a Hitachi H-7000 transmission electron microscope, equipped with 4K×4K Gatan digital camera (Gatan, Inc., Pleasanton, CA).
Western blots
Whole-cell lysates from tissue culture cells were prepared using SDS-PAGE sample buffer supplemented with benzonase nuclease (Sigma, E1014) and boiled for 5 min. Xenopus embryo extracts were prepared as previously described (Edens and Levy, 2014b). Proteins were separated on SDS-PAGE gels (4–20% gradient) and transferred onto PVDF membrane. Membranes were blocked in Odyssey PBS Blocking Buffer (Li-Cor, 927-40000). The primary antibodies used were mouse anti-NTF2 at 1:500 (Sigma, N9527), DM1A mouse anti-α-tubulin at 1:2000 (Santa Cruz Biotechnology, sc-32293), and mouse anti-actin at 1:200 (Abgent, AM1965b). The secondary antibody was IRDye-680RD-conjugated anti-mouse-IgG used at 1:20,000 (Li-Cor 925-68070). Blots were scanned on a Li-Cor Odyssey CLx instrument and band quantification was performed with ImageStudio. For a given sample, NTF2 band intensity was normalized to the actin or tubulin signal.
Tissue culture
NTF2 was cloned from a plasmid consisting of NTF2 in pRSET B obtained from Karsten Weis (Lane et al., 2000) into pEmCherry-C2 (a derivative of pEGFP-C2, a gift from Anne Schlaitz) at EcoRI/KpnI (pDL23). Primary radial growth phase melanoma cells lines (WM35, WM3211, WM1789) were obtained from the Wistar repository through the Coriell Institute. The normal melanocyte cell line (AG21857) was obtained from the NIA Cell Culture Repository through the Coriell Institute. HeLa cells stably expressing GFP-tagged histone H2B were a gift from Jay Gatlin (University of Wyoming, Laramie, WY), and MRC-5 normal human lung fibroblast cells were a gift from Jason Gigley (University of Wyoming, Laramie, WY). Cells were transiently transfected and visualized as previously described (Jevtic et al., 2015). Images were acquired and nuclear sizes quantified as described above in the ‘Microscopy and image quantification’ section.
Nuclear-to-cell volume ratio measurements
Nuclei were isolated from tissue culture cells using the EZ Prep Nuclei Isolation Kit (Sigma-Aldrich, NUC-101). Cells were grown to 90% confluency, washed with cold PBS and harvested in Nuclei EZ lysis buffer using a cell scraper. After a 5-min incubation on ice, cells were collected by centrifugation and resuspended in lysis buffer. This was repeated a total of three times, and the final pellet of nuclei was resuspended in 200 µl Nuclei EZ storage buffer. Successful isolation of nuclei was verified by staining a small aliquot of nuclei with Hoechst 33342. Nuclei were stained with Trypan Blue, and nuclear diameters were quantified using a Countess Automated Cell Counter (Invitrogen). To measure cell size, cells were treated with trypsin for 5 min and collected in growth medium. After an additional 5-min incubation in growth medium, cells were stained with Trypan Blue, and the diameters of viable cells were quantified using a Countess Automated Cell Counter (Invitrogen). Cell and nuclear volumes were extrapolated from the diameter measurements of roughly spherical cells and nuclei. Nuclear-to-cell volume ratios were calculated by dividing nuclear volume by cell volume.
Tissue microarrays
A hematoxylin and eosin (H&E)-stained tissue microarray (ME1004cs, US Biomax) was digitized using an Aperio Slide Scanner. Nuclear diameter was determined with Aperio ImageScope software, and nuclear cross-sectional area was measured with ImageJ. To quantify NTF2 levels, an unstained ME1004cs microarray was deparaffinized in xylene, followed by sequential washes in 100%, 95%, and 70% ethanol. The slide was equilibrated in PBS, and antigen retrieval was performed in 0.01 M sodium citrate buffer pH 6.0 at 95°C for 15 min. The slide was then incubated with primary mouse anti-NTF2 antibody at 1:200 (Sigma, N9527) for 1 h at room temperature, washed with PBS, and incubated for 1 h at room temperature with Alexa-Fluor-568-conjugated donkey anti-mouse-IgG secondary antibody (Molecular Probes, A10037) at 1:1000. Hoechst 33342 staining (20 µg/ml) was performed for 5 min. The slide was overlaid with Vectashield (Vector Labaratories) and a coverslip and sealed with nail polish. Images were acquired at the same exposure time. For each tissue sample, images were acquired for five randomly selected regions. For each image, NTF2 staining intensities were measured using ImageJ for five or more equally sized regions and corrected for the background signal. For each tissue sample, more than 20 NTF2 staining intensity measurements were performed, normalized based on the area of the region quantified, and averaged.
Statistics
For each coverslip, at least 50, and usually >800, nuclei were quantified. Averaging and statistical analysis were performed for independently repeated experiments. Where noted, nuclear area and intensity measurements were normalized to controls. Two-tailed Student's t-tests assuming equal variances were performed in Excel (Microsoft) to evaluate statistical significance. The P-values, number of independent experiments and error bars are denoted in the figure legends.
Acknowledgements
We thank Don Jarvis for use of the Countess Automated Cell Counter and Karen White for constructive comments on the manuscript. We also thank Rebecca Heald for support in the early stages of this research.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
L.D.V., P.J., and D.L.L. designed the study and wrote the paper. D.L.L. constructed plasmids, synthesized mRNA for microinjections, and purified recombinant proteins. L.D.V. performed tissue culture experiments. P.J. performed Xenopus experiments. Z.Z. performed TEM of NPCs. B.A.S. quantified nuclei on tissue microarrays. All authors analyzed the results and approved the final version of the manuscript.
Funding
This work was supported by the National Institutes of Health [grant number R15GM106318 to D.L.L.]; and the American Cancer Society [grant number RSG-15-035-01-DDC to D.L.L.]. L.D.V. was supported by a graduate assistantship from Wyoming IDeA Networks for Biomedical Research Excellence [grant number NIH/NIGMS P20GM103432]. P.J. was supported by a graduate assistantship from the University of Wyoming Agricultural Experiment Station. Deposited in PMC for release after 12 months.
References
- Abdalla F., Boder J., Markus R., Hashmi H., Buhmeida A. and Collan Y. (2009). Correlation of nuclear morphometry of breast cancer in histological sections with clinicopathological features and prognosis. Anticancer Res. 29, 1771-1776. [PubMed] [Google Scholar]
- Adam S. A., Sengupta K. and Goldman R. D. (2008). Regulation of nuclear lamin polymerization by importin alpha. J. Biol. Chem. 283, 8462-8468. 10.1074/jbc.M709572200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R., Ribbeck K., Akin D., Kent H. M., Feldherr C. M., Görlich D. and Stewart M. (1999). Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J. Mol. Biol. 293, 579-593. 10.1006/jmbi.1999.3166 [DOI] [PubMed] [Google Scholar]
- Bayliss R., Leung S. W., Baker R. P., Quimby B. B., Corbett A. H. and Stewart M. (2002). Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J. 21, 2843-2853. 10.1093/emboj/cdf305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. C. and Forbes D. J. (2006). In vitro study of nuclear assembly and nuclear import using Xenopus egg extracts. Methods Mol. Biol. 322, 289-300. 10.1007/978-1-59745-000-3_20 [DOI] [PubMed] [Google Scholar]
- Chan Y.-H. M. and Marshall W. F. (2010). Scaling properties of cell and organelle size. Organogenesis 6, 88-96. 10.4161/org.6.2.11464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.-H. M. and Marshall W. F. (2012). How cells know the size of their organelles. Science 337, 1186-1189. 10.1126/science.1223539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K. H., Factor R. E. and Ullman K. S. (2012). The nuclear envelope environment and its cancer connections. Nat. Rev. Cancer 12, 196-209. 10.1038/nrc3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson W. D., Kent H. M. and Stewart M. (1996). Separate binding sites on nuclear transport factor 2 (NTF2) for GDP-Ran and the phenylalanine-rich repeat regions of nucleoporins p62 and Nsp1p. J. Mol. Biol. 263, 517-524. 10.1006/jmbi.1996.0594 [DOI] [PubMed] [Google Scholar]
- Clarkson W. D., Corbett A. H., Paschal B. M., Kent H. M., McCoy A. J., Gerace L., Silver P. A. and Stewart M. (1997). Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran. J. Mol. Biol. 272, 716-730. 10.1006/jmbi.1997.1255 [DOI] [PubMed] [Google Scholar]
- Conklin E. (1912). Cell size and nuclear size. J. Exp. Embryol. 12, 1-98. 10.1002/jez.1400120102 [DOI] [Google Scholar]
- Dahl E., Kristiansen G., Gottlob K., Klaman I., Ebner E., Hinzmann B., Hermann K., Pilarsky C., Durst M., Klinkhammer-Schalke M. et al. (2006). Molecular profiling of laser-microdissected matched tumor and normal breast tissue identifies karyopherin alpha2 as a potential novel prognostic marker in breast cancer. Clin. Cancer Res. 12, 3950-3960. 10.1158/1078-0432.CCR-05-2090 [DOI] [PubMed] [Google Scholar]
- Dangou J. M., Kiss R., DePrez C., Jeannot M. C., Fastrez R., Pasteels J. L. and Verhest A. (1993). Heterogeneity of DNA ploidy, proliferation index and nuclear size in human colorectal carcinomas. Anal. Quant. Cytol. Histol. 15, 23-31. [PubMed] [Google Scholar]
- Dey P. (2010). Cancer nucleus: morphology and beyond. Diagn. Cytopathol. 38, 382-390. 10.1002/dc.21234 [DOI] [PubMed] [Google Scholar]
- Edens L. J. and Levy D. L. (2014a). Size scaling of subcellular organelles and structures in Xenopus laevis and Xenopus tropicalis. In Xenopus Development (ed. Kloc M. and Kubiak J. Z.), pp. 325-345. Hoboken, NJ: John Wiley & Sons, Inc. [Google Scholar]
- Edens L. J. and Levy D. L. (2014b). cPKC regulates interphase nuclear size during Xenopus development. J. Cell Biol. 206, 473-483. 10.1083/jcb.201406004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens L. J., White K. H., Jevtic P., Li X. and Levy D. L. (2013). Nuclear size regulation: from single cells to development and disease. Trends Cell Biol. 23, 151-159. 10.1016/j.tcb.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Feldherr C., Akin D. and Moore M. S. (1998). The nuclear import factor p10 regulates the functional size of the nuclear pore complex during oogenesis. J. Cell Sci. 111, 1889-1896. [DOI] [PubMed] [Google Scholar]
- Gambus A., Khoudoli G. A., Jones R. C. and Blow J. J. (2011). MCM2–7 form double hexamers at licensed origins in Xenopus egg extract. J. Biol. Chem. 286, 11855-11864. 10.1074/jbc.M110.199521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluz O., Wild P., Meiler R., Diallo-Danebrock R., Ting E., Mohrmann S., Schuett G., Dahl E., Fuchs T., Herr A. et al. (2008). Nuclear karyopherin alpha2 expression predicts poor survival in patients with advanced breast cancer irrespective of treatment intensity. Int. J. Cancer 123, 1433-1438. 10.1002/ijc.23628 [DOI] [PubMed] [Google Scholar]
- Goehring N. W. and Hyman A. A. (2012). Organelle growth control through limiting pools of cytoplasmic components. Curr. Biol. 22, R330-R339. 10.1016/j.cub.2012.03.046 [DOI] [PubMed] [Google Scholar]
- Goldberg M. W., Rutherford S. A., Hughes M., Cotter L. A., Bagley S., Kiseleva E., Allen T. D. and Clarke P. R. (2000). Ran alters nuclear pore complex conformation. J. Mol. Biol. 300, 519-529. 10.1006/jmbi.2000.3891 [DOI] [PubMed] [Google Scholar]
- Gonzalez Y., Meerbrey K., Chong J., Torii Y., Padte N. N. and Sazer S. (2009). Nuclear shape, growth and integrity in the closed mitosis of fission yeast depend on the Ran-GTPase system, the spindle pole body and the endoplasmic reticulum. J. Cell Sci. 122, 2464-2472. 10.1242/jcs.049999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y., Iwabuchi M., Ohsumi K. and Kimura A. (2013). Intranuclear DNA density affects chromosome condensation in metazoans. Mol. Biol. Cell 24, 2442-2453. 10.1091/mbc.E13-01-0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M., Mori C., Kojidani T., Bunai F., Hori T., Fukagawa T., Hiraoka Y. and Haraguchi T. (2009). Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate tetrahymena. Curr. Biol. 19, 843-847. 10.1016/j.cub.2009.03.055 [DOI] [PubMed] [Google Scholar]
- Jevtic P. and Levy D. L. (2014). Mechanisms of nuclear size regulation in model systems and cancer. Adv. Exp. Med. Biol. 773, 537-569. 10.1007/978-1-4899-8032-8_25 [DOI] [PubMed] [Google Scholar]
- Jevtic P. and Levy D. L. (2015). Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Curr. Biol. 25, 45-52. 10.1016/j.cub.2014.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtic P., Edens L. J., Vuković L. D. and Levy D. L. (2014). Sizing and shaping the nucleus: mechanisms and significance. Curr. Opin. Cell Biol. 28, 16-27. 10.1016/j.ceb.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtic P., Edens L. J., Li X., Nguyen T., Chen P. and Levy D. L. (2015). Concentration-dependent effects of nuclear lamins on nuclear size in Xenopus and mammalian cells. J. Biol. Chem. 290, 27557-27571. 10.1074/jbc.M115.673798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent H. M., Clarkson W. D., Bullock T. L. and Stewart M. (1996). Crystallization and preliminary X-ray diffraction analysis of nuclear transport factor 2. J. Struct. Biol. 116, 326-329. 10.1006/jsbi.1996.0049 [DOI] [PubMed] [Google Scholar]
- Ladouceur A.-M., Dorn J. F. and Maddox P. S. (2015). Mitotic chromosome length scales in response to both cell and nuclear size. J. Cell Biol. 209, 645-652. 10.1083/jcb.201502092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane C. M., Cushman I. and Moore M. S. (2000). Selective disruption of nuclear import by a functional mutant nuclear transport carrier. J. Cell Biol. 151, 321-332. 10.1083/jcb.151.2.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D. L. and Heald R. (2010). Nuclear size is regulated by importin alpha and Ntf2 in Xenopus. Cell 143, 288-298. 10.1016/j.cell.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D. L. and Heald R. (2012). Mechanisms of intracellular scaling. Annu. Rev. Cell Dev. Biol. 28, 113-135. 10.1146/annurev-cellbio-092910-154158 [DOI] [PubMed] [Google Scholar]
- Levy D. L. and Heald R. (2015). Biological scaling problems and solutions in amphibians. Cold Spring Harb. Perspect. Biol. 8, pii:a019166 10.1101/cshperspect.a019166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg L. G. (1992). Nuclear DNA ploidy in mammary carcinomas; using nuclear size as co-parameter reveals more complex patterns. Anal. Cell Pathol. 4, 389-394. [PubMed] [Google Scholar]
- Ly T., Ahmad Y., Shlien A., Soroka D., Mills A., Emanuele M. J., Stratton M. R. and Lamond A. I. (2014). A proteomic chronology of gene expression through the cell cycle in human myeloid leukemia cells. Elife 3, e01630 10.7554/elife.01630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman S. K., Guan T., Bednenko J., Wodrich H. and Gerace L. (2002). Influence of cargo size on Ran and energy requirements for nuclear protein import. J. Cell Biol. 159, 55-67. 10.1083/jcb.200204163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid A. S. and Weis K. (2006). Nuclear transport is becoming crystal clear. Chromosoma 115, 98-109. 10.1007/s00412-005-0043-3 [DOI] [PubMed] [Google Scholar]
- Malone C. D., Falkowska K. A., Li A. Y., Galanti S. E., Kanuru R. C., LaMont E. G., Mazzarella K. C., Micev A. J., Osman M. M., Piotrowski N. K. et al. (2008). Nucleus-specific importin alpha proteins and nucleoporins regulate protein import and nuclear division in the binucleate Tetrahymena thermophila. Eukaryot. Cell 7, 1487-1499. 10.1128/EC.00193-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. F. (2012). Organelle size control systems: from cell geometry to organelle-directed medicine. Bioessays 34, 721-724. 10.1002/bies.201200043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerzon M., Gao Z., Liu J., Wu J.-C., Malone C. J. and Starr D. A. (2009). Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Dev. Biol. 327, 433-446. 10.1016/j.ydbio.2008.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto H., Isobe H., Akita H., Ishiguro A., Endo T., Inoue S. and Kawakami Y. (1992). The flow cytometric nuclear-DNA content, tumor-origin, nuclear size and prognosis in squamous-cell lung-cancer. Int. J. Oncol. 1, 325-329. 10.3892/ijo.1.3.325 [DOI] [PubMed] [Google Scholar]
- Morrison J., Yang J.-C., Stewart M. and Neuhaus D. (2003). Solution NMR study of the interaction between NTF2 and nucleoporin FxFG repeats. J. Mol. Biol. 333, 587-603. 10.1016/j.jmb.2003.08.050 [DOI] [PubMed] [Google Scholar]
- Mossbacher U., Knollmayer S., Binder M., Steiner A., Wolff K. and Pehamberger H. (1996). Increased nuclear volume in metastasizing “thick” melanomas. J. Invest. Dermatol. 106, 437-440. 10.1111/1523-1747.ep12343580 [DOI] [PubMed] [Google Scholar]
- Neumann F. R. and Nurse P. (2007). Nuclear size control in fission yeast. J. Cell Biol. 179, 593-600. 10.1083/jcb.200708054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop P. D. and Faber J. (1967). Normal Table of Xenopus laevis (Daudin). Amsterdam: North-Holland Publishing Company. [Google Scholar]
- Paschal B. M. and Gerace L. (1995). Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J. Cell Biol. 129, 925-937. 10.1083/jcb.129.4.925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshkin L., Wühr M., Pearl E., Haas W., Freeman R. M. Jr., Gerhart J. C., Klein A. M., Horb M., Gygi S. P. and Kirschner M. W. (2015). On the Relationship of Protein and mRNA Dynamics in Vertebrate Embryonic Development. Dev. Cell 35, 383-394. 10.1016/j.devcel.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafelski S. M. and Marshall W. F. (2008). Building the cell: design principles of cellular architecture. Nat. Rev. Mol. Cell Biol. 9, 593-602. 10.1038/nrm2460 [DOI] [PubMed] [Google Scholar]
- Ribbeck K., Lipowsky G., Kent H. M., Stewart M. and Görlich D. (1998). NTF2 mediates nuclear import of Ran. EMBO J. 17, 6587-6598. 10.1093/emboj/17.22.6587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick G. and Macara I. G. (2005). A systems analysis of importin-{alpha}-{beta} mediated nuclear protein import. J. Cell Biol. 168, 1027-1038. 10.1083/jcb.200409024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulov L., Gruber R., Cohen I. and Harel A. (2011). A dominant-negative form of POM121 binds chromatin and disrupts the two separate modes of nuclear pore assembly. J. Cell Sci. 124, 3822-3834. 10.1242/jcs.086660 [DOI] [PubMed] [Google Scholar]
- Smith A., Brownawell A. and Macara I. G. (1998). Nuclear import of Ran is mediated by the transport factor NTF2. Curr. Biol. 8, 1403-1406. 10.1016/S0960-9822(98)00023-2 [DOI] [PubMed] [Google Scholar]
- Sørensen F. B. (1996). Quantitative analysis of nuclear size for prognosis-related malignancy grading. In Advances in Oncobiology (ed. Heppner G. and Bittar E. E.), pp. 221-255. Greenwich, CT: JAI Press, Inc. [Google Scholar]
- Sørensen F. B., Bichel P. and Jakobsen A. (1992). DNA level and stereologic estimates of nuclear volume in squamous cell carcinomas of the uterine cervix. A comparative study with analysis of prognostic impact. Cancer 69, 187-199. [DOI] [PubMed] [Google Scholar]
- Stewart M. (2007). Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195-208. 10.1038/nrm2114 [DOI] [PubMed] [Google Scholar]
- Tamura K. and Hara-Nishimura I. (2011). Involvement of the nuclear pore complex in morphology of the plant nucleus. Nucleus 2, 168-172. 10.4161/nucl.2.3.16175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Fukao Y., Iwamoto M., Haraguchi T. and Hara-Nishimura I. (2010). Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22, 4084-4097. 10.1105/tpc.110.079947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theerthagiri G., Eisenhardt N., Schwarz H. and Antonin W. (2010). The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J. Cell Biol. 189, 1129-1142. 10.1083/jcb.200912045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Velthoven R., Petein M., Oosterlinck W. J., Zandona C., Zlotta A., Van der Meijden A. P. M., Pasteels J.-L., Roels H., Schulman C. and Kiss R. (1995). Image cytometry determination of ploidy level, proliferative activity, and nuclear size in a series of 314 transitional bladder cell carcinomas. Hum. Pathol. 26, 3-11. 10.1016/0046-8177(95)90108-6 [DOI] [PubMed] [Google Scholar]
- Vukovic L. C., Jevtic P., Edens L. J. and Levy D. L. (2016). New insights into mechanisms and functions of nuclear size regulation. Int. Rev. Cell Mol. Biol. 322, 1-60. 10.1016/bs.ircmb.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Walters A. D., Bommakanti A. and Cohen-Fix O. (2012). Shaping the nucleus: factors and forces. J. Cell Biochem. 113, 2813-2821. 10.1002/jcb.24178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Stenkvist B. G. and Tribukait B. (1992). Morphometry of nuclei of the normal and malignant prostate in relation to DNA ploidy. Anal. Quant. Cytol. Histol. 14, 210-216. [PubMed] [Google Scholar]
- Wang C.-I., Wang C.-L., Wang C.-W., Chen C.-D., Wu C.-C., Liang Y., Tsai Y.-H., Chang Y.-S., Yu J.-S. and Yu C.-J. (2010). Importin subunit alpha-2 is identified as a potential biomarker for non-small cell lung cancer by integration of the cancer cell secretome and tissue transcriptome. Int. J. Cancer 128, 2364-2372. 10.1002/ijc.25568 [DOI] [PubMed] [Google Scholar]
- Webster M., Witkin K. L. and Cohen-Fix O. (2009). Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 122, 1477-1486. 10.1242/jcs.037333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K., Ryder U. and Lamond A. I. (1996). The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 15, 1818-1825. [PMC free article] [PubMed] [Google Scholar]
- Wilson E. B. (1925). The karyoplasmic ratio. In The Cell in Development and Heredity, pp. 727-733. New York: The Macmillan Company. [Google Scholar]
- Zink D., Fischer A. H. and Nickerson J. A. (2004). Nuclear structure in cancer cells. Nat. Rev. Cancer 4, 677-687. 10.1038/nrc1430 [DOI] [PubMed] [Google Scholar]