Abstract
Tumor-derived exosomes (TEXs) are emerging as a new type of cancer biomarker. TEXs are membrane-bound, virus-size vesicles of endocytic origin present in all body fluids of cancer patients. Based on the expanding albeit incomplete knowledge of their biogenesis, secretion by tumor cells and cancer cell-specific molecular and genetic contents, TEXs are viewed as promising, clinically-relevant surrogates of cancer progression and response to therapy. Preliminary proteomic, genetic and functional profiling of tumor cell-derived or cancer plasma-derived exosomes confirms their unique characteristics. Alterations in protein or nucleic acid profiles of exosomes in plasma of cancer patients responding to therapies appear to correlate with clinical endpoints. However, methods for TEX isolation and separation from the bulk of human plasma-derived exosomes are not yet established and their role as biomarkers remains to be confirmed. Further development and validation of TEXs as noninvasive, liquid equivalents of tumor biopsies are necessary to move this effort forward.
Keywords: Tumor-derived exosomes, TEX, cancer, non-invasive biomarkers, cancer prognosis, TEX isolation, molecular profiling, protein cargo, nucleic acid content
1. Introduction
Cancer is a major public health problem in the United States and the world. Epidemiological data indicate that one of three women and one in two men in the United States are likely to develop cancer over their lifetime [1]. While the incidence of some cancers, e.g., lung and colorectal cancers, has significantly decreased in recent years, that of the other solid tumors, including melanoma, has increased [2,3]. Developments in cancer therapies, early cancer diagnosis and general awareness of risk factors for cancer have clearly improved survival. However, there remains an urgent need for the introduction of new more effective and less invasive surrogate markers which could guide early diagnosis, development of therapies, choice of therapeutic strategies for individual patients and accurate estimates of prognosis. Also, reliable surrogate biomarkers for evaluations of response to cancer therapies are needed.
While numerous disease-related surrogate endpoints exist, including histopathology markers (dysplasia, hyperplasia, carcinoma in situ [CIS], tumor stage/grade, metastasis), serum biomarkers, such as CEA, PSA, CA-125, or clinical endpoints such as disease-free survival (DFS) or time to progression (TTP), they do not reliably and consistently correlate with each other or with patients’ responses to therapy and outcome. There are also mechanistic surrogate endpoints which emerge from pre-clinical studies as potentially promising determinants of molecular signaling that might impact clinical responses or survival. While often featured in the biomarker literature, these mechanistic surrogate markers seldom pass the validation tests performed in sufficiently large cohorts of patients with similar demographic and clinicopathological features. To date, few, if any validated cancer biomarkers predictive of clinical response or outcome have emerged. Biomarkers are also very much needed in the drug development arena, where the time frame for advancing a drug from the research lab to the clinic is estimated at about 15 years, and where a reliable marker could substantially reduce this time frame. Not surprisingly, a search for intermediate cancer biomarkers of response or survival continues at a rapid pace, and in the era of personalized medicine research, there is a great interest in identifying biomarkers of therapeutic outcome or response.
The advent of novel therapies for cancer, specifically immunotherapies [4,5] has further increased the need for more reliable biomarkers of prognosis and response to therapy. The choice of immune therapies for patients with cancer who are most likely to achieve clinical benefits is largely dependent on the predictive value of the available biomarkers. Unfortunately, these are few and poorly defined, as exemplified, e.g., by variable expression levels on tumor cells of PD-L1, the ligand whose presence has been considered critical for a successful anti-PD-L1 Ab blockade [6]. However, clinical responses to the antibody blockade of PD-L1 do not appear to correlate to the ligand expression levels on the tumor, and the search for other biomarkers of response to this immune therapy continues [7]. Given the largely unfulfilled expectations for the predictive or even diagnostic usefulness of most available cancer biomarkers, the current emphasis on finding and validating biomarkers that would meet the required criteria is a priority.
What should be expected of a cancer biomarker and how can its performance be determined? The so called REMARK criteria have been formulated and are listed in the literature as guidelines for biomarker studies [8]. Briefly, a new biomarker has to be validated in prospective (and not retrospective) studies of adequate size and statistical power. These studies should include a unique cohort of patients in whom the biomarker correlates with disease activity and the known (if any) molecular factors predictive of survival. The biomarker should have a defined molecular mechanism of biological activity, and the data in support of its validity have to be based on thorough specimen collection, assay results confirming specificity, sensitivity, reproducibility, robustness as well as statistical rigor and on a stringent patient follow-up. In addition, the assays for biomarkers to be used clinically should be simple, inexpensive and lend themselves readily to high through-put technologies. These are by no means trivial requirements, and they emphasize the difficulties associated with the field of biomarker discovery.
A great deal of discussion has centered on the usefulness and validity of serum/plasma biomarkers vs tissue biomarkers in cancer [9]. Body fluids are readily available, while human tissue specimens are not, and serial specimens needed for monitoring of responses to therapy are only available by sampling of body fluids. However, only tumor specimens provide accurate information about the tumor microenvironment (TME), and serum/ plasma are much less useful, although ascites, pleural effusions or saliva may be more informative. In this context, the discovery that exosomes present in all body fluids of patients with cancer carry a molecular cargo that reflects the profile of tumor cells and perhaps of other components of the TME has created considerable excitement in the biomarker field.
The objective of this review is to consider the evidence in support of the potential role of tumor-derived exosomes as biomarkers which in the near future might facilitate monitoring of cancer progression and its outcome.
2. Definition, biogenesis and characteristics of exosomes
Exosomes are virus-size membrane-bound vesicles secreted by normal as well as malignant cells, and they are present in all body fluids [10,11]. Since the late 1970s, it has been known that various cells can release extracellular vesicles (EVs), which carry membrane-tethered as well as intravesicular molecules and deliver them to distant cellular targets [12–15]. EVs vary widely in size, molecular content and biological activities [16]. Apoptotic bodies are the largest type of EVs (1,000–5,000 nm), while exosomes are the smallest with the diameter of 30–120 nm [17]. Microvesicles, an intermediate-sized EVs, result from “pinching off,” or “blebbing,” of the cellular membrane into vesicles ranging in diameter from 200–1,000 nm. Among EVs, exosomes occupy a unique position primarily because of their biogenesis [17]. It begins when the plasma membrane buds inward, forming an endosome. An early endosome matures to a late endosome. As the late endosome membrane buds inward, and closes, it forms intraluminal vesicles, and endosome converts into a multivesicular body (MVB) [17,18]. MVBs contain multiple vesicles bound by inverted endosomal membranes enclosing bits of cytoplasm [19,20]. When MVBs containing pools of future exosomes fuse with the plasma membrane, vesicles are released into extracellular spaces. Exosome formation and release are ATP-dependent, and thus exosomes are products of live cells. Exosomes differ from other EVs not only by their small size and distinct biogenesis but also by other characteristic properties such as morphology, buoyant density on sucrose gradients and distinctive surface protein profiles [21]. The molecular cargo of the exosome membrane is of special interest because it is enriched in the components derived from the plasma membrane of the parent cell. It also contains endosomal markers, which is taken as evidence for the endocytic origin of exosomes [17].
The vesicular content of exosomes includes nucleic acids, enzymes, cytokines as well as various soluble factors, and it reflects the cytoplasmic content of the parent cell [20,22]. Exosome membranes are enriched in tetraspanins which are organized into tetraspanin-enriched domains (TEMs) and are thought to play a key role in exosome biogenesis [23,24]. Tetraspanins such as CD9, CD63, CD37, CD53, CD81, CD83 and CD151 are widely used as exosome markers, although exosomes derived from different cell types may carry only some, not all, of these tetraspanins [25]. Exosomes also carry components of the endosomal sorting complex responsible for transport (ESCRT) and various accessory molecules such as ALIX and TSG101 [26]. These are also often used as exosomal markers [26]. The ESCRT complex is involved in sorting of cellular components into exosomes and in exosome release from parental cells [26]. This biogenesis process consists of a coordinated series of steps involving many molecules and is executed by all cells secreting exosomes, as recently reviewed in a greater detail [17]. The mechanistic insights of this process are still being investigated.
The evidence allowing for a distinction to be made between exosomes and microvesicles is muted at best. It remains unclear whether exosomes derived from MVBs or microvesicles formed by “blebbing” of plasma membrane (hence, called ectosomes) carry molecular cargos that are more representative of the parent cell and thus potentially more useful as biomarkers. Further, a possibility of exosome aggregation into larger vesicles or, conversely, microvesicles splitting into smaller vesicles, makes exosome isolation and functional characterization both difficult as well as problematic. Nevertheless, given that exosome biogenesis involves an active rather than passive series of cellular events, it seems reasonable to consider exosomes as preferable “representatives” of a cellular phenotype and genotype of the parent cell. Based on this premise, exosomes are emerging as biomarkers that carry potentially useful information about parental cells.
a. Tumor-derived exosomes
While exosome secretion occurs under physiologic conditions, and all cells are capable of their release, tumor cells are avid exosome producers. This may be because in a hypoxic environment of tumors, stressed tumor cells would be likely to produce more exosomes [27,28]. Tumor-derived exosomes (TEXs) have been reported to carry a cargo of molecules and factors able to transfer information from the parent tumor cell to other cells present within and outside the tumor microenvironment [29,30]. TEX appear to have properties distinct from those of exosomes secreted by normal cells and have been reported to mediate immune suppression as well as immune activation [31–33]. Exosome fractions obtained from plasma of cancer patients are enriched in various immunosuppressive molecules, including death receptor ligands such as FasL, PD-L1, TRAIL and inhibitory cytokines, IL-10 and TGF-β1, as well as well as PGE2 [34,35]. In contrast to exosomes derived from normal cells, human tumors produce exosomes that have been shown to induce apoptosis of activated CD8+ T cells, promote differentiation and functions of regulatory T cells (Treg) and interfere with dendritic cell (DC) differentiation, favoring expansion of myeloid-derived suppressor cells (MDSC) [34–36]. Importantly, in addition to their immunosuppressive cargo, TEX also carry tumor-associated antigens, a variety of co-stimulatory proteins and the MHC molecules, all of which provide them with the capability to stimulate immune responses [33,37,38]. The dual signaling capability, both immuno-stimulatory and immuno-inhibitory, places TEX in a special category as potential biomarkers of cancer progression and of immune response to the progressing tumor.
In addition to immunomodulation, TEX mediate a variety of pro-tumorigenic effects [39]. They promote tumor growth, sustain autocrine loops and modify functions of stromal cells [39]. Considerable literature exists describing TEX as carriers of oncogenic signals or active oncogenes from tumor to normal cells and supporting the role of TEX in neoplastic transformation [29,40–42]. Thus, in their capacity to favor tumor growth, TEX or “oncosomes” may serve as putative diagnostic tools or therapeutic targets.
b. Biological functions of exosomes
Exosomes appear to be involved in a broad variety of biological functions (Table 1). In fact, various names given to exosomes based on their functions, such as oncosomes, tolerosomes, argosomes, etc. [40], reflect their involvement in many physiological and pathological events. Exosomes which carry membrane and cytosolic components of the parent cell are excellent vehicles for information transfer. The surface of exosomes is decorated by the parent cell-specific proteins (including various receptors and ligands) as well as endosome-derived proteins, exposing these components to the milieu outside the parent cell. This provides a mechanism for exosome interactions with recipient cells [43–46]. The exosomal cargo components are biologically active, forming molecular arrays which, upon interaction with and transport to the recipient cell, alter its functional attributes [47,48]. In this way, exosomes can play a key role in a wide variety of physiological processes. Multiple cell types have been reported to secrete exosomes, including all types of hematopoietic cells, fibroblasts, vascular endothelial cells, intestinal epithelial cells, neuronal cells and others [49–53]. Exosomes produced by normal cells regulate normal physiological functions. In contrast, infected, transformed or otherwise altered cells appear to secrete exosomes which mediate quite different functions.
Table 1.
Biological functions attributed to tumor-derived exosomes (TEX)
| References | |
|---|---|
| Tumor progression | |
| Autocrine effects on tumor cells: Proliferation, survival, migration | 16, 32, 44 |
| Stimulation of angiogenesis in the TME | 32, 44, 129 |
| Extracellular matrix remodeling | 61, 62 |
| Education and re-programming of the bone marrow to a pro-metastatic niche | 54,55 |
| Metastasis | 54,55 |
| Suppression of host anti-tumor immunity | 57, 58, 59, 60 |
| Horizontal transfer of oncogenic mutations from tumor to normal cells | 39, 40, 54 |
| Intercellular communication and target cell reprogramming (TEX as “signalosomes”) | |
| Transfer of proteins, lipids and nucleic acids (mRNA, miRNA, DNA) from tumor to normal cells: alterations in phenotype and functions | 39, 40–48 |
| Thrombogenesis: promotion of coagulation | 130 |
| Angiogenesis: vessel formation at sites distant from tumor | 131, 132, 133 |
| Hematopoietic cell differentiation | 63, 64, 67 |
| Immune regulation | |
| Immunosuppression of tumor-specific responses | 57–60 |
| Immune stimulation of tumor-specific responses | 36, 37, 38 |
| Drug resistance | |
| Removal of toxic drugs from the cytosol | 65 |
| Exchange of drug transporters | 66 |
Cancer cells actively secrete masses of exosomes that are distributed throughout the body and become a part of the tumor-driven communication network. Table 1 lists functions attributed to tumor-derived exosomes (TEX). TEX can manipulate the tumor microenvironment as well as distant tissue sites to promote tumor progression and tumor metastasis [54,55]. The participation of exosomes in metastatic niche formation was elegantly demonstrated in vivo by Peinado et al [54]. Exosomes produced by mouse melanoma cells facilitated cancer metastasis by altering the bone marrow milieu and the vascular compartment, preparing or educating tissue sites for acceptance of metastatic cells and promoting their growth [54,56]. TEX were shown to carry molecules and signals that disable immune cells or cause death of activated immune cells, thus preventing them from exercising antitumor functions and promoting tumor escape from the host immune system [10,32,34,35,57–60]. TEX have been shown to participate in angiogenesis [61,62], hematopoietic cell development and differentiation [63,64] as well as resistance of tumor cells to drugs [65,66]. TEX also modulate various cellular and intercellular signaling pathways that are implicated in cancer promotion, such as the TGF-β, the Wnt-β-catenin and the Notch pathways [63,64,67] and the pro-tumorigenic effects of TEX seem to predominate in the tumor microenvironment. Exosome secretion by tumor cells was found to be regulated by the p53 protein [68] and PTEN, which regulates the PI3K-AKT signaling, was found to be carried in TEX and to mediate phosphatase activity in recipient cells [69,70]. These examples indicate that tumor cells utilize exosomal signaling as a major aspect of pro-tumorigenic intracellular communication.
While TEX are purported to carry information imparted to them by the parent tumor cell to other tumor or normal cells, the composition of this cargo depends on the sorting/packaging machinery (ESCRT) and its accessory molecules [26]. The efficient delivery of this cargo to recipient cells is critical for information transfer. If sorting via the ESCRT pathway in the parental cell is specific, as suggested above, released exosomes are targeted to pre-determined cellular destinations [20]. However, an alternative hypothesis, initially formed when exosomes were first described, is that exosomes are simply discarded, unwanted cellular debris [71]. This view of exosomes does not entirely fit with the biological effects they are described to mediate. Biological effects of TEX are orchestrated at the level of recipient cells and appear to specify definite functional alterations that follow exosome transfer and are distinct from those mediated by exosomes produced by normal cells [44,72]. At present, signals and molecular mechanisms of exosome interactions with various target cells are not entirely clear.
Depending on the nature of the target cell, exosomes may be readily or not so readily internalized. Exosomes can transfer materials to target cells via three ways: receptor-mediated uptake, direct fusion with cell membrane or phagocytosis [73]. Phagocytic cells, such as macrophages or dendritic cells (DC) rapidly take up exosomes [74]. Other cells, e.g., cultured human brain microvascular endothelial cells co-incubated with green fluorescent protein (GFP)-labeled exosomes, were shown to internalize them readily, so that green nanovesicles were seen in the cytosol within hours of co-culture [44]. But it appears that other cell types, e.g., human lymphocytes, at least resting lymphocytes, do not readily internalize exosomes [Muller L, Whiteside TL, unpublished data]. In T cells, uptake of TEX seems to depend on receptor-mediated uptake as previously reported [75]. Exosomes co-incubated with human T cells deliver signals to surface receptors present on the T cells, initiate a Ca2+ flux and alter expression levels of mRNAs coding for various immunomodulatory proteins, which ultimately results in changes of expression levels in CD69, a surface activation protein, in the recipient cells [76]. Thus, in T cells and other recipient cells, signals delivered by exosomes alter the cellular transcriptome, and these changes have functional consequences after being translated into proteins. Mechanisms responsible for exosome-induced mRNA and protein alterations in recipient cells are only partly understood.
Functions attributed to exosomes are numerous and quite varied. It is unclear whether all or only subsets of TEX mediate these functions. It also seems unlikely that all TEX would be programmed to carry information for all functions listed in Table 1. Therefore, a hypothesis was introduced in support of selective packaging of information in exosomes and targeted delivery of messages to cells in or outside the tumor microenvironment [24]. In this scenario, exosomes designated to prepare, e.g., a distant lymph node for metastases, would deliver a message distinct from that carried by other TEX. To date, there is no evidence in support of this hypothesis, and multi-functionality of TEX remains an unexplained and somewhat perplexing puzzle. In fact, the actual biological role of the entire exosome system remains unknown.
3. Methods for exosome isolation
In recent years, many different technologies have been introduced for the isolation of exosomes from supernatants of cultured cells or from body fluids. The lack of consensus for selection of the best method is a major hurdle in exosome research. The proposed methods have been recently comprehensively reviewed, with a detailed commentary on their respective advantages and disadvantages [77–79]. Most of these methods were developed using exosomes in supernatants of cultured cells. Here, special attention is directed to the methods being developed for the isolation of TEX from body fluids of cancer patients. To be able to utilize exosomes as cancer biomarkers, their origin, morphological integrity, molecular and genetic content and functional attributes have to be preserved, and their recovery devoid of losses due to sample processing. This is a critical aspect of the development that will ultimately determine whether TEX meet the criteria required for cancer biomarkers. With this rationale in mind, TEX isolation assumes a predominant place in the process of biomarker development.
a. Exosome isolation from tumor cell supernatants
Supernatants of tumor cell lines grown in culture are frequently used for exosome isolation, because all exosomes are derived from the same cellular source and because a relatively simple chemical composition of most culture media facilitates isolation of exosomes devoid of ‘contaminating’ proteins, lipids and sugars. Human or animal sera used as supplements for cell cultures are ultracentrifuged prior to their use to remove “contaminating” extracellular vesicles. Nearly all studies of TEX reported to date in the literature were performed using supernatants of cultured tumor cells, e.g., Kasumi-1 (AML) or PCI-13 (HNSCC) cell lines in the author’s laboratory [76,80]. Supernatants of cell lines undergo differential centrifugation first at low and then higher speeds to remove cell fragments and large EVs. Next, exosomes are pelleted by ultracentrifugation at 100,000×g for 2–3h, re-suspended in PBS and prepared for the subsequent enrichment on continuous sucrose density gradients [77]. This method yields single exosomes banding at the sucrose density of 1.13–1.19g/mL [50,81]. However, low levels of exosome recovery, probably due to a loss of aggregated exosomes, which sediment to the bottom of the gradient, and the low-throughput nature of this methodology limit its utility. This “conventional” isolation procedure has been modified by incorporating ultrafiltration with a 0.22μ filter immediately after differential centrifugation followed by size exclusion chromatography (SEC) on a Sepharose 2B column and then high-speed ultracentrifugation [76]. SEC, first introduced by Taylor et al [82] allows for the recovery of “cleaner” exosomes in the void volume [77], as unwanted “impurities” elute in later fractions. A more recent modification utilizes mini-SEC columns packed with Sepharose 2BL (the bed volume is 10 mL), which may be homemade [83] or purchased and which isolate morphologically-intact and biologically-active exosomes from supernatants of tumor cells. Mini-SEC columns replace ultracentrifugation with a simple, one-step, highly reproducible scaled-down size-exclusion chromatography, yielding early fractions enriched in exosomes [83].
b. Isolation of exosomes from body fluids
Exosome isolation from plasma is more complex than that from supernatants of cultured cells. First, plasma contains a mix of exosomes derived from many different cells in varying proportions, so that the source of exosomes present in plasma is unknown. Second, plasma exosomes are ‘coated’ with proteins, lipids, glycoproteins or glycolipids likely to cause their aggregation and a potential loss upon centrifugation. Third, isolation of exosomes derived from a specific cell type, such as tumor cells, is a considerable problem, because the desired exosomal fraction, e.g., that containing TEX, represents only a small part of the total comprised largely of exosomes originating from erythrocytes and platelets [49,84–86]. Therefore, special methods are required for recovery of TEX from body fluids of patients with cancer.
The use of human plasma vs serum for exosome isolation has been a subject of much discussion because of the possibility for exosome losses from sera due to the clot formation. However, in our hands, plasma or serum serve as equally good sources of circulating exosomes based on the recovery, purity, morphology and biological function [76]. Exosome isolation from human fresh vs frozen (after low-speed centrifugation) plasma has also been evaluated [76]. This is an important issue as most exosome studies performed today are retrospective and utilize specimens frozen/thawed for months if not years [84]. Few if any prospective studies of plasma-derived exosomes have been conducted so far because of a need for clinical results to be available for correlations with exosome molecular or functional profiles. However, the use of banked frozen/thawed plasma or serum specimens for exosome isolation presents some problems. When plasma is frozen immediately after low-speed centrifugation, larger vesicles that are left behind as well as some exosomes are disrupted or damaged during freezing and release nucleic acids and proteins. We showed by TEM that even the differentially-centrifuged frozen/thawed plasma contains numerous string-like aggregates, which surround exosomes [76]. Removal of these aggregates by enzymatic treatments with DNAse, RNAse or hyaluronidase in part reduced their presence but tended to aggregate exosomes. Thus, attempts at improving exosome recovery from frozen/thawed plasma specimens by enzymatic treatments were neither productive nor cost effective. Probably, fresh platelet-depleted plasma would be the best source of exosomes for biomarker studies. Frozen plasma centrifuged at low then higher (10,000 × g to 15,000 × g) speeds and ultrafiltrated using a bacterial (0.22μ) filter before banking would be the second best. After thawing of plasma, a second ultrafiltration greatly reduces “contamination” with larger EVs, subcellular fractions, protein aggregates, protein-nucleic acid aggregates or plasma proteins [76]. Differential centrifugation of plasma without ultrafiltration is not adequate for the removal of these “contaminants.” After ultrafiltration, the use of above-described mini-SEC columns yields fractions which are highly enriched in relatively “clean” exosomes [83]. Coomassie blue staining of PAGE gels loaded with the min-SEC-isolated plasma-derived exosomes indicated minimal levels of contamination with immunoglobulins [83]. The recovery, quality and functionality of circulating exosomes purified from such plasma or serum specimens were found to be adequate for further exosome characterization [76,83].
Once exosomes are isolated, they are either immediately used for studies or stored for future use. In our hands, freezing of freshly isolated exosomes (i.e., after their recovery on mini-SEC columns or by ultracentrifugation) does not alter their morphology and does not seem to impair their function, as also previously reported [87,88]. The exosome resistance to freezing is probably due to the more balanced surface to volume ratio, as compared to much larger cells or larger vesicles. Thus, if isolated exosomes cannot be used within a few days, they can be aliquoted and stored at −80°C for future use.
There are numerous commercially-available technologies for exosome isolation. They are often designed to monitor either exosomal proteins, lipids or nucleic acids but almost never all of the above simultaneously. These methods do not differentiate exosomes from other EVs. While recovery of nucleic acids from EVs or exosomes appears to be relatively reliable, other components of the exosomal cargo may be readily lost during isolation due to damages of the exosomal membrane or stripping of components by solvents. Some procedures might lead to the concentration of contaminating non-exosomal materials, others might recover only a fraction of total exosomes. These artifacts can skew the molecular profiles of exosomes and obscure their significance as biomarkers. In view of potential isolation artifacts, it is only reasonable to aim for the method that isolates morphologically-intact, non-aggregated exosomes with the defined diameter of 30–120 nm, that serve as a reliable source of proteins, lipids, and nucleic acids and that mediate expected biological functions. In the author’s laboratory, the mini-SEC method was found to measure up to these criteria and, therefore, it is currently used exclusively for exosome isolation and purification from banked specimens of human plasma. These, of course, are total exosome fractions, and further manipulations are necessary to separate exosomes produced by tumor cells from those produced by other, normal cells.
c. Isolation of TEX from plasma of cancer patients
Plasma or other body fluids of patients with cancer contain exosomes produced by tumor cells and by non-tumor, normal cells. To study TEX and confirm their unique molecular and functional properties, it is first necessary to separate them from larger EVs (e.g., by mini-SEC) and then from non-tumor-derived exosomes and to do so without incurring losses in TEX recovery. Studies of exosomes produced by cultured tumor cells showed that molecular profiles of these vesicles are tumor-like and are distinct from those of normal cells [see Table 2]. Based on these comparative in vitro data, exosomes circulating in plasma of cancer patients are expected to carry a unique set of membrane-embedded molecules, which mimic those present in the membrane of tumor cells. If this is true, then TEX could be distinguished and separated from non-tumor-derived exosomes in the same plasma by beads charged with antibodies specific for tumor-associated antigens (TAA) carried by TEX [89]. Thus, monoclonal antibodies (mAbs) specific for the TAA expressed or overexpressed on the surface of tumor but not normal cells are necessary for immunocapture of TEX. This will require a judicial selection of mAbs for each tumor type.
Table 2.
Tumor associated proteins in EVs/exosomes derived from human tumor cell linesa
| Not cell-type specificb | EV cell source | Biological effects | Reference |
| Tetraspanins: CD9, CD63, CD81 | Normal or tumor cells | Exosome sorting | 134, 135 |
| Heat shock proteins: Hsp70, HSP90 | Various tumor cells | Exosome antigenicity | 54,136 |
| Biogenesis associated: TSG-101, ALIX | Normal or tumor cells | Exosome formation | 26, 136 |
| Membrane transport: anexins, flotellin, caveolin-1, Rab GTPase | Normal or tumor cells | Exosome release | 137, 138, 139 |
| Immune cell apoptosis FasL, TRAIL, galectin-9 | Various tumor cells | Immune suppression by exosomes | 32, 34, 35 124, 140 |
| Cancer cell-associatedc | Tumor type | Pro-tumor effects | Reference |
| Amplified oncogenes (K-ras mut) | CRC | Oncogene transfer | 40–44 |
| EGFRVIII mut | GBM | Growth | 141, 142 |
| IDH1, IDH2 mut | GBM, meduloblastoma | Growth, invasion | 141, 143, 144 |
| Amphiregulin | BRCA, CRC | Growth, invasion | 145 |
| Survivin | Inhibits apoptosis | 146 | |
| GD2 ganglioside | NB | Immune suppression | 146 |
| FGF-2 | Meduloblastoma | Growth | 143 |
| LMP-1 | NPC | Signal Transduction | 148 |
| MUC-1 | BRCA | Growth, invasion | 149 |
| Del-1 | Mesothelioma | Angiogenesis | 143 |
| EpCAM | CRC | Growth, invasion | 96 |
| TGF-β | GBM, BRCA | Immune Suppression | 142 |
| Mesothelioma | Stroma remodeling | 61, 62 | |
| VEGF, TIMP 1, MMP-9 | GBM | Growth, invasion, stroma | 44 |
Abbreviations: GBM, glioblastoma multiforme; med bl, medulloblastoma; BRCA, breast carcinoma; NB, neuroblastoma; NPC, nasopharyngeal carcinoma; CRC, colorectal cancer.
EVs were isolated from supernatants of tumor cell lines by various methods, without discrimination of larger EVs from smaller exosomes.
The proteins listed are present in EVs isolated from tumor cells at higher levels compared to those observed in EVs obtained from normal cells.
As a “proof of principle,” an immunoaffinity-based capture method for CD34+ exosomes from plasma of AML patients was developed [80]. Immunocaptured CD34+ exosomes were separated from non-captured CD34neg exosomes, dissociated from the beads, recovered and analyzed for protein levels, numbers of nanoparticles by NanoSight or q-Nano and morphology by TEM. Their non-captured counterparts were analyzed in parallel. The presence of tetraspanins, used as “markers” of exosomes [25], and of leukemia-associated antigens or immunoregulatory molecules of interest, such as IL-10, TGF-β, FasL, PD-1, PDL-1, CD39, CD73, HSP70, HSP90 was studied by Western blots. The ability of TEX to suppress immune cell activities was measured in flow cytometry-based functional assays [76,90,91]. The recovery of TEX calculated in µg protein/mL of the input plasma varied between AML patients, but was adequate for subsequent analyses by or protein immune arrays or qRT-PCR for selected sets of genes. The studies showed that in CD34+ AML, blast-derived CD34+ exosomes not only carried a distinctive molecular cargo but were also functionally different from non-blast exosomes in assays measuring NK-cell activation [76].
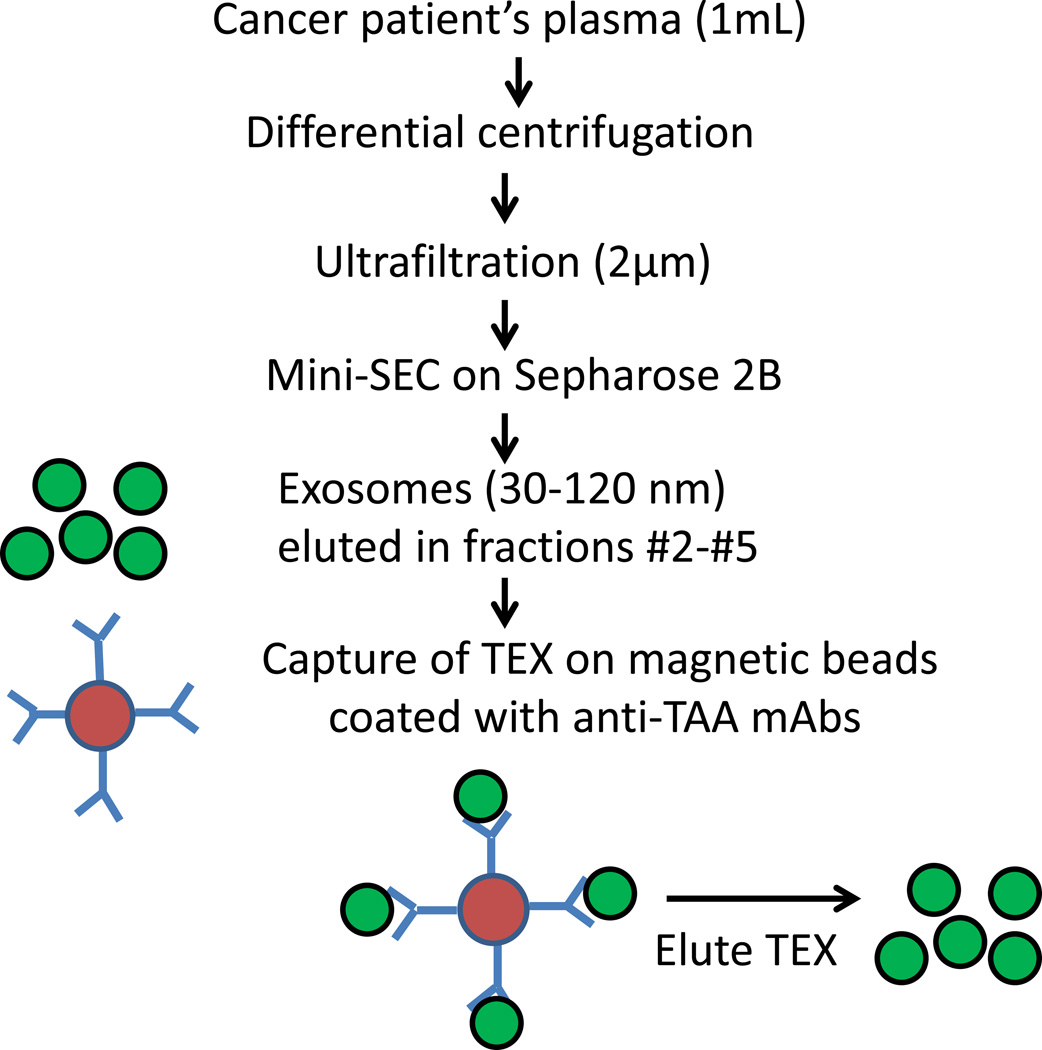
Immunocapture method can be implemented using several different TAA-reactive mAbs to acquire TEX from plasma of patients with cancer (Figure 2). For example, to capture melanoma TEX, a combination of mAbs recognizing CSGP4, also known as the high molecular weight melanoma associated antigen (HMWMAg), and GM3 mAb recognizing NeuGc ganglioside can be used for capture of TEX [92,93]. For HNSCC, capture experiments can be performed, e.g., with anti-survivin mAb in combination with PRAME-specific mAb [94,95]. Antibodies to EPCAM have been used to capture TEX in plasma of patients with colon carcinoma [96]. While these TAA may be present on non-tumor cells, they are overexpressed in the tumor and presumably are carried by TEX. In attempting immunocapture of TEX from patient’s plasma, it is advisable to check by Western blots whether exosomes are strongly positive for a tumor marker of choice.
Figure 2.
The strategy for isolation of TEX from plasma of a patient with cancer. Initially, a one-step mini-SEC is used to isolate total plasma exosomes. Next, TEX are captured on mAb-coated magnetic beads. The mAb selected for immune capture of TEX is specific for a tumor-associated antigen (TAA) overexpressed on tumor cells. Captured TEX are eluted from beads using acidified buffer. Following capture and elution, TEX retain morphologic integrity and mediate biological functions [83].
In plasma of cancer patients, total exosome fractions were found to be significantly enlarged relative to those in NC plasma [76,90,91]. This was especially evident in patients with advanced cancers [76]. Not only total protein levels but also the content of TEX in the total exosome fraction based on expression of selected markers might vary depending on the disease stage and therapy used [90]. In situations when cancer therapy is effective, the TEX content might decrease, so that immune capture of TEX gives negative results. Obviously, this is critically important information that impacts significance of TEX as biomarkers of response to therapy. However, to interpret this change correctly, reliable, reproducible and consistent methodology for TEX monitoring has to be in place. Thus, the selection and establishment of methods for TEX isolation is the major barrier in advancing TEX to the biomarker status.
A special advantage of TEX immunocapture is that it provides an opportunity for studies of both TEX and their non-tumor-derived counterparts. This might provide useful insights into changes induced by the tumor or TEX in normal cells present in the tumor microenvironment. It is possible that exosomes produced by normal cells residing in the tumor milieu might prove to be as informative cancer biomarkers as TEX in respect to tumor progression.
4. Evaluation of exosome molecular and genetic profiles
Molecular profiling of exosomes or TEX isolated from human body fluids can be accomplished by a variety of methods. Mass spectrometry is favored by many investigators [97], but, as discussed, it is especially prone to difficulties with plasma-derived exosomes, where “contaminating” plasma proteins might mask genuine exosome-associated components. Removal of “contaminants” with low pH buffers or solvents does not seem to ameliorate the problem and might impair exosome integrity. In our hands, mass spectrometry results of exosome or TEX enzymatic digests were deficient in components which in Western blots and functional assays represent biologically-important exosome cargos. Some of the cargo components that are of special interest such as cytokines or enzymes are not seen by mass spectrometry and may have to be amplified by antibodies to be detected and measured. It appears that proteomics by mass spectrometry may not be the method of choice for the analysis of tumor-associated proteins carried by exosomes isolated from plasma of cancer patients. Many investigators have switched to mass spectrometry of exosomes recovered from urine in order to bypass the problems encountered with profiling of plasma-derived exosomes [98]. The downside of this strategy is that exosomes isolated from urine, having been “filtered” by kidneys may be only partly and not fully representative of the total population of TEX. Ab-based protein arrays or qRT-PCR for mRNA expression levels of selected groups of genes have been successfully used for molecular and genetic profiling of exosomes [44]. The protein arrays are especially appealing because of the signal amplification they offer. The disadvantage is that both these technologies require substantial quantities of exosomes for analysis.
a. Total protein levels of plasma-derived exosomes
Protein levels in total exosome fractions isolated from plasma are significantly higher in patients with cancer than in healthy donors [54,91]. Determined by the standard protein assays and calculated in µg protein/mL plasma, exosomal protein levels may exceed values of 150µg protein/mL plasma relative to e.g., 10µg protein/mL plasma in NC [91]. Further, protein levels in exosomal fractions isolated from patients with cancer were reported to correlate with disease activity, tumor grade, tumor stage, response to therapy and even survival [54,90,91]. In AML patients undergoing standard of care chemotherapy, exosomal protein levels were reduced following induction chemotherapy concomitantly with the blast reduction in the bone marrow [90]. During consolidation therapy with high-dose cytarabine, exosomal protein levels were increased relative to those measured post induction therapy, reaching a mean level similar to that seen at diagnosis in some patients [90]. Exosomal protein levels in plasma of AML patients who achieved long-term remission were not significantly different from those seen with exosomes from plasma of healthy donors [90]. These data suggest that in AML, protein levels of plasma-derived exosomes might serve as a measure of leukemic blast persistence after chemotherapy and potentially as predictors of disease relapse.
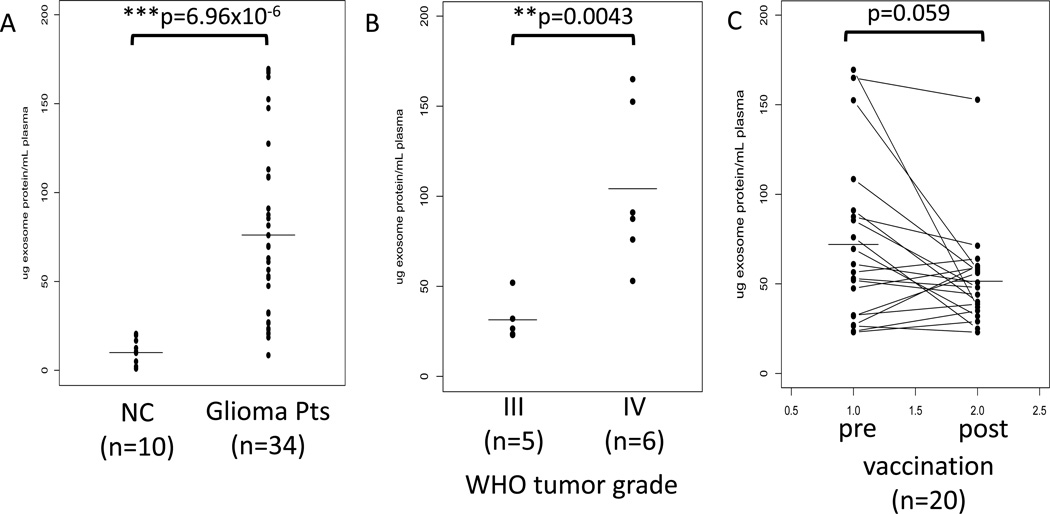
In patients with recurrent malignant glioma (n=20) who participated in a phase I/II dendritic cell/peptide-based vaccination trial at the author’s institution, protein levels were measured in total exosomal fractions obtained from plasma collected prior to and after vaccination [99]. Exosomal protein levels positively correlated (p<0.0043) with the WHO tumor grade at diagnosis (Figure 3). These protein levels rapidly decreased after vaccination, in many albeit not all patients. The observed pre- to post-vaccine decreases in exosomal protein levels correlated with immunological and clinical responses of the patients, an indication that exosomal protein levels might serve as a measure of post-vaccination immune responses and potentially as a predictive marker of clinical response to the vaccine [99]. It should be emphasized that it may not be TEX but rather immune cell-derived exosomes that serve as markers of immune responses to the vaccine.
Figure 3.
Protein content (normalized to µg/mL plasma) of exosomes isolated from plasma specimens of patients with glioma by differential centrifugation, ultrafiltration, SEC and ultracentrifugation. In A, mean protein levels are higher in patients than NC. In B, patients with stage IV disease have higher protein levels in exosome fractions than those with stage III disease. In C, protein levels in exosomes isolated prior to and after vaccination with a DC-based peptide vaccine. The data shown in A, B and C are reproduced from Muller L, Muller-Haegele S, Mitsuhashi M, Gooding W, Okada H, Whiteside TL, Exosomes isolated from plasma of glioma patients enrolled in a vaccination trial reflect anti-tumor immune activity and might predict survival, Oncoimmunology, 4 (6), e1008347 (2015) [99] by permission of Taylor & Francis LLC.
These two examples added to the data reported by others [54] suggest that a simple measurement of protein content in plasma-derived exosomes might have a predictive value in cancer. Recent data also indicate that not only total protein levels but also the individual protein content in exosomes can provide useful prognostic information. While examining one exosomal protein in lieu of a protein profile appears to be hardly acceptable, in combination with functional analysis it might be informative. For example, TGF-β1, a factor known to inhibit natural killer (NK)-cell cytotoxicity and promote differentiation and expansion of regulatory T cells (Treg), were found to be significantly elevated in exosomes isolated from plasma of AML patients [90]. AML exosomes isolated before, during or after chemotherapy carried different forms of TGF-β1 (pro-peptide, LAP or an active, mature form) in distinctly different proportions. After their solubilization/acidification, these exosomes were shown to contain different levels of active TGF-β1 which down-regulated cytotoxicity mediated by NK cells [90]. These data suggest that TGF-β1 expression levels in exosomes reflect TGF-β1 activation and utilization in AML, directly linking the presence of TGF-β1+ exosomes to immune cell suppression commonly seen in AML [91]. We studied functional relevance of the different forms of TGF-β1 seen in AML exosomes by Western blotting and found that NKG2D down-regulation on NK cells was only mediated by exosomes carrying mature TGF-β1 and that the content of mature TGF-β1 correlated to the degree of suppression these exosomes induced [90]. Alterations in levels of the pro-peptide, LAP and mature TGF-β1 in exosomes during therapy and in remission could influence NK-cell functions and/or Treg generation in AML patients. These results illustrate the potential impact of individual exosome components on immune cell functions in AML and thus on disease progression.
b. Protein profiling in plasma-derived exosomes
Given that increased protein levels and the enrichment in physiologically-relevant tumor-associated proteins in circulating exosomes have been reported to correlate with cancer progression and outcome [54,90,100,101], there is a great deal of interest in profiling the molecular content exosomes in plasma of cancer patients. The results of proteomics available in the EXOCarta (now Vesiclepedia) databases indicate that exosomes carry a wide variety of biologically important molecules such as oncogenic proteins, receptors, receptor ligands, enzymes, cytokines and factors modulating cellular behavior [102,103]. To a large extent, the efforts of cataloguing exosome-associated proteins have been limited to studies of EVs/exosomes isolated from supernatants of tumor cell lines. These studies made no distinction between EVs and exosomes and, as summarized in several recent reviews, the data defining molecular signatures of “tumor-derived exosomes” were obtained by analysis of total EV fractions isolated by various procedures. Nevertheless, these data indicate that EVs isolated from tumor cell supernatants have a defined protein and lipid signature which consists of conserved as well as tissue/cell type-specific sets of molecules [39,104]. Table 2 provides a short list of proteins reported to be carried by EVs originating from various cultured tumor cells. The list illustrates the presence of selected, well-known tumor-associated proteins in EV fractions. The protein signatures of these EVs produced by different tumor cells are different from each other and from signatures of EVs derived from normal cells, suggesting “cancer cell-type specificity” of the cargo.
Few reports of proteomics results for EVs/exosomes isolated from plasma of cancer patients are available. Reports on EVs/exosomes isolated from urine are more frequent [98,105]. Table 3 lists examples of the proteins found in EVs obtained from body fluids of patients with cancer. This is a selected list illustrating the presence of molecules known to be associated with cancer, its progression and/or response to therapy. Their presence in the EV/exosome cargo implies that these vesicles could play a potential role as cancer biomarkers. It has been suggested that TEX might serve as a “liquid biopsy” that could be non-invasively and repeatedly measured in body fluids of cancer patients without a need for the tumor biopsy [101,106,107]. This intriguing concept has yet to be validated by extensive studies of plasma-derived exosomes and the parent tumor cells in situ performed in parallel. Another hypothesis predicts that TEX mimic the molecular content of the parent tumor cell more precisely than do total plasma-derived exosomes. This concept was discussed above and is an important part of ongoing efforts to qualify exosomes as future cancer biomarkers. However, the role of TEX as a more “faithful” molecular surrogates of tumor cells then total EV/exosome fractions remains speculative. Few data are currently available that demonstrate actual application of TEX to diagnosis, prognosis or patient management [90,99]. To move the field forward, it is necessary to reach a rapid consensus on technical issues surrounding exosome and TEX isolation to be followed by retrospective and then prospective studies correlating TEX with cancer progression and response to therapies. Future studies comparing predictive values of TEX with those of total EV/exosomes in body fluids of cancer patients will determine which population of vesicles is to be considered as a better tumor surrogate and merit further development as a non-invasive cancer biomarker.
Table 3.
Tumor-associated proteins in EVs/exosomes isolated from body fluids of patients with various cancersa
| Plasma/serumb | Tumor type | Pro-tumor effects | Reference |
|---|---|---|---|
| EGFRVIII mut | GBM | Growth | 141, 142 |
| Survivin | PROCA | Apoptosis inhibited | 146 |
| PTEN | PROCA | Signaling | 152 |
| Claudin-4 | OVA CA | Invasion | 153 |
| TYRP2, VLA-4, HSP70 | Melanoma | Growth progression | 54 |
| PSA | PROCA | Growth, progression | 152 |
| EGFR | NSCLC | Signaling, growth | 135 |
| Caveolin, CD63 | Melanoma | 139 | |
| Galectin-9 | NPC | Immune suppression | 140 |
| LMP-1 | NPC | Signaling | 154 |
| FasL | HNSCC | Immune suppression | 34, 35, 155 |
| Met (phosphor) | Melanoma | Oncogene transfer | 54 |
| KRAS | PANCCA | Oncogene transfer | 55 |
| CEA | CRC, NSCLC | Growth, invasion | 135 |
| TGF-β | AML, OV CA | Immune suppression | 90, 156 |
| Urinec | |||
| PSA | PRO CA | Growth, progression | 157 |
| PSMA | PRO CA | Growth, progression | 158 |
| MMP-9 | RCC | Stroma remodeling | 159 |
| Carbonic anhydrase | RCC | Stroma remodeling | 159 |
| α6-integrin | BLCA | Growth, invasion | 160 |
| MUC-1 | BLCA | Growth, invasion | 160 |
| LRG-1 | NSCLC | Cell adhesion | 161 |
| Basigin | CRC, BLCA | Migration, invasion | 162 |
| Salivad | |||
| Dipeptidyl peptidase IV (CD26) | Normal donors | 163 | |
| Alix, TSg101, HSP70, IgA, CD26 | Normal donors | 164 | |
| 63 different proteins, including annexin, keratin, actin, Igs and S100 | Normal donors | 165 | |
EVs/exosomes were isolated from body fluids of patients with cancer. EVs were not discriminated from exosomes.
The data are selected from published reports to illustrate the range of tumor-associated proteins carried by EVs/exosomes.
Selected proteins carried by urine exosomes are listed based on published data.
Only exosomes from saliva of normal donors were studied to date
c. mRNA and miRNA expression levels in plasma-derived exosomes
The presence in exosome cargo of mRNAs and microRNAs (miRNAs) and their transfer via exosomes to recipient cells was first demonstrated by Valadi and colleagues in 2007 [43]. Since then, numerous reports have confirmed that tumor-derived EVs carried abundant miRNAs also called oncomirs [100,108–110]. These small, non-coding RNA molecules were also abundant in EVs/exosomes isolated from plasma or other body fluids of patients with all types of cancer [110]. More recent data show that miRNA content is limited in exosomes and that larger vesicles contain higher miRNA levels [111]. It is now clear that miRNAs delivered to recipient cells regulate gene expression in target cells by either repressing the translation or causing the degradation of multiple-target mRNAs, depending on the cellular context. The horizontal transfer of miRNA to target cells leads to functional alterations with consequences that impact on tumor progression and metastasis [110]. With this in mind, much attention has been given to exosome-bound miRNAs and their potential role as genetic biomarkers of cancer [109–112]. The data presented in Table 4 indicate that miRNA signatures for EVs/exosomes derived from plasma of patients with different cancers are distinct. However, it has been shown that miRNA signatures in EVs/exosomes do not correspond to those in the parent tumor cells [43,44,113]. This suggests that a sorting process in the parent cell selects and actively loads miRNAs into EVs and that miRNA content of EVs is specified by the tumor cell. Further, pre-miRNAs packaged into exosomes can be converted to mature mi-RNAs. Very recent studies show that breast Ca-associated exosomes contain the RISC-Loading Complex which enables them to process pre-miRNAs into mature miRNAs and that the elements necessary for this maturation process, including DICER and CD43 are present in the exosomes [42]. This means that horizontally-transferred cancer exosomes can induce rapid silencing of mRNAs in target cells, re-programming their transcriptome and converting normal epithelial cells to tumor cells in a DICER-dependent manner [42]. Thus, tumor-derived exosomes emerge as pre-programmed carriers of miRNAs, whose profile might provide important insights into the status of the parent cancer cells.
Table 4.
Tumor-associated miRNAs in EVs/exosomes isolated from human body fluidsa
| Cancer type | miRNAs | Reference |
|---|---|---|
| OvCA | miR-21, miR-141, miR-200a, miR-200b, miR-200c, miR-203, miR-205, miR-2014 | 100, 107 |
| CerCA | miR-21, miR-146a | 117 |
| CRC | Let-7a, miR-1224, miR-150, miR-1246, miR-21, miR-23a miR-223 | 114 |
| GBM | miR-21, miR-320, miR-574-3p | 166, 167 |
| NSCLC | miR-151a, miR-30a-3p, miR-154-3p, miR-100, miR-629, miR-2006-5p | 107, 108, 168 |
| PROCA | miR-107, miR-1306, miR-141, miR-181a-2, and 8 others | 129, 169, 170 |
| RCC (stem cells) | miR-200c, miR-92, miR-141, miR-196, miR-29a, miR-29c, miR-650, miR-151 | 129 |
| HCC | miR-584, miR-517c, miR-378, and 8 others | 171 |
| AMLb | Let-7a, miR-9, miR-99b, miR-150, miR-155, miR-191, miR-223 | 172 |
EVs/exosomes were isolated from patients’ body fluids by various methods, and exosomes were not discriminated from EVs. The table presents a selected list of data to illustrate the diversity of miRNA contents in vesicles isolated from human body fluids.
miRNAs detected in supernatants of an AML cell line Molm-14.
Abbreviations: OVCA, ovarian carcinoma; CerCA, cervical carcinoma; GBM, glioblastoma; NSCLC, non-small cell lung cancer; PROCA, prostate carcinoma; RCC, renal cell carcinoma; AML, acute myeloid leukemia.
Work by Taylor and Gercel-Taylor identified the first cancer-specific miRNA signature in exosomes derived from plasma of patients with ovarian Ca [100]. In this study, circulating TEX immunocaptured by using anti-EpCAM Abs were shown to carry 8 miRNAs that discriminated ovarian Ca from a benign disease (Table 4). Similarly, exosomes in plasma of CRC patients carried a distinct miRNA signature [114]. Other studies have shown that exosomes in plasma of certain cancer types, including glioblastoma [44], lung [115] and breast [116] cancers, have distinct miRNA profiles (Table 4). Specifically, certain miRNA species, such as miR-21, were found to be highly expressed in exosomes from plasma of ovarian Ca, glioblastoma, breast Ca and pancreatic Ca (Table 4). Further, miR-21 expression levels appeared to correlate with the disease presence, progression and response to therapy [100,117]. In NSCLC, exosomal miRNA correlated with disease free survival and overall survival [118]. The data regarding unique expression of miRNA in tumor-derived exosomes in different cancers are very promising, but more research is needed to standardize the methodologies and increase the sample sizes in an effort to validate the use of exosomal miRNA signatures as cancer biomarkers. At present, tumor-associated circulating miRNAs in plasma seem to be more widely evaluated as promising cancer biomarkers than exosomal miRNAs.
Less is known about the importance of exosomal mRNA as a potential biomarker of the cancer presence or progression. Skog et al reported that exosomes derived from clinical glioblastoma (GBM) samples contained over 4,700 distinct mRNA species and provided evidence for possible selective packaging of mRNA within exosomes, as also discussed above [44]. Hong et al detected over 11,000 distinct mRNAs in exosomes derived from supernatants of colorectal cell line SW480 [119]. Although less abundant than miRNAs, many of the mRNAs carried by exosomes are known to be involved in critical cellular activities such as cell cycle regulation, cell division, chromosome segregation, migration and angiogenesis [44,119]. Importantly, in several of the GBM patients, exosomes carried mRNA for the mutant version of EGFR, EGFRvIII, suggesting that exosomes in patients’ plasma might serve as biomarkers for non-invasive detection of glioblastoma [44]. Our own recent data with exosomes isolated from the pre-post vaccination plasma of patients with gliomas in the phase I/II clinical trial mentioned above showed that: (a) exosome fractions yielded sufficient mRNA for qRT-PCR to measure expression levels of 24 immunoregulatory genes in paired pre-post vaccine exosomes; (b) a significant decrease after vaccination in expression levels of 4 genes seen in exosomes from many albeit not all patients; and (c) the down-regulated genes (IL-8, TGF-β, TIMP-1 and ZAP70) are known to be related to angiogenesis, immune regulation and clinical outcome in gliomas [99]. These changes discriminated immunological responders from non-responders to the vaccine. Further, the observed changes in mRNA expression levels of the four genes were related to clinical responses, as 3 patients who responded immunologically to the vaccine are still alive at 64 months after vaccination. When the hazard ratios for the 4 genes were calculated, only ΔCt for IL-8 expression showed trend toward significance at the HR = 1.56 with p<0.070 for overall survival and HR = 1.48 with p<0.057 for time to progression [99]. Overall, this small retrospective study showed that the assessment of mRNA expression levels in exosomes isolated from plasma of glioma patients receiving immune-based therapy was worthwhile. Ours was the first study in which protein and mRNA expression levels for immune-related genes in plasma exosomes were shown to correlate with glioma patients’ response to vaccination therapy [99]. This confirmed the potential usefulness of exosomes as predictors of clinical and immunological responses to the vaccine in patients with advanced glioma despite the fact that total exosomal fractions rather than TEX were evaluated in this study. More extensive examination of transcripts carried by TEX in cancer patients undergoing immune therapies seems to be the next step in the development of exosomal mRNA profiles as surrogates of response.
5. TEX as markers of anti-tumor immune responses
Immune cell dysfunction is commonly seen in patients with cancer, and it is magnified as cancer progresses [120]. Human tumors use a variety of strategies to protect themselves from immune intervention by the host, executing a tumor escape [121]. This process of tumor escape characterizes all human malignancies, although the mechanisms tumors employ to implement it may vary [120,121]. We and others have shown that circulating exosomes of patients with cancer carry a variety of immunosuppressive molecules, including FasL, TGF-β, IL-10, TRAIL, PGE2, the checkpoint inhibitory receptors and their ligands as well as enzymes responsible for adenosine production [35,91,122,123]. Because of this cargo, tumor-derived exosomes exert profound inhibitory effects on anti-tumor immunity in patients with cancer [120,124]. TEX not only directly interfere with anti-tumor functions of T cells and NK cells, they also promote differentiation and proliferation of Treg and MDSC [35,91,125–127]. We have suggested that the TEX immunosuppressive profile could serve as a measure of tumor-induced immune dysfunction and tumor progression. This rationale is based on recent data indicating that the extent of immune dysfunction in cancer reflects tumor progression and correlates with outcome [5,128]. If so, then TEX could serve as surrogate markers of immune dysfunction in cancer and, by extension, of unfavorable prognosis. Also, a change to a less immunosuppressive profile in TEX could serve as a measure of response to therapy. We are testing this concept using exosomes isolated from plasma of patients with AML, melanoma and HNSCC. Our preliminary unpublished data suggest that the presence of immunosuppressive proteins in TEX correlates with clinical endpoints and response to therapy.
6. Expert commentary
Tumor-derived exosomes (also called TEX), present in all body fluids of patients with cancer, are currently considered as potentially promising candidates for a new class of biomarkers for diagnosis, prognosis and therapeutic responses in cancer. As the knowledge of biogenesis, molecular content and biological attributes of exosomes and TEX increases, so does the interest in their potential as cancer biomarkers. There are compelling reasons for this interest. The field of oncology has been in need of biomarkers that could fulfill the criteria not only for assay specificity, sensitivity, precision, reproducibility or robustness, but above all, for clinical relevance. The rapid development of new cancer drugs in the last decade requires reliable surrogate markers for guidance. As new therapeutic options are being offered to cancer patients, selection of those who are likely to benefit from therapy becomes a key issue. The ability to predict outcome and evaluate response to therapy are necessary for clinicians to optimize drug delivery. Whether exosomes or TEX, emerging as novel and promising biomarkers can satisfy these various requirements is speculative at best.
Applications of TEX analysis to diagnosis, monitoring, prognosis or response to therapy in cancer are rare. Nevertheless, exosomes, and especially TEX, present an attractive alternative to the existing cancer biomarkers and are moving to the forefront of biomarker discovery efforts. TEX are ubiquitous, enriched in body fluids of cancer patients relative to healthy controls, membrane bound with the content that is not vulnerable to degradation and mimics that of parent tumor cells. They are stable and small enough to cross the blood-brain barrier. The cargo of TEX comprising proteins, lipids, glycans and nucleic acids (both DNAs and RNAs) allows for monitoring of their molecular or genetic profiles and for establishing tumor-specific signatures. TEX have the potential of serving as a “liquid biopsy” which can be non-invasively and repeatedly acquired and studied in the course of disease or therapy, thereby eliminating the need for a surgical or needle tumor biopsy. Another advantage is that TEX carry biologically-active components, including enzymes, ligands and soluble factors, which make it possible to in vitro or in vivo study TEX for effects they (and by extension parent tumor cells) mediate upon interacting with target cells. Perhaps of greatest interest, however, are reports that signatures of exosomes in cancer patients’ body fluids change to inform about the presence of disease, its progression and response to therapy as well as outcome. These reports are largely retrospective, are few in number and obtained with small patient cohorts. Nevertheless, they raise hope for establishing exosomes as future cancer biomarkers.
Although the advantages offered by exosomes are unique and in many respects exceed the attributes of conventional circulating cancer biomarkers, there are also problems with exosomes, and especially with TEX. Chief among these problems is the heterogeneity of EVs in the body fluids of cancer patients. Exosomes are arbitrarily defined based on their size, cellular origin and functional properties as a subset of EVs. TEX are a subpopulation of exosomes released by tumor cells. The lack of established methods for separation of exosomes from EVs and for isolation of TEX is the second major barrier. As a result, the identity of EV populations evaluated in various studies remains elusive, and the available data do not discriminate TEX from other EVs. It is unclear whether TEX upon their isolation from plasma will be better or worse biomarkers than total plasma EVs. Also, it is not at all clear which of the multiple exosome components or signatures (i.e., the molecular genetic or functional signature) reported in the literature is likely to correlate better with clinical endpoints or prognosis. These barriers have to be eliminated before any significant progress can be made in developing exosomes as cancer biomarkers. As for TEX, providing they will pass the initial test for representing the tumor better than total exosome fractions or EVs, validation of their status as a “liquid tumor biopsy” will require laborious comparisons with relevant tumors in situ. Many hurdles will have to be overcome before exosomes or TEX will be ready for prime time as cancer biomarkers. Nevertheless, the available data suggest that efforts required for further development and validation of exosome-based strategies may be worthwhile.
7. Five year view
The last five years have seen a remarkable and rapid progress in bringing TEX to the attention of the scientific and clinical communities. Although few in numbers, results linking molecular or genetic profiles of TEX with prognosis and response to therapy are promising. There is little doubt that further development will extend and strengthen the existing evidence in favor of exosomes, including TEX. A consensus regarding methods for the isolation and characterization of these nanovesicles from cancer patients’ body fluids is likely to provide a novel, more effective, efficient and high-throughput platform for measuring exosomes in patient’s plasma. The concept of TEX as a “liquid biopsy’ will be extensively investigated. Prospective clinical trials will test the predictive role of TEX in cancer. Advantages will be taken of TEX abilities to suppress or activate immune cells in order to gage the potential of the tumor to modulate the immune system. With the improved understanding of the role exosomes play in the entire process of carcinogenesis, assays will be developed to interrogate the TEX potential to regulate all of the pathways that lead to tumor progression, including growth, migration, angiogenesis, silencing of immune anti-tumor responses, metastasis and oncogenic transformation. This will require numerous technical innovations and rigorous clinical testing. However, at the end of the rainbow will be a highly sophisticated, biologically- and clinically-sound approach to using tumor-derived exosomes as surrogate markers of tumor progression, outcome and response to therapy that will outpace and replace biomarkers in use today.
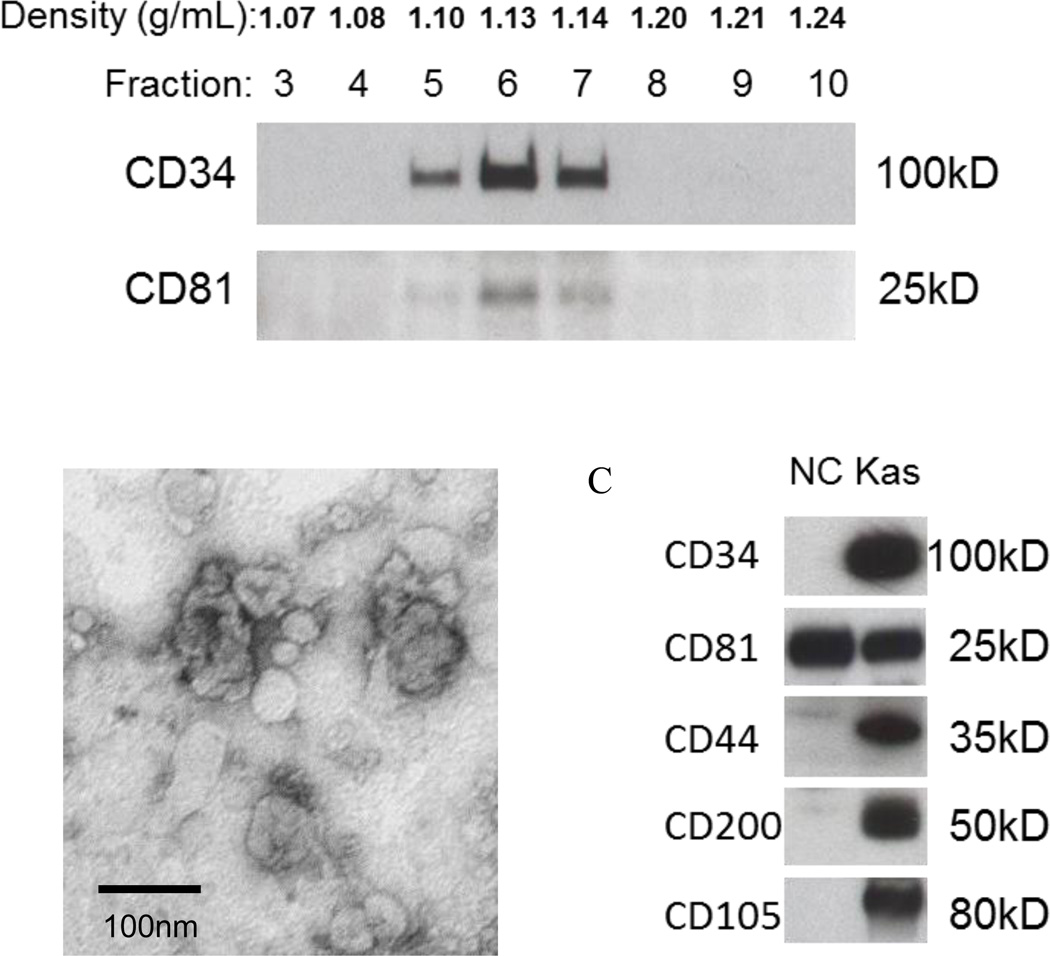
Figure 1.
Characteristics of exosomes isolated from the supernatant of Kasumi-1 (AML) cell line using differential centrifugation, ultrafiltration, size-exclusion chromatography and ultracentrifugation were studied. In A, following ultracentrifugation, exosomes were floated on a continuous sucrose density gradient, and the collected gradient fractions were tested by Western blots for CD34 and CD81. CD34+ and CD81+ exosomes were recovered at the sucrose density of 1.10–1.14 g/mL. In B, transmission electron microscopy of negatively-stained Kasumi-1 exosomes. In C, Western blots of Kasumi-1 exosomes (Kas) and exosomes isolated from plasma of a normal donor (NC). Each lane was loaded with 10µg exosomal protein. Reproduced from Hong CS, Muller L, Boyiadzis M, Whiteside TL, Isolation and characterization of CD34+ blast-derived exosomes in acute myeloid leukemia. PLoS One, 9(8), e103310 (2014) [80].
Key Issues.
Exosomes are the smallest type (30–120 nm) of extracellular vesicles (EVs) present in human body fluids.
Tumor-derived exosomes (TEX), a subset of exosomes, are avidly produced by tumor cells and accumulate in cancer patients’ plasma.
Pre-clinical studies suggest that TEX mediate intercellular communication and are involved in a wide spectrum of other biological activities.
TEX carry a cargo of proteins, lipids, glycans and nucleic acids, including mRNA, miRNA and DNA, which may be tumor-specific.
Molecular and genetic components of TEX are biologically active in vitro and in vivo in murine cancer models.
TEX have a potential of serving as a “liquid biopsy” which can be non-invasively monitored in patient’s plasma.
So far, TEX analysis has not been clinically applied to evaluate prognosis or responses to therapy in cancer.
The lack of consensus on exosome and TEX isolation methods has been a major barrier in TEX development as cancer biomarkers.
In the near future, intense efforts will be made to establish the role of TEX as surrogate markers of tumor progression, outcome and response to therapy.
Acknowledgments
The author was in part supported by the NIH grant R0-1 CA168-62.
Footnotes
Financial & competing interests disclosure
The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as:
* of interest
** of considerable interest
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Research UK. Melanoma statistics and outlook. 2014 Available from: http://www.cancerresearchuk.org/about-cancer/type/melanoma/treatment/mela. [Google Scholar]
- 3.Kohler BA, Sherman RL, Howlader N, et al. Annual report to the nation on the status of cancer, 1975–2011, featuring incidence of breast cancer subtypes by race/ethnicity, poverty, and state. J Natl Cancer Inst. 2015;107(6) doi: 10.1093/jnci/djv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parish CR. Cancer immunotherapy: the past, the present and the future. Immunol Cell Biol. 2003;81(2):106–113. doi: 10.1046/j.0818-9641.2003.01151.x. [DOI] [PubMed] [Google Scholar]
- 5.Beatty GL, Gladney WL. Immune escape mechanisms as a guide for cancer immunotherapy. Clin Cancer Res. 2015;21(4):687–692. doi: 10.1158/1078-0432.CCR-14-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther. 2015;14(4):847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 7.Mahoney KM, Freeman GJ, McDermott DF. The Next Immune-Checkpoint Inhibitors: PD-1/PD-L1 Blockade in Melanoma. Clin Ther. 2015 doi: 10.1016/j.clinthera.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97(16):1180–1184. doi: 10.1093/jnci/dji237. **This article specifies requirements for acceptable evaluations of cancer biomarkers. It is a must read for all investigators working with prospective cancer biomarkers.
- 9.Shi Q, Mandrekar SJ, Sargent DJ. Predictive biomarkers in colorectal cancer: usage, validation, and design in clinical trials. Scand J Gastroenterol. 2012;47(3):356–362. doi: 10.3109/00365521.2012.640836. [DOI] [PubMed] [Google Scholar]
- 10.Taylor DD, Gercel-Taylor C. Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Semin Immunopathol. 2011;33(5):441–454. doi: 10.1007/s00281-010-0234-8. [DOI] [PubMed] [Google Scholar]
- 11.Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011;9:86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33(3):967–978. doi: 10.1016/0092-8674(83)90040-5. *One of the first reports (together with refs. 13–15) on the involvement of EVs in transfer of biological materials from one cell to another.
- 13.Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97(2):329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101(3):942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262(19):9412–9420. [PubMed] [Google Scholar]
- 16.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19(2):43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Brinton LT, Sloane HS, Kester M, Kelly KA. Formation and role of exosomes in cancer. Cell Mol Life Sci. 2015;72(4):659–671. doi: 10.1007/s00018-014-1764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Marks MS, Heijnen HF, Raposo G. Lysosome-related organelles: unusual compartments become mainstream. Curr Opin Cell Biol. 2013;25(4):495–505. doi: 10.1016/j.ceb.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. *This exceptionally well-written current review provides a balanced view of EVs, their classification and their evolving importance in biology.
- 21.Gould SJ, Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64(3):676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 23.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6(10):801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 24.Rana S, Zoller M. Exosome target cell selection and the importance of exosomal tetraspanins: a hypothesis. Biochem Soc Trans. 2011;39(2):559–562. doi: 10.1042/BST0390559. [DOI] [PubMed] [Google Scholar]
- 25.Andreau Z, Yanez-Mo M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:1–12. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Colombo M, Moita C, van Niel G, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126(Pt 24):5553–5565. doi: 10.1242/jcs.128868. **This paper provides mechanistic information about the ESCRT system and its role in exosome biogenesis.
- 27.Park JE, Tan HS, Datta A, et al. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics. 2010;9(6):1085–1099. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Nedawi K, Meehan B, Micallef J, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 30.Dutta S, Warshall C, Bandyopadhyay C, Dutta D, Chandran B. Interactions between exosomes from breast cancer cells and primary mammary epithelial cells leads to generation of reactive oxygen species which induce DNA damage response, stabilization of p53 and autophagy in epithelial cells. PLoS One. 2014;9(5):e97580. doi: 10.1371/journal.pone.0097580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wieckowski E, Whiteside TL. Human tumor-derived vs dendritic cell-derived exosomes have distinct biologic roles and molecular profiles. Immunol Res. 2006;36(1–3):247–254. doi: 10.1385/IR:36:1:247. [DOI] [PubMed] [Google Scholar]
- 32. Andreola G, Rivoltini L, Castelli C, et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med. 2002;195(10):1303–1316. doi: 10.1084/jem.20011624. *One of the first reports on the potential role of tumor-derived microvesicles in immune suppression involving the Fas/FasL pathway.
- 33.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14(3):195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wieckowski EU, Visus C, Szajnik M, Szczepanski MJ, Storkus WJ, Whiteside TL. Tumor-derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor-reactive activated CD8+ T lymphocytes. J Immunol. 2009;183(6):3720–3730. doi: 10.4049/jimmunol.0900970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JW, Wieckowski E, Taylor DD, Reichert TE, Watkins S, Whiteside TL. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res. 2005;11(3):1010–1020. [PubMed] [Google Scholar]
- 36.Xiang X, Poliakov A, Liu C, et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int J Cancer. 2009;124(11):2621–2633. doi: 10.1002/ijc.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bobrie A, Thery C. Exosomes and communication between tumours and the immune system: are all exosomes equal? Biochem Soc Trans. 2013;41(1):263–267. doi: 10.1042/BST20120245. [DOI] [PubMed] [Google Scholar]
- 38.Gabrielsson S, Scheynius A. Exosomes in immunity and cancer--friends or foes? Semin Cancer Biol. 2014;28:1–2. doi: 10.1016/j.semcancer.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Saleem SN, Abdel-Mageed AB. Tumor-derived exosomes in oncogenic reprogramming and cancer progression. Cell Mol Life Sci. 2015;72(1):1–10. doi: 10.1007/s00018-014-1710-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rak J, Guha A. Extracellular vesicles--vehicles that spread cancer genes. Bioessays. 2012;34(6):489–497. doi: 10.1002/bies.201100169. *This review summarizes the role of EVs in transporting oncogenes and oncogenic signals from tumor to normal cells.
- 41.Demory Beckler M, Higginbotham JN, Franklin JL, et al. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12(2):343–355. doi: 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289(7):3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. **This report provides evidence for exosomes serving as vehicles for transfer of mRNA and micro-RNA between cells.
- 44. Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. **This paper shows that EVs transport RNA and protein from glioblastoma to receipient cells which induced alterations in their cellular behavior and could serve as cancer biomarkers.
- 45.Subra C, Grand D, Laulagnier K, et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J Lipid Res. 2010;51(8):2105–2120. doi: 10.1194/jlr.M003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Record M, Subra C, Silvente-Poirot S, Poirot M. Exosomes as intercellular signalosomes and pharmacological effectors. Biochem Pharmacol. 2011;81(10):1171–1182. doi: 10.1016/j.bcp.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 47.Martins VR, Dias MS, Hainaut P. Tumor-cell-derived microvesicles as carriers of molecular information in cancer. Curr Opin Oncol. 2013;25(1):66–75. doi: 10.1097/CCO.0b013e32835b7c81. [DOI] [PubMed] [Google Scholar]
- 48.Atay S, Godwin AK. Tumor-derived exosomes: A message delivery system for tumor progression. Commun Integr Biol. 2014;7(1):e28231. doi: 10.4161/cib.28231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 50.Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183(3):1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blanchard N, Lankar D, Faure F, et al. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J Immunol. 2002;168(7):3235–3241. doi: 10.4049/jimmunol.168.7.3235. [DOI] [PubMed] [Google Scholar]
- 52.Lachenal G, Pernet-Gallay K, Chivet M, et al. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol Cell Neurosci. 2011;46(2):409–418. doi: 10.1016/j.mcn.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 53.van Niel G, Raposo G, Candalh C, et al. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology. 2001;121(2):337–349. doi: 10.1053/gast.2001.26263. [DOI] [PubMed] [Google Scholar]
- 54. Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. **This is an important study demonstrating the in vivo role of melanoma-derived exosomes in educating the bone marrow microenvironment and converting it to the pro-metastatic milieu.
- 55.Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17(6):816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71(11):3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 57.Iero M, Valenti R, Huber V, et al. Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ. 2008;15(1):80–88. doi: 10.1038/sj.cdd.4402237. [DOI] [PubMed] [Google Scholar]
- 58.Huber V, Fais S, Iero M, et al. Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: role in immune escape. Gastroenterology. 2005;128(7):1796–1804. doi: 10.1053/j.gastro.2005.03.045. [DOI] [PubMed] [Google Scholar]
- 59. Whiteside TL. Immune modulation of T-cell and NK (natural killer) cell activities by TEXs (tumour-derived exosomes) Biochem Soc Trans. 2013;41(1):245–251. doi: 10.1042/BST20120265. **This paper provides evidence for immunosppressive effects mediated by tumor-derived exosomes interacting in vitro with various human immune cells.
- 60.Valenti R, Huber V, Iero M, Filipazzi P, Parmiani G, Rivoltini L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 2007;67(7):2912–2915. doi: 10.1158/0008-5472.CAN-07-0520. [DOI] [PubMed] [Google Scholar]
- 61.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70(23):9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 62.Webber JP, Spary LK, Sanders AJ, et al. Differentiation of tumour-promoting stromal myofibroblasts by cancer exosomes. Oncogene. 2015;34(3):290–302. doi: 10.1038/onc.2013.560. [DOI] [PubMed] [Google Scholar]
- 63.Gross JC, Chaudhary V, Bartscherer K, Boutros M. Active Wnt proteins are secreted on exosomes. Nat Cell Biol. 2012;14(10):1036–1045. doi: 10.1038/ncb2574. [DOI] [PubMed] [Google Scholar]
- 64.Luga V, Zhang L, Viloria-Petit AM, et al. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151(7):1542–1556. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 65.Safaei R, Larson BJ, Cheng TC, et al. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther. 2005;4(10):1595–1604. doi: 10.1158/1535-7163.MCT-05-0102. [DOI] [PubMed] [Google Scholar]
- 66.Corcoran C, Rani S, O'Brien K, et al. Docetaxel-resistance in prostate cancer: evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS One. 2012;7(12):e50999. doi: 10.1371/journal.pone.0050999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beloribi S, Ristorcelli E, Breuzard G, et al. Exosomal lipids impact notch signaling and induce death of human pancreatic tumoral SOJ-6 cells. PLoS One. 2012;7(10):e47480. doi: 10.1371/journal.pone.0047480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66(9):4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- 69.Putz U, Howitt J, Doan A, et al. The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Sci Signal. 2012;5(243):ra70. doi: 10.1126/scisignal.2003084. [DOI] [PubMed] [Google Scholar]
- 70.Putz U, Mah S, Goh CP, Low LH, Howitt J, Tan SS. PTEN secretion in exosomes. Methods. 2015;77–78:157–163. doi: 10.1016/j.ymeth.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 71.Johnstone RM, Mathew A, Mason AB, Teng K. Exosome formation during maturation of mammalian and avian reticulocytes: evidence that exosome release is a major route for externalization of obsolete membrane proteins. J Cell Physiol. 1991;147(1):27–36. doi: 10.1002/jcp.1041470105. [DOI] [PubMed] [Google Scholar]
- 72.Abd Elmageed ZY, Yang Y, Thomas R, et al. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells. 2014;32(4):983–997. doi: 10.1002/stem.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3 doi: 10.3402/jev.v3.24641. *This paper summarizes the existing knowledge of the mechanisms responsible for the uptake of EVs by various cells.
- 74.Feng D, Zhao WL, Ye YY, et al. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11(5):675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- 75.Nolte-'t Hoen EN, Buschow SI, Anderton SM, Stoorvogel W, Wauben MH. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood. 2009;113(9):1977–1981. doi: 10.1182/blood-2008-08-174094. [DOI] [PubMed] [Google Scholar]
- 76.Muller L, Hong CS, Stolz DB, Watkins SC, Whiteside TL. Isolation of biologically-active exosomes from human plasma. J Immunol Methods. 2014 doi: 10.1016/j.jim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Taylor DD, Shah S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods. 2015 doi: 10.1016/j.ymeth.2015.02.019. *A commentary on methods for isolation of EVs from supernatants or body fluids written by one of the first investigators attempting to isolate and characterize these vesicles using column chromatography.
- 78.Brownlee Z, Lynn KD, Thorpe PE, Schroit AJ. A novel "salting-out" procedure for the isolation of tumor-derived exosomes. J Immunol Methods. 2014;407:120–126. doi: 10.1016/j.jim.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saenz-Cuesta M, Arbelaiz A, Oregi A, et al. Methods for extracellular vesicles isolation in a hospital setting. Front Immunol. 2015;6:50. doi: 10.3389/fimmu.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hong CS, Muller L, Boyiadzis M, Whiteside TL. Isolation and characterization of CD34+ blast-derived exosomes in acute myeloid leukemia. PLoS One. 2014;9(8):e103310. doi: 10.1371/journal.pone.0103310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3(Unit 3):22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 82.Taylor DD, Doellgast GJ. Quantitation of peroxidase-antibody binding to membrane fragments using column chromatography. Anal Biochem. 1979;98(1):53–59. doi: 10.1016/0003-2697(79)90704-8. [DOI] [PubMed] [Google Scholar]
- 83.Hong CS, Funk S, Muller L, Boyiadzis M, Whiteside TL. Isolation of biologically active and morphologically intact exosomes from human plasma. 2015 doi: 10.3402/jev.v5.29289. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Witwer KW, Buzas EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gyorgy B, Paloczi K, Kovacs A, et al. Improved circulating microparticle analysis in acid-citrate dextrose (ACD) anticoagulant tube. Thromb Res. 2014;133(2):285–292. doi: 10.1016/j.thromres.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 86.van der Meel R, Krawczyk-Durka M, van Solinge WW, Schiffelers RM. Toward routine detection of extracellular vesicles in clinical samples. Int J Lab Hematol. 2014;36(3):244–253. doi: 10.1111/ijlh.12247. [DOI] [PubMed] [Google Scholar]
- 87.Sokolova V, Ludwig AK, Hornung S, et al. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf B Biointerfaces. 2011;87(1):146–150. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 88.Jayachandran M, Miller VM, Heit JA, Owen WG. Methodology for isolation, identification and characterization of microvesicles in peripheral blood. J Immunol Methods. 2012;375(1–2):207–214. doi: 10.1016/j.jim.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tauro BJ, Greening DW, Mathias RA, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56(2):293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 90. Hong CS, Muller L, Whiteside TL, Boyiadzis M. Plasma exosomes as markers of therapeutic response in patients with acute myeloid leukemia. Front Immunol. 2014;5:160. doi: 10.3389/fimmu.2014.00160. **This report is one of the first linking the plasma protein levels of the exosomal fractions and the exosome molecular content with therapeutic responses in patients with AML.
- 91.Szczepanski MJ, Szajnik M, Welsh A, Whiteside TL, Boyiadzis M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-beta1. Haematologica. 2011;96(9):1302–1309. doi: 10.3324/haematol.2010.039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y, Sabbatino F, Wang X, Ferrone S. Detection of chondroitin sulfate proteoglycan 4 (CSPG4) in melanoma. Methods Mol Biol. 2014;102:523–535. doi: 10.1007/978-1-62703-727-3_28. [DOI] [PubMed] [Google Scholar]
- 93.Rodriguez M, Llanes L, Perez A, Perez R, Vazquez AM. Generation and characterization of an anti-idiotype monoclonal antibody related to GM3(NeuGc) ganglioside. Hybrid Hybridomics. 2003;22(5):307–314. doi: 10.1089/153685903322538836. [DOI] [PubMed] [Google Scholar]
- 94.Pickhard A, Grober S, Haug AK, et al. Survivin and pAkt as potential prognostic markers in squamous cell carcinoma of the head and neck. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;117(6):733–742. doi: 10.1016/j.oooo.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 95.Szczepanski MJ, Whiteside TL. Elevated PRAME expression: what does this mean for treatment of head and neck squamous cell carcinoma? Biomark Med. 2013;7(4):575–578. doi: 10.2217/bmm.13.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taylor DD, Gercel-Taylor C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front Genet. 2013;4:142. doi: 10.3389/fgene.2013.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kalra H, Adda CG, Liem M, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 2013;13(22):3354–3364. doi: 10.1002/pmic.201300282. [DOI] [PubMed] [Google Scholar]
- 98.Nilsson J, Skog J, Nordstrand A, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100(10):1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Muller L, Muller-Haegele S, Mitsuhashi M, Gooding W, Okada H, Whiteside TL. Exosomes isolated from plasma of glioma patients enrolled in a vaccination trial reflect anti-tumor immune activity and might predict survival. Oncoimmunology. 2015;4(6):e1008347. doi: 10.1080/2162402X.2015.1008347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 101.Santiago-Dieppa DR, Steinberg J, Gonda D, Cheung VJ, Carter BS, Chen CC. Extracellular vesicles as a platform for 'liquid biopsy' in glioblastoma patients. Expert Rev Mol Diagn. 2014;14(7):819–825. doi: 10.1586/14737159.2014.943193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim DK, Kang B, Kim OY, et al. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kalra H, Simpson RJ, Ji H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10(12):e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lasser C. Exosomes in diagnostic and therapeutic applications: biomarker, vaccine and RNA interference delivery vehicle. Expert Opin Biol Ther. 2015;15(1):103–117. doi: 10.1517/14712598.2015.977250. [DOI] [PubMed] [Google Scholar]
- 105.Nawaz M, Camussi G, Valadi H, et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat Rev Urol. 2014;11(12):688–701. doi: 10.1038/nrurol.2014.301. [DOI] [PubMed] [Google Scholar]
- 106.Roberson CD, Atay S, Gercel-Taylor C, Taylor DD. Tumor-derived exosomes as mediators of disease and potential diagnostic biomarkers. Cancer Biomark. 2010;8(4–5):281–291. doi: 10.3233/CBM-2011-0211. [DOI] [PubMed] [Google Scholar]
- 107.Rolfo C, Castiglia M, Hong D, et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta. 2014;1846(2):539–546. doi: 10.1016/j.bbcan.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 108.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 109.Henderson MC, Azorsa DO. The genomic and proteomic content of cancer cell-derived exosomes. Front Oncol. 2012;2:38. doi: 10.3389/fonc.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 111.Chevillet JR, Kang Q, Ruf IK, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111(41):14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zocco D, Ferruzzi P, Cappello F, Kuo WP, Fais S. Extracellular vesicles as shuttles of tumor biomarkers and anti-tumor drugs. Front Oncol. 2014;4:267. doi: 10.3389/fonc.2014.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nolte-'t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, t Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40(18):9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9(4):e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rosell R, Wei J, Taron M. Circulating MicroRNA Signatures of Tumor-Derived Exosomes for Early Diagnosis of Non-Small-Cell Lung Cancer. Clin Lung Cancer. 2009;10(1):8–9. doi: 10.3816/CLC.2009.n.001. [DOI] [PubMed] [Google Scholar]
- 116.Corcoran C, Friel AM, Duffy MJ, Crown J, O'Driscoll L. Intracellular and extracellular microRNAs in breast cancer. Clin Chem. 2011;57(1):18–32. doi: 10.1373/clinchem.2010.150730. [DOI] [PubMed] [Google Scholar]
- 117.Liu J, Sun H, Wang X, et al. Increased exosomal microRNA-21 and microRNA-146a levels in the cervicovaginal lavage specimens of patients with cervical cancer. Int J Mol Sci. 2014;15(1):758–773. doi: 10.3390/ijms15010758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Silva J, Garcia V, Zaballos A, et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J. 2011;37(3):617–623. doi: 10.1183/09031936.00029610. [DOI] [PubMed] [Google Scholar]
- 119.Hong BS, Cho JH, Kim H, et al. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics. 2009;10:556. doi: 10.1186/1471-2164-10-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Whiteside TL. Immune responses to cancer: are they potential biomarkers of prognosis? Front Oncol. 2013;3:107. doi: 10.3389/fonc.2013.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Whiteside TL. Induced regulatory T cells in inhibitory microenvironments created by cancer. Expert Opin Biol Ther. 2014:1–15. doi: 10.1517/14712598.2014.927432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schuler PJ, Saze Z, Hong CS, et al. Human CD4(+) CD39(+) regulatory T cells produce adenosine upon co-expression of surface CD73 or contact with CD73(+) exosomes or CD73(+) cells. Clin Exp Immunol. 2014;177(2):531–543. doi: 10.1111/cei.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Szajnik M, Czystowska M, Szczepanski MJ, Mandapathil M, Whiteside TL. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg) PLoS One. 2010;5(7):e11469. doi: 10.1371/journal.pone.0011469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Filipazzi P, Burdek M, Villa A, Rivoltini L, Huber V. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin Cancer Biol. 2012;22(4):342–349. doi: 10.1016/j.semcancer.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 125.Clayton A, Mitchell JP, Court J, Linnane S, Mason MD, Tabi Z. Human tumor-derived exosomes down-modulate NKG2D expression. J Immunol. 2008;180(11):7249–7258. doi: 10.4049/jimmunol.180.11.7249. [DOI] [PubMed] [Google Scholar]
- 126.Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007;67(15):7458–7466. doi: 10.1158/0008-5472.CAN-06-3456. [DOI] [PubMed] [Google Scholar]
- 127.Abusamra AJ, Zhong Z, Zheng X, et al. Tumor exosomes expressing Fas ligand mediate CD8+ T-cell apoptosis. Blood Cells Mol Dis. 2005;35(2):169–173. doi: 10.1016/j.bcmd.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 128.Chang S, Kohrt H, Maecker HT. Monitoring the immune competence of cancer patients to predict outcome. Cancer Immunol Immunother. 2014;63(7):713–719. doi: 10.1007/s00262-014-1521-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Grange C, Collino F, Tapparo M, Camussi G. Oncogenic micro-RNAs and Renal Cell Carcinoma. Front Oncol. 2014;4:49. doi: 10.3389/fonc.2014.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Geddings JE, Mackman N. Tumor-derived tissue factor-positive microparticles and venous thrombosis in cancer patients. Blood. 2013;122(11):1873–1880. doi: 10.1182/blood-2013-04-460139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Al-Nedawi K, Meehan B, Kerbel RS, Allison AC, Rak J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc Natl Acad Sci U S A. 2009;106(10):3794–3799. doi: 10.1073/pnas.0804543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nazarenko I, Rana S, Baumann A, et al. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010;70(4):1668–1678. doi: 10.1158/0008-5472.CAN-09-2470. [DOI] [PubMed] [Google Scholar]
- 133.Zhuang G, Wu X, Jiang Z, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31(17):3513–3523. doi: 10.1038/emboj.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.van Niel G, Charrin S, Simoes S, et al. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell. 2011;21(4):708–721. doi: 10.1016/j.devcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jakobsen KR, Paulsen BS, Baek R, Varming K, Sorensen BS, Jorgensen MM. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles. 2015;4:26659. doi: 10.3402/jev.v4.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Baietti MF, Zhang Z, Mortier E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14(7):677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 137.Ostrowski M, Carmo NB, Krumeich S, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1):19–30. doi: 10.1038/ncb2000. sup pp 11–13. [DOI] [PubMed] [Google Scholar]
- 138.Hsu C, Morohashi Y, Yoshimura S, et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010;189(2):223–232. doi: 10.1083/jcb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Logozzi M, De Milito A, Lugini L, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4(4):e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Klibi J, Niki T, Riedel A, et al. Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood. 2009;113(9):1957–1966. doi: 10.1182/blood-2008-02-142596. [DOI] [PubMed] [Google Scholar]
- 141.Shao H, Chung J, Balaj L, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18(12):1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Graner MW, Alzate O, Dechkovskaia AM, et al. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009;23(5):1541–1557. doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Epple LM, Griffiths SG, Dechkovskaia AM, et al. Medulloblastoma exosome proteomics yield functional roles for extracellular vesicles. PLoS One. 2012;7(7):e42064. doi: 10.1371/journal.pone.0042064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Redzic JS, Ung TH, Graner MW. Glioblastoma extracellular vesicles: reservoirs of potential biomarkers. Pharmgenomics Pers Med. 2014;7:65–77. doi: 10.2147/PGPM.S39768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Higginbotham JN, Demory Beckler M, Gephart JD, et al. Amphiregulin exosomes increase cancer cell invasion. Curr Biol. 2011;21(9):779–786. doi: 10.1016/j.cub.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Khan S, Bennit HF, Turay D, et al. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer. 2014;14:176. doi: 10.1186/1471-2407-14-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Marimpietri D, Petretto A, Raffaghello L, et al. Proteome profiling of neuroblastoma-derived exosomes reveal the expression of proteins potentially involved in tumor progression. PLoS One. 2013;8(9):e75054. doi: 10.1371/journal.pone.0075054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Flanagan J, Middeldorp J, Sculley T. Localization of the Epstein-Barr virus protein LMP 1 to exosomes. J Gen Virol. 2003;84(Pt 7):1871–1879. doi: 10.1099/vir.0.18944-0. [DOI] [PubMed] [Google Scholar]
- 149.Staubach S, Razawi H, Hanisch FG. Proteomics of MUC1-containing lipid rafts from plasma membranes and exosomes of human breast carcinoma cells MCF-7. Proteomics. 2009;9(10):2820–2835. doi: 10.1002/pmic.200800793. [DOI] [PubMed] [Google Scholar]
- 150.Mears R, Craven RA, Hanrahan S, et al. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics. 2004;4(12):4019–4031. doi: 10.1002/pmic.200400876. [DOI] [PubMed] [Google Scholar]
- 151.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73(10):1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 152.Mizutani K, Terazawa R, Kameyama K, et al. Isolation of prostate cancer-related exosomes. Anticancer Res. 2014;34(7):3419–3423. [PubMed] [Google Scholar]
- 153.Li J, Sherman-Baust CA, Tsai-Turton M, Bristow RE, Roden RB, Morin PJ. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer. 2009;9:244. doi: 10.1186/1471-2407-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Houali K, Wang X, Shimizu Y, et al. A new diagnostic marker for secreted Epstein-Barr virus encoded LMP1 and BARF1 oncoproteins in the serum and saliva of patients with nasopharyngeal carcinoma. Clin Cancer Res. 2007;13(17):4993–5000. doi: 10.1158/1078-0432.CCR-06-2945. [DOI] [PubMed] [Google Scholar]
- 155.Bergmann C, Strauss L, Wieckowski E, et al. Tumor-derived microvesicles in sera of patients with head and neck cancer and their role in tumor progression. Head Neck. 2009;31(3):371–380. doi: 10.1002/hed.20968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Szajnik M, Derbis M, Lach M, et al. Exosomes in Plasma of Patients with Ovarian Carcinoma: Potential Biomarkers of Tumor Progression and Response to Therapy. Gynecol Obstet (Sunnyvale) 2013;(Suppl 4):3. doi: 10.4172/2161-0932.S4-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mitchell PJ, Welton J, Staffurth J, et al. Can urinary exosomes act as treatment response markers in prostate cancer? J Transl Med. 2009;7:4. doi: 10.1186/1479-5876-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Principe S, Jones EE, Kim Y, et al. In-depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. Proteomics. 2013;13(10–11):1667–1671. doi: 10.1002/pmic.201200561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Raimondo F, Morosi L, Corbetta S, et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol Biosyst. 2013;9(6):1220–1233. doi: 10.1039/c3mb25582d. [DOI] [PubMed] [Google Scholar]
- 160.Welton JL, Khanna S, Giles PJ, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9(6):1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li Y, Zhang Y, Qiu F, Qiu Z. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis. 2011;32(15):1976–1983. doi: 10.1002/elps.201000598. [DOI] [PubMed] [Google Scholar]
- 162.Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep. 2012;28(5):1551–1558. doi: 10.3892/or.2012.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ogawa Y, Kanai-Azuma M, Akimoto Y, Kawakami H, Yanoshita R. Exosome-like vesicles with dipeptidyl peptidase IV in human saliva. Biol Pharm Bull. 2008;31(6):1059–1062. doi: 10.1248/bpb.31.1059. [DOI] [PubMed] [Google Scholar]
- 164.Xiao H, Wong DT. Proteomic analysis of microvesicles in human saliva by gel electrophoresis with liquid chromatography-mass spectrometry. Anal Chim Acta. 2012;723:61–67. doi: 10.1016/j.aca.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 165.EVpedia. [cited 7 October, 2014]; Available from http://evpedia.info/ [Google Scholar]
- 166.Roth P, Wischhusen J, Happold C, et al. A specific miRNA signature in the peripheral blood of glioblastoma patients. J Neurochem. 2011;118(3):449–457. doi: 10.1111/j.1471-4159.2011.07307.x. [DOI] [PubMed] [Google Scholar]
- 167.Manterola L, Guruceaga E, Gallego Perez-Larraya J, et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol. 2014;16(4):520–527. doi: 10.1093/neuonc/not218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8(9):1156–1162. doi: 10.1097/JTO.0b013e318299ac32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bryant RJ, Pawlowski T, Catto JW, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106(4):768–774. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Brase JC, Johannes M, Schlomm T, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128(3):608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 171.Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54(4):1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Huan J, Hornick NI, Shurtleff MJ, et al. RNA trafficking by acute myelogenous leukemia exosomes. Cancer Res. 2013;73(2):918–929. doi: 10.1158/0008-5472.CAN-12-2184. [DOI] [PubMed] [Google Scholar]